Abstract
Recent evidence suggests human embryonic stem (ES) and induced pluripotent stem (iPS) cell lines have differences in their epigenetics marks and transcriptomes, yet the impact of these differences on subsequent terminally differentiated cells is less well understood. Comparison of purified, homogeneous populations of somatic cells derived from multiple independent human iPS and ES lines will be required to address this critical question. Here, we report a differentiation protocol based on embryonic development that consistently yields large numbers of endothelial cells (EC) derived from multiple human ES or iPS cells. Mesoderm differentiation of embryoid bodies was maximized and defined growth factors were used to generate KDR+ EC progenitors. Magnetic purification of a KDR+ progenitor subpopulation resulted in an expanding, homogeneous pool of ECs that expressed EC markers and had functional properties of ECs. Comparison of the transcriptomes revealed limited gene expression variability between multiple lines of human iPS–derived ECs, or between lines of ES– and iPS–derived ECs. These results demonstrate a method to generate large numbers of pure human EC progenitors and differentiated ECs from pluripotent stem cells, and suggest individual lineages derived from human iPS cells may have significantly less variance than their pluripotent founders.
Medical Subject Headings: Endothelial Cells, Stem Cells, Induced Pluripotent Stem Cells, Cell Differentiation
Introduction
Induced pluripotent stem (iPS) cells have recently emerged as a promising alternative to embryonic stem cells for modeling disease and developing therapeutics [1]. While seemingly equivalent, many recent studies suggest subtle differences between these two pluripotent cell types, including specific culture-derived genetic abnormalities, propensity to differentiate towards a specific lineage, and somatic epigenetic memory [2]. In contrast, the impact of the putative differences between iPS and ES cells on fully differentiated cell states after in vitro directed differentiation has not been explored to the same extent, especially in human cell types where cell-type specific reporter lines have been difficult to generate. Well-defined cell surface markers make endothelial cells (ECs) a useful platform to systematically profile a homogeneous cell population derived from pluripotent stem cells without the requirement for reporter cell lines.
The endothelium is a monolayer that invests the luminal surface of all blood and lymphatic vessels. ECs composing this diaphanous film of tissue modulate the growth and reactivity of the underlying smooth muscle, control the interaction of the vessel wall with circulating blood elements, and regulate vascular responses to hemodynamic forces [3]. Transplanted human ES or iPS derived ECs lead to increased function and vascularization in multiple animal disease models including hind limb perfusion and myocardial infarction in addition to stably carrying blood up to 150 days after transplantation with no safety issues as yet reported [4–7].
Several methods for generating ECs from human pluripotent stem cells have been reported. Original embryoid body differentiation methods supplemented with high VEGF generated 5–8% CD31 positive cells after two weeks in culture [8, 9]. Recent improvements claim efficiencies from 15–57% CD31 positive cells by day 14, however, these methods have been difficult to consistently replicate across multiple pluripotent stem cell lines either because of protocol complexity, batch variation in required reagents, or other unexplained factors [10, 11]. Independent pluripotent stem cell lines may require optimization of conditions for each cell line due to inherent variation amongst lines [12]. Most importantly, a comprehensive genome-wide analysis of gene expression variability in human ECs, or any other specific human lineage, among multiple different ES or iPS lines has not been reported [13, 14].
Here, we report a differentiation protocol that recapitulates normal development and consistently yielded large numbers of relatively pure ECs derived from multiple independent human ES or iPS cell lines. Comprehensive profiling of this well-defined cell population revealed remarkably few gene expression differences between ECs derived from multiple hiPSCs or hESCs, as well as ECs derived with different differentiation protocols. These findings suggest that differentiated cell types derived from hES and hiPS cells lines, and from multiple hiPS cell lines, may have limited transcriptome variance, increasing the likelihood of successful disease-modeling using iPS-based technology.
Materials and Methods
Human PSC Culture and Differentiation into Endothelial Cells
hESCs (H1, H7, and H9) and hiPSCs (iPS1, iPS2, and iPS3) were cultured according to WiCell Protocols under feeder-free conditions on matrigel-coated plates in mTeSR®1 (Stem Cell Technologies, Vancouver, BC) in a hypoxic environment (5% CO2, 5% O2). hiPSC lines were all derived by reprogramming fibroblasts with four factors (Oct4, Sox2, Klf4, and c-Myc) and were fully characterized for pluripotency. iPS1 is the iPSC line K23F (Shinya Yamanaka & Kiichiro Tomoda, unpublished), iPS2 is 3S5F [15], and iPS3 is Huf5 [6, 16]. To induce differentiation, hESCs and hiPSCs were detached with Dispase (Gibco, Carlsbad, CA) and scraped with a cell lifter and then placed into StemPro-34 (Invitrogen) supplemented with 10 ng/mL pen/strep, 2 mM L-glutamine, 150 mg/mL Transferrin, 1 mM ascorbic acid, and 4×10−4 M monothioglycerol (MTG) (Sigma, St. Louis, MO). All cytokines, including human-bFGF, human-Activin A, human-BMP4 (R&D Systems, Minneapolis, MN) and human-VEGF (Calbiochem, Gibbstown, NJ), were added at the indicated time points and concentrations. Pluripotent stem cells were cultured in a hypoxic environment (5% CO2, 5% O2, 90% N2) until D6 of differentiation. After D6, MACS was performed according to the manufacturers protocol and KDR+ cells were plated on fibronectin (Sigma) in a normoxic environment (5% CO2, 20% O2, 75% N2) in ECM medium (ScienCell, Carlsbad, CA) with 5% FBS, pen/strep and ECGS (10 mg/ml BSA, 10 mg/ml apo-transferrin, 5 mg/ml insulin, 10 ng/ml EGF, 2 ng/ml bFGF, 2 ng/ml VEGF, 2 ng/ml IGF-1, 1 mg/ml hydrocortisone and 30 ng/mL retinoic acid). KDR+ cells were allowed to grow to 80–90% confluence and were then split 1:3 using 0.05% trypsin. Control differentiations consisted of EB formation followed by addition of 10 ng/mL VEGF from D1 to D14 and then CD31+ cell sorting on D14.
Flow Cytometry and Magnetic Cell Sorting
Accutase (Millipore, Bellerica, MA) was used to generate single-cell suspensions from EBs plated overnight on fibronectin. The cells were stained with KDR-PE-conjugated antibodies (R&D Systems) and FACS sorted on a FACS ARIA 2 or MACS sorted on a midi column (Miltenyi Biotec, Auburn, CA) according to the manufacturer’s protocol. CD31 antibodies conjugated to magnetic beads (Miltenyi Bio) were used with midi columns to further purify cultures contaminated with more than 10% non-ECs. Characterization of fully differentiated endothelial cells was determined by PECAM-1–488-conjugated primary antibody (CD31, R&D Systems) or VE-cadherin-PE-conjugated antibody (CD144; R&D Systems) staining. EPHB2 staining was performed with recombinant mouse EphB4 Fc chimera protein (R&D Systems). Fluorescent intensity was determined for 10,000 cells total and percentages shown in all Figures are the percent of live single cells that fall within the gate shown. Gates were determined using a negative control (isotype specific IgG) stained cell population.
Immunocytochemical Staining
To characterize the phenotype of ECs, purified cells were stained with antibodies against endothelial markers, including PECAM-1 (CD31, R&D Systems), VE-cadherin (CD144; R&D Systems), endothelial nitric oxide synthase (eNOS; BD Pharmingen, San Diego, CA, USA), von Willebrand factor (vWF; Abcam, Cambridge MA). Briefly, the cells were fixed with paraformaldehyde (4%), permeabilized with Triton X-100 (0.1%), and blocked with either normal goat or donkey serum (1%) for 30 minutes, followed by overnight incubation with the primary antibodies at 4°C. The cells were washed with 1× PBS and incubated with Alexa Fluor-488 or -594 secondary antibodies for 1 hour at room temperature. Cells were washed with 1× PBS, and the nuclei were stained with Hoechst 33342 dye (Invitrogen, Carlsbad, CA).
Acetylated LDL Uptake
Cells were incubated with Dil-labeled Ac-LDL (10 µg/ml; Invitrogen) for 4 hours at 37°C. After incubation, the cells were washed with 1× PBS before being visualized and photographed under a fluorescent microscope.
In Vitro Matrigel Assay
Cells (2.5 × 105) were seeded on 24-well plates pre-coated with growth factor-reduced Matrigel (BD Discovery Labware, Bedford, MA) and incubated for 24 hours at 37°C. Images were taken using a light microscope.
In Vivo Matrigel Injection
To determine if hiPSC– and hESC–derived ECs formed functional blood vessels in vivo, a Matrigel plug assay was performed using immunodeficient SCID mice. Matrigel was mixed with bFGF (50 ng/ml; Peprotech, Rocky Hill, NJ) and ECs (5×105). The mixture was subcutaneously injected into SCID mice. After 14 days, the animals were euthanized and the Matrigel plugs were removed. Paraffin-embedded matrigel sections were stained with human CD31. The number of capillaries formed by the ECs was counted under 200× magnification from 6–7. All animals were treated in accordance with an approved protocol from the Stanford Administrative Panel on Laboratory Animal Care.
