Abstract
Fragile-X Syndrome (FXS) is a multi-organ disease leading to mental retardation, macro-orchidism in males, and premature ovarian insufficiency in female carriers. FXS is also a prominent monogenic disease associated with autism spectrum disorders (ASD). FXS is typically caused by the loss of FRAGILE X-MENTAL RETARDATION 1 (FMR1) expression, which encodes for the RNA-binding protein (RBP), FMRP. We report the discovery of distinct RNA recognition elements (RREs) that correspond to the two independent RNA binding domains of FMRP, and the binding sites within the mRNA targets for wild-type and I304N mutant FMRP isoforms and its paralogs, FXR1 and FXR2. RRE frequency, ratio, and distribution determine target mRNA association with FMRP. Among highly-enriched targets, we identified many genes involved in ASD and demonstrate that FMRP affects their protein levels in cell culture, mice, and human brain. Unexpectedly, we discovered that these targets are also dysregulated in Fmr1-/- mouse ovaries, showing signs of premature follicular overdevelopment. These results indicate that FMRP targets shared signaling pathways across different cellular contexts. As it is become increasingly appreciated that signaling pathways are important to FXS and ASD, our results here provide a molecular guide towards the pursuit of novel therapeutic targets for these neurological disorders.
INTRODUCTION
Most clinical cases of FXS are a result of a hyper-expansion and methylation of CGG repeats within the promoter of FMR1, leading to a loss of its expression1-3. The FMR1 RBP family has three members, FMR1, FXR1, and FXR2, which possess two centrally located KH domains and a C-terminal arginineglycine (RG)-rich region implicated in mRNA binding 4-7. FMR1 encodes for multiple protein isoforms, but is predominantly expressed as a 69 kDa protein (isoform 7)8,9. Isoform (iso) 1 and six other alternative splice variants include exon 12, with iso1 encoding the full-length protein (71 kDa). Exon 12 insertion lengthens the second KH (KH2) RBD, possibly influencing FMR1 RNA-binding specificity or affinity. The I304N mutation, described in a FXS patient, is also located in the KH2 and is reported to attenuate association with RNA and polysomes10-12. FMR1 proteins are implicated in various RNA processes including RNA subcellular localization by facilitating nucleo-cytoplasmic shuttling13 and association with motor proteins14-16. FMR1 proteins were also suggested to mediate translational regulation12,17. Given the critical role of FMR1 in human cognition and premature ovarian insufficiency18,19, intensive efforts towards the identification of its RNA targets have been employed, with the goal that their discovery would shed light upon the array of related disorders and provide options for molecular therapy18-26. No precise RRE has been defined and very few bona fide mRNA targets are confirmed27.
RESULTS AND DISCUSSION
To identify the binding sites of FMR1 family proteins (Fig. 1a and Supplementary Fig. 1), we first compared photocrosslinking methods28-30 using stable FLAG-HA FMR1 iso7 expressing HEK293 cells (Fig. 1b), as these cells and human brain share 90% of expressed genes based on a comparison of existing RNAseq datasets31-33 (Supplementary Fig. 2). The difference in FMR1 levels between the experimental system and brain is 1.3 fold, as calculated using RNAseq data and the quantitated expression of FMR1 in our stable cells. We found that 4SU PAR-CLIP provided the highest yield of crosslinked RNAs, and used this approach for all FMR1 family proteins (Fig. 1c). cDNA libraries were generated after PAR-CLIP and Illumina-sequenced (Supplementary Table 1). Genome-aligned reads were grouped by PARalyzer34 to identify segments of RNA that represented peaks of T-to-C conversion, termed binding sites. PARalyzer separated closely spaced binding sites connected by overlapping reads and yielded a median RNA segment length of 33 nt (Supplementary Fig. 3). FMRP iso1 and 7 bound to approx. 80,000-100,000 sites, of which > 85% mapped to ~6,000 mRNAs (Supplementary Tables 1, 2, and https://fmrp.rockefeller.edu). FXR1 and FXR2 protein binding sites are comprised within FMRP binding sites with an overlap of 95% (Supplementary Table 3).
