Abstract
We present an attractive new system for the specific and sensitive detection of the malaria causing Plasmodium parasites. The system relies on isothermal conversion of single DNA cleavage-ligation events catalyzed specifically by the Plasmodium enzyme topoisomerase I to micrometer sized products detectable at the single-molecule level. Combined with a droplet-microfluidics Lab-on-a-Chip platform, this design allowed for sensitive, specific and quantitative detection of all human malaria causing Plasmodium species in single drops of unprocessed blood with a detection limit of less than one parasite/μL. Moreover, the setup allowed for detection of Plasmodium parasites in non-invasive saliva samples from infected patients. During recent years malaria transmission has declined worldwide and with this the number of patients with low-parasite density has increased. Consequently, the need for accurate detection of even a few parasites is becoming increasingly important for the continued combat against the disease. We believe that the presented droplet-microfluidics platform, which has a high potential for adaptation to point-of-care setups suitable for low-resource settings may contribute significantly to meet this demand. Moreover, potential future adaptation of the presented setup for the detection of other microorganisms may form the basis for the development of a more generic platform for diagnosis, fresh water- or food quality control or other purposes within applied or basic science.
Keywords: Droplet-microfluidics, Malaria, Diagnosis, Enzyme Activity Detection, Rolling Circle Amplification, Lab-on-a-Chip
By definition, enzymes convert substrate molecules into distinct products without themselves being consumed in the process. Consequently, highly sensitive detection of biomolecules can be achieved by enzymatic signal enhancement. In line with this, polymerase driven PCR amplification of nucleotide sequences presently provides the most sensitive way to detect biomarkers. For some applications such as detection of disease causing pathogens, where time-to-readout is a major concern, a drawback of PCR is the need for thermal cycling. This hampers quantification and requires specialized equipment unavailable to most physicians, which in turn delays onset of treatment considerably.1–3 Therefore, detection of pathogens by isothermal nucleotide amplification, which poses little requirement on equipment and follows linear reaction kinetics is an attractive solution.3, 4 One such technique is Rolling-Circle-Amplification (RCA), by which a circular DNA template is converted to a ~103 tandem repeat Rolling-Circle-Product (RCP) that can easily be visualized at the single-molecule level.5, 6 RCA has successfully been employed to detect nucleic-acid sequences7, 8, proteins9, 10 or small-molecules.11 However, the disadvantage of most published methods is the requirement of extensive sample preparation, and the formation of only one or few RCPs per target molecule.
Here we present the specific circularization of a RCA substrate by the essential DNA-cleaving enzyme topoisomerase I (pTopI) from the human malaria causing Plasmodium parasites. This allows specific, quantitative and highly sensitive detection of the malaria causing genus, Plasmodium, in small volumes of unprocessed and crude clinical samples using the Rolling-Circle-Enhanced-Enzyme-Activity-Detection (REEAD) system in combination with a droplet-microfluidics Lab-on-a-Chip platform.14 This is to our knowledge the first example of a diagnostic test based directly on detection of endogenous enzyme activity. Moreover, it is the first report of a REEAD setup capable of distinguishing between typeIB topoisomerases in a genus specific manner for a diagnostic purpose. In contrast to other detection strategies the presented Plasmodium-specific REEAD-on-a-Chip system enjoys a “double amplification” advantage since each target enzyme (pTopI) generates numerous DNA circles for signal-enhancement by RCA in addition to the uniform mixing and fast reaction kinetics imposed by droplet-microfluidics, making the setup highly sensitive and yet quantitative in nature.
RESULTS AND DISCUSSION
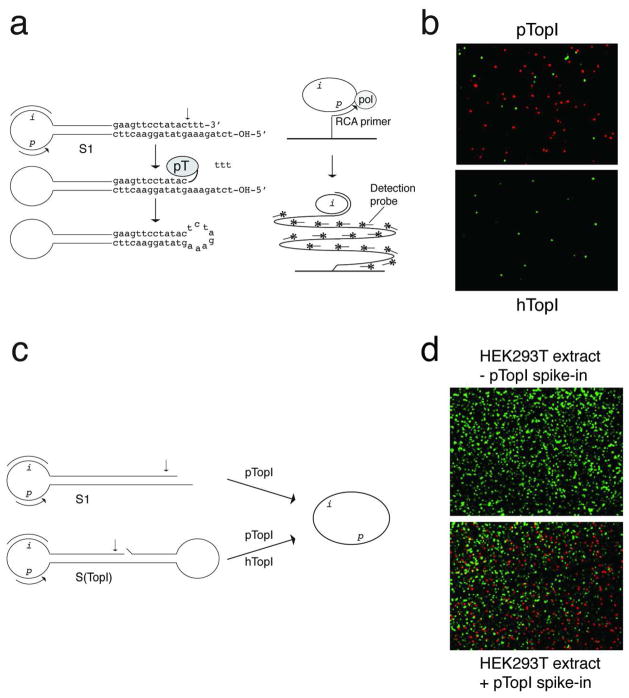
Like human topoisomerase I (hTopI), pTopI belongs to the typeIB topoisomerase family15, which introduces transient single-strand breaks in double-stranded DNA. Cleavage results in a covalent enzyme-DNA intermediate allowing religation of the generated nick.15 To investigate if different DNA recognition by pTopI and hTopI would allow the design of a pTopI-specific REEAD substrate, the cleavage pattern of P. falciparum topoisomerase I (used as a model for pTopI for the REEAD substrate design) and hTopI were compared using standard assays.16 This analysis demonstrated that pTopI can cleave DNA close to a 3′-end and ligate a protruding 5′-end of the non-scissile DNA strand, which hTopI cannot17 (Supplementary Figure S1). Based on this, we designed five different REEAD substrates, which each fold into a hairpin structure containing a probe- and primer annealing sequence in the single-stranded loop-region and a potential cleavage-ligation site for pTopI in the double-stranded stem-region. These substrates were tested for reactivity with pTopI and cross-reactivity with hTopI using the basic REEAD setup.12 All tested substrates were converted to closed circles by pTopI to serve as templates for solid support RCA and visual readout by hybridization of red fluorescent probes as described in12. The reaction is schematically illustrated in Figure 1a and the results shown in Figure 1b and in Supplementary Figure S2. None of the substrates reacted with hTopI (data not shown, and Figure 1b). Of the substrates tested, S1 (shown in Figure 1a) was the most efficient substrate for pTopI and chosen for further experiments (see Supplementary Figure S2).
