Abstract
Embryonic stem cells dynamically fluctuate between phenotypic states, as defined by expression levels of genes such as Nanog, while remaining pluripotent. The dynamic phenotype of stem cells is in part determined by gene expression control and dictated by various signaling pathways and transcriptional regulators. We sought to define the activities of two TGF-beta-related signaling pathways, Bone morphogenetic protein (BMP) and Nodal signaling, in modulating mouse embryonic stem cell heterogeneity in undifferentiated culture conditions. Both BMP and Nodal signaling pathways were seen to be active in distinct Nanog subpopulations, with subtle quantitative differences in activity. Pharmacological and genetic modulation of BMP or Nodal signaling strongly influenced the heterogeneous state of undifferentiated ES cells, as assessed by dynamic expression of Nanog reporters. Inhibition of Nodal signaling enhanced BMP activity, which through the downstream target Id factors, enhanced the capacity of ES cells to remain in the Nanog-high epigenetic state. The combined inhibition of Nodal and BMP signaling resulted the accumulation of Nanog-negative cells, even in the presence of LIF, uncovering a shared role for BMP and Nodal signaling in maintaining Nanog expression and repression of differentiation. These results demonstrate a complex requirement for both arms of TGF-beta-related signaling to influence the dynamic cellular phenotype of undifferentiated ES cells in serum-based media, and that differing subpopulations of ES cells in heterogeneous culture have distinct responses to these signaling pathways. Several pathways, including BMP, Nodal, and FGF signaling, have important regulatory function in defining the steady-state distribution of heterogeneity of stem cells.
Keywords: Embryonic stem cells, heterogeneity, BMP, Nodal, Nanog
Introduction
Embryonic stem (ES) cells are derived from the inner cell mass of the early blastocyst and maintain the capacity to differentiate to various cell types of the adult in vivo and in vitro1. These remarkable features of long-term self-renewal and pluripotency have made mouse and human ES cells valuable models to study mechanisms of self-renewal and differentiation. To remain in the undifferentiated state, ES cells require specific culture conditions, which regulate transcriptional regulatory networks to maintain pluripotency2. Undifferentiated mouse ES cells show highly variable expression of some components of the pluripotency program, including transcriptional regulators such as Nanog, Zfp42 (Rex-1), Brachyury (T), and Dppa3 (Stella)3–10. Mouse epiblast stem cells11 and undifferentiated human ES cells12, 13 also exhibit variable levels of many genes.
In pluripotent cells, heterogeneity is due to temporal heterogeneity, in which stem cells dynamically fluctuate between phenotypic states, as defined by expression levels of genes such as Nanog in mouse ES cells. When cells of a particular state are purified and replated, the cells will eventually re-establish heterogeneous populations5, 7; ES cells interconvert between these pluripotent states while still not committed to differentiate. Thus heterogeneity results from a complex dynamic equilibrium of cell subpopulations with distinct gene expression levels. Heterogeneity may be an important phenotype in stem cell populations, to allow cells to respond to differentiation cues while still remaining otherwise undifferentiated14.
The dynamic expression of Nanog and its role in pluripotency suggests that this factor may act as both a ‘marker’ and a ‘maker’ of heterogeneous subpopulations. Substantial data has shown that the divergent homeobox gene Nanog is an important component of the core self-renewal machinery15–18 and participates in the regulation of genes associated with the undifferentiated phenotype. Purified Nanog-high cells have a significantly increased capacity for integration into embryos when injected into blastocysts5; Nanog-low cells in contrast do not readily integrate to form chimeric mice. Heterogeneity is thus a fundamental aspect of pluripotent stem cells and is intrinsically linked to the self-renewal machinery that controls stem cell identity. An understanding of how heterogeneity arises and is regulated will provide critical information on how self-renewal and pluripotency are maintained in different stem cell types.
The heterogeneous status of ES cells in culture can be influenced by altering the FGF signaling pathway. Inhibition of autocrine FGF-MEK-ERK signaling greatly enhances the Nanog-high phenotypic state18–21. These data are among the first observations to suggest that the heterogeneous phenotype of mouse ES cells is not an entirely random process but is heavily influenced by the active signaling pathways of undifferentiated cell culture; heterogeneity is thus a regulated process. Thus the dynamic phenotype of stem cells is in part determined by gene expression control and dictated by various signaling pathways, transcriptional regulators, and chromatin marks. The complexity of the gene regulatory pathways controlling the core pluripotency program suggests other pathways likely also are involved in heterogeneity, but are not characterized.
In this report, we sought to define the activities of two TGF-beta-related signaling pathways, Bone morphogenetic protein (BMP) and Nodal signaling, in modulating mouse embryonic stem cell heterogeneity in undifferentiated culture conditions. The Nodal signaling pathway has known roles in controlling pluripotency of human ES cells22, 23. Although Nodal is important in regulating proliferation of mouse ES cells24, a role of this signaling pathway in stem cell self-renewal and homeostasis has not been determined. Our previous studies demonstrated that autocrine Nodal signaling modulates BMP signaling pathway in undifferentiated ES cells25, and BMP signaling plays a critical role in maintaining the undifferentiated state of mouse ES cells26. In this work we show that modulation of BMP or Nodal signaling strongly influences the heterogeneous state of undifferentiated ES cells, as assessed by dynamic expression of Nanog. Inhibition of Nodal signaling acts to enhance BMP activity, which through the downstream target Id factors, enhances the capacity of ES cells to remain in the Nanog-high epigenetic state. Our efforts also uncovered a shared role for BMP and Nodal signaling in maintaining Nanog expression and repression of differentiation. These data indicate the BMP and Nodal signaling pathways have essential and complex roles in controlling stem cell self-renewal, and that differing subpopulations of ES cells in heterogeneous culture have distinct responses to these signaling pathways. These results suggest multiple pathways have regulatory function to define the spectrum of dynamic phenotypes of stem cells in culture.
Material and Methods
Mouse ES cell culture
Experiments utilized E14Tg2A (E14) ES cells, TNG ES cells5, and BNG ES cells (described below). ES cells were maintained as described previously25 on gelatin-coated or fibroblast co-cultured plates. Cells were grown in serum-based ES cell media: DMEM, 15% FBS, penicillin-streptomycin, L-glutamine, non-essential amino acids, beta-mercaptoethanol, and 103 units/ml LIF. In serum-free experiments, FBS was replaced with knockout serum replacement (KOSR; Invitrogen). For specific studies, ES cells were treated with 10 ng/ml Activin (R&D), 10 ng/ml BMP4 (R&D), 5 µM SB431542 (Sigma), 100 nM LDN193189 (Stemgent), 1uM PD0325901 (Stemgent), SB505124 (Sigma), Noggin (R&D), and 1ug/mL doxycycline (Sigma). Time periods for treatments are indicated for each experiment.
Transgenesis of ES cell lines
To generate BAC-Nanog-GFP (BNG) ES cells, Nanog-GFP BAC27 was introduced into Ainv15 ES cells28 or AInv15-Smad7 ES cells25 via lipofection, modified from previous protocols29. In brief, ES cells were plated onto four wells of a 24 well gelatin coated plate at a concentration of 10^5 cells per well. After 24 hr culture, 1.0ug of supercoiled BAC DNA was transfected into each well (Lipofectamine, Invitrogen). The next day the cells were plated onto resistant feeders and hygromycin selection (150ug/ml) was initiated 24 hr later. Expanded clones were characterized for GFP expression and response to small molecules via flow cytometry. Id1 coding sequences were introduced into the doxycycline responsive cassette of BNG cells via CRE-mediated recombination as described25.