Endothelial Cell Migration Assay
EC migration was measured using BD Biocoat Angiogenesis System and the manufacturer’s protocol. Briefly, a cell suspension of 4.0 × 10 cells/ml was prepared for each EC sample and added into 24-well plate inserts. The insert plates were incubated for 24 hours at 37°C, 5% CO2. The inserts were removed and transferred to another plate containing calcein AM solution, and the plates were incubated for 90 minutes at 37°C, 5% CO2. A fluorescence plate reader with bottom-reading capabilities at excitation/emission wavelengths of 494/517 nm was used to quantify fluorescence of the cells that successfully migrated. The fold migration was calculated by the fluorescence value of the treated cells over the untreated cells
Nitric Oxide Measurement
Production of nitric oxide was determined using HPLC measurement of nitrite in media after 24 hrs of cell conditioning. Following media collection, the cells were lysed with RIPA buffer containing protease inhibitors and the protein concentration for each cell type was measured using BCA assay to adjust nitrite levels for cell number.
RNA Extraction, Microarray, and qRT-PCR validation
All cells were washed with PBS and placed in Trizol (Invitrogen). Total RNA was collected using phenol/chloroform extraction with phase-lock tubes (5 Prime, Gaithersburg, MD). The RNA was cleaned and DNAse digested on RNeasy columns (Qiagen, Valencia, CA). RNA was amplified, fragmented, and labeled using NuGEN Technologies Applause Plus kits and the manufacturer's instructions. Labeled samples were then hybridized to Affymetrix Human Gene 1.0 ST GeneChips. The GeneChips were then washed and stained on Affymetrix 450 Fluidics Stations and scanned on an Affymetrix 3000 Scanner according to standard protocols. Validation of a selected set of differentially expressed genes was performed by quantitative reverse transcriptase-PCR (qRT-PCR) using standard Taq-Man primer sets (Applied Biosystems) on an ABI-7900 HT.
Bioinformatics
Microarrays were normalized to control for array-specific effects with the Affymetrix Power Tools software using Robust Multi-Array normalization. For statistical analyses, all probe sets in which none of the groups had an average log2 intensity greater than 3.0 were removed. This is a standard cutoff, under which the expression levels are indistinguishable from background. All array data are available from Gene Expression Omnibus (accession number GSE37631). Linear models were fitted for each gene using the limma package (Smyth 2008) in R/Bioconductor. Moderated t-statistics and the associated P-values were calculated. P-values were adjusted for multiple testing by controlling for false-discovery rate (FDR, which is the expected percentage of falsely-declared differentially expressed genes among the set of all declared differentially expressed genes) using the Benjamini-Hochberg method. Differential expression was defined using a threshold of 2–4-fold change as indicated and a P-value of < 0.05. Hierarchical cluster analysis was performed on normalized array values with Cluster 3.0 with Euclidean distance and single linkage for >10,000 genes and average linkage for smaller gene sets. Heatmaps were generated with Java TreeView. GO Analysis was completed using GO Elite with default settings.
Statistical Analysis
All experiments were performed at least three times unless otherwise stated. All statistical analysis was completed using GraphPad Prism software. Two-way analysis was completed with two-tailed unpaired t tests with a 95% confidence interval. Multiple comparison tests were performed by One-way ANOVA with Bonferroni post test with Alpha = 0.05 (95% confidence interval).
Results
Generation of KDR+ precursors
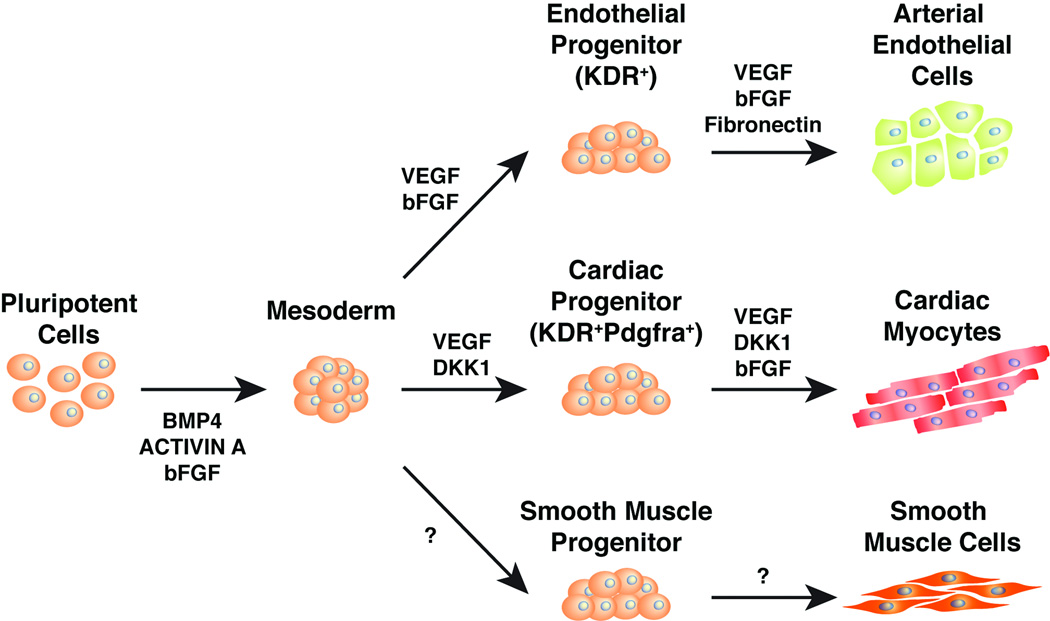
Large numbers of cardiac myocytes have been efficiently generated from mouse and human stem cells by inducing pre-cardiac mesoderm cells marked by KDR and PDGFR-a cell surface receptors, then biasing toward the cardiac muscle fate [17, 18]. Since the pre-cardiac mesoderm cells are multipotent and give rise to endothelial and smooth muscle cells in addition to cardiac myocytes, we searched for approaches to bias these precursors into the EC lineage [19].
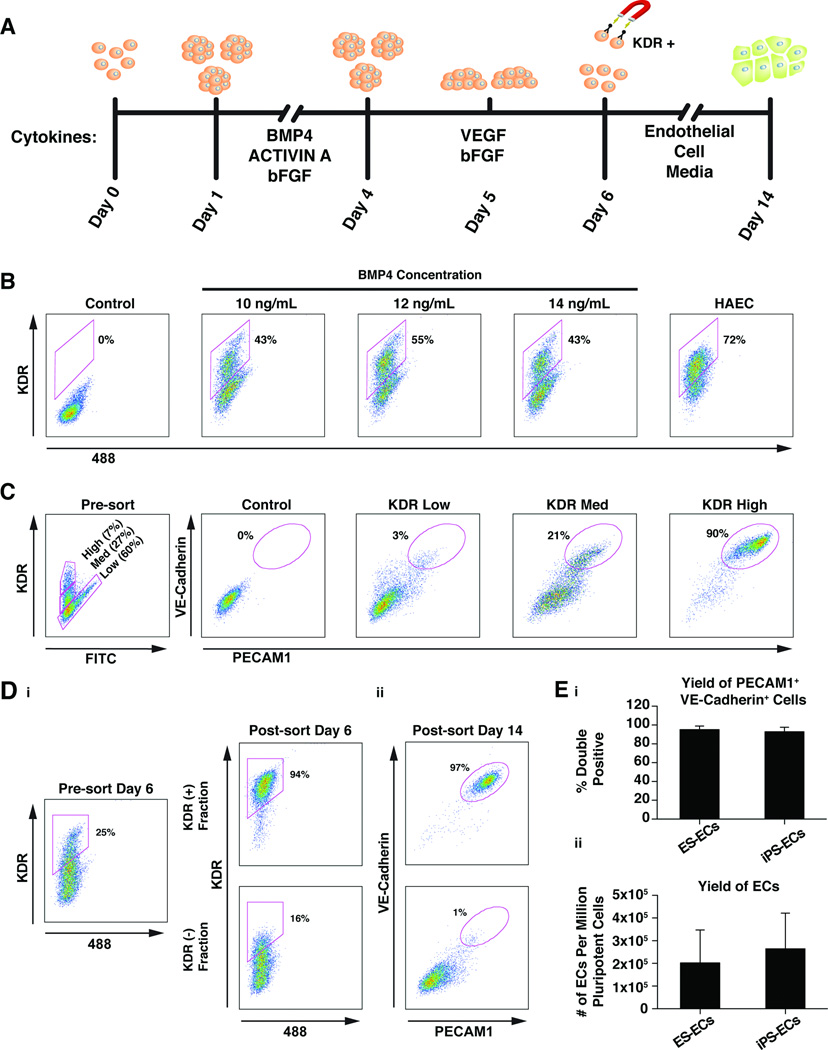
hESCs and hiPSCs were grown in feeder-free conditions and aggregated to form embryoid bodies (EBs). KDR+ cells were induced from day 1–4 (D1–D4) (stage 1) with defined medium containing bFGF (5 ng/mL), Activin A (6 ng/mL) and a range of BMP4 (4–14 ng/mL) in a manner similar to a reported protocol for directed differentiation of ESCs into cardiac myocytes [18] (Fig. 1A). We attempted to further differentiate pre-cardiac mesoderm into endothelial precursors from D4–D5 (stage 2) with VEGF (10 ng/mL) and bFGF (10 ng/mL), two cytokines critically important for angiogenesis [20, 21]. At D5.4, EBs were plated on fibronectin-coated plates which promote attachment and proliferation of endothelial cells [22]. On D6, single-cell suspensions were generated using Accutase and subsequently stained for KDR. Expression of KDR ultimately becomes restricted to the endothelial lineage [19, 20], so we tested a range of BMP4 concentrations (BMP 4–14 ng/ml) (Fig. S1A) for the ability to generate KDR+ cells at D6. BMP4 administered at 12 ng/mL from D1–4 resulted in maximal KDR expression (55%) at D6, as detected by flow cytometry (Fig. 1B). This concentration of BMP4 was used for subsequent differentiations of all pluripotent cell lines, though optimization for each line may further maximize EC yield. In order to determine the level of commitment to the endothelial lineage at D6 we used flow cytometric analysis to profile the surface markers PECAM1 (CD31) and VE-Cadherin (CD144), which are highly expressed in mature human ECs. While 44% of the cells were KDR+, only 12% or 13% of the cells were CD31+ or CD144+, respectively (Fig. S1B).