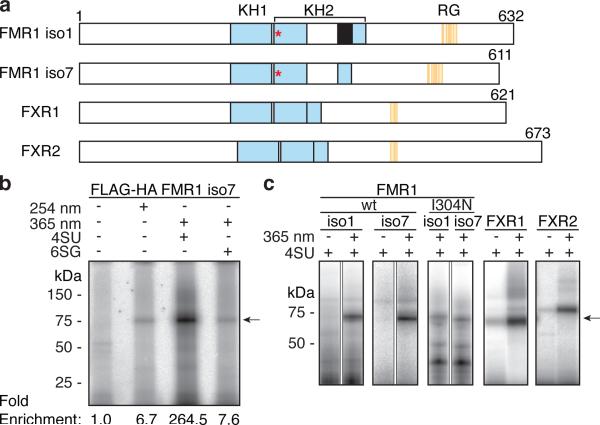
Figure 1. PAR-CLIP of FMR1 family proteins.
a, FMR1 family proteins comprise two type-I KH domains (cyan). FMRP isoform 1 and 7 (iso1 and 7) vary by the presence of exon 12 (black) within KH2. The I304N mutation is located within the KH2 domain (red asterisk). The RG-rich region (orange bars) is also implicated in RNA-binding. The lengths of proteins in amino acids are indicated. We established stable inducible cell lines expressing FLAG-HA epitope-tagged wt and I304N mutants of FMR1 (iso1 and 7), and its homologs FXR1 and 247. b, RNA-FMRP crosslinking comparing CLIP (254 nm) to 4SU-or 6SG-PAR-CLIP (365 nm). RNA-radiolabeled FLAG immunoprecipitates (IPs) of lysates from crosslinked FLAG-HA-FMRP-iso7-expressing HEK293 cells were separated by SDS-PAGE. The migrations of protein mass standards are indicated. Enrichment of radiolabelled RNA covalently bound to FLAG-HA-FMRP (arrow) was determined after normalizing by Western blot analysis (not shown). c, 4SU-PAR-CLIP of FMR1 family proteins analogous to (b).
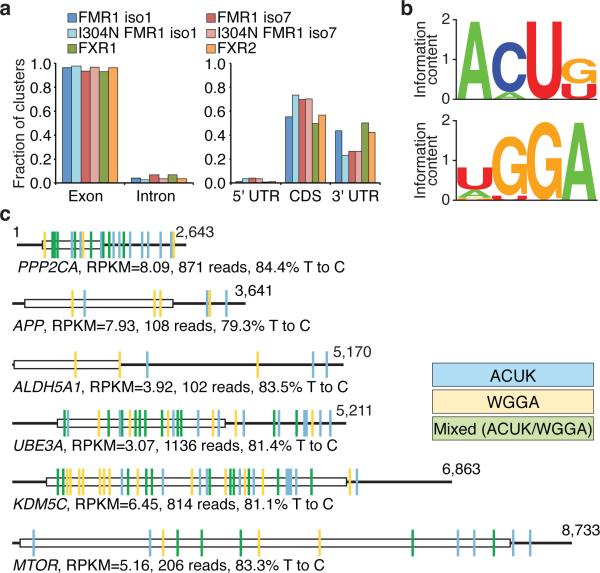
Nearly all mRNA binding sites were located in exons (>90%) (Fig. 2a) and distributed between CDS and 3'UTR (>95%, total) with slightly more CDS sites, similar to distributions seen for other cytoplasmic RBPs28. The computational sequence analysis method cERMIT35 revealed two major RREs, ACUK and WGGA (K=G/U, and W=A/U) (Fig 2b, Supplementary Fig. 4). Together, ACUK and WGGA RREs were found in ≥50% of mRNA binding sites in iso1 and iso7; they occurred exclusively or together within the same binding site (Fig. 2c). Remaining binding sites typically contained close derivatives of either RRE.
Figure 2. Analysis of FMR1 family protein mRNA binding sites.
a, Distribution of binding sites within mRNA targets of the FMR1 protein family. b, Two major RREs were inferred from FMRP iso1 and iso7 binding sites. c, Distribution of FMRP binding sites, color-coded based on cERMIT-inferred RREs, across representative targets. Open boxes and thick lines indicate CDS and UTRs, respectively; numbers indicate nucleotide number.