Fluorescent cell sorting and analysis
ES cells were trypsinized to a single cell suspension and analyzed by BD FACSAria (cell sorting) and BD LSRII flow cytometer (cell analysis) for GFP expression. A convention was established for sorting and analyzing subpopulations of BNG ES cells based on the profile of GFP expression of unsorted cells (Fig. 1B). The GFP-medium and GFP-high populations were determined by gating 30–35% of the cells from the peak of the GFP distribution. Cells expressing higher levels of GFP than the GFP-high subpopulation were classified as GFP-very high; whereas cells with expression less than GFP-medium cells but above background were identified as GFP-low cells. Analysis of control E14 ES cells were used as a negative control for flow analysis and sorting. After cell sorting, subpopulations were analyzed to confirm purity of the population.
Figure 1. Reporter models of Nanog heterogeneity in ES cells.
ES cell cultures from TNG (A) and BNG ES cells (B) were sorted according to GFP expression levels. C) Gene expression of sorted BNG ES cells showed differential expression of pluripotency and differentiation markers. D) GFP expression and cell morphology were analyzed in FACS-purified BNG cells.
RNA analysis
RNA was isolated (Qiagen), and cDNA was synthesized (Invitrogen) following manufacturer’s instructions. TaqMan primer sets with the 7500 Real Time PCR system (Applied Biosystems) were used for quantitative real-time PCR analysis. Upon request, specific ABI TaqMan Primer/Probe assay identification numbers are available.
Protein analysis
Cells were pelleted and then lysed with radioimmune precipitation assay buffer supplemented with Halt Protease and Phosphatase Inhibitor Cocktails (Pierce). Protein samples were separated on BioRad Tris-HCl gels, and blots were probed with primary antibodies for pSmad2 (Millipore), Smad2 (Zymed Laboratories Inc.), pSmad1/5/8 (Cell Signaling), Smad1 (Zymed Laboratories Inc.), GAPDH (Santa Cruz Biotechnology), pERK (Cell Signaling), Nanog (BD Pharmingen), and Id1 (Santa Cruz) and incubated overnight at 4 °C with appropriate secondary antibodies. SuperSignal West Pico Chemiluminescence (Pierce) was used to detect the western blots.
Fluorescent Imaging
Reporter ES cells were sorted to GFP-high, GFP-low, or GFP-negative populations. Sorted cells were plated on chamber slides pre-coated with gelatin and cultured from 24 to 72 hours. Images were taken to visualize GFP fluorescence at 24, 48, and 72 hours. Hoechst 33342 dye was used in some experiments to visualize nuclei of live cells.
Clonogenicity assay
Sorted cells were plated at a low density (1000 cells per 10cm plate) onto gelatin-coated plates and allowed to grow for 6 days. Colony number and morphology were determined via microscopic examination.
Proliferation assay
Unsorted cells were plated onto gelatin-coated wells of a 24 well plate in quadruplicate and allowed to grow for 72 hrs. Cells were then lysed and total cell number in each well was indirectly determined using a fluorescence dye to quantify DNA content (Invitrogen).
Statistical analysis
Analysis of RNA was conducted in triplicate samples. Results were graphed to display means ± standard deviation. Student t-tests were performed to determine statistical significance of the data with a p value of < 0.05 considered statistically significant.
Results
Reporter models to monitor Nanog heterogeneity in ES cells
To monitor the dynamic expression of Nanog, two mouse ES cell lines harboring Nanog-GFP reporters were analyzed in detail. The TNG cell line harbors a GFP reporter targeted to the endogenous Nanog locus5. Additionally, a Nanog-GFP reporter BAC27 was introduced into AINV15 cells28, resulting in the BNG ES cell line. Both ES cell lines demonstrated distinct populations as assessed via flow cytometric analysis (Fig. 1A,B), with a small population of cells with undetectable GFP levels. Cells were sorted to negative, low, medium, high, and very-high GFP subpopulations. Via western and real-time expression analysis, these subpopulations of cells exhibited quantitatively distinct levels of Nanog expression (Fig. 1A,B, Supplemental Fig. 2). GFP-low TNG cells exhibited approximately 40% levels of Nanog transcript levels compared to Nanog-high cells (Fig. 1B), demonstrating the GFP reporter accurately reflects endogenous Nanog expression. Differences between the Nanog-GFP BAC reporter and the Nanog-GFP targeted allele reporter are minor and likely reflect subtle differences in the sensitivity of the GFP to report lower levels of Nanog. Thus GFP expression accurately reflected endogenous Nanog gene expression in both cell models.
To monitor the dynamic expression, subpopulations of cells were sorted on the basis of GFP expression and replated (Fig. 1D). After 72 hours, the sorted cells exhibited varied expression levels of the reporter. Specifically, GFP-negative sorted TNG cells exhibited some GFP expression, whereas GFP-positive sorted cells displayed some cells which lacked GFP expression. BNG cells initially sorted for high levels of GFP exhibited a tight ES colony morphology, with clearly defined borders. GFP-low cells had a more flattened appearance (Fig. 1D), but after several days of culture, cell clumps with tighter morphologies were apparent. These data demonstrate that the Nanog-BAC reporter models generated for this analysis accurately reflect endogenous Nanog expression and exhibit heterogeneous and dynamic expression.
To further characterize the gene activity of the subpopulations, the expression of several genes were monitored in sorted BNG cells. Several genes associated with stem cell self-renewal exhibited varied levels of expression within the Nanog expressing subpopulations. Higher levels of expression of Klf4 and Stella (Dppa3; Fig. 1C) were associated with high levels of Nanog; Klf4 levels were approximately 3-fold and 4-fold higher in GFP-high cells compared to GFP-low and GFP-negative cells, respectively. Tet1 and Tet2 were also seen to exhibit highly variable levels of expression. These loci encode proteins with roles in converting methylated DNA to 5-hydroxymethylcytosine, with functions in differentiation and pluripotency30, 31. Importantly, other pluripotency factors, such as Oct4 (Pou5f1) did not exhibit substantial variable expression levels in the Nanog-GFP subpopulations, consistent with previous studies5, 7 In contrast, genes such as Brachyury and Fgf8 were substantially higher in GFP-low subpopulations. Other markers associated with differentiation, such as the early neurectoderm marker Pax6, did not exhibit differential expression, suggesting the heterogeneous expression of some markers is not a generic phenomenon of differentiation. Indeed, Brachyury has previously been shown to be expressed at low levels in many ES cell lines and exhibits heterogeneous expression3. Thus, although the reporters reflect heterogeneous Nanog expression, a substantial number of other genes associated with pluripotency also exhibit variable levels of expression in these GFP subpopulations. These data are indicative of a complex network of dynamic gene expression in undifferentiated ES cells.