Figure 1.
KDRhigh cells differentiate into mature endothelial cells. (A): Endothelial cell differentiation protocol. Human ES or iPS cells were induced to form cardiac mesoderm using BMP, Activin A, and bFGF. Embryoid bodies were placed into conditions promoting endothelial cell formation at day 4, plated on fibronectin at day 5 and then KDRhigh endothelial precursor cells were magnetically sorted on day 6. The precursors were plated in endothelial cell maintenance conditions and expanded until day 14 when they were frozen. (B): Flow cytometric analysis of the effects of various BMP4 concentrations on KDR expression. The control population consisted of cells labeled with isotype control antibodies (IgG). The experiment was repeated three times and representative data from one experiment is displayed. (C): Analysis of the EC differentiation potential of three cell sub-populations with varying KDR expression levels. Three populations were sorted by FACS at day 6 and then analyzed for expression of the mature human endothelial cell-surface markers VE-Cadherin and PECAM1 on day 14. The control population consisted of cells labeled with isotype control antibodies (IgG). The experiment was repeated three times and representative data from one experiment is displayed. (D): i. Analysis of KDR expression before and after magnetic sorting of KDRhigh cells at day 6. ii. Two KDR fractions (+ and −) were replated after magnetic sorting and reanalyzed on day 14 for expression of VE-Cadherin and PECAM1. The experiment was repeated three times and representative data from one experiment is displayed. (E): i. Mean percentage of cells expressing both PECAM1 and VE-Cadherin at Day 14 of differentiation from ES or iPS cells from 4 different cell lines in 4 separate experiments for each group. Error bars represent standard deviation. Mean values were not statistically different when compared with two-tailed unpaired t test. ii. Mean yield of ECs from 1 million pluripotent stem cells from 4 different cell lines in 4 separate experiments for each group. Error bars represent standard deviation. Mean values were not statistically different when compared with two-tailed unpaired t test.
Endothelial Cell Differentiation Potential of KDR+ Populations
Cells expressing KDR at D6 could be divided into three sub-populations based on KDR expression level: KDRlow, KDRmed, KDRhigh. To investigate their potentials to differentiate into ECs, the three sub-populations were sorted by fluorescence activated cell sorting (FACS) and re-plated on fibronectin-coated plates in EC medium commonly used to maintain primary EC lines. We used PECAM1 (CD31) and VE-Cadherin (CD144) cell surface staining at D14 to monitor and optimize the EC differentiation. The KDRhigh population resulted in greater than 90% CD31/CD144 double positive ECs (CD31+/CD144+) (Fig. 1C). The KDRmed or KDRlow populations generated 20% or 3% CD31+/CD144+ ECs, respectively. Interestingly, a mixture of KDRhigh/med cells resulted in greater than 90% CD31+/CD144+ ECs, while a mixture of KDRmed/low cells generally resulted in fewer than 10% CD31+/CD144+ ECs (data not shown). This may indicate that the KDRmed sub-population is a plastic precursor cell responsive to non-cell autonomous differentiation cues from surrounding cells. The KDRlow fraction’s inhibition of EC differentiation may represent a dominant effect over the KDRhigh population or may be due to more rapid proliferation of the KDRlow derivatives that then become the dominant cell type.
Isolating large numbers of KDRhigh/med cells that could ultimately become ECs required a more efficient method than FACS, which is time consuming and destructive to cells needing further culture and expansion. Therefore, we employed magnetic cell sorting (MACS) to isolate KDRhigh/med cells using a combination of a KDR antibody conjugated to Phycoerythrin (PE) and an anti-PE antibody conjugated to a magnetic bead. We purified KDRhigh/med cells by MACS and quantified the KDR-expressing cells by FACS from the pre- and post-magnetic sort populations. By MACS at D6, we recovered a KDR+ population that was 94% KDRhigh/med cells (Fig. 1D). The percent KDRhigh/med of the total cell population varied widely between cell lines and between differentiations of the same cell line with a range of 6.78 to 72.7% (Table 1).
Table 1.
Summary of KDRhigh/med yield on day 6 of differentiation.
| Pluripotent Cell Line | KDRhigh/med |
|---|---|
| H1 | 6.78 |
| H1 | 12.8 |
| H7 | 43.8 |
| H9 | 33.6 |
| H9 | 43.4 |
| H9 | 44.8 |
| H9 | 59.1 |
| H9 | 76.2 |
| iPS2 | 9.79 |
| iPS2 | 19.6 |
| iPS2 | 30.5 |
| iPS2 | 72.7 |
Data are displayed as percentage of total cell population staining positive for KDR at d6 of differentiation. Duplicate lines indicate independent differentiations done on separate days. Percentages from same cell line are arranged in ascending order.
Once replated on fibronectin in EC medium, the KDRhigh/med endothelial precursor cells had a high proliferative capacity and were allowed to grow to 80–90% confluence and then were split 1:3 by using 0.05% trypsin. Endothelial cells round up and can be tapped off of the plate before other contaminating cell types release from the culture dish when using 0.05% trypsin, which helped maintain nearly pure populations of CD31+/CD144+ ECs. All differentiations with adequate enrichment of KDRhigh/med cells, regardless of starting pre-sort KDRhigh/med percentage, resulted in 95% ± 3.7 CD31+/CD144+ cells at day 14 for ES cells and 93% ± 4.8 for iPS cells (Fig. 1D and E). The CD31+/CD144+ ECs were frozen at D14 and could be thawed and expanded again. The yield of ECs varied among batches and lines, but ranged from 1.5 × 105 to 4.0 × 105 cells per million pluripotent cells and correlated with the proliferation rate of the KDRhigh/med cells after sorting rather than the percentage of KDRhigh/med cells at the time of sorting (Fig. 1E). It should be noted that this calculation is based on the number of starting ES cells on day 0 and not the number of cells present at D1 after aggregation (~50% of starting population) as was reported in James et al., 2010.