Figure 1. Development of REEAD for detection of pTopI.
a, shows the REEAD setup exemplified by the pTopI reaction with S1. pTopI mediated cleavage-ligation converts S1 to a single-stranded DNA circle, which templates RCA initiated from a glass slide-coupled primer hybridizing to “p” of S1. Unreacted S1 cannot template RCA. The generated RCPs are visualized microscopically upon hybridization of fluorescent probes annealing to “i-regions” of RCPs. An arrow indicates the cleavage site for pTopI. Grey ellipses labeled “pT” or “pol” denote pTopI or Phi29 polymerase, respectively. b, example of the microscopic views obtained upon incubation of S1 with pTopI (top panel) or hTopI (bottom panel) followed by REEAD. RCPs originating from circularized S1 and spike-in control circles were visualized by rhodamine (red)- and FITC (green) labeled probes, respectively. c, illustrates S1 and S(TopI) used in the experiments shown in d. d, typical microscopic view showing the result of REEAD analyses of nuclear extracts from HEK293T cells without (top panel) or with pTopI spike-in (bottom panel). RCPs originating from circularized S1 or S(TopI) were visualized by rhodamine (red)- or FITC (green) labeled probes.
The specificity of S1 in crude biological samples was addressed using nuclear extract from human HEK293T cells with or without spike-in purified pTopI enzyme. Besides S1, the S(TopI) substrate, previously demonstrated to detect hTopI in cell extracts12, was added to the reaction mixtures as a positive control for the experimental procedure (Figure 1c). In all examined microscopic views red signals corresponding to single RCPs matching circularized S1 were observed only upon addition of pTopI spike-in. Green signals originating from circularized S(TopI) were observed in both samples (Figure 1d). This demonstrated the specificity of S1 for pTopI in a background of the human cell-nucleus content.
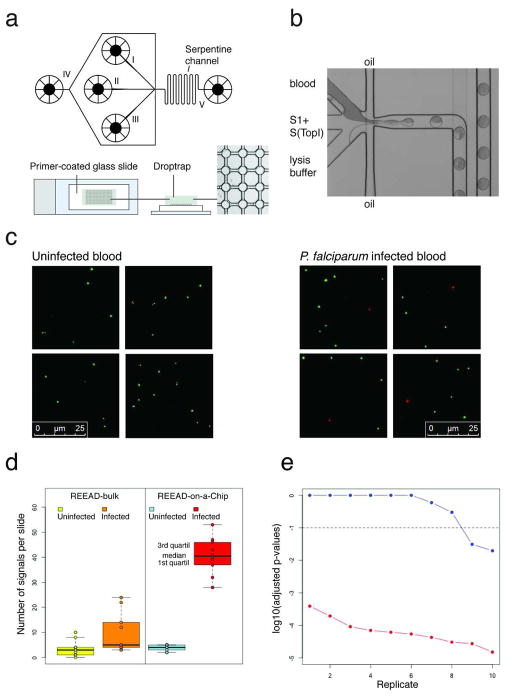
To address the performance of pTopI-specific REEAD in detecting Plasmodium in clinically relevant samples, S1 and S(TopI) were reacted with extracts from blood samples from uninfected patients or from a P. falciparum-infected patient (patient #1, Supplementary Table S3), where the latter was diluted with uninfected blood to a parasite density of 1,000 parasites/μL. The need for extensive sample preparation poses an obstacle for many diagnostic tests.3 We therefore aimed for a simple protocol without extensive sample preparation and suitable for analyses of small volumes (≤50 μL) of blood. We tested two procedures. In one procedure, named “REEAD-bulk”, extracts were prepared by mixing blood with a low-salt lysis buffer before addition of substrates. In the other procedure, named “REEAD-on-a-Chip”, REEAD was combined with a droplet-microfluidics Lab-on-a-Chip platform18 to allow confinement of blood, substrates and the low-salt lysis buffer in pL droplets in which the reaction took place (Figure 2a and 2b). Subsequently, droplets were retained in a drop-trap device and exsiccated onto a primer-coated glass slide to allow RCA12, 14 (Figure 2a, lower panel). Examples of the microscopic views obtained by this procedure are shown in Figure 2c. Green hTopI-generated signals were observed after analyses of either infected- or uninfected blood confirming successful reaction procedures. Red pTopI-specific signals were observed only after analyses of blood from patient #1.