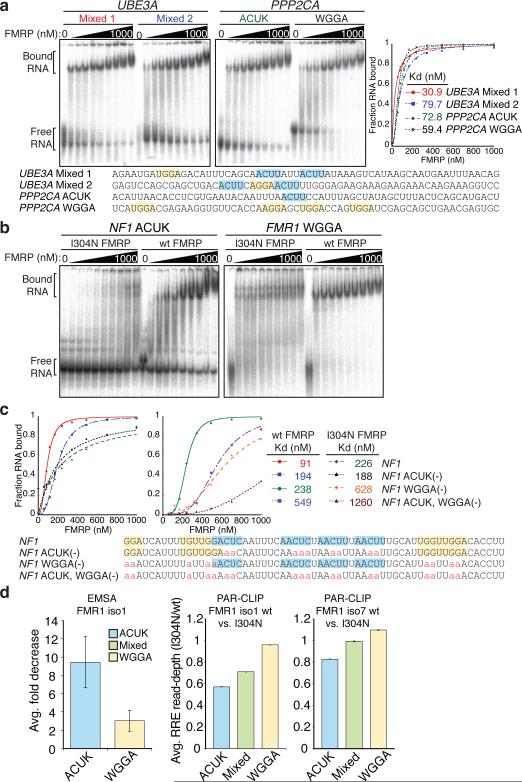
We performed electrophoretic mobility shift assays (EMSAs) to test RNAs representing FMRP binding sites using recombinant FMRP purified from Sf9 cells (Supplementary Fig. 5 and 6). FMRP target sites were selected based on whether they contained ACUK, WGGA or Mixed (ACUK/WGGA) RREs (Supplementary Table 4). Testing RNAs of various lengths, we found that oligonucleotide lengths of ≥45 nt were required to observe gel shifts and reach dissociation constants below 0.5 μM, suggesting RNA backbone contacts outside the RREs contribute to the association in vitro. WGGA-containing RNAs exhibited the widest range and strongest affinities, generally correlating with the number of RREs within a PARalyzer-defined binding site. An RNA segment containing nine WGGAs bound almost 2 orders of magnitude tighter than those containing one WGGA, whereas binding of ACUK-containing RNAs varied only 5-fold. EMSAs using RNAs representing target sites within PPP2CA, and UBE3A are shown (Fig. 3a).
Figure 3. RNA binding assays using natural FMRP target sites containing ACUK and WGGA RREs, and the effect of a KH2 mutation to its target RNA spectrum.
a, EMSAs of RNAs representing UBE3A or PPP2CA binding sites containing various RREs. Binding curves and constants are shown. The sequences of the RNAs are provided with WGGA (yellow) and ACUK RREs (cyan) highlighted. b, Impact of KH2 mutation in FMRP on target sites containing ACUK versus WGGA RREs. The RNA affinities of wt and I304N FMRP iso1 were compared using binding sites in NF1 (ACUK) and FMR1 (WGGA). c, Binding curves of wt and I304N FMRP for an RNA segment representing a mixed RRE binding site in NF1, and several mutant sequence versions (ACUK (-), WGGA (-), and ACUK, WGGA (-)). d, Comparison of FMRP iso1 affinity for RRE type in EMSAs and FMRP iso1 and 7 wt and I304N PAR-CLIP libraries. Error bars in EMSA summary represent s.d., n= 9 (ACUK), or 8 (WGGA). The ratio of sequence reads aligned to each RRE binding site was calculated between wt and I304N FMRP PAR-CLIP libraries. The average sequence-depth ratio of wt over I304N binding site, per RRE-type, are shown. Error bars in the read-depth analyses represent the avg. min and max values across all subsampled mutant libraries (n= 14 and 26 for iso 1 and 7, respectively).
We also tested sites in APP and FMR1 mRNA (Supplementary Fig. 7), two previously identified mRNA targets. APP was originally discovered as an FMRP target based on a predicted G-quartet region7 although the actual site was subsequently identified in vitro at an upstream segment36, which was identified here (APP Site 1) as containing WGGA. FMRP targets its own mRNA though it was an association only observed in vitro5,6,37 and in immunoprecipitates38.
Recombinant I304N iso1 FMRP showed a ~10-fold average decrease in its affinity towards ACUK-containing RNAs compared to wt FMRP iso1 (Fig. 3b and Supplementary Table 5). We characterized binding to wt and mutant RNA sequences derived from an NF1 mixed RRE site (Fig. 3c). I304N FMRP bound wt and NF1 ACUK(-) RNAs similarly, whereas wt FMRP showed a two-fold reduction in affinity for NF1 ACUK(-) RNA. Additionally, wt FMRP bound NF1 ACUK, WGGA(-) RNA similar to I304N FMRP for NF1 WGGA(-) RNA. Our results indicate that the KH2 domain associates with the ACUK RRE. Since FMRP associates with mRNAs at more than one binding site, its association at multiple sites will impact the final regulatory effect. We compared the distribution of RREs in I304N FMRP vs. wt FMRP PAR-CLIPs and found a transcriptome-wide depletion in the recovery of sequence reads for ACUK binding sites for both I304N isoforms, consistent with the biochemistry (Fig. 3d). Interestingly, the RRE fractional distribution of I304N FMRP iso1 was similar to wt FMRP iso7, suggesting that alteration of the KH2 domain by mutation or exon insertion affects binding-site selectivity. Taken together, the biochemical and PAR-CLIP results with I304N FMRP indicate that we identified the natural target sites of the FMRP KH2 domain.