Biological and Functional Characterization
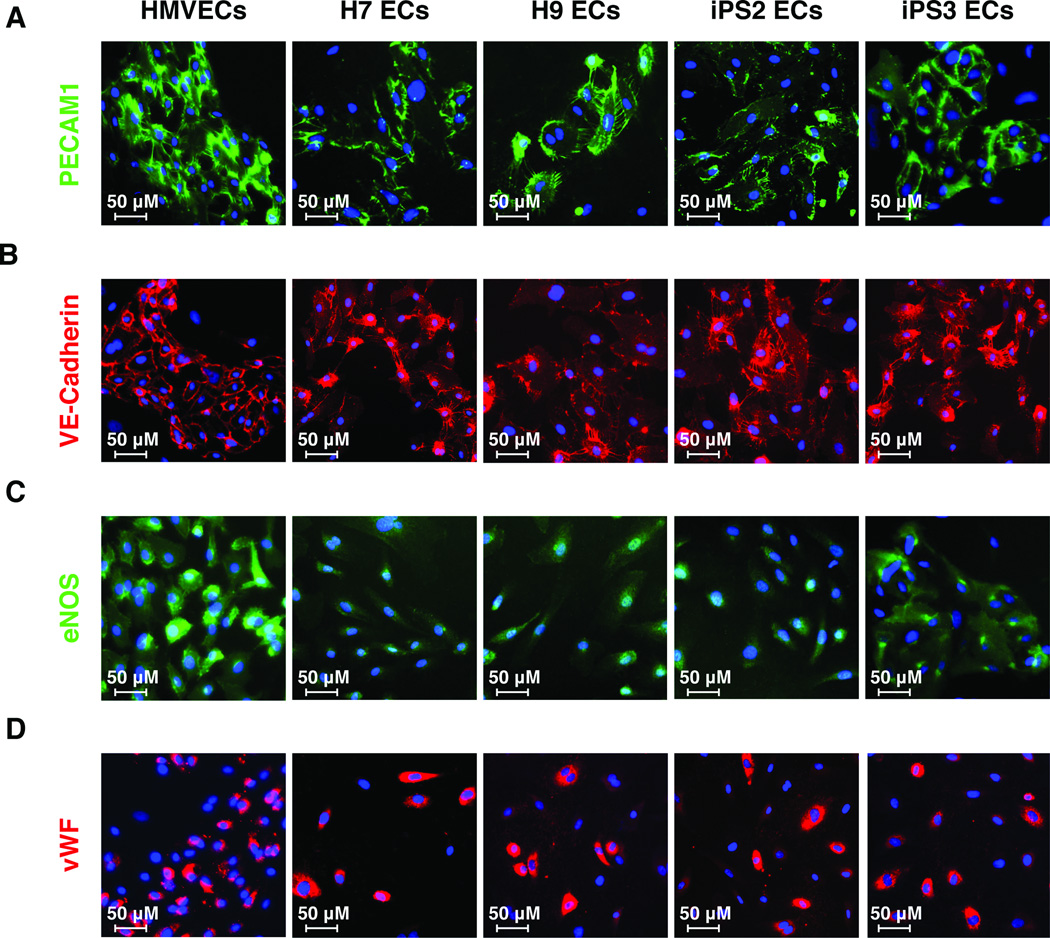
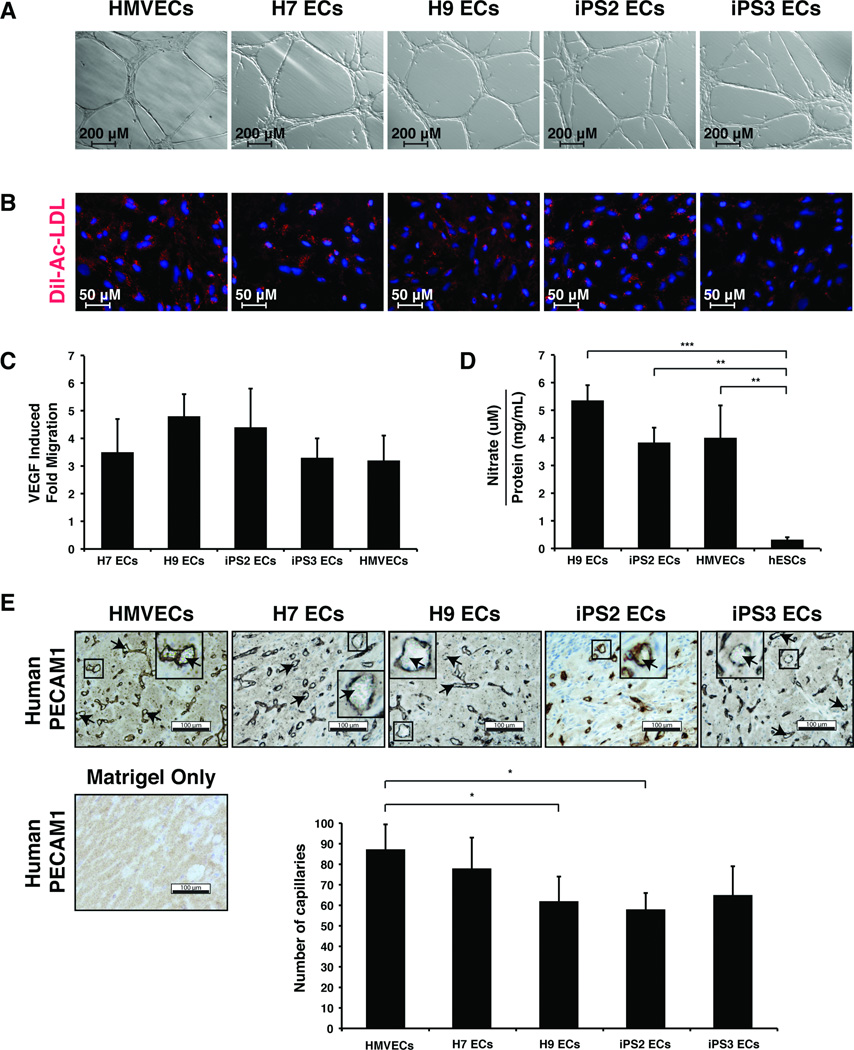
CD31+/CD144+ cells were derived from various hiPS (iPS1, iPS2, iPS3) and hES cell lines (H1, H7, H9) and used for genome-wide gene expression analysis. Two lines of each EC type (hiPS or hES) were randomly selected for full characterization and were subjected to a comprehensive series of standard endothelial characterization tests, including immunostaining, fluorescent acetylated LDL (Dil-Ac-LDL) uptake assays, and Matrigel assays. Both hiPS-ECs and hES-ECs developed cobblestone morphology and stained positive for the endothelial markers PECAM-1 (CD31), VE-cadherin (CD144), endothelial nitric oxide (eNOS), and von Willebrand factor (vWF) (Fig. 2, Fig. S2). These cells also formed vascular network-like structures when placed on Matrigel (Fig. 3A) and were able to incorporate 1,1'-dioctadecyl-3,3,3',3'-tetramethlyindocarbocyanine perchlorate-acetylated-low density lipoprotein (Dil-Ac-LDL) (Fig. 3B). hiPS-ECs and hES-ECs also migrated in response to VEGF (Fig. 3C). When compared to a hESC line, all EC lines tested produced significantly higher levels of nitric oxide, which is a fundamental determinant of vascular homeostasis (Fig. 3D).
Figure 2.
Morphology and protein expression of endothelial cells derived from pluripotent stem cells. Immunofluorescent staining for PECAM1 (A), VE-Cadherin (B), eNOS (C), and vWF (D) in ECs derived from pluripotent stem cells at day 21 of differentiation protocol. Control cells are human microvascular endothelial cells (HMVECs). Nuclei were visualized with DAPI (blue).
Figure 3.
Functional characterization of endothelial cells derived from pluripotent stem cells. (A): Bright-field microscopy of Matrigel tube formation assay showing in vitro angiogenesis potential. (B): Fluorescent microscopy of Dil-Ac-LDL uptake (red) and nuclei (blue) in endothelial cells derived from pluripotent stem cells. (C): Quantification of endothelial cell migration induced by VEGF. The graph shows fold migration, which refers to migration rate when stimulated with VEGF divided by migration rate without VEGF. All measurements were performed in triplicate and were not statistically different from one other using one-way ANOVA with Bonferroni's multiple comparison post hoc test. (D): Quantification of nitric oxide production. Cells were grown for 24 hr. in serum free media in the presence of 50ng/mL VEGF at which time nitrite concentration was measured in the conditioned media by HPLC. The nitrite concentration was adjusted for cell number by measuring the protein concentration of the cell lysate, resulting in arbitrary units of nitric oxide production. All measurements were performed using one-way ANOVA with Bonferroni's multiple comparison post hoc test (* P< 0.05, ** P< 0.01, *** P< 0.001). (E): Bright field microscopy of representative sections from in vivo Matrigel plug angiogenesis assay. Human PECAM1 staining is shown in brown. Blood cells are indicated (arrowheads). Scale bars represent 100 µm. Quantification of average capillary number from 6 fields of view per cell line is displayed ± standard deviation. Statistical significance was determined using one-way ANOVA with Bonferroni's multiple comparison post hoc test (* P< 0.05). No human PECAM1 positive cells or capillaries were observed in animals injected with matrigel only and this condition was excluded from subsequent graphing and statistical analysis.
An in vivo Matrigel plug assay was used to evaluate the angiogenic capabilities of CD31+/CD144+ pluripotent cell derived ECs. When ECs are mixed with Matrigel and injected subcutaneously in a mouse, they organize into functional capillaries that link with the endogenous circulatory system and carry red blood cells. All of the hiPS-ECs and hES-ECs were able to form functional blood capillaries in a similar fashion to primary ECs, as indicated by the robust staining for human CD31 and the presence of blood cells within some lumens (Fig. 3E).
Gene Expression Analysis of Pluripotent Cell-Derived Endothelial Cells
Global gene expression of the pluripotent cell–derived endothelial cells (ES/iPS) and primary human ECs was profiled by microarrays and 32 genes with the highest differential gene expression between various groups were subsequently analyzed by qRT-PCR (Figures S4, S5, S6, S7). Human aortic ECs (HAECs), human saphenous vein ECs (HSVECs), and human lymphatic ECs (HLECs) were chosen as representative primary cell lines of the three major EC types of the body: arterial, venous, and lymphatic. The primary ECs were grown in vitro in EC medium until passage 3, while still actively dividing, and the cells were collected for total RNA extraction. D14 ES- or iPS-derived ECs were thawed and matured until D21, at which point cell proliferation had decreased, as expected. All lines were collected for RNA at full confluence when the ECs were contact inhibited and displayed cobblestone morphology.
We compared three human ES–derived ECs (H1, H7, and H9 ECs) to three genetically independent human iPS–derived EC lines (iPS1, iPS2, and iPS3 ECs), as well as one of the same iPS cell line differentiated on two separate days (iPS2 Day X and iPS2 Day Y). In addition, we compared ES- and iPS-derived EC lines differentiated with two independent methods (H9 EC-A, H9 EC-B, iPS3 EC-A, and iPS3 EC-B). Method A represents the new differentiation method described here and Method B is an embryoid body + VEGF method.