Figure 2. Optimization of REEAD for detection of Plasmodium in blood samples from infected patients.
a, The microfluidic lab-on-a-chip device used for REEAD-on-a-Chip. Top panel, schematic illustration of the microfluidic channel device. Sample to be analyzed, lysis buffer and S1 and S(TopI) were loaded into the system in three different channels (marked I, II, and III). By competition with oil (fed by channel IV) the three different components were confined in pL droplets, lead via a channel system to the outlet (V) and subsequently confined in the drop-trap device. The serpentine channel, ensuring mixing of droplet content, is indicated on the figure. Lower panel, shows the drop-trap device. Droplets were confined in cavities at the intersections in the drop-trap (right panel), and exsiccated onto a primer-coated glass slide (left and middle panel) to support RCA. b, a light-microscopic view of the microfluidic platform. In this example blood, substrates and lysis buffer were loaded into the three different channels and confined in oil surrounded pL droplets. c, shows four representative segments from typical microscopic views obtained from analyzing blood from uninfected or P. faliciparum infected (1,000 parasites/μL) patients using the REEAD-on-a-Chip setup. RCPs originating from circularized S1 or S(TopI) were visualized by rhodamine (red)- or FITC (green) labeled probes. d, boxplot representing the results of the experiment comparing the REEAD-bulk and the REEAD-on-a-Chip setups. The left panel displays the number of pTopI signals/slide obtained with the REEAD-bulk setup (yellow for the uninfected blood samples and orange for the infected replicates). The right panel represents the results for the REEAD-on-a-Chip setup (blue for uninfected blood samples and red for the infected replicates). e, FDR-adjusted p-values for testing each replicate of the infected blood against the reference uninfected samples (in a log10 scale) for the REEAD-on-a-Chip (in red) and the REEAD-bulk (in blue) setups. The replicates are sorted in a decreasing order of p-values. The dashed horizontal line represents a threshold for malaria detection at a significance level of 10%. The p-values for the replicates obtained by the REEAD-on-a-Chip setup vary in the range from 3.85×10−04 to 1.51×10−05 and the three smaller p-values for the replicates of the REEAD-bulk setup are 0.2963, 0.0309 and 0.0196.
To compare the performance of the REEAD-bulk and REEAD-on-a-Chip procedures, 10 replicates of the analysis of the P. falciparum infected blood sample described above and a reference panel of 10 blood samples from different uninfected patients (#6–#15, Supplementary Table S3) were analyzed using each of the two procedures. Subsequently, all the pTopI-specific signals generated on the microscopic slides were counted. As indicated in the boxplot shown in Figure 2d, the median of the counts obtained for the positive replicates differed significantly from the median of the counts obtained for the uninfected samples when using the REEAD-on-a-Chip setup (p-value=8.39×10−5, Wilcoxon test). In contrast, only marginally significant differences between positive and negative replicates were observed when using the REEAD-bulk protocol (p-value=0.01227, n= 20). Figure 2e shows the results of contrasting each replicate of the infected blood determination with the replicates of the uninfected blood (pooled). As evident from the plot only 2 out of 10 replicates were detected positive using REEAD-bulk (p-values 0.0309 and 0.0196), while all the 10 replicates of the infected sample were detected positive using the REEAD-on-a-Chip setup (p-values ranging from 3.85×10−4 to 1.51×10−5). Finally, the data from the REEAD-bulk setup present signs of over-dispersion (variance/mean ratios equal to 2.79 and 6.69 for uninfected samples and the infected replicates, respectively, and false-discovery-rate (FDR) adjusted p-values equal to 0.00594 and 0.00004 for the uninfected samples and the infected replicates, respectively, using parametric conditional resampling test for over-dispersion of counts). No evidence of over-dispersion was found in the data obtained with the REEAD-on-a-Chip setup (adjusted p-values given by 0.94285 and 0.297573 for the uninfected samples and infected replicates, respectively).
Microfluidics is characterized by many intrinsic advantages, such as low sample consumption and short processing time. However, the superior performance of the microfluidic setup observed in this study most probably results from a confined reaction in the pL droplets, where each droplet represents an individual micro-reactor. This offers an efficient mixing of reagents resulting in improved extraction and faster reaction kinetics.18 This is consistent with the absence of detectable over-dispersion in the repeated assays using REEAD-on-a-Chip in contrast to the over-dispersion found when using REEAD-bulk. Over-dispersion is associated with the presence of additional variation sources19, 20 and only extraction and circularization differed between the two setups. Based on the observations described above we continued our investigations using the “REEAD-on-a-Chip” setup.
The performance of REEAD-on-a-Chip on field samples was tested in a trial including 20 blood samples from patients infected in west Africa or India with a parasite density varying from <50 to 275,000 parasites/μL of P. falciparum (#17–#26, #29, #30, #32–#36), P. vivax (#27, #28), or P. ovale (#21, #31) and 11 blood samples from uninfected patients (#6–#16) (Supplementary Table S4). Only sporadic signals were observed in the 11 uninfected samples (mean=4 signals/slide, SE=1.3) and no significant differences between the expected numbers of signals of those samples were detected (p-value 0.9045, n=31, Supplementary Table S4). Therefore, in this trial an infected sample was reported positive when the total number of pTopI signals detected was statistic significantly larger than the expected number of signals for the uninfected reference samples (for α < 1.9×10−5 which is equivalent to ≥17 signals per slide, Supplementary Table S4). According to this criterion the test identified all the Plasmodium positive samples, regardless parasite species and density.