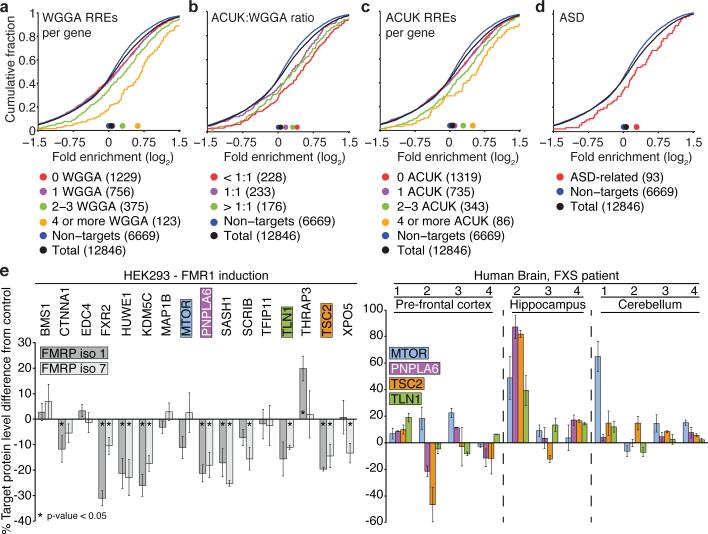
To rank FMRP targets, we measured their enrichment in RIP-chip, which would indicate stable interactions. 3,593 PAR-CLIP-identified targets showed enrichment by RIP-chip, of which 939 genes were two- to six-fold enriched; 646 transcripts were two-fold enriched but not identified as PAR-CLIP targets. We used binding-site information obtained by PAR-CLIP to infer the salient features for stable association in RIP-Chip (Fig. 4, Supplementary Fig. 8, and Supplementary Table 6). Increasing frequency of WGGA- and ACUK-containing elements led to greater RIP-chip enrichment, in agreement with in vitro affinity measurements. On average, top targets contained more RRE binding sites (18 per transcript) compared to the least-enriched targets (13 per transcript). Top-ranked targets had a CDS:3'UTR binding site ratio of 3.7:1 compared to bottom-ranked targets that had 1:2. Transcripts with ACUK:WGGA RRE ratios <1 were the most significantly enriched population. Importantly, we identified 93 genes independently implicated in ASD among the highly-enriched FMRP targets (Fig. 4d and Supplementary Table 7), which is striking given the relationship of FXS with ASD20,21,39.
Figure 4. RRE-dependent enrichment criteria for FMRP association with mRNAs.
a, RIP-chip experiments were performed using FLAG-HA-FMRP iso1. a-d, Cumulative distribution fraction plots of FMRP targets based on indicated criteria. Transcripts were grouped and color-coded based on indicated bins. Non-targets are mRNA transcripts with zero PAR-CLIP binding sites, although detectable in the array; total is the sum of non-targets and PAR-CLIP identified targets detectable by RIP-chip. d, Enrichment of ninety-three PAR-CLIP identified ASD-related target genes. e, Immunoblot densitometry analysis of top-ranking FMRP targets from RIP-chip and PAR-CLIP analyses in HEK293 and human brain. In cell culture, target protein expression differences of indicated proteins were determined upon induction of FMR1 iso1 or 7 expression. Similarly, relative protein expression was measured using lysates prepared from indicated brain regions of four FXS patients, compared to age/sex/anatomic-matched controls. Error bars represent s.e.m., with n = 2-11 (depending on protein measured and whether the sample was HEK293 or brain lysate). PABPC1 protein level served as loading and ratio control as it was a gene with PAR-CLIP binding sites but showed no RIP-chip enrichment (-0.10 LFE).
Enrichment of RNAs in RIP-chip depends on the saturation of target sites with FMRP. To estimate the yield of saturation, protein copy number, target mRNA copies, and the number of binding sites within those transcripts have to be accounted. We determined endogenous and co-expressed FLAG-HA-tagged FMR1 protein copy numbers to be approx. 60,000 and 70,000 molecules per cell, respectively, using quantitative Western blotting and reference recombinant protein. Each cell contains 20 pg total RNA, of which 4% are the approx. 1 Mio mRNA molecules/cell. Considering their relative abundance based on HEK293 RNAseq, we estimate that an equal distribution of FMRP would occupy 6% of binding sites/cell among its 6,000 target transcripts, or up to 20% if FXR1 and 2 protein estimates are included. Since PAR-CLIP-identified targets had varying enrichment, the association of FMRP with them is not uniform. The ~3,500 transcripts enriched in RIP-chip are estimated to have at least 18% FMRP binding site occupancy, whereas top-ranked 900 genes (two-fold enriched) potentially exhibit 78%. The presence of multiple binding sites within targets suggests that multiple FMR1 family proteins bind each transcript to influence their regulation.