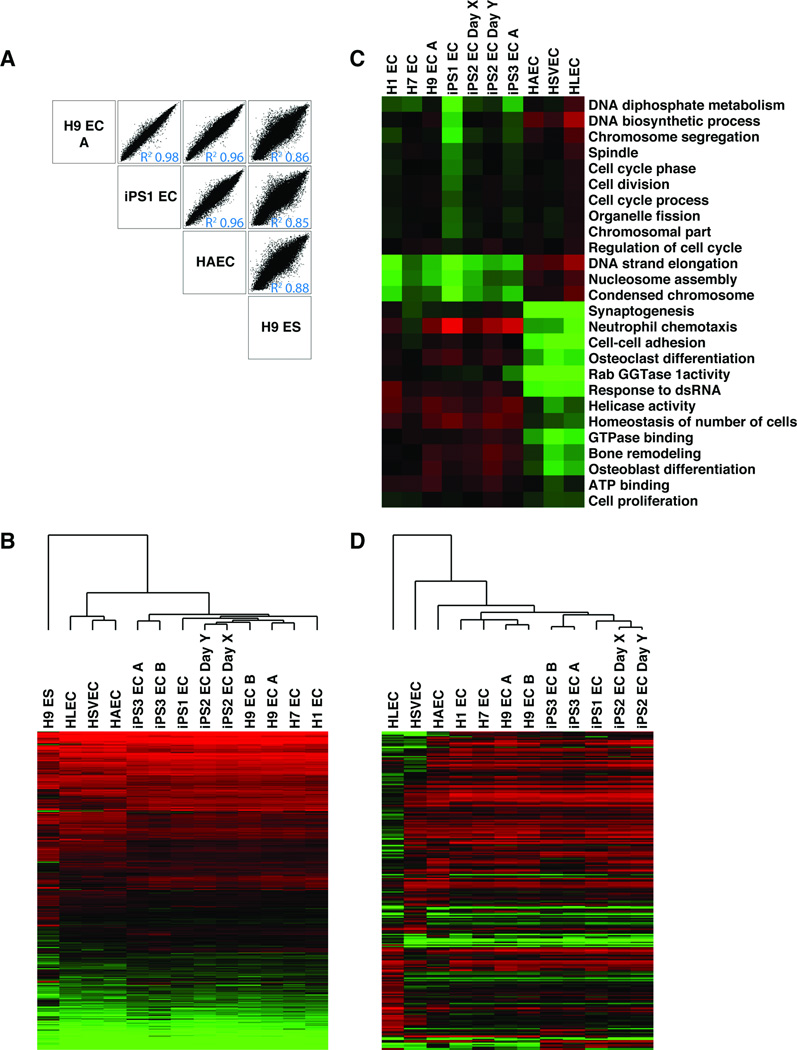
When comparing array log2 intensity values from all ESC– and iPSC–derived ECs pair wise, Pearson’s R2 values were >0.97 for all pairs (Fig. 4A and Fig S2A). Clustering analysis of all genes with twofold or greater difference in gene expression between any two samples clearly revealed a single cluster containing all three primary ECs and a second cluster that included all ES– or iPS–derived ECs (Fig. 4B). Genes differentially expressed between these two clusters included mainly those involved in cell cycle and adhesion, likely reflecting the nature of the 6 day primary culture vs. longer-term differentiation of the pluripotent stem cells (day 21).
Figure 4.
Global gene expression in endothelial cells derived from multiple pluripotent stem cell lines and primary endothelial cells. (A): Log2 intensity plots of representative samples from four groups of cells: ESC-ECs (H9 EC-A), iPS-ECs (iPS1-EC), Primary ECs (HAEC), and ESCs (H9 ES). (B): Heat map and clustering analysis of genes displaying at least two fold differential expression between any sample (15,740 genes). Scale extends from 2.5 to 13. Red indicates log2 intensity >7.0, and green <7.0. iPS2 EC Day X and iPS2 EC Day Y indicate the same iPSC line differentiated in two independent experiments. iPS3 EC A and iPS3 EC B refer to the same iPSC line differentiated by two different methods. (C): Heat map of GO analysis terms that differentiate pluripotent-cell derived ECs from primary ECs. Genes with threefold difference in expression between pluripotent cell derived ECs and primary ECs and P value < 0.05 were selected (968 genes) and GO Elite was used to group them by GO category with an adjusted P value < 0.05. Red indicates average log2 intensity of genes in each category that are higher than 8.75, and green lower than 8.75. (D): Heat Map and clustering analysis of genes with at least four fold differential expression between any two of the primary endothelial cell lines, not considering X and Y chromosome genes (240 genes). Red indicates log2 intensity >7.0 and Green <7.0. All Array data available from Gene Expression Omnibus (accession number GSE37631).
Importantly, when comparing ECs from the same pluripotent cell line differentiated in two separate experiments (iPS2 Day X ECs and iPS2 Day Y ECs) or ECs from the same pluripotent cell line differentiated with two separate methods (iPS3 EC-A:iPS3 EC-B or H9 EC-A:H9 EC-B), differences in gene expression were very small: Pearson’s R = 0.99 for all comparisons and only 7 protein coding genes were differentially regulated by more than 2 fold between the two different methods (Fig. S3A and Table S1). Expression of cell cycle genes were similar under all conditions and protocols.
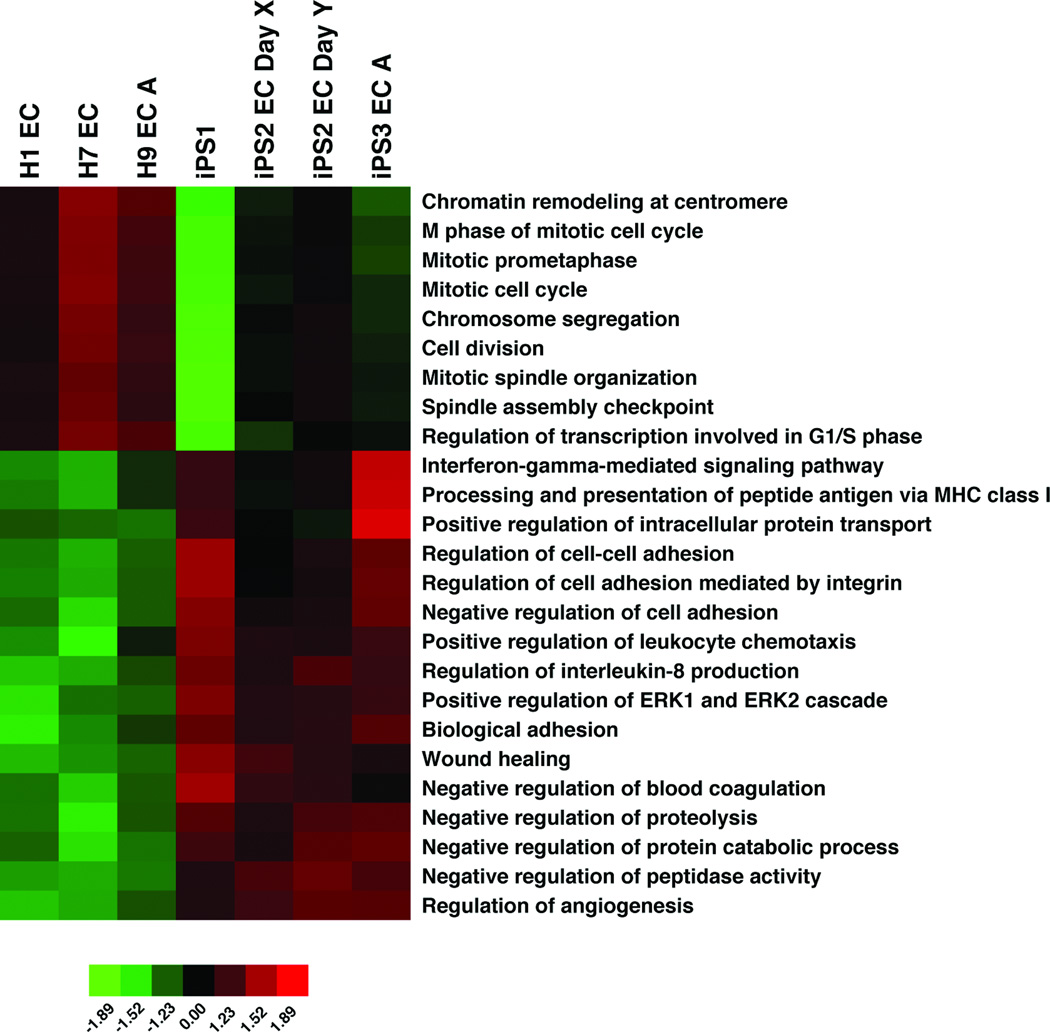
When comparing ES– and iPS–derived endothelial cells, only 17 protein coding genes were differentially expressed by more than threefold and 147 genes showed greater than twofold differences (P value ≤0.05) (Tables S2 and S3 and Fig. S4). Genes upregulated in iPS-ECs showed significant overlap with genes reported to be upregulated in iPS derived beating clusters or iPS cells in an undifferentiated state when compared to ES counterparts, whereas genes downregulated in iPS-ECs did not show overlap with genes downregulated specifically in iPS cells or iPS derived beating clusters compared to their ES counterparts (Table S4) [23–25]. When analyzed with GO-Elite, iPS-ECs had reduced expression of genes involved in cell division/cell cycle and higher expression levels of genes involved in cell adhesion, chemotaxis, and proteolysis compared to ES-ECs, potentially indicating a difference in growth and senescence (Fig. 5 and Table S5).
Figure 5.
Gene ontology differences between ES-ECs and iPS ECs. Heat map of GO analysis terms that differentiate iPS-ECs from ES-ECs. Genes with twofold difference in expression between iPS-ECs and ES-ECs and P value < 0.05 were selected (147 genes) and GO Elite was used to group them by GO category with an adjusted P value < 0.05.
To calculate the difference in gene expression variance across all genes between hES– and hiPS–derived ECs, the non-parametric Wilcoxon signed rank test was used. This test calculates and determines which group of ECs (hES– or hiPS–derived) has higher variance in gene expression for each individual gene on the microarray (28,439 genes). Then, each group is assigned a percentage corresponding to the number of individual genes across the entire microarray that are more variable. If hES or hiPS cells have a fundamental difference in their ability to generate endothelial cells or a certain type of endothelial cells, then one might expect global gene expression to be more highly variable between different lines in one of the types of pluripotent derived ECs. For example, if hiPS cells stochastically have incomplete epigenetic remodeling during reprogramming and thus more heterogeneity between pluripotent lines, one might expect more variation in expression when comparing each gene individually across the transcriptome. 54% of the genes showed higher variance in hiPS–ECs, while 46% were more variant in hESC–ECs, indicating iPS– and ES–derived ECs have similar variance in gene expression and implying a lack of fundamental difference between the ability of hES and iPS cells to generate ECs.