TGF-beta-related signaling is active in Nanog subpopulations
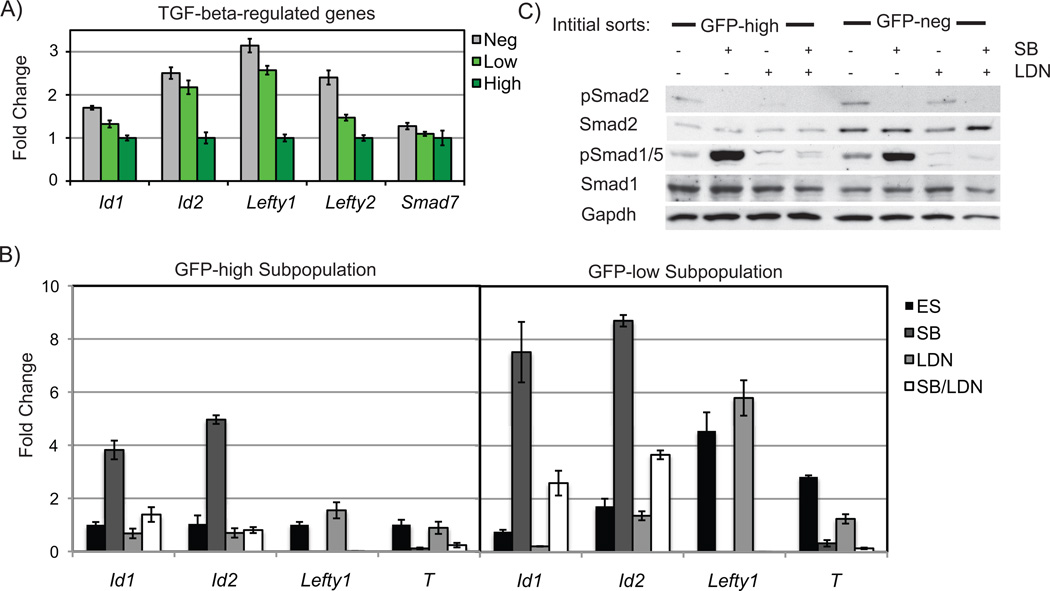
Our previous studies characterized a function for autocrine Nodal signaling in modulating the intracellular activity of BMP in undifferentiated mouse ES cells25. Given the activity of BMP signaling in maintaining ES cell self-renewal26, 32, we wished to determine roles of both Nodal and BMP signaling in stem cell heterogeneity. We first examined the expression of downstream targets of the Nodal and BMP signaling pathways in Nanog subpopulations, including Id genes (BMP targets) and the Nodal signaling targets Lefty1, Lefty2, Brachyury (T), and Fgf825, 26. These genes exhibited variable levels of expression in the Nanog subpopulations of BNG cells (Fig. 2A) and TNG ES cells (Supplemental Fig. 1), with higher levels of these factors associated with the lower expression of Nanog. One exception was Smad7, which is regulated in part by Nodal signaling, but exhibited similar levels of expression in all GFP subpopulations. Quantitative differences in the activity of BMP and Nodal signaling in different subpopulations was confirmed when the intracellular activities were examined via western analysis of the activated Smads. In sorted subpopulations of TNG ES cells, GFP-negative cells had slightly higher levels of phospho-Smad2 and phospho-Smad1/5 than the GFP-positive cells (Fig. 2C).
Figure 2. Activity of TGF-beta signaling in Nanog subpopulations.
A) Expression of TGFbeta regulated genes was lower in GFP-high than GFP-low cells. B) GFP-low and GFP-high cells had similar expression changes to Nodal and BMP signaling inhibition. C) TNG cells were treated in an identical manner and protein was analyzed for levels of phosph-Smad2 and phospho-Smad1/5 activity.
The sorted Nanog subpopulations were analyzed to compare their responses to TGF-beta signaling modulation. Two specific pharmacological inhibitors were used to target BMP or Nodal signaling. LDN193189 (LDN), inhibits BMP type I receptors ALK2/333, and SB431542 (SB) targets the Activin type I receptors ALK4/5/734, 35. Inhibition of BMP signaling reduced pSmad1/5 levels in GFP-positive and GFP-negative expressing TNG cells (Fig. 2C). In response to Nodal inhibition, both GFP-positive and GFP-negative populations decreased phosphorylation of Smad2 and drastically increased phosphorylation of Smad1/5, consistent with Nodal signaling acting as a negative regulator of BMP signaling via Smad7 regulation25. Decreased expression of Lefty1 and T was observed when Nodal signaling was inhibited, and Id1 and Id2 expression was enhanced (Fig. 2B), although quantitative differences in the magnitude of response was observed in different subpopulations. For example, SB treatment of GFP-low cells induced Id1 expression 10-fold, while GFP-high cells responded with a 4-fold upregulation. Significant expression of some targets of the Nodal signaling pathway, such as T, were largely restricted to the Nanog-low population. Inhibition of BMP signaling repressed Id1 expression to a higher degree in GFP-low cells compared to GFP-high cells. Dual inhibition of Nodal and BMP significantly decreased the SB-mediated Id induction in both subpopulations. Signaling modulation via other antagonists (Nodal antagonist SB505124 or the BMP antagonist Noggin) demonstrated similar findings (Supplemental Fig. 3). Thus, both BMP and Nodal signaling are active in distinct Nanog subpopulations, with subtle quantitative differences; Nanog-high subpopulations have lower intracellular activity of both pathways. Importantly, the regulation of BMP signaling via Nodal signaling25 is maintained in all ES cell subpopulations.
TGF-beta signaling modulates dynamic heterogeneity
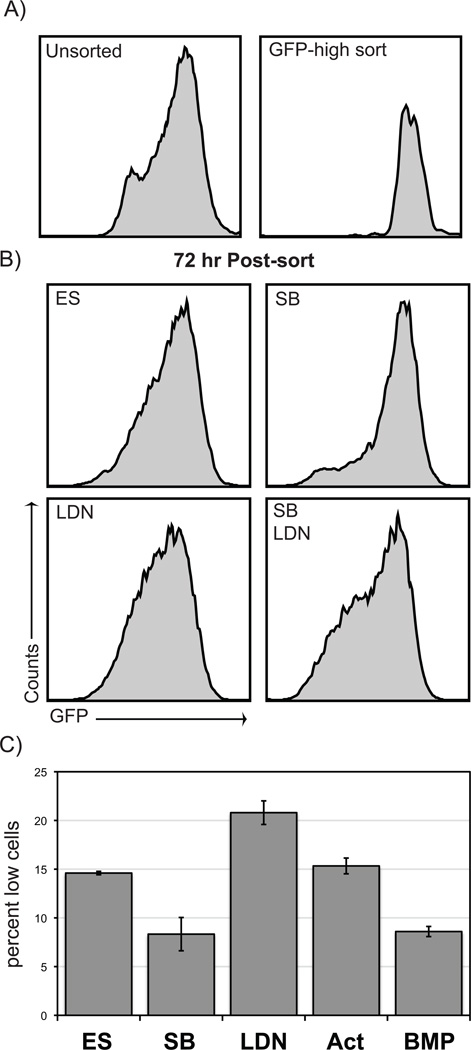
We wished to determine if altered BMP or Nodal signaling modulates the capacity of ES cell subpopulations to dynamically alter their phenotype. To monitor the dynamic transition of ES cells, subpopulations of Nanog-GFP cells were purified, replated for further culture, and then analyzed for the resulting heterogeneity via flow analysis. Nanog-high ES cells were purified and replated in SB supplemented media for 72 hours and then flow analyzed. In control cultures, cells that were initially GFP-high exhibited broader distribution of Nanog expressing subpopulations (Fig. 3B), with significant number of GFP-medium and -low expressing populations. In contrast, Nodal inhibition by SB significantly enhanced the capacity of GFP-high cells to remain in the GFP-high phenotypic state. SB treated cells displayed a reduction in cells of the GFP-low phenotypic state (decreased GFP-low cells by 43% compared to control; Fig. 3B; Supplemental Fig. 4). Similar, and even more striking, results were seen when the purified GFP-low subpopulation was analyzed (Fig. 4A). Cultures of GFP-low cells in SB supplemented media resulted in 47% more GFP-high and -very high cells compared to control conditions. Treatment of purified GFP-high cells with another Nodal pharmacological inhibitor, SB505124, also decreased the percentage of cells that transitioned to a GFP-low state (Supplemental Fig. 3). Treatment of cells with recombinant activin A did not result in a significant effect on heterogeneity (Fig. 3C). This result is consistent with our previous data showing that treatment with recombinant activin has a modest effect on Nodal downstream targets25, likely due to the signaling pathway being nearly saturated from autocrine production of Nodal. These results indicate that autocrine Nodal signaling influences the dynamic heterogeneity of undifferentiated ES cells, acting to direct ES cells toward the Nanog-low phenotypic state.