To assess the impact of FMR1 binding sites on mRNA stability, siRNA knockdown of FMR1 or the FMR1 family was performed and mRNA expression profiles were analysed by microarray. We found no evidence for FMR1 affecting target mRNA abundance (data not shown).
A panel of FMR1-targets were selected based on enrichment in RIP-Chip, low-to-intermediate expression in RNAseq, similar abundance in human brain (using published microarray31 and RNAseq datasets32,33) and with documented neurological and human disease relevance, then analysed them by quantitative Western blot to determine their protein levels as a function of FMRP expression (Fig. 4e). FXR2, HUWE1, KDM5C, and MTOR protein levels, among others, showed up to 30% reduction in protein levels upon expression of FMR1, in HEK293. We analysed lysates prepared from human postmortem brains. Four FXS brains (Supplementary Fig. 9) were available with age/sex/anatomic-matched controls from pre-frontal cortical, hippocampus, and cerebellar regions. While only four of eight antibodies yielded quantifiable bands in brain lysates, we observed a general trend of elevated target protein expression levels in FXS brains. This is the inverse of FMR1 overexpression effects in HEK293, and consistent with FMRP affecting the protein levels of its mRNA targets.
The mRNA targets identified here are from a human transcriptome where the vast majority of genes are comparably expressed in human brain (Supplementary Fig. 2). We discovered ASD-related and numerous other genes implicated in neuronal disorders associated with FXS and validated representatives by EMSA, RIP-chip, and immunoblot. We found genes involved in Angelman (AS), Prader-Willi, Rett, and Cornelia de Lange Syndromes. Interestingly, the ASD and AS-associated gene UBE3A ubiquitinates ARC and SACS140; ARC is a well-known target and here we identify SACS1 as a targeted transcript. These findings potentially provide the molecular link to tie together elements of clinically overlapping disorders, principally setting a molecular target framework for characterizing the connections between FXS and its associated phenotypes.
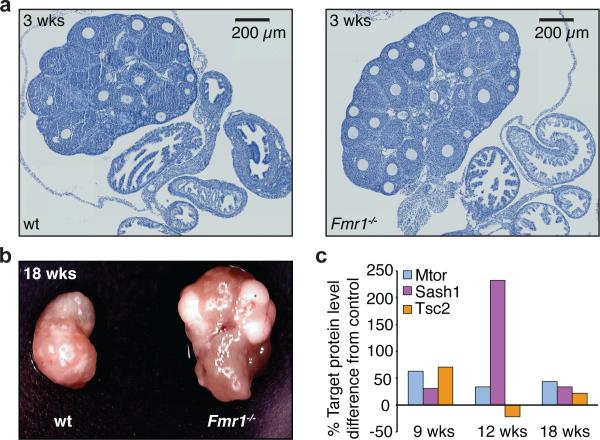
Although FX-related diseases are primarily considered CNS disorders, at least two other target organs are affected, the testes and ovaries. We reasoned that changes in FMR1 expression lead to dysregulation of largely overlapping sets of targets shared across all affected organs. Thus dysregulated genes and pathways in brain might also contribute to phenotypes in testes and ovary. We therefore examined the ovaries of Fmr1-/- mice41 since CNS and macroorchidism phenotypes were reported, yet ovary development had largely been under-investigated. Ovaries from Fmr1-/- mice were markedly larger by 3 weeks post-birth compared to wt controls (Fig. 5a-b). At 12 and 18 wks post-birth, KO ovaries were 22% and 72% larger by mass compared to age-matched controls, respectively. Importantly, we found increased protein levels of Tsc2, Sash1 and Mtor (Fig. 5c). As it is independently known that the Mtor pathway can regulate ovarian development, it is tempting to conclude that increased activity, in the absence Fmr1 expression, contributes to enlarged ovaries histologically consistent with precocious follicular development. These observations suggest that Fmr1-KO mice have the potential to model FXS-related premature ovarian insufficiency in which it remains unclear whether elevated FMR1 mRNA or the observed reduction of FMRP protein itself is causative in female carriers affected by this disease.
Figure 5. Ovarian phenotype in Fmr1-/- mice.
Ovaries from wt and Fmr1-/- female mice were harvested at 3, 9, 12, and 18 wks and processed for histological (a), morphological (b), and quantitative western analyses (c). a, By 3 wks of age, histological staining (hematoxylin) of sectioned ovaries show greater than expected number of follicles compared to wt. b, Ovaries from 18 wk old Fmr1-/- mice are larger than wt and exhibit prominent cysts consistent with corpus luteal development. c, Lysates were prepared from 9, 12, and 18 wk ovaries from two different wt and KO mice each, and analysed by quantitative Western using Mtor, Sash1, and Tsc2 antibodies. As in human samples, Pabpc1 was used for normalization.