The limited gene expression variation between the ES– or iPS–derived ECs allowed us to group them together for further comparisons with primary culture of ECs. Pairwise comparisons between ES– or iPS–derived ECs and primary ECs resulted in a range of Pearson’s R2 values of 0.92–0.97. Grouping all of the pluripotent cells together and comparing them to primary ECs, we found 839 genes with at least threefold change up or down (P value ≤0.05) (Table S6). When analyzed with GO Elite, ES– and iPS–derived ECs had reduced expression of genes involved in cell division/cell cycle and higher expression levels of genes involved in integrin-mediated cell adhesion and chemotaxis compared to primary ECs, including CDK2, CDK6, FAT1, RGS5 AND PCDHB5 (Fig. 4C and Fig. S5). This difference likely reflects the fact that primary cells were analyzed after only 3 passages and 6 days of culture, while pluripotent cell-derived ECs were passaged 7–8 times and cultured for 21 days prior to analysis; both cell types are known to decrease their proliferative rate with prolonged culture
Limiting the comparisons to endothelial-specific genes [26], the ES– or iPS–derived ECs and primary cells all expressed high levels of KDR, PECAM1, CDH5 (VE-cadherin), LAMA4, MCAM, and THBS1. ES– or iPS–derived ECs expressed lower levels of vWF, PRKACA, and ROBO4 and higher levels of PLSCR4 when compared to primary ECs (Fig. S3B). qRT-PCR showed lower expression levels of Nitrtic Oxide Synthase 3 and Von Willebrand Factor and higher expression of VEGF receptor 2 in pluripotent derived ECs compared to primary ECs (Fig. S6). Using a set of known EC type–specific genes to perform cluster analysis, ES– and iPS–derived ECs lacked expression of canonical lymphatic specific genes PROX1, PDPN, and SOX18 (Fig. S3C). Cluster analysis indicated ES– and iPS–derived ECs were most similar to arterial cells in vitro. To perform an unbiased cluster analysis, the top 250 genes differentially expressed between the three primary endothelial cell types were identified and genes with X and Y chromosome locations were removed to control for sex of primary cell source. Then, gene expression analysis and clustering was extended to the ES– or iPS–derived ECs. This impartial clustering analysis also indicated ES– or iPS–derived ECs, by the method described here, are most similar to primary arterial cells in vitro (Fig. 4D, Fig. S7). In addition, FACS analysis showed that 86% of pluripotent cell-derived ECs were positive for expression of the arterial EC marker, EPHB2, although the levels of VEGF in the media could also induce EPHB2 expression in venous derived cells (Fig. S1C).
Discussion
This study describes a simple method for differentiating human pluripotent stem cells into ECs amenable to multiple pluripotent cell lines, which revealed limited gene expression variability among multiple human iPS- and ES-derived ECs. In this method, stem cells were guided through cell fate decisions mimicking their EC development in vivo, and the intermediate progenitors could be isolated, unlike previously described approaches. This advancement provides a platform for investigating novel aspects of human EC fate commitment, including chromatin remodeling and gene expression changes. Furthermore, the limited variance in gene expression of a defined human iPS-derived cell type supports the potential for successful detection of meaningful transcriptome alterations in iPS-based disease models.
Of the EC subtypes, ECs derived using this differentiation method most closely resembled arterial cells, as previously reported for in vitro derived ECs [27]. Future functional studies such as monocyte adhesion assays could further define the EC subtype. The medium used for the growth and expansion of the KDR+ precursors contains relatively high concentration of human-VEGF, which promotes the arterial fate by inducing NOTCH signaling [28]. Immature ECs show plasticity and can switch between arterial and venous fates [29]. Further maturation into fully differentiated arterial cells may require higher levels of VEGF, cAMP, or induction of NOTCH signaling by shear stress. Based on our knowledge of EC development in vivo, NOTCH signaling inhibition or retinoic acid treatment might induce ECs to venous or lymphatic fates, respectively [28, 30]. Isolation of significant numbers of human KDR+ endothelial precursors, uniquely enabled by the approach we describe, will permit interrogation of early EC-type fate determination through high-throughput screening or candidate approaches, taking cues from zebrafish and mouse development.
The gene expression of the pluripotent derived ECs resembles the primary derived ECs, though they express lower levels of NOS3 and vWF than the primary cells, which may indicate incomplete maturation. Importantly, the in vivo Matrigel plug angiogenesis assays indicate that the endothelial cells produced from ES or iPS derived sources are capable of organizing into functional capillaries that link with the endogenous circulatory system and carry red blood cells, though further studies are necessary to determine if the cells from this protocol can functionally rescue a disease model such as hind limb ischemia or myocardial infarction.
This study is the first comprehensive analysis of ECs derived from multiple hESC and hiPSC lines. The wide variation in percent KDRhigh/med cells derived from the same pluripotent cell line during separate differentiations is quite surprising and seems to be influenced by the state of pluripotent cells in combination with reagent variability, which is a major problem for directed differentiation protocols. This variation may impede interrogation of the role genetic mutations play in affected and unaffected iPS cell differentiation potential and requires the use of multiple cell lines from each group. It seems that the health and purity of the starting ES or iPS cell line on the first day of differentiation played a larger role in the success of the differentiation than the passage number of the pluripotent cell line, as some of the most successful differentiations were from ES cells with a passage number over 200. Fortunately, after purification using a cell surface marker, the resulting cell populations were seemingly more homogeneous, allowing for more accurate comparisons between different pluripotent cell lines. Our data indicate some significant disparity in gene expression between pluripotent cell–derived ECs and primary ECs. Many of these differences are related to cell cycle and adhesion. One possible explanation is that the primary cells were collected at passage 3 (day 6 of culture) to minimize in vitro gene expression changes and, thus, were in a more proliferative state. In contrast, the pluripotent cell–derived ECs were grown to day 21 (7–8 passages), and their proliferation rates had greatly decreased by that time, indicating a shift towards a senescent state, as expected. Senescent primary ECs at high passage numbers display higher levels of integrin β4 signaling, similar to our pluripotent cell–derived ECs, indicating a shared characteristic of senescence at high passage number between the two EC types [31]. The addition of the TGFβ-inhibitory molecule, SB431542, during EC differentiation seems to promote cell proliferation and inhibit senescence and could be a useful tool for expanding pure populations of ECs [11].
Our data indicate that despite gene expression heterogeneity between undifferentiated hESC and hiPSC lines as a whole [32], ECs derived from both pluripotent cell types are very similar. While there are some iPS-specific upregulated genes, most of the gene expression differences between ES-ECs and iPS-ECs were related to cell cycle and adhesion. These gene programs seem to be mutually exclusive in our data sets and most likely reflect differences between proliferation genes (high cell cycle, low adhesion) and senescence genes (low cell cycle, high adhesion) that would be expected in endothelial cells maintained in long-term culture. In addition, the high concordance of gene expression between ECs derived from the same pluripotent cell line via separate experiments or differentiation methods indicates that in a well-defined differentiated cell type, such as CD31+/CD144+ ECs, gene expression differences are most likely due to the inherent differentiation capacity of the originating cell line. These findings are relevant as we consider bioengineering vascularized tissues or creating vascular grafts for ischemic repair using hiPSCs. In addition, given the limited variability, ECs derived from patient-specific iPSCs should be useful in modeling diseases caused by defects in EC differentiation or maintenance, enabling the identification of molecular mechanisms underlying such ailments. Successful modeling of endothelial diseases with limited gene expression “noise” will enable drug-screening strategies using iPSC–derived ECs to identify novel therapeutics.
Supplementary Material
Figure S1. Extended analysis of EC differentiation. (A): Flow cytometric analysis of the effects of various BMP4 concentrations on KDR expression on day 6 of differentiation. (B): Flow cytometric analysis of the mature EC differentiation state on day 6 before KDR+ cell sorting using PECAM and VE-Cadherin. The control population consisted of cells labeled with isotype control antibodies. (C): Flow cytometric analysis for EPHB2 staining on PECAM / VE-Cadherin double positive cells on day 14. HAEC, HSVEC, and HLEC were used as positive and negative controls and H9 ECs were differentiated from ES cells.
Table S3. Differential Gene Expression iPS-ECs vs. ES-ECs. Twofold Cutoff (P<0.05) - 147 Protein Coding Genes.
Table S4. iPS-EC Specific Gene Signatures Compared to Three Other Reports of iPS Specific Gene Signatures (Gupta et al., Chin et al., Marchetto et al.). Comparison of Chin et al. done with late iPS vs ES cell data set. Green= iPS-EC Gene Expression Fold Change Matches Direction of Fold Change in Report Cited, Red cells= iPS-EC Gene Expression Fold Change is Opposite to Direction of Fold Change in Report Cited.
Table S5. Summary of Top 25 GO Categories for Genes Differentially Regulated by Twofold or Greater Between iPS-ECs and ES-ECs Including Gene Lists for Each GO Category.
Table S6. Differential Gene Expression pcdECs vs. Primary ECs. Threefold Cutoff (P<0.05) – 839 Protein Coding Genes
Figure S2. Control immunofluorescence staining. (A) Staining of positive control (HMVECs) and negative control (H1 ES) with PECAM1 and eNOS primary antibodies (green). The same alexa-488 secondary was used to visualize both primary antibodies. Negative control no primary wells were stained with secondary antibody only. (B) Staining for VE-Cadherin or vWF (red). The same alexa-594 secondary was used to visualize both primary antibodies. (C) Dil-Ac-LDL alexa 594 uptake (red) in positive control (HMVEC) or negative control (H1 ES) cells. Negative control cells received no Alexa 594 conjugated LDL. Nuclei in all panels visualized with DAPI (blue).