In addition to P. falciparum, P. vivax and P. ovale, which were detected in the test described above, the REEAD system was able to detect infections with P. knowlesi and P. malariae (Supplementary Table S5). This is consistent with the fact that the biomarker, pTopI, is an essential enzyme that is conserved among Plasmodium species and suggests that REEAD-on-a-Chip may be developed into a highly sensitive and specific pan-malaria test. Moreover, the essential functions of the TopI cleavage-ligation15 reaction argues against the occurrence of Plasmodium strains undetectable by REEAD-on-a-Chip. This is the case for many of the common commercially available Rapid Diagnostic Tests (RDTs) specific for biomarkers such as HRP2, which detect P. falciparum only and are further hampered in regions were the hrp2 genes are deleted from large proportions of the parasites.2
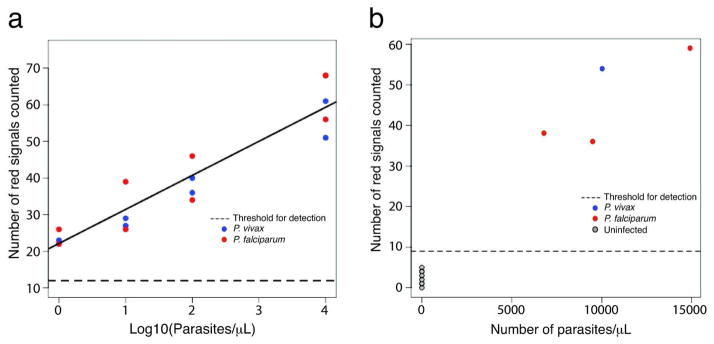
To address if REEAD-on-a-Chip is suitable for quantitative determination of parasites and to calculate the detection limit, serial dilutions of blood samples with a known parasite density (Supplementary Table S3) from individuals infected with P. falciparum (#1) or P. vivax (#5) were analyzed and the results compared to those obtained using a reference panel of blood samples from 11 uninfected patients (#6–#16). The microfluidic setup could detect both Plasmodium species in the studied range of 1–10,000 parasites/μL. Note that this does not indicate an upper limit for detection (see Supplementary Table S4). The expected number of signals increased linearly with the logarithm of the number of parasites at a rate of 9.3 units per 10-fold increase in the number of parasites/μL (Figure 3a). This regression yields an estimated detection limit of the current REEAD-on-a-Chip protocol of 0.06 parasites/μL (zM level) (see methods). Note that the best PCR protocol demonstrates a detection limit of 0.01–1 parasites/μL. However, such sensitivity is achieved only after extensive sample preparation and/or concentration procedures, which often requires larger sample volume and involves several centrifugation steps.21 This is redundant for the REEAD-on-a-Chip procedure.
Figure 3. REEAD-on-a-Chip analyses of Plasmodium infected blood or saliva samples.
a, graphic representation of a quantitative analyses of blood samples from patient #1 (P. falciparum), or #5 (P. vivax) diluted by uninfected blood to a parasite density of 10,000, 100, 10 or 1 parasite/μL. The total numbers of pTopI-specific signals on the individual slides are plotted as a function of Log10 to numbers of parasites/μL in the individual samples. Red and blue dots show the results of two repeated analyses of the P. falciparum and P. vivax samples, respectively. As a reference a panel of blood samples from 11 uninfected patients (#6–#16) were analyzed and used to determine an approximate threshold for detection with a significance level of α=10−3 (dotted line). b, samples of saliva from three P. falciparum (red dots), one P. vivax (blue dot), and eight uninfected (grey dots) individuals were analyzed using the REEAD-on-a-Chip setup. The total numbers of pTopI-specific signals observed on each slide are plotted as a function of parasite density observed in the blood from the infected patients. The dotted line shows an approximate threshold for detection with a significance level of α=10−3.
Plasmodium parasites have been reported present in saliva from infected patients.22 Since diagnosis based on non-invasive samples such as saliva poses obvious advantages we therefore analyzed saliva samples obtained from eight uninfected (#9–#16), and four P. falciparum or P. vivax (#2–#5) infected individual(s) (Supplementary Table S3). All infected samples tested positive (Figure 3b) suggesting that REEAD-on-a-Chip will allow for parasite-based diagnosis of malaria using non-invasive saliva samples. Clearly, this would impact the possibilities of disease surveillance worldwide especially when it comes to identifying asymptomatic carriers, which is of particular importance in elimination programs.1
CONCLUSION
In conclusion the presented REEAD-on-a-Chip setup allowed highly sensitive detection of Plasmodium infection in small volumes of blood or saliva samples. In terms of detection limit (less than one parasite/μL) the assay out-competes state-of-the-art light-microscopy (detection limit; 10–200 parasites/μL1, 2) and RDTs (require parasitaemia ≥200 parasite/μL). Compared to PCR, which is the only method with a sensitivity matching that of REEAD-on-a-Chip, the latter assay presents the advantages of being directly quantitative in nature and requiring no sample preparation. Indeed, to obtain a detection limit comparable to REEAD-on-a-Chip, PCR requires extensive sample preparation21 and is under typical use not substantially more sensitive than light-microscopy.2 Moreover, the REEAD-on-a-Chip setup is easy-to-operate, and requires no thermal cycling making it adaptable to point-of-care testing even at low-resource settings. This of course requires an unambiguous and automated read-out format as an alternative to the presented microscopic readout. This can be obtained, for example, by transforming the biological events to electrical signals, which has been demonstrated feasible for vast numbers of electrochemical DNA sensors and requires only low power.23 Moreover, the pump-system used for droplet microfluidics can be simplified by a pumpless syringe based inlet system24, which is currently being designed. Even in the current proof-of-concept setup time-to-readout (~2.5 hours) and prize (~2 USD per sample) of the REEAD-on-a-Chip is acceptable for routine use and is expected considerably reduced once a point-of-care test is fully developed.