Figure 3. Nanog subpopulation dynamics in response to TGF-beta modulation.
A,B) GFP-high sorted cells were cultured and then flow analyzed for GFP expression. SB treatment significantly decreased the number of GFP-low cells. In contrast, both LDN treatment and SB+LDN cotreatment increased the number of GFP-low cells. C) Graph depicts triplicate samples.
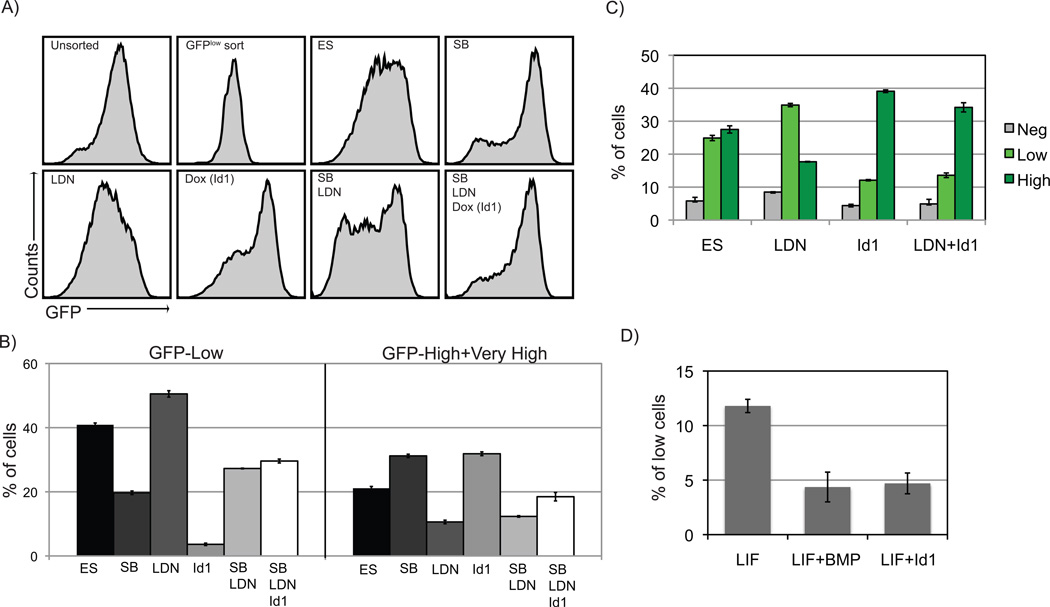
Figure 4. Id1 overexpression.
A) Id1 overexpression significantly enhanced the percentage of GFP-high cells and significantly attenuated the effects of SB+LDN treatment. B) Graphs summarize data from triplicate experiments. C) Id1 overexpression decreased the effects of LDN treatment on Nanog-GFP expression. D) In KOSR, Id1 induced a similar reduction of GFP-low cells compared to recombinant BMP stimulation.
Inhibition of BMP signaling had a dramatically different effect on Nanog heterogeneity compared to Nodal inhibition. Using either the LDN pharmacological inhibitor or the recombinant BMP antagonist Noggin, both GFP-low and GFP-high subpopulations of BNG ES cells demonstrated an increased propensity to shift towards the Nanog-low expressing phenotype. (Fig. 3 & 4 and Supplemental Fig. 3 & 4). Supplementation of media with recombination BMP4 resulted in the dramatic enhancement of maintaining the Nanog-high state (Fig. 3C); this response was as effective as SB treatment. These data demonstrate that BMP signaling is important for maintaining the Nanog-high state in serum-based media, with BMP and Nodal signaling having opposing activities in regulating heterogeneity.
To determine the hierarchy of effects of Nodal or BMP signaling in ES cell heterogeneity, both pathways were inhibited by simultaneous treatment with both SB and LDN. Sorted BNG cells grown in the combination of SB and LDN exhibited a profile of heterogeneity similar to LDN treatment alone (Fig. 3B, 4B). The effect of Nodal inhibition via SB on heterogeneity, namely the increase in the GFP-high subpopulations, was eliminated in the combined treatment of SB and LDN. These results suggest that the SB-induced propensity to maintain Nanog-high populations is a BMP-dependent process.
Forced expression of Id1 enhances Nanog-high state
The Id factors are prominent targets of BMP signaling36, and are critical downstream components of the self-renewal activity of BMP signaling in mouse ES cells26. Nodal inhibition enhances BMP activity and Id expression in undifferentiated ES cells25 (Fig. 2B). We wished to determine if enhancement of ES cells to remain in the GFP-high phenotypic state in response to Nodal inhibition is due to enhanced BMP signaling and associated increase in Id expression. A doxycycline-inducible Id1 transgene was targeted into BNG cells via CRE-mediated transgenesis28 to generate the BNG-Id1 cell line. Treatment of BNG-Id1 cells with doxycycline (dox) induced an approximate 20-fold enhancement of Id1 expression (Supplemental Fig. 5). Sorted GFP-low BNG-Id1 cells were plated in combinations of SB, LDN, and dox supplemented culture media. After three-day treatment of dox, BNG-Id1 ES cells had 2.2-fold fewer GFP-low cells along with a concominant increase in cells with high and very high Nanog-GFP expression in comparison to cells grown in ES media (Fig. 4A and B), and, closely mimicking the effects of SB treatment. Treatment of BNG-Id1 ES cells with dox also largely ablated the effects of LDN treatment (Fig. 4C). Thus, under basal ES media conditions, forced Id1 expression directs cells toward the Nanog-high state.
Components of fetal bovine serum are known to activate the BMP signaling pathway. To further study the roles of Id1 in Nanog heterogeneity under conditions without BMP agonists, BNG-Id1 cells were grown in knock-out serum replacement (KOSR) media. GFP-high sorted BNG-Id1 cells grown in KOSR media exhibited an almost 3-fold decrease in Nanog-low cells when BMP was added to the media (Fig. 4D). When dox was added to induce Id1 expression in KOSR media, the proportion of GFP-low cells were identical to BMP treatment (Fig. 4D). These results strongly suggest that the Id factors are critical downstream targets of BMP signaling in regulating heterogeneity. The enhanced capacity of ES cells to remain in the Nanog-high epigenetic state when Nodal signaling is inhibited is thus due to enhanced BMP activity and resulting increase in Id gene expression.