Figure S3. Extended microarray analysis related to Fig. 4. (A): Log2 intensity plots of all samples from four groups of cells; ESC ECs (H1, H7, H9, H9 cont.), iPSC ECs (iPS1, iPS2A, iPS2B, iPS3, iPS3 Cont.), primary ECs (HAEC, HSVEC, HLEC) and ESCs (H9 ES). Red points are between group comparisons. Values in lower left are Pearson’s R values of log transformed values. (B): Heat map and clustering analysis of genes specifically expressed in endothelial cells. Scale extends from 2.5 to 13. Red indicates log2 intensity higher than 7.5, and green lower than 7.5. (C): Heat map and clustering analysis of genes culled from the literature that are specifically expressed in different endothelial cell subtypes. Overlapping bars indicate genes expressed in two of the three primary cell lines. Scale extends from 2.5 to 13. Red indicates log2 intensity higher than 7.5, and green lower than 7.5.
Figure S4. Microarray data validation by qRT-PCR of genes differentially expressed between ES-ECs and iPS-ECs by at least 4 fold. Average gene expression from three biological replicates is plotted and error bars show standard deviation. Primary EC bar is average of HAECs, HSVECs and HLECs. RNA samples from H9 hES cells (ES), day 6 H9 differentiated cells (KDR−), or endothelial precursors (KDR+), are included for reference. Statistical significance was determined using one-way ANOVA with Bonferroni's multiple comparison post hoc test(* P< 0.05, ** P< 0.01, *** P< 0.001, **** P< 0.0001).
Figure S5. Microarray data validation by qRT-PCR of genes differentially expressed between primary-ECs and pluripotent derived-ECs (ES-ECs and iPS-ECs). Average gene expression from three biological replicates is plotted and error bars show standard deviation. RNA samples from H9 hES cells (ES), day 6 H9 differentiated cells (KDR−), or endothelial precursors (KDR+), are included for reference but were not included in the statistical analysis because of the null hypothesis being tested, i.e., that there is no difference in expression between primary ECs and pluripotent derived ECs. Statistical significance was determined using one-way ANOVA with Bonferroni's multiple comparison post hoc test(* P< 0.05, ** P< 0.01, **** P< 0.0001).
Figure S6. EC specific genes expressed in indicated cell types. Average gene expression from three biological replicates is plotted and error bars show standard deviation. Statistical significance was determined using one-way ANOVA with Bonferroni's multiple comparison post hoc test(* P< 0.05, ** P< 0.01, *** P< 0.001, **** P< 0.0001).
Figure S7. Microarray data validation by qRT-PCR of genes differentially expressed between (A) Arterial (B) Venous and (C) Lymphatic EC subtypes. Grey bars show the mean expression level from each of the primary EC types and error bars show technical standard deviation as a reference to compare the pluripotent derived ECs. Black bars show the average of three biological replicates of each EC type (ES or iPS derived) and error bars show standard deviation. RNA samples from H9 hES cells (ES), day 6 H9 differentiated cells (KDR−), or endothelial precursors (KDR+), are included for reference but were not included in the statistical analysis. ES-ECs and iPS ECs were compared with students t test and no genes showed statistical differences in expression between these two pairs.
Table S1. Differential Gene Expression Method A vs Method B. Twofold Cutoff (P<0.05)- 7 Protein Coding Genes.
Table S2. Differential Gene Expression iPS-ECs vs. ES-ECs. Threefold Cutoff (P<0.05) - 18 Protein Coding Genes.
Figure 6.
Model depicting the directed differentiation of human pluripotent stem cells into three mesodermally derived somatic cell types important for cardiovascular function.
ACKNOWLEDGEMENTS
We are grateful to the Srivastava lab and Gladstone community for useful discussions. We thank A. Holloway and A. Williams from the Gladstone Bioinformatics core, S. Elmes from UCSF Laboratory for Cell Analysis, and the Gladstone Genomics and Stem Cell Cores for technical expertise. We also thank Shinya Yamanaka and Kiichiro Tomoda for the kind gift of control iPS cell lines 1 and 2. Finally, we are grateful for editorial help from G. Howard and manuscript preparation by B. Taylor.
D.S. was supported by grants from NHLBI/NIH, L.K. Whittier Foundation, William Younger Family Foundation, Eugene Roddenberry Foundation, the California Institute for Regenerative Medicine (CIRM), and by NIH/NCRR grant (C06RR018928) to the Gladstone Institutes. M.W. was supported by a pre-doctoral fellowship from the American Heart Association. K.N.I. was supported by AHA Scientist Development Grant. J.P.C. and R.A.H. were supported by grants from the National Institutes of Health (RC2HL103400, 1U01HL100397, U01HL099775), the California Tobacco Related Disease Research Program of the University of California (18XT-0098), The Wallace H. Coulter Translational Research Grant Program, and the Stanford Cardiovascular Institute.
Footnotes
Author contributions: M.P.W.: conception and design, provision of study material or patients, collection and/or assembly of data, data analysis and interpretation, writing and critical revision of manuscript; R.A.J.: collection and/or assembly of data, data analysis and interpretation, manuscript writing and critical revision of manuscript; L.L.: collection and/or assembly of data: K.N.I.: conception and design, critical revision of manuscript; J.P.C.: financial support, critical revision of manuscript, experimental design; D.S.: conception and design, financial support, final approval of manuscript
Conflicts of Interest: D.S. is a member of the Scientific Advisory Board of iPierian Inc. and RegeneRx Pharmaceuticals.
REFERENCES
- 1.Robinton DA, Daley GQ. The promise of induced pluripotent stem cells in research and therapy. Nature. 2012;481:295–305. doi: 10.1038/nature10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leeper NJ, Hunter AL, Cooke JP. Stem Cell Therapy for Vascular Regeneration: Adult, Embryonic, and Induced Pluripotent Stem Cells. Circulation. 2010;122:517–526. doi: 10.1161/CIRCULATIONAHA.109.881441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cooke JP. Flow, NO, atherogenesis. Proc Natl Acad Sci USA. 2003;100:768–770. doi: 10.1073/pnas.0430082100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho SW, Moon SH, Lee SH, et al. Improvement of Postnatal Neovascularization by Human Embryonic Stem Cell-Derived Endothelial-Like Cell Transplantation in a Mouse Model of Hindlimb Ischemia. Circulation. 2007;116(21):2409–2419. doi: 10.1161/CIRCULATIONAHA.106.687038. [DOI] [PubMed] [Google Scholar]
- 5.Wang ZZ, Au P, Chen T, et al. Endothelial cells derived from human embryonic stem cells form durable blood vessels in vivo. Nat Biotechnol. 2007;25(3):317–318. doi: 10.1038/nbt1287. [DOI] [PubMed] [Google Scholar]
- 6.Rufaihah AJ, Huang NF, Jame S, et al. Endothelial Cells Derived From Human iPSCS Increase Capillary Density and Improve Perfusion in a Mouse Model of Peripheral Arterial Disease. Arterioscler Thromb Vasc Biol. 2011;31(11):e72–e79. doi: 10.1161/ATVBAHA.111.230938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Descamps B, Emanueli C. Vascular differentiation from embryonic stem cells: Novel technologies and therapeutic promises. Vascular Pharmacology. 2012;56(5–6):267–279. doi: 10.1016/j.vph.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 8.Nourse MB, Halpin DE, Scatena M, et al. VEGF induces differentiation of functional endothelium from human embryonic stem cells: implications for tissue engineering. Arterioscler Thromb Vasc Biol. 2010;30:80–89. doi: 10.1161/ATVBAHA.109.194233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kane NM, Xiao Q, Baker AH, et al. Pluripotent stem cell differentiation into vascular cells: A novel technology with promises for vascular re(generation) Pharmacology And Therapeutics. 2011;129:29–49. doi: 10.1016/j.pharmthera.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 10.Kane NM, Meloni M, Spencer HL, et al. Derivation of endothelial cells from human embryonic stem cells by directed differentiation: analysis of microRNA and angiogenesis in vitro and in vivo. Arterioscler Thromb Vasc Biol. 2010;30:1389–1397. doi: 10.1161/ATVBAHA.110.204800. [DOI] [PubMed] [Google Scholar]
- 11.James D, Nam H-S, Seandel M, et al. Expansion and maintenance of human embryonic stem cell-derived endothelial cells by TGFbeta inhibition is Id1 dependent. Nat Biotechnol. 2010;28:161–166. doi: 10.1038/nbt.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kattman SJ, Witty AD, Gagliardi M, et al. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 13.Li Z, Hu S, Ghosh Z, Han Z, Wu JC. Functional characterization and expression profiling of human induced pluripotent stem cell- and embryonic stem cell-derived endothelial cells. Stem Cells And Development. 2011;20:1701–1710. doi: 10.1089/scd.2010.0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tatsumi R, Suzuki Y, Sumi T, et al. Simple and highly efficient method for production of endothelial cells from human embryonic stem cells. Cell Transplant. 2011;20:1423–1430. doi: 10.3727/096368910X547444. [DOI] [PubMed] [Google Scholar]