We believe REEAD-on-a-Chip may provide the basis for a highly valuable pan-malaria test, which can complement state-of-the-art methods for detection of low-parasitaemia cases. Such a test is needed as malaria transmission declines. With this, diagnosis of low-parasitaemia cases becomes increasingly more important for individual case management as well as for disease surveillance and for eradication programs in semi-immune populations, where identification and treatment of all (including asymptomatic) infected individuals are important determinants of success.1 Even in its present form, REEAD-on-a-Chip requires no specialized skills, and may be of direct use at community hospitals in non-endemic regions where lack of experience among technicians currently results in misdiagnoses and reporting delays.2 Also, since the candidate enzyme, pTopI, is a potential new drug target in the combat against multi-drug resistant malaria25, 26, the presented REEAD-on-a-Chip would be an attractive setup for high-throughput drug screening. Finally, with the potential future development of REEAD substrates specific to other relevant enzyme activities the presented REEAD-on-a-Chip setup may form the basis for a more generic platform for nano-medicine or other purposes within applied or basic science.
METHODS
REEAD substrates, primers and probes
All oligonucleotides were purchased from DNA Technology A/S. The sequences of the oligonucleotides are shown in Supplementary Information.
Enzyme expression and purification
The P. falciparum TopI gene (PlasmoDB accession number PFE0520c)27 was codon optimized (by GeneArt) for expression in Saccharomyces cerevisiae. The optimized gene was PCR amplified and cloned into the pYES2.1/V5-His-TOPO vector (Invitrogen). A positive clone was identified by sequencing and the generated plasmid pPT100 was transformed into the yeast S. cerevisiae top1Δ strain RS190 (a kind gift from R. Sternglanz, State University of New York, USA) according to standard procedures. pTopI was expressed and purified as previously described for hTopI.28 hTopI was expressed and purified as previously described.28 The protein concentrations were estimated from Coomassie blue-stained SDS-polyacrylamide gels by comparison to serial dilutions of BSA.
Cell culture and nuclear extract preparation
Human embryonic kidney HEK293T cells were cultured in Dulbecco’s Modified Eagle Medium (GIBCO) supplemented with 10% fetal bovine serum (FBS) (GIBCO), 100 units/mL penicillin and 100 mg/mL streptomycin (Invitrogen). Cells were incubated in a humidified incubator (5% CO2/95% air atmosphere at 37°C). Cells were harvested with 0.5% Trypsin-EDTA (GIBCO). Media was discarded and the cells washed in Phosphate-Buffered Saline (1xPBS) prior to nuclear extraction performed as previously described.13 The cell extracts were used for REEAD directly or spiked with purified pTopI prior to REEAD.
Bulk preparation of extracts from blood samples
40 μL of ice-cold lysis buffer (10 mM Tris-HCL pH 7.5, 0.5 mM EDTA, 1 mM DTT, 1 mM PMSF, 0.2% Tween 20) was added and carefully mixed (by vortexing) with 8 μL of 37° C blood (uninfected or Plasmodium infected) and incubated on ice for 15 min.
REEAD analyses of bulk extracted blood (REEAD-bulk)
Circularization reactions were carried out in 40 μL reaction volumes containing a divalent cation depleted buffer (10 mM Tris-HCl, pH 7.5, 0.5 mM EDTA, 250 mM NaCl, 1 mM DTT, 1 mM PMSF) supplemented with REEAD substrate(s) as stated in the text. Standard concentrations S(TopI): 67 nM and S1: 167 nM. Reactions were initiated by the addition of the purified enzymes (hTopI or pTopI), cell extracts, or 10 μL bulk blood extraction. Incubation was carried out for 30 min. at 37°C.