Elevated signal transduction activity of the Mtor pathway42,43 was reported in Fmr1 KO-mice, and was attributed to Fmrp targeting of Pik3ca mRNA. Indeed, PAR-CLIP identified several FMRP binding sites within the PIK3CA transcript. However, we find that it is a less-enriched RIP-chip target compared to MTOR and TSC2, whose protein levels appear regulated in an FMRP-dependent manner. Interestingly, recent evidence demonstrated that Tsc mutant mice44-46, which have increased mTOR activity, had impaired mGluR-LTD and protein synthesis compared to Fmr1-/- mice; crossing Tsc2+/- with Fmr1-/y mice corrected the phenotypes44. Given our results we suspect that in Tsc2+/-, Fmr1-/y mice, Tsc2 and Mtor protein levels (among others) were elevated, correcting the balance of protein expression and leading to the reversal of phenotypes observed. The reported model44 by which separate pools of mRNA are differentially regulated by partially convergent pathways in FMRP (in)dependent ways, remains unclear. This is in part because FMRP associates with transcripts of ERK pathway components as well. Therapeutic targeting of the MTOR pathway has become an important goal – but must be further guided by additional functional analysis, particularly of FMRP targets upstream and downstream of MTOR and interconnected signaling pathways (Supplementary Fig. 10). Combined, our validation work in Fmr1-KO mouse ovaries and in human brain demonstrate that the effect of FMRP binding to specific target genes identified in cell culture is extensible to physiologically relevant contexts.
METHODS SUMMARY
Methods are described in greater detail within Supplementary Information. FVB129P2 Fragile-X mice were a kind gift from Dr. Suzanne Zukin (Albert Einstein College of Medicine). Gateway plasmids (Invitrogen) generated in this study will be deposited in addgene.org. FlpIn T-Rex HEK293 (Invitrogen) inducible-stable cell lines were generated per manufacturer's instrutions. The titers, source and use of antibodies used in this study are listed in Supplementary Information. PAR-CLIP was performed essentially as described, except that the second RNase T1 digestion was omitted following the IP. Recombinant wt and mutant FMRP iso1 proteins were purified using baculoviral expression system (Invitrogen). Electrophoretic mobility shift assays and Western blots were quantified using ImageGauge (Fuji). Parameters of computation analyses are described in Supplementary Information and within the relevant sections within https://fmrp.rockefeller.edu/. Relevant datasets, including raw data are available at https://fmrp.rockefeller.edu/ and GEO (GSE39686).
Supplementary Material
ACKNOWLEDGEMENTS
Human tissue was obtained from the NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland, Baltimore, MD. We would like to thank the following members of the Tuschl lab for their support and assistance: Greg Wardle, Dr. Neil Renwick, and Dr. Iddo Ben-Dov. We would like to thank Dr. Jack Keene for his invaluable advice throughout the project. We would like to acknowledge Dr. Mohsen Khorshid, Dr. Lukas Burger, and Dr. Mihaela Zavolan for analyzing PAR-CLIP data at the initial stages of the project and discussions. We would like to thank the MSKCC in-situ core for their assistance with the mouse histology. Finally, we would like to express our appreciation for all members of the Tuschl laboratory for their assistance and collegiality. This work was supported, in part, by the following agencies: NIH/NCRR/RU CCTS (M.A., UL1RR024143), Simons Foundation Autism Research Initiative (T.T., CEN5300891) and NIH (T.T., R01 MH080442).
Footnotes
CONTRIBUTIONS
M.A designed, executed, supervised, and interpreted experiments. N.M., P.B., and D.L.C. carried out the sequence alignment, annotation, and PARalyzer pipeline. N.M. and P.B. performed the computational analysis on the RIP-chip. M.A., J.B.M., and J.N. purified FMRP proteins, performed the EMSAs, and carried out the quantitative Westerns and analyses. M.A. and M.M. performed the RIP-chips. S.D. assisted in the Illumina sequencing of all PAR-CLIP libraries. M.H. helped in the initial PAR-CLIP experiments. C.L. and Z.W. carried out and analyzed mouse experiments. U.O. supervised computational efforts. T.T. supervised and helped in the design of experiments. M.A. and T.T. wrote the manuscript.
COMPETING FINANCIAL INTEREST
T.T. is co-founder and scientific advisor to Alnylam Pharmaceuticals and Regulus Therapeutics.