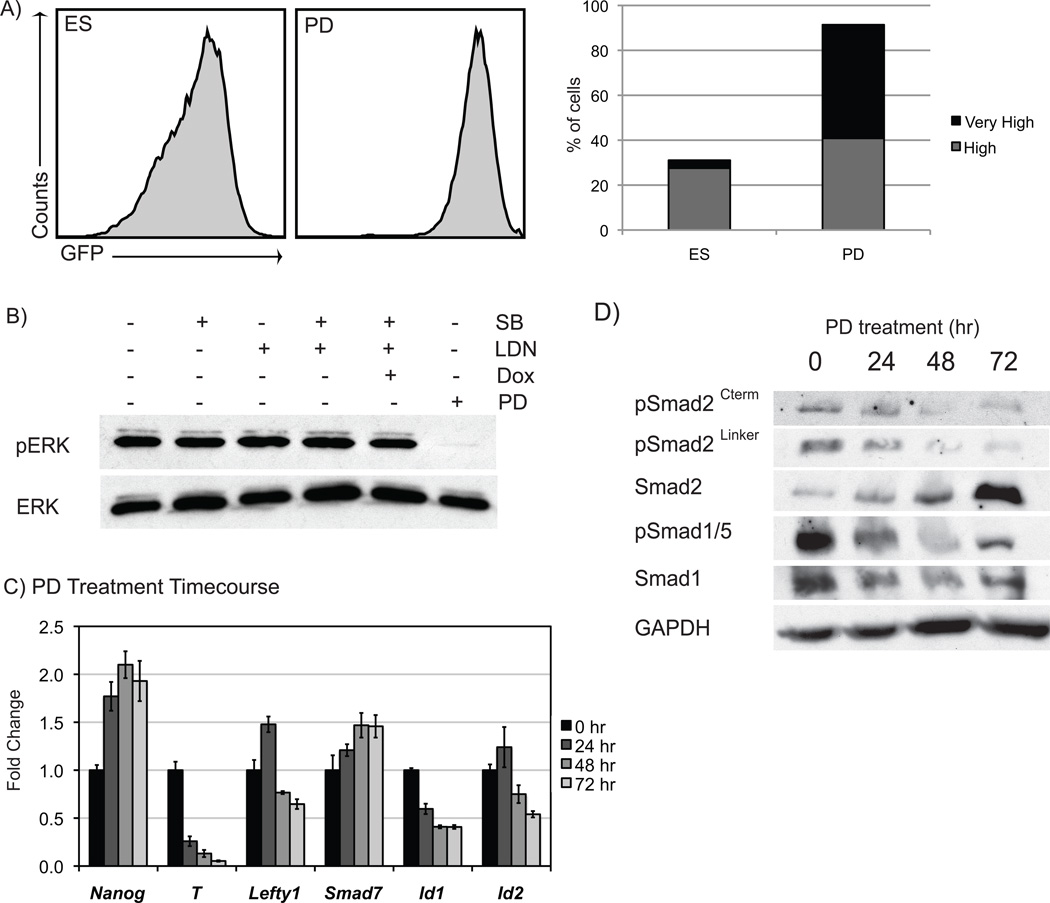
Inhibition of FGF4/MEK signaling attenuates Smad phosphorylation and dramatically increases Nanog-high phenotypic state
FGF signaling plays an important role in modulating ES cell heterogeneity via its regulation of the MEK signaling pathway20, 21. PD0325901 (PD), a potent and selective non-competitive inhibitor of MEK137, inhibits FGF signaling and is a crucial component of the 2i cocktail shown to maintain undifferentiated mouse ES cells38. GFP-high BNG ES cells treated with PD in serum-based media for 72 hours displayed a dramatic increase in the number of GFP-high and -very high cells (Fig. 5A). PD treatment increased the number of GFP-very high cells to 51%, compared to the 4% of GFP-very high cells in control ES media cultures. Nanog mRNA was also elevated approximately 2-fold by PD treatments (Fig. 5C). To determine if FGF signaling is modulated in response to Nodal or BMP signaling, we examined the downstream signaling pathway via western analysis. Phospho-ERK expression was greatly diminished by PD treatment but was unaffected by SB or LDN treatments (Fig. 5B). These data suggest that the effects of Nodal or BMP signaling on heterogeneity either occur independent of FGF signaling, or are downstream of ERK in FGF-MEK signaling.
Figure 5. Inhibition of FGF/MEK signaling.
A) PD treatment enhanced GFP-high and -very high cells. B) SB and LDN treatments did not affect levels of pERK while PD treatment vastly reduced pERK. Inhibition of FGF/MEK signaling reduced the phosphorylation of Smad2 and Smad1/5 (C) and decreased the expression of many TGF-beta-related genes (D).
To analyze the effects of MEK inhibition on Nodal and BMP signaling, a PD treatment time course was conducted, and RNA and protein samples were analyzed. Corresponding with a high level of Nanog expression, Brachyury (T) expression was decreased by PD treatment (Fig. 5C). And while inhibitory Smad7 levels increased, other markers of Nodal and BMP activity (Lefty1 and the Id genes) decreased transcript expression. Treatment with PD also diminished Smad phosphorylation of both arms of the TGF-beta pathway (Fig. 5D). Smad1/5 showed decreased phosphorylation after 24 hours, and significant decreases in the phosphorylation of Smad2 were demonstrated after 48 hours. Multiple Smad2 serine residues decreased phosphorylation after MEK1 inhibition; the Ser465/467 residue in the C terminal region and the Ser245/250/255 in the linker region showed loss of phosphorylation. These data are consistent with decreased Nodal and BMP activity associated with the GFP-high and -very high phenotypic state, and suggesting that FGF4 signaling provides a permissive environment for Nodal and BMP activity.
Nodal and BMP signaling share function in regulating Nanog expression and inhibiting differentiation
A consistent increase in GFP-negative cells was observed in response to combined treatment of SB and LDN, most strikingly in Nanog-low purified subpopulations (Fig. 4A and B). These results suggest that Nodal and BMP signaling may have an additional shared function in keeping ES cells from entering the GFP-negative epigenetic state. To examine this overlap in function, we generated a BNG ES cell line with a dox-inducible Smad7 cassette to enhance Smad7 expression and impede both BMP and Nodal signaling24. Dox-treated BNG-Smad7 cells increased Smad7 levels 3-fold relative to cells grown in media alone (Supplemental Fig. 5).
BNG-Smad7 cells were purified into GFP-high and GFP-low subpopulations and treated for three days in different media conditions to modulate Nodal and BMP signaling. Treatment with dox or with SB+LDN significantly increased the population of GFP-negative cells in both GFP-low and -high sorted cells (Fig. 6A and B). Triple treatment with SB+LDN+dox induced the most dramatic increase of GFP-negative cells, particularly in the GFP-low sorted cells, which resulted in over 45% of the cell population becoming GFP-negative. A significant reduction in proliferation was observed when both Nodal and BMP signaling are inhibited (Supplemental Fig. 6C). The increase in GFP-negative cells in response to combined inhibition of Nodal and BMP signaling was also observed in serum-free media. After 5 days, BNG-Smad7 cells grown in KOSR supplemented with media lacking BMP but including LIF, SB, LDN, and dox inhibited an 8.7-fold increase in Nanog-negative cells compared to cells grown in LIF and BMP-supplemented media (Supplemental Fig. 6A). These data suggest that the combined strong inhibition of Nodal and BMP signaling results the accumulation of Nanog-negative cells, even in the presence of LIF.
Figure 6. TGF-beta inhibition decreases Nanog.
In GFP-high (A) and GFP-low (B) sorted ES cells, combined inhibition of Nodal and BMP signaling increased the percentage of GFP-negative cells. C,D) Under SB+LDN+Smad7 growth conditions, GFP-negative cells with differentiated morphologies were apparent (see bracket). RNA analysis showed Nanog expression after 24 hours (E) and differentiation markers after 6 days (F).
We examined endogenous Nanog expression levels in response to short term inhibition of Nodal and BMP signaling in unsorted cells. Single treatments of SB or LDN did not change Nanog expression after 24 hours (Fig. 6D); however, when SB and LDN were co-administered, Nanog expression dropped to approximately 80% that of untreated cells. Strong inhibition of both Nodal and BMP signaling via triple treatment of dox, SB, and LDN further reduced Nanog mRNA levels in both serum-based media (Fig. 6D) and in serum free media (Supplemental Figure 6D). When GFP-high BNG-Smad7 ES cells were grown in 72 hr culture in SB+LDN+dox, striking cell morphologies were observed (Fig. 6C). In addition to cells with coherent undifferentiated morphologies, a substantial number of cells lacking GFP expression were observed with flattened morphology, either with either a squamous epithelial phenotype or a loosely coherent mesenchymal phenotype. These phenotypes are indicative of highly differentiated cells, which were rarely apparent in untreated cells. This highly differentiated morphology was also preponderant in SB+LDN+dox treated cells in a clonal outgrowth assay. After 6 days of inhibition of both Nodal and BMP signaling, single plated cells demonstrated a significant reduction in capacity for clonal outgrowth, with SB+LDN+dox-treated cells displaying a x reduction in clone number. Many of the resulting colonies grown in SB+LDN+dox media consisted solely of cells with a highly differentiated morphology lacking Nanog expression (46%; Fig. 6D, Supplemental Fig. 6B), a phenotype rarely observed in clones grown in untreated media. The Nanog-low sorted cells were more sensitive to SB+LDN+dox treatment compared to Nanog-high cells; Nanog-low cells had a significantly reduced capacity for clonogenicity and a dramatically increased number of clones consisting of differentiated cells in response to SB+LDN+dox (Supplemental Fig. 6B).