REEAD-on-a-Chip analyzes of blood or saliva samples
The microfluidic setup consists of two devices: a flow-focusing droplet generator and a drop-trap. Both devices were fabricated by conventional soft lithography techniques29, casting and curing the PDMS prepolymer on a SU-8 3025 (MicroChem) master of a channel height at around 25 μm. PDMS prepolymer (Sylgard 184) was prepared in a 10:1 (base: curing agent) ratio and cured at 65°C for 1 hour. Prior to the experiments, the channels were wetted with oil/surfactant (PFPE-based Surfactant made essentially as described in30) for at least 15 min.. Two syringe pumps (Harvard Apparatus) were used to control the flow rates of oil/surfactant and reagents independently, forming monodisperse water-in-oil droplets at a frequency of 0.8–1.5 kHz. The droplet volume and generation frequency were controlled by the flow rate ratio, determined by the competition between continuous phase (carrier fluid (HFE-7500 fluorocarbon oil (3M): the oil/surfactant, flow rate 15 μL/min.) and disperse phase (aqueous reagents: blood/saliva, lysis buffer and REEAD substrates, flow rate 2.5 μL/min.). Blood or saliva, lysis buffer (10 mM Tris-HCL pH 7.5, 0.5 mM EDTA, 1 mM DTT, 1 mM PMSF, 0.2% Tween 20), and REEAD substrates (final concentration in the droplets: S(TopI): 67 nM, S1: 167 nM) were loaded in each their channel in the microfluidic device and droplet generation initiated. The generated droplets were harvested in Eppendorf tubes. 5 μL of the droplets were placed on a primer-printed glass slide prepared as described below. The PDMS drop-trap was gently placed on top of the glass slide containing droplets. The geometry of the drop-trap was designed according to the size of generated droplets. The droplets were left to exsiccate and the REEAD substrates hybridized to the primers (see REEAD – microscopic readout), before the drop-trap was removed.
REEAD – microscopic readout
A 5′-amine-conjugated primer was coupled to CodeLink Activated Slides (SurModics) according to the manufacturer’s description. 5 μL circularization reaction sample was hybridized to the immobilized primers for 30 min. at RT (22–25°C). Washing, RCA, hybridization of probes, and microscopic visualization were performed as previously described.12 The quantification of pTopI-specific signals was performed by counting all red signals on the microscopic slides.
Statistical analyses
The performance of the REEAD-on-a-Chip and the REEAD-bulk protocols were compared in an experiment where 10 replicates of the analyses of a P. falciparum infected blood sample containing 1,000 parasites/μL and a panel of 10 blood samples of different uninfected patients were analyzed using each of the two protocols. The total numbers of pTopI-signals observed on the entire surface of a microscopic slide containing RCPs resulting from 5 μL of circularization-mixture/droplets were determined for each of these assays. The following statistical procedures were applied for the data generated with each protocol. First, a one-sided Wilcoxon test was used to compare the median of the observations from the infected blood with the median of the observations of the uninfected blood. The over-dispersion of each group of replicates (infected and uninfected with each protocol) was tested using a parametric conditional resampling test for over-dispersion of counts (see19 page 142). A sequence of generalized linear models for Poisson over-dispersed counts19 were used to analyze this experiment, since the replicates obtained with the REEAD-bulk protocol showed significant over-dispersion. Next, we tested whether the uninfected replicates could be pooled (defining suitable nested models and using F tests with p-values obtained by non-parametric bootstrap19 with 10,000 bootstrap repetitions, to avoid use of asymptotic inference) obtaining the p-values 0.545 and 0.512 for the REEAD-on-a-Chip and the REEAD-bulk protocols respectively. We then inspected each replicate of the infected blood sample by contrasting it with the pooled observations of the uninfected blood samples (Wald test). After applying a false discovery rate correction (FDR) for multiple testing, we obtained the p-values reported in Figure 2e.
The possibility of using the REEAD-on-a-Chip protocol for quantitative determinations of the density of parasites was addressed by mounting an experiment involving four dilutions of blood samples with a known parasite density (densities of 1, 10, 100 and 10,000 parasites/μL) from individuals infected with P. falciparum (#1) or P. vivax (#5) and two replicates, yielding 2×4×2=16 observations. A sequence of generalized linear model defined with the Poisson distribution and the identity link was used to study the results of this experiment.20 We showed that a model assuming that the expected number of pTopI-signals/slide, say E(S), is linearly related to the logarithm of the number of parasites/μL, log10 (P), more precisely,
| (1) |
where α and β are parameters to be estimated. Comparing a succession of nested models by the likelihood-ratio-tests we found that there are no statistical evidences of inhomogeneity (p-value=0.4138, n=16), lack of adjustment of the linear relationship described in equation (1) (one response-curve per species, p-value=0.9196, n=16) and differences between the response-curve of each of the two species (p-value=0.3513, n=16). The estimated value of the parameters of the model defined by (1) are α̂ = 22.1 (SE=1.9) and β̂ = 9.3 (SE=1.0), moreover β differs significantly from zero (p-value=2×10−16), i.e. the number of pTopI-signals/slide increases significantly with the number of parasites and there are no evidences in our experiment of differences in this increase pattern between P. falciparum and P. vivax. The detection limit was estimated by predicting with the regression (1) the number of parasites P such that the number of signals S attained a threshold for detection obtained with the panel of uninfected reference samples. Using a significance level of α = 10−3 the threshold for detection is 11 signals/slide, yielding an estimate of the detection limit of .
The samples of saliva were analyzed with the REEAD-on-a-Chip protocol combined with fluorescent-microscope counting of the total number of pTopI-signals/slide. Using suitably defined Poisson generalizer linear models20, no evidences of differences between the expected numbers of pTopI-signals/slide among the uninfected control samples were found (bootstrap p-value 0.139). The total number of pTopI-signals/slide from samples of saliva from infected patients were then compared to the distribution of the number of pTopI-signals/slide of the control uninfected samples following a procedure similar to the described above.