REFERENCES CITED
- 1.Verkerk AJ, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 2.Kremer EJ, et al. Mapping of DNA instability at the fragile X to a trinucleotide repeat sequence p(CCG)n. Science (New York, N.Y. 1991;252:1711–1714. doi: 10.1126/science.1675488. [DOI] [PubMed] [Google Scholar]
- 3.Vincent A, et al. Abnormal pattern detected in fragile-X patients by pulsed-field gel electrophoresis. Nature. 1991;349:624–626. doi: 10.1038/349624a0. [DOI] [PubMed] [Google Scholar]
- 4.Siomi H, Siomi MC, Nussbaum RL, Dreyfuss G. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell. 1993;74:291–298. doi: 10.1016/0092-8674(93)90420-u. [DOI] [PubMed] [Google Scholar]
- 5.Ashley CTJ, Wilkinson KD, Reines D, Warren ST. FMR1 protein: conserved RNP family domains and selective RNA binding. Science (New York, N.Y. 1993;262:563–566. doi: 10.1126/science.7692601. [DOI] [PubMed] [Google Scholar]
- 6.Brown V, et al. Purified recombinant Fmrp exhibits selective RNA binding as an intrinsic property of the fragile X mental retardation protein. J. Biol. Chem. 1998;273:15521–15527. doi: 10.1074/jbc.273.25.15521. [DOI] [PubMed] [Google Scholar]
- 7.Darnell JC, et al. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 8.Ashley CT, et al. Human and murine FMR-1: alternative splicing and translational initiation downstream of the CGG-repeat. Nat. Genet. 1993;4:244–251. doi: 10.1038/ng0793-244. [DOI] [PubMed] [Google Scholar]
- 9.Verkerk AJ, et al. Alternative splicing in the fragile X gene FMR1. Hum. Mol. Genet. 1993;2:399–404. doi: 10.1093/hmg/2.4.399. [DOI] [PubMed] [Google Scholar]
- 10.De Boulle K, et al. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat. Genet. 1993;3:31–35. doi: 10.1038/ng0193-31. [DOI] [PubMed] [Google Scholar]
- 11.Siomi H, Choi M, Siomi MC, Nussbaum RL, Dreyfuss G. Essential role for KH domains in RNA binding: impaired RNA binding by a mutation in the KH domain of FMR1 that causes fragile X syndrome. Cell. 1994;77:33–39. doi: 10.1016/0092-8674(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 12.Feng Y, et al. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol. Cell. 1997;1:109–118. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- 13.Fridell RA, Benson RE, Hua J, Bogerd HP, Cullen BR. A nuclear role for the Fragile X mental retardation protein. EMBO J. 1996;15:5408–5414. [PMC free article] [PubMed] [Google Scholar]
- 14.Ling SC, Fahrner PS, Greenough WT, Gelfand VI. Transport of Drosophila fragile X mental retardation protein-containing ribonucleoprotein granules by kinesin-1 and cytoplasmic dynein. Proc. Nat. Acad. Sci. 2004;101:17428–17433. doi: 10.1073/pnas.0408114101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davidovic L, et al. The fragile X mental retardation protein is a molecular adaptor between the neurospecific KIF3C kinesin and dendritic RNA granules. Hum. Mol. Genet. 2007;16:3047–3058. doi: 10.1093/hmg/ddm263. [DOI] [PubMed] [Google Scholar]
- 16.Dictenberg JB, Swanger SA, Antar LN, Singer RH, Bassell GJ. A Direct Role for FMRP in Activity-Dependent Dendritic mRNA Transport Links Filopodial-Spine Morphogenesis to Fragile X Syndrome. Dev. Cell. 2008;14:926–939. doi: 10.1016/j.devcel.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corbin F, et al. The fragile X mental retardation protein is associated with poly(A)+ mRNA in actively translating polyribosomes. Hum. Mol. Genet. 1997;6:1465–1472. doi: 10.1093/hmg/6.9.1465. [DOI] [PubMed] [Google Scholar]
- 18.Oostra BA, Willemsen R. FMR1: a gene with three faces. Biochim. Biophys. Acta. 2009;1790:467–477. doi: 10.1016/j.bbagen.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Rubeis S, Fernández E, Buzzi A, Di Marino D, Bagni C. Molecular and cellular aspects of mental retardation in the Fragile X syndrome: from gene mutation/s to spine dysmorphogenesis. Adv. Exp. Med. Biol. 2012;970:517–551. doi: 10.1007/978-3-7091-0932-8_23. [DOI] [PubMed] [Google Scholar]
- 20.Gross C, Berry-Kravis EM, Bassell GJ. Therapeutic strategies in fragile X syndrome: dysregulated mGluR signaling and beyond. Neuropsychopharmacology. 2012;37:178–195. doi: 10.1038/npp.2011.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hagerman R, Lauterborn J, Au J, Berry-Kravis E. Fragile X syndrome and targeted treatment trials. Results Probl Cell Differ. 2012;54:297–335. doi: 10.1007/978-3-642-21649-7_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darnell JC, et al. FMRP Stalls Ribosomal Translocation on mRNAs Linked to Synaptic Function and Autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krueger DD, Bear MF. Toward fulfilling the promise of molecular medicine in fragile X syndrome. Annu. Rev. Med. 2011;62:411–429. doi: 10.1146/annurev-med-061109-134644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfeiffer BE, Huber KM. The state of synapses in fragile X syndrome. Neuroscientist. 2009;15:549–567. doi: 10.1177/1073858409333075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belmonte MK, Bourgeron T. Fragile X syndrome and autism at the intersection of genetic and neural networks. Nature neuroscience. 2006;9:1221–1225. doi: 10.1038/nn1765. [DOI] [PubMed] [Google Scholar]