- 15.Nakamura K, Salomonis N, Tomoda K, Yamanaka S, Conklin BR. G(i)-coupled GPCR signaling controls the formation and organization of human pluripotent colonies. Plos One. 2009;4:7780. doi: 10.1371/journal.pone.0007780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Byrne JA, Nguyen HN, Reijo Pera RA. Enhanced generation of induced pluripotent stem cells from a subpopulation of human fibroblasts. Plos One. 2009;4:7118. doi: 10.1371/journal.pone.0007118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kattman SJ, Huber TL, Keller GM. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell. 2006;11:723–732. doi: 10.1016/j.devcel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 18.Yang L, Soonpaa MH, Adler ED, et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- 19.Yamashita J, Itoh H, Hirashima M, et al. Flk1-positive cells derived from embryonic stem cells serve as vascular progenitors. Nature. 2000;408:92–96. doi: 10.1038/35040568. [DOI] [PubMed] [Google Scholar]
- 20.Olsson A-K, Dimberg A, Kreuger J, Claesson-Welsh L. VEGF receptor signalling - in control of vascular function. Nat Rev Mol Cell Biol. 2006;7:359–371. doi: 10.1038/nrm1911. [DOI] [PubMed] [Google Scholar]
- 21.Cross MJ, Claesson-Welsh L. FGF and VEGF function in angiogenesis: signalling pathways, biological responses and therapeutic inhibition. Trends Pharmacol Sci. 2001;22:201–207. doi: 10.1016/s0165-6147(00)01676-x. [DOI] [PubMed] [Google Scholar]
- 22.Grant DS, Kleinman HK, Martin GR. The role of basement membranes in vascular development. Ann N. Y. Acad Sci. 1990;588:61–72. doi: 10.1111/j.1749-6632.1990.tb13197.x. [DOI] [PubMed] [Google Scholar]
- 23.Gupta MK, Illich DJ, Gaarz A, et al. Global transcriptional profiles of beating clusters derived from human induced pluripotent stem cells and embryonic stem cells are highly similar. Bmc Dev Biol. 2010;10:98. doi: 10.1186/1471-213X-10-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chin MH, Mason MJ, Xie W, et al. Induced Pluripotent Stem Cells and Embryonic Stem Cells Are Distinguished by Gene Expression Signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchetto MCN, Yeo GW, Kainohana O, et al. Reh TA, editor. Transcriptional Signature and Memory Retention of Human-Induced Pluripotent Stem Cells. Plos One. 2009;4:e7076. doi: 10.1371/journal.pone.0007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho M, Yang E, Matcuk G, et al. Identification of endothelial cell genes by combined database mining and microarray analysis. Physiol Genomics. 2003;13:249–262. doi: 10.1152/physiolgenomics.00186.2002. [DOI] [PubMed] [Google Scholar]
- 27.Yamamizu K, Matsunaga T, Uosaki H, et al. Convergence of Notch and - catenin signaling induces arterial fate in vascular progenitors. The Journal Of Cell Biology. 2010;189:325–338. doi: 10.1083/jcb.200904114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lanner F, Sohl M, Farnebo F. Functional arterial and venous fate is determined by graded VEGF signaling and notch status during embryonic stem cell differentiation. Arterioscler Thromb Vasc Biol. 2007;27:487–493. doi: 10.1161/01.ATV.0000255990.91805.6d. [DOI] [PubMed] [Google Scholar]
- 29.Oliver G, Srinivasan RS. Endothelial cell plasticity: how to become and remain a lymphatic endothelial cell. Development. 2010;137:363–372. doi: 10.1242/dev.035360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marino D, Dabouras V, Brändli AW, Detmar M. A role for all-trans-retinoic acid in the early steps of lymphatic vasculature development. J Vasc Res. 2011;48:236–251. doi: 10.1159/000320620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun C, Liu X, Qi L, et al. Modulation of vascular endothelial cell senescence by integrin β4. J Cell Physiol. 2010;225:673–681. doi: 10.1002/jcp.22262. [DOI] [PubMed] [Google Scholar]
- 32.Bock C, Kiskinis E, Verstappen G, et al. Reference maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Extended analysis of EC differentiation. (A): Flow cytometric analysis of the effects of various BMP4 concentrations on KDR expression on day 6 of differentiation. (B): Flow cytometric analysis of the mature EC differentiation state on day 6 before KDR+ cell sorting using PECAM and VE-Cadherin. The control population consisted of cells labeled with isotype control antibodies. (C): Flow cytometric analysis for EPHB2 staining on PECAM / VE-Cadherin double positive cells on day 14. HAEC, HSVEC, and HLEC were used as positive and negative controls and H9 ECs were differentiated from ES cells.
Table S3. Differential Gene Expression iPS-ECs vs. ES-ECs. Twofold Cutoff (P<0.05) - 147 Protein Coding Genes.
Table S4. iPS-EC Specific Gene Signatures Compared to Three Other Reports of iPS Specific Gene Signatures (Gupta et al., Chin et al., Marchetto et al.). Comparison of Chin et al. done with late iPS vs ES cell data set. Green= iPS-EC Gene Expression Fold Change Matches Direction of Fold Change in Report Cited, Red cells= iPS-EC Gene Expression Fold Change is Opposite to Direction of Fold Change in Report Cited.
Table S5. Summary of Top 25 GO Categories for Genes Differentially Regulated by Twofold or Greater Between iPS-ECs and ES-ECs Including Gene Lists for Each GO Category.
Table S6. Differential Gene Expression pcdECs vs. Primary ECs. Threefold Cutoff (P<0.05) – 839 Protein Coding Genes
Figure S2. Control immunofluorescence staining. (A) Staining of positive control (HMVECs) and negative control (H1 ES) with PECAM1 and eNOS primary antibodies (green). The same alexa-488 secondary was used to visualize both primary antibodies. Negative control no primary wells were stained with secondary antibody only. (B) Staining for VE-Cadherin or vWF (red). The same alexa-594 secondary was used to visualize both primary antibodies. (C) Dil-Ac-LDL alexa 594 uptake (red) in positive control (HMVEC) or negative control (H1 ES) cells. Negative control cells received no Alexa 594 conjugated LDL. Nuclei in all panels visualized with DAPI (blue).
Figure S3. Extended microarray analysis related to Fig. 4. (A): Log2 intensity plots of all samples from four groups of cells; ESC ECs (H1, H7, H9, H9 cont.), iPSC ECs (iPS1, iPS2A, iPS2B, iPS3, iPS3 Cont.), primary ECs (HAEC, HSVEC, HLEC) and ESCs (H9 ES). Red points are between group comparisons. Values in lower left are Pearson’s R values of log transformed values. (B): Heat map and clustering analysis of genes specifically expressed in endothelial cells. Scale extends from 2.5 to 13. Red indicates log2 intensity higher than 7.5, and green lower than 7.5. (C): Heat map and clustering analysis of genes culled from the literature that are specifically expressed in different endothelial cell subtypes. Overlapping bars indicate genes expressed in two of the three primary cell lines. Scale extends from 2.5 to 13. Red indicates log2 intensity higher than 7.5, and green lower than 7.5.
Figure S4. Microarray data validation by qRT-PCR of genes differentially expressed between ES-ECs and iPS-ECs by at least 4 fold. Average gene expression from three biological replicates is plotted and error bars show standard deviation. Primary EC bar is average of HAECs, HSVECs and HLECs. RNA samples from H9 hES cells (ES), day 6 H9 differentiated cells (KDR−), or endothelial precursors (KDR+), are included for reference. Statistical significance was determined using one-way ANOVA with Bonferroni's multiple comparison post hoc test(* P< 0.05, ** P< 0.01, *** P< 0.001, **** P< 0.0001).
Figure S5. Microarray data validation by qRT-PCR of genes differentially expressed between primary-ECs and pluripotent derived-ECs (ES-ECs and iPS-ECs). Average gene expression from three biological replicates is plotted and error bars show standard deviation. RNA samples from H9 hES cells (ES), day 6 H9 differentiated cells (KDR−), or endothelial precursors (KDR+), are included for reference but were not included in the statistical analysis because of the null hypothesis being tested, i.e., that there is no difference in expression between primary ECs and pluripotent derived ECs. Statistical significance was determined using one-way ANOVA with Bonferroni's multiple comparison post hoc test(* P< 0.05, ** P< 0.01, **** P< 0.0001).
Figure S6. EC specific genes expressed in indicated cell types. Average gene expression from three biological replicates is plotted and error bars show standard deviation. Statistical significance was determined using one-way ANOVA with Bonferroni's multiple comparison post hoc test(* P< 0.05, ** P< 0.01, *** P< 0.001, **** P< 0.0001).
Figure S7. Microarray data validation by qRT-PCR of genes differentially expressed between (A) Arterial (B) Venous and (C) Lymphatic EC subtypes. Grey bars show the mean expression level from each of the primary EC types and error bars show technical standard deviation as a reference to compare the pluripotent derived ECs. Black bars show the average of three biological replicates of each EC type (ES or iPS derived) and error bars show standard deviation. RNA samples from H9 hES cells (ES), day 6 H9 differentiated cells (KDR−), or endothelial precursors (KDR+), are included for reference but were not included in the statistical analysis. ES-ECs and iPS ECs were compared with students t test and no genes showed statistical differences in expression between these two pairs.
Table S1. Differential Gene Expression Method A vs Method B. Twofold Cutoff (P<0.05)- 7 Protein Coding Genes.
Table S2. Differential Gene Expression iPS-ECs vs. ES-ECs. Threefold Cutoff (P<0.05) - 18 Protein Coding Genes.