Supplementary Material
Acknowledgments
We are thankful to technicians Noriko Y. Hansen, Erik Hagen Nielsen, Liv Papanikolaou, Anette Olesen, and Dorthe Mejdahl Lind for technical assistance during this project. We also thank Dr. Eigil Kjeldsen for providing negative samples for the project, Dr. Bent Deleuran for contributing with important equipment, and Dr. Felicie F. Andersen for preparing some of the figures in the Supplementary materials. This work was supported by NIH (HL89764), NSF EEC-0425626, the Danish Research Councils (11-116325/FTP) and (11-105736/FSS), Karen Elise Jensen’s Foundation, Dagmar Marshall’s Foundation, Dir. Einar Hansen og Vera Hansen’s Foundation, The Harboe Foundation, The Augustinus Foundation, Louis Hansen’s Foundation, The Hørslev Foundation, Fabrikant Einar Willumsen’s Foundation, Købmand Sven Hansen & hustru Ina Hansen’s Foundation, Dir. Emil Hertz & hustru Inger Hertz’ Foundation, Civilingeniør Frode Nygaard’s Foundation, Kong Christian den Tiende’s Foundation, The Gangsted Foundation, KU’s Foundation for Cancer research, Ludvig og Franciska Andersen’s Foundation, Arvid Nilsson’s Foundation, Frimodt-Heineke’s Foundation, the Aase and Ejnar Danielsens Foundation, the Minister Erna Hamiltons Foundation for Science and Art, the Apolodoro Plausonius Foundation, and the Italian association for cancer research AIRC.
Footnotes
Supporting Information Available: Figures of the purification and DNA cleavage ability of pTopI and hTopI, tests of specific DNA substrates for pTopI together with overviews of patient samples and DNA oligoes are available free of charge via the Internet at http://pubs.acs.org.
REFERENCES AND NOTES
- 1.McMorrow ML, Aidoo M, Kachur SP. Malaria Rapid Diagnostic Tests in Elimination Settings--Can They Find the Last Parasite? Clin Microbiol Infect. 2011;17:1624–1631. doi: 10.1111/j.1469-0691.2011.03639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bronzan RN, McMorrow ML, Kachur SP. Diagnosis of Malaria: Challenges for Clinicians in Endemic and Non-Endemic Regions. Mol Diagn Ther. 2008;12:299–306. doi: 10.1007/BF03256295. [DOI] [PubMed] [Google Scholar]
- 3.McNerney R, Daley P. Towards a Point-of-Care Test for Active Tuberculosis: Obstacles and Opportunities. Nat Rev Microbiol. 2011;9:204–213. doi: 10.1038/nrmicro2521. [DOI] [PubMed] [Google Scholar]
- 4.Lutz S, Weber P, Focke M, Faltin B, Hoffmann J, Muller C, Mark D, Roth G, Munday P, Armes N, et al. Microfluidic Lab-on-a-Foil for Nucleic Acid Analysis based on Isothermal Recombinase Polymerase Amplification (RPA) Lab Chip. 2010;10:887–893. doi: 10.1039/b921140c. [DOI] [PubMed] [Google Scholar]
- 5.Lizardi PM, Huang X, Zhu Z, Bray-Ward P, Thomas DC, Ward DC. Mutation Detection and Single-Molecule Counting using Isothermal Rolling-Circle Amplification. Nat Genet. 1998;19:225–232. doi: 10.1038/898. [DOI] [PubMed] [Google Scholar]
- 6.Nallur G, Luo C, Fang L, Cooley S, Dave V, Lambert J, Kukanskis K, Kingsmore S, Lasken R, Schweitzer B. Signal Amplification by Rolling Circle Amplification on DNA Microarrays. Nucleic Acids Res. 2001;29:E118. doi: 10.1093/nar/29.23.e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smolina I, Miller NS, Frank-Kamenetskii MD. PNA-based Microbial Pathogen Identification and Resistance Marker Detection: An Accurate, Isothermal Rapid Assay based on Genome-Specific Features. Artif DNA PNA XNA. 2010;1:76–82. doi: 10.4161/adna.1.2.13256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larsson C, Koch J, Nygren A, Janssen G, Raap AK, Landegren U, Nilsson M. In Situ Genotyping Individual DNA Molecules by Target-Primed Rolling-Circle Amplification of Padlock Probes. Nat Methods. 2004;1:227–232. doi: 10.1038/nmeth723. [DOI] [PubMed] [Google Scholar]
- 9.Konry T, Smolina I, Yarmush JM, Irimia D, Yarmush ML. Ultrasensitive Detection of Low-Abundance Surface-Marker Protein using Isothermal Rolling Circle Amplification in a Microfluidic Nanoliter Platform. Small. 2011;7:395–400. doi: 10.1002/smll.201001620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang L, Fung CW, Cho EJ, Ellington AD. Real-Time Rolling Circle Amplification for Protein Detection. Anal Chem. 2007;79:3320–3329. doi: 10.1021/ac062186b. [DOI] [PubMed] [Google Scholar]
- 11.Cho EJ, Yang L, Levy M, Ellington AD. Using a Deoxyribozyme Ligase and Rolling Circle Amplification to Detect a Non-Nucleic Acid Analyte, ATP. J Am Chem Soc. 2005;127:2022–2023. doi: 10.1021/ja043490u. [DOI] [PubMed] [Google Scholar]