Gene expression was examined in cells with combined Nodal and BMP inhibition for 6 days in both serum-based media and serum-free media (Fig. 6F and Supplemental Figure 6D). SB+LDN+dox treatment mildly enhanced the expression of neural (Pax6) and trophectoderm (Gata2) markers. The upregulation of Gata2 is consistent with previous observations of SB effects on ES cells25. However, substantial increases in genes associated with mesendodermal differentiation were observed in response to combined Nodal and BMP inhibition, including an 8-fold increase in FoxA2 and 6-fold increase in the trophectoderm/posterior streak marker Cdx2. Thus a variety of differentiation markers are derepressed in response to combined loss of Nodal and BMP activity in undifferentiated ES cells. These data point to a previously undescribed and shared role for exogenous BMP and autocrine Nodal signaling in regulating Nanog expression and maintaining the undifferentiated state of mouse ES cells when cultured in LIF.
Given that BMP signaling regulates Id gene expression, we wished to determine if Id factors are important components of the combined role of Nodal and BMP in regulating Nanog expression. To test this, the BNG-Id1 cell line was used to force Id expression in the context of Nodal and BMP inhibition via SB and LDN. After 6 days, combined treatment of SB +LDN lowered Nanog expression (Fig. 6E). Analysis of BNG-Id1 cells revealed that forced expression of Id1 only partially enhanced the reduced expression of Nanog in response to SB and LDN after 24 hours (Fig. 6E). Expression of Id1 was thus not sufficient to raise Nanog levels to basal expression, suggesting that Id-independent mechanisms exist by which Nodal and BMP signaling pathways cooperate to maintain Nanog expression.
Discussion
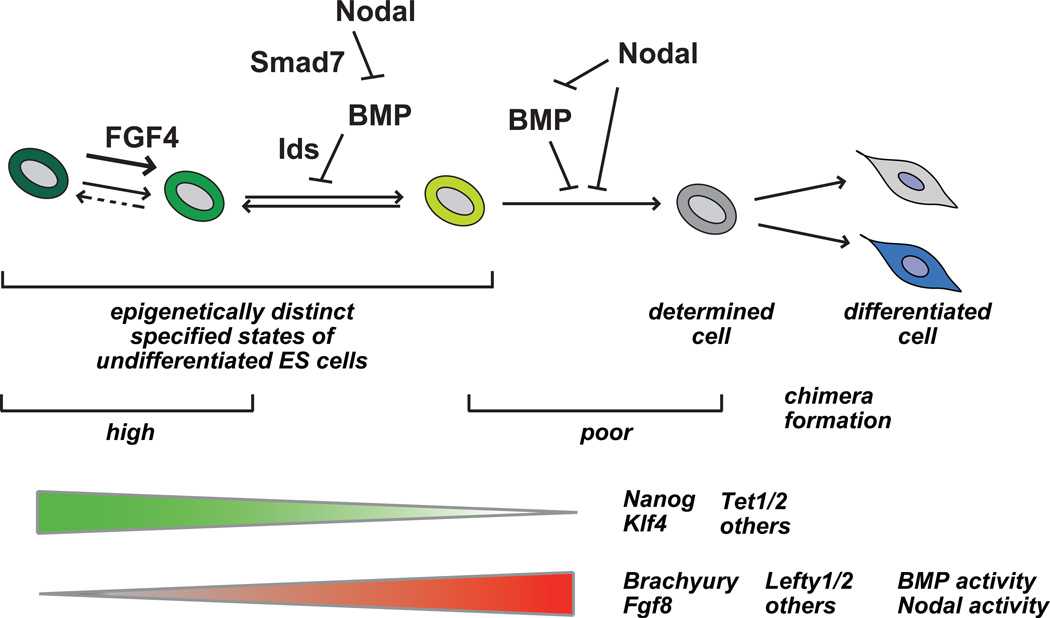
In this report we sought to characterize the role of the TGF-beta-related signaling pathways on dynamic heterogeneity. Using two Nanog reporter models, our results show a substantial number of genes with variable expression associated with Nanog heterogeneity, including several downstream components of Nodal and BMP signaling. These studies demonstrate that modulation of either Nodal or BMP signaling substantially influences the dynamic phenotypic transition of ES cells as assessed by Nanog expression. Our results point to a regulatory cascade of autocrine Nodal signaling modulating BMP activity, which through the regulation of Id gene expression, influences the dynamic expression of Nanog in ES cells (Fig. 7) in serum-based media. These data also indicate a novel shared requirement for Nodal and BMP signaling to maintain the Nanog-low state. These observations, in combination with other studies21, denote a central role for the intracellular activities of BMP, Nodal, and FGF signaling in defining the steady-state distribution of heterogeneity of undifferentiated ES cells in culture. Thus heterogeneity is a regulated process, under the control of several extrinsic and intrinsic signals.
Figure 7. Model for Nodal and BMP function in regulating dynamic heterogeneity of mouse ES cells in serum-based culture.
See text for details.
Distinct activities of Nodal and BMP are observed in differing subpopulations of undifferentiated mouse ES cells. These functional differences are observed by the direct targets of Nodal signaling, which are distinct in Nanog-high and –low subpopulations. For example, Lefty1 and Lefty2 are major direct targets of Nodal in both Nanog-high and -low subpopulations (Fig, 2B). The essential role of Nodal in Nanog-high cells is largely limited to its regulation of BMP signaling. However, in Nanog-low cells, Nodal signaling is more complex, and is essential for the Nanog-low restricted expression of Brachyury. Additionally, Nodal is responsible for the repression of several markers of differentiation25 (Fig. 6F) in the Nanog-low population. Our work has uncovered a context-dependent shared role for Nodal and BMP signaling in the inhibition of differentiation, an essential activity which is restricted to Nanog-low cells. Thus the activities and essential functions of Nodal and BMP signaling are context dependent, in that the Nanog epigenetic state determines distinct activities of these pathways. Future work will need to be done to examine the differences in the these phenotypic states, perhaps at the level of chromatin architecture, which influence the regulation of Nodal and BMP targets.
The combined inhibition of BMP and Nodal signaling results in a highly differentiated cell morphology and upregulation of genes associated with ectoderm and mesoderm differentiation, which is more evident in Nanog-low cells (Fig. 6). When both pathways are inhibited, an immediate reduction of Nanog transcript levels are also observed within 24 hr (Fig. 6D), suggesting that the regulation of Nanog expression is responsible for this observed differentiation. The observation that Nanog-low cells are more sensitive to the combined loss of Nodal and BMP signaling is consistent with this possible mechanism. The mechanism by which Nodal and BMP share function to maintain Nanog expression and repress differentiation will be an important topic for later studies. It remains to be determined if these pathways have a shared role for in the direct transcriptional regulation of Nanog expression via Smad factors, or if this regulation is indirect. BMP signaling may directly regulate Nanog gene expression in mouse ES cells, as the SMAD1 protein has been found to be localized to genomic locations near the Nanog locus39, although the functional consequences of this localization have not been examined. Studies in human ES cells have indicated a role for Nodal signaling in directly regulating NANOG expression via Smad2/3 complexes22. The Nodal target Brachyury has also been shown to regulate Nanog expression3, providing a possible indirect mechanism of control by Nodal signaling. There is little precedence for shared functionality of these distinct arms of the TGF-beta superfamily; thus a mechanistic understanding of this shared activity will be of interest to the TGF-beta signaling community.