- 26.Brown V, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- 27.Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hafner M, et al. Transcriptome-wide Identification of RNA-Binding Protein and MicroRNA Target Sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Licatalosi DD, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ascano M, Hafner M, Cekan P, Gerstberger S, Tuschl T. Identification of RNA-protein interaction networks using PAR-CLIP. WIREs RNA. 2012;3:159–177. doi: 10.1002/wrna.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Nat. Acad. Sci. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kishore S, et al. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat. Meth. 2011;8:559–564. doi: 10.1038/nmeth.1608. [DOI] [PubMed] [Google Scholar]
- 34.Corcoran DL, et al. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol. 2011;12:R79. doi: 10.1186/gb-2011-12-8-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Georgiev S, et al. Evidence-ranked motif identification. Genome Biol. 2010;11:R19. doi: 10.1186/gb-2010-11-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Westmark CJ, Malter JS. FMRP mediates mGluR5-dependent translation of amyloid precursor protein. PLoS Biol. 2007;5:e52. doi: 10.1371/journal.pbio.0050052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schaeffer C, et al. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 2001;20:4803–4813. doi: 10.1093/emboj/20.17.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ceman S, Brown V, Warren ST. Isolation of an FMRP-associated messenger ribonucleoprotein particle and identification of nucleolin and the fragile X-related proteins as components of the complex. Mol. Cell. Biol. 1999;19:7925–7932. doi: 10.1128/mcb.19.12.7925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhakar AL, Dölen G, Bear MF. The Pathophysiology of Fragile X (and What It Teaches Us about Synapses). Annu Rev Neurosci. 2012 doi: 10.1146/annurev-neuro-060909-153138. doi:10.1146/annurev-neuro-060909-153138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greer PL, et al. The Angelman Syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell. 2010;140:704–716. doi: 10.1016/j.cell.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fmr1 knockout mice: a model to study fragile X mental retardation. The Dutch-Belgian Fragile X Consortium. Cell. 1994;78:23–33. [PubMed] [Google Scholar]
- 42.Gross C, et al. Excess phosphoinositide 3-kinase subunit synthesis and activity as a novel therapeutic target in fragile X syndrome. J Neurosci. 2010;30:10624–10638. doi: 10.1523/JNEUROSCI.0402-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma A, et al. Dysregulation of mTOR signaling in fragile X syndrome. J Neurosci. 2010;30:694–702. doi: 10.1523/JNEUROSCI.3696-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Auerbach BD, Osterweil EK, Bear MF. Mutations causing syndromic autism define an axis of synaptic pathophysiology. Nature. 2011;480:63–68. doi: 10.1038/nature10658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bateup HS, Takasaki KT, Saulnier JL, Denefrio CL, Sabatini BL. Loss of Tsc1 in vivo impairs hippocampal mGluR-LTD and increases excitatory synaptic function. J Neurosci. 2011;31:8862–8869. doi: 10.1523/JNEUROSCI.1617-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chévere-Torres I, et al. Metabotropic glutamate receptor-dependent long-term depression is impaired due to elevated ERK signaling in the ΔRG mouse model of tuberous sclerosis complex. Neurobiol. Dis. 2012;45:1101–1110. doi: 10.1016/j.nbd.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spitzer J, et al. PAR-CLIP (Photoactivatable Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation) - a step-by-step protocol to the transcriptome-wide identification of binding sites of RNA-binding proteins. Meth. Enzymol. 2010 doi: 10.1016/B978-0-12-420120-0.00008-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Landthaler M, et al. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA (New York, N.Y. 2008;14:2580–2596. doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.