- 12.Stougaard M, Lohmann JS, Mancino A, Celik S, Andersen FF, Koch J, Knudsen BR. Single-Molecule Detection of Human Topoisomerase I Cleavage-Ligation Activity. ACS Nano. 2009;3:223–233. doi: 10.1021/nn800509b. [DOI] [PubMed] [Google Scholar]
- 13.Andersen FF, Stougaard M, Jorgensen HL, Bendsen S, Juul S, Hald K, Andersen AH, Koch J, Knudsen BR. Multiplexed Detection of Site Specific Recombinase and DNA Topoisomerase Activities at the Single Molecule Level. ACS Nano. 2009;3:4043–4054. doi: 10.1021/nn9012912. [DOI] [PubMed] [Google Scholar]
- 14.Juul S, Ho YP, Koch J, Andersen FF, Stougaard M, Leong KW, Knudsen BR. Detection of Single Enzymatic Events in Rare or Single Cells using Microfluidics. ACS Nano. 2011;5:8305–8310. doi: 10.1021/nn203012q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Champoux JJ. DNA Topoisomerases: Structure, Function, and Mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 16.Gentry AC, Juul S, Veigaard C, Knudsen BR, Osheroff N. The Geometry of DNA Supercoils Modulates the DNA Cleavage Activity of Human Topoisomerase I. Nucleic Acids Res. 2011;39:1014–1022. doi: 10.1093/nar/gkq822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christiansen K, Svejstrup AB, Andersen AH, Westergaard O. Eukaryotic Topoisomerase I-Mediated Cleavage Requires Bipartite DNA Interaction. Cleavage of DNA Substrates Containing Strand Interruptions Implicates a Role for Topoisomerase I in Illegitimate Recombination. J Biol Chem. 1993;268:9690–9701. [PubMed] [Google Scholar]
- 18.Ho YP, Grigsby CL, Zhao F, Leong KW. Tuning Physical Properties of Nanocomplexes Through Microfluidics-Assisted Confinement. Nano Lett. 2011;11:2178–82. doi: 10.1021/nl200862n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davison AC, Hinkley D. Bootstrap Methods and their Application (8th printing) Cambridge: Cambridge Series in Statistical and Probabilistic Mathematics; 2006. [Google Scholar]
- 20.McCullagh P, Nelder J. Generalized Linear Models. 2. Boca Raton: Chapman and Hall/CRC; 1989. [Google Scholar]
- 21.Mahajan B, Zheng H, Pham PT, Sedegah MY, Majam VF, Akolkar N, Rios M, Ankrah I, Madjitey P, Amoah G, et al. Polymerase Chain Reaction-based Tests for Pan-Species and Species-Specific Detection of Human Plasmodium Parasites. Transfusion. 2012 doi: 10.1111/j.1537-2995.2011.03541.x. [DOI] [PubMed] [Google Scholar]
- 22.Putaporntip C, Buppan P, Jongwutiwes S. Improved Performance with Saliva and Urine as Alternative DNA Sources for Malaria Diagnosis by Mitochondrial DNA-based PCR Assays. Clin Microbiol Infect. 2011;17:1484–1491. doi: 10.1111/j.1469-0691.2011.03507.x. [DOI] [PubMed] [Google Scholar]
- 23.Drummond TG, Hill MG, Barton JK. Electrochemical DNA sensors. Nat Biotechnol. 2003;21:1192–1199. doi: 10.1038/nbt873. [DOI] [PubMed] [Google Scholar]
- 24.Abate AR, Weitz DA. Syringe-Vacuum Microfluidics: A Portable Technique to Create Monodisperse Emulsions. Biomicrofluidics. 2011;5:14107. doi: 10.1063/1.3567093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia-Estrada C, Prada CF, Fernandez-Rubio C, Rojo-Vazquez F, Balana-Fouce R. DNA Topoisomerases in Apicomplexan Parasites: Promising Targets for Drug Discovery. Proc Biol Sci. 2010;277(1689):1777–17787. doi: 10.1098/rspb.2009.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reguera RM, Redondo CM, Gutierrez de Prado R, Perez-Pertejo Y, Balana-Fouce R. DNA Topoisomerase I from Parasitic Protozoa: a Potential Target for Chemotherapy. Biochim Biophys Acta. 2006;1759:117–131. doi: 10.1016/j.bbaexp.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 27.Aurrecoechea C, Brestelli J, Brunk BP, Dommer J, Fischer S, Gajria B, Gao X, Gingle A, Grant G, Harb OS, et al. PlasmoDB: a Functional Genomic Database for Malaria Parasites. Nucleic Acids Res. 2009;37(Database issue):D539–543. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lisby M, Olesen JR, Skouboe C, Krogh BO, Straub T, Boege F, Velmurugan S, Martensen PM, Andersen AH, Jayaram M, et al. Residues Within the N-terminal Domain of Human Topoisomerase I Play a Direct Role in Relaxation. J Biol Chem. 2001;276:20220–20227. doi: 10.1074/jbc.M010991200. [DOI] [PubMed] [Google Scholar]
- 29.Qin D, Xia Y, Whitesides GM. Soft Lithography for Micro- and Nanoscale Patterning. Nat Protoc. 2010;5:491–502. doi: 10.1038/nprot.2009.234. [DOI] [PubMed] [Google Scholar]
- 30.Holtze C, Rowat AC, Agresti JJ, Hutchison JB, Angile FE, Schmitz CH, Koster S, Duan H, Humphry KJ, Scanga RA, et al. Biocompatible Surfactants for Water-in-Fluorocarbon Emulsions. Lab Chip. 2008;8:1632–1639. doi: 10.1039/b806706f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.