The self-renewal activity of BMP signaling in mouse ES cells functions via regulation of Id signaling26. Our results show that the control of heterogeneity off undifferentiated ES cells is also due to its regulation of the Id factors, as the upregulation of Id1 enhances the capacity of ES cells to remain or attain the Nanog-high state. We have observed an increased activity of BMP signaling in the Nanog-low subpopulations (Fig. 2), which may suggest a potential feedback response to maintain the undifferentiated state and to direct Nanog-low cells toward the Nanog-high state. The biochemical activity of the Id factors in BMP-dependent functions of both self-renewal and Nanog heterogeneity are not known. Id factors are well-known antagonists of basic-helix-loop-helix (bHLH) transcription factors40, and function by binding to bHLH proteins and inhibit their capacity for heterodimerization and DNA binding. It remains to be determined if Id factors antagonize bHLH factors in stem cell self-renewal. Non-bHLH binding partners of Id factors have been reported as well41,42, suggesting other potential protein interaction partners. Biochemical identification of Id binding partners in undifferentiated ES cells will be required to understand the role of these factors in pluripotency.
In serum-based media, Nanog-very high cells represent a fairly rare cell type (5%), suggesting a strict upper limit to Nanog expression levels in standard culture conditions in serum. Importantly, inhibition of FGF-MEK signaling with PD greatly enhances the capacity of ES cells to exist in the Nanog-very high state. These data strongly indicate that autocrine FGF signaling via expression of FGF421 is a major limiter of ES cells to exist in the phenotypic state characterized by very-high levels of Nanog. Inhibition of FGF-MEK signaling results in a delayed reduction of both Nodal and BMP intracellular signaling (Fig. 5). FGF signaling thus provides a permissive environment for Nodal and BMP signaling in undifferentiated ES cells. We also observe relatively reduced BMP and Nodal signaling in unmanipulated Nanog-high epigenetic state compared to Nanog-low cells (Fig. 2). These data suggest that the reduced BMP and Nodal signaling in response to PD treatment is indirect, and are a reflection of the phenotypic status of the Nanog-very high state. Previous studies have indicated a negative regulatory role for NANOG protein in modulating BMP signaling via direct interaction with SMAD13, suggesting a potential mechanism in which increased Nanog expression in response to PD may modulate BMP and possibly Nodal activity.
Computational models have attempted to explain heterogeneity as a readout of ‘transcriptional noise’, in which small stochastic changes in gene expression give rise to variable expression levels of some genes and concomitant phenotypic changes43. These models generally do not take into account the known complexity of how critical genes such as Nanog are regulated3, 44–47. The data presented here, along with other studies, demonstrate that many genes in fact show heterogeneous expression patterns in ES cells (Fig. 7), including a variety of genes with roles in stem cell self-renewal. These observations strongly suggest that the dynamic equilibrium of the pluripotent phenotype is established by numerous controls, likely to involve networks of self-renewal factors, as well as multiple signaling pathways that impinge upon these transcriptional networks. Heterogeneity is thus a fundamental aspect of pluripotent stem cells and is intrinsically linked to the self-renewal factors that control the stem cell identity. The capacity for ES cells to exhibit distinct molecular signatures but similar differentiation potential has been observed in expression profiles of cells grown in serum-based media and in ‘2i’ conditions48, and are consistent with observations of the complex molecular differences present in subpopulations of heterogeneous populations.
Supplementary Material
TNG cells were sorted to GFP-positive and GFP-negative subpopulations. A) Gene expression between the subpopulations was analyzed after 24 hours. Sorted GFP-positive and GFP-negative cells were treated with SB, LDN, or SB+LDN. Cultures were reanalyzed for GFP expression after 3 (B) or 6 (C) days. D) Graphs depict triplicate samples.
A) BNG cells were sorted to subpopulations. They were flow analyzed to determine GFP expression. B) Expression of Nanog and T in BNG GFP-subpopulations after 24 hours. C) Nanog protein levels from subpopulations immediately after sorting. D) Mean fluorescence readings from Nanog-GFP subpopulations. E) Nanog and T expression correlate.
A, B) Comparison of LDN to Noggin and of SB431542 to SB505124 for T, Lefty and Id factors. C) Representative flow profiles after 3 days of treatment. D) Percent of GFP-low cells 3 days after treatment.
BNG ES cells were treated with SB, LDN, and PD for 3 days. Cells were analyzed by flow to determine level of Nanog-GFP expression.
A) Dox treatment of AINV-BNG Id1 cells increased Id1 expression over 20-fold. B) Protein analysis also revealed a striking increase in Id1 protein. C) Dox treatment of AINV-Smad7 increased Smad7 expression by approximately 3-fold.
A) The percentages of GFP-neg and GFP-low cells of BNG-Smad7 GFP-high sorted cells is increased with SB+LDN and SB+LDN+Smad7 treatments in serum-free media. B. Reduced colony outgrowth and increased differentiation in clonogenicity assay in serum media. C) Lower cell number in response to SB+LDN+dox after 72 hr as assessed by DNA content. D) Gene expression analysis demonstrated increased expression of Foxa2, Pax6, and Cdx2 following SB+LDN and SB+LDN+Smad7 treatments in serum-free media.
Acknowledgements
We thank the KUMC Flow Cytometry Core Facility for assistance in cell sorting and analysis and Stan Fernald in the KUMC Imaging Core for assistance in microscopy. The TNG ES cell line was kindly provided by Ian Chambers, and the Nanog-GFP BAC was kindly provided by Christoph Schaniel.
Research support:
This work was supported by a National Institutes of Health postdoctoral training grant (KEG, T32HD007455) and a National Institutes of Health COBRE program project grant (JLV, P20RR024214).
Footnotes
Author contributions:
Katherine E. Galvin-Burgess: Conception and design, Collection and/or assembly of data, Data analysis and interpretation, Manuscript writing
Emily D. Travis: Collection and/or assembly of data
Kelsey E. Pierson: Collection and/or assembly of data
Jay L. Vivian: Conception and design, Financial support, Collection and/or assembly of data, Data analysis and interpretation, Manuscript writing, Final approval of manuscript
The authors declare no conflict of interest.
References
- 1.Smith AG. Embryo-derived stem cells: of mice and men. Annu Rev Cell Dev Biol. 2001;17:435–462. doi: 10.1146/annurev.cellbio.17.1.435. [DOI] [PubMed] [Google Scholar]
- 2.Ohtsuka S, Dalton S. Molecular and biological properties of pluripotent embryonic stem cells. Gene Ther. 2008;15:74–81. doi: 10.1038/sj.gt.3303065. [DOI] [PubMed] [Google Scholar]
- 3.Suzuki A, Raya A, Kawakami Y, et al. Nanog binds to Smad1 and blocks bone morphogenetic protein-induced differentiation of embryonic stem cells. Proc Natl Acad Sci U S A. 2006;103:10294–10299. doi: 10.1073/pnas.0506945103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suzuki A, Raya A, Kawakami Y, et al. Maintenance of embryonic stem cell pluripotency by Nanog-mediated reversal of mesoderm specification. Nat Clin Pract Cardiovasc Med. 2006;3(Suppl 1):S114–S122. doi: 10.1038/ncpcardio0442. [DOI] [PubMed] [Google Scholar]
- 5.Chambers I, Silva J, Colby D, et al. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- 6.Singh AM, Hamazaki T, Hankowski KE, et al. A heterogeneous expression pattern for Nanog in embryonic stem cells. Stem Cells. 2007;25:2534–2542. doi: 10.1634/stemcells.2007-0126. [DOI] [PubMed] [Google Scholar]
- 7.Hayashi K, Lopes SM, Tang F, et al. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell. 2008;3:391–401. doi: 10.1016/j.stem.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toyooka Y, Shimosato D, Murakami K, et al. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development. 2008;135:909–918. doi: 10.1242/dev.017400. [DOI] [PubMed] [Google Scholar]
- 9.Vieyra DS, Rosen A, Goodell MA. Identification and characterization of side population cells in embryonic stem cell cultures. Stem Cells Dev. 2009;18:1155–1166. doi: 10.1089/scd.2008.0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka TS. Transcriptional heterogeneity in mouse embryonic stem cells. Reprod Fertil Dev. 2009;21:67–75. doi: 10.1071/rd08219. [DOI] [PubMed] [Google Scholar]
- 11.Han DW, Tapia N, Joo JY, et al. Epiblast stem cell subpopulations represent mouse embryos of distinct pregastrulation stages. Cell. 2010;143:617–627. doi: 10.1016/j.cell.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 12.Stewart MH, Bosse M, Chadwick K, et al. Clonal isolation of hESCs reveals heterogeneity within the pluripotent stem cell compartment. Nat Methods. 2006;3:807–815. doi: 10.1038/nmeth939. [DOI] [PubMed] [Google Scholar]
- 13.Hough SR, Laslett AL, Grimmond SB, et al. A continuum of cell states spans pluripotency and lineage commitment in human embryonic stem cells. PLoS One. 2009;4:e7708. doi: 10.1371/journal.pone.0007708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graf T, Stadtfeld M. Heterogeneity of embryonic and adult stem cells. Cell Stem Cell. 2008;3:480–483. doi: 10.1016/j.stem.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 15.Boiani M, Scholer HR. Regulatory networks in embryo-derived pluripotent stem cells. Nat Rev Mol Cell Biol. 2005;6:872–884. doi: 10.1038/nrm1744. [DOI] [PubMed] [Google Scholar]
- 16.Loh YH, Wu Q, Chew JL, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 17.Kim J, Chu J, Shen X, et al. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chambers I, Colby D, Robertson M, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 19.Hamazaki T, Kehoe SM, Nakano T, et al. The Grb2/Mek pathway represses Nanog in murine embryonic stem cells. Mol Cell Biol. 2006;26:7539–7549. doi: 10.1128/MCB.00508-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lanner F, Rossant J. The role of FGF/Erk signaling in pluripotent cells. Development. 2010;137:3351–3360. doi: 10.1242/dev.050146. [DOI] [PubMed] [Google Scholar]
- 21.Lanner F, Lee KL, Sohl M, et al. Heparan sulfation-dependent fibroblast growth factor signaling maintains embryonic stem cells primed for differentiation in a heterogeneous state. Stem Cells. 2010;28:191–200. doi: 10.1002/stem.265. [DOI] [PubMed] [Google Scholar]
- 22.Xu RH, Sampsell-Barron TL, Gu F, et al. NANOG is a direct target of TGFbeta/activin-mediated SMAD signaling in human ESCs. Cell Stem Cell. 2008;3:196–206. doi: 10.1016/j.stem.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vallier L, Mendjan S, Brown S, et al. Activin/Nodal signalling maintains pluripotency by controlling Nanog expression. Development. 2009;136:1339–1349. doi: 10.1242/dev.033951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogawa K, Saito A, Matsui H, et al. Activin-Nodal signaling is involved in propagation of mouse embryonic stem cells. J Cell Sci. 2007;120:55–65. doi: 10.1242/jcs.03296. [DOI] [PubMed] [Google Scholar]
- 25.Galvin KE, Travis ED, Yee D, et al. Nodal signaling regulates the bone morphogenic protein pluripotency pathway in mouse embryonic stem cells. J Biol Chem. 2010;285:19747–19756. doi: 10.1074/jbc.M109.077347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ying QL, Nichols J, Chambers I, et al. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- 27.Schaniel C, Li F, Schafer XL, et al. Delivery of short hairpin RNAs--triggers of gene silencing--into mouse embryonic stem cells. Nat Methods. 2006;3:397–400. doi: 10.1038/nmeth0506-397. [DOI] [PubMed] [Google Scholar]
- 28.Kyba M, Perlingeiro RC, Daley GQ. HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell. 2002;109:29–37. doi: 10.1016/s0092-8674(02)00680-3. [DOI] [PubMed] [Google Scholar]
- 29.Magin-Lachmann C, Kotzamanis G, D'Aiuto L, et al. In vitro and in vivo delivery of intact BAC DNA -- comparison of different methods. J Gene Med. 2004;6:195–209. doi: 10.1002/jgm.481. [DOI] [PubMed] [Google Scholar]
- 30.Wu H, D'Alessio AC, Ito S, et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473:389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koh KP, Yabuuchi A, Rao S, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qi X, Li TG, Hao J, et al. BMP4 supports self-renewal of embryonic stem cells by inhibiting mitogen-activated protein kinase pathways. Proc Natl Acad Sci U S A. 2004;101:6027–6032. doi: 10.1073/pnas.0401367101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu PB, Deng DY, Lai CS, et al. BMP type I receptor inhibition reduces heterotopic [corrected] ossification. Nat Med. 2008;14:1363–1369. doi: 10.1038/nm.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inman GJ, Nicolas FJ, Callahan JF, et al. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol Pharmacol. 2002;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- 35.Laping NJ, Grygielko E, Mathur A, et al. Inhibition of transforming growth factor (TGF)-beta1-induced extracellular matrix with a novel inhibitor of the TGF-beta type I receptor kinase activity: SB-431542. Mol Pharmacol. 2002;62:58–64. doi: 10.1124/mol.62.1.58. [DOI] [PubMed] [Google Scholar]
- 36.Hollnagel A, Oehlmann V, Heymer J, et al. Id genes are direct targets of bone morphogenetic protein induction in embryonic stem cells. J Biol Chem. 1999;274:19838–19845. doi: 10.1074/jbc.274.28.19838. [DOI] [PubMed] [Google Scholar]
- 37.Bain J, Plater L, Elliott M, et al. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ying QL, Wray J, Nichols J, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen X, Xu H, Yuan P, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 40.Benezra R, Davis RL, Lockshon D, et al. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 41.Lasorella A, Iavarone A, Israel MA. Id2 specifically alters regulation of the cell cycle by tumor suppressor proteins. Mol Cell Biol. 1996;16:2570–2578. doi: 10.1128/mcb.16.6.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yates PR, Atherton GT, Deed RW, et al. Id helix-loop-helix proteins inhibit nucleoprotein complex formation by the TCF ETS-domain transcription factors. EMBO J. 1999;18:968–976. doi: 10.1093/emboj/18.4.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalmar T, Lim C, Hayward P, et al. Regulated fluctuations in nanog expression mediate cell fate decisions in embryonic stem cells. PLoS Biol. 2009;7:e1000149. doi: 10.1371/journal.pbio.1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodda DJ, Chew JL, Lim LH, et al. Transcriptional regulation of nanog by OCT4 and SOX2. J Biol Chem. 2005;280:24731–24737. doi: 10.1074/jbc.M502573200. [DOI] [PubMed] [Google Scholar]
- 45.Wu da Y, Yao Z. Isolation and characterization of the murine Nanog gene promoter. Cell Res. 2005;15:317–324. doi: 10.1038/sj.cr.7290300. [DOI] [PubMed] [Google Scholar]
- 46.Pereira L, Yi F, Merrill BJ. Repression of Nanog gene transcription by Tcf3 limits embryonic stem cell self-renewal. Mol Cell Biol. 2006;26:7479–7491. doi: 10.1128/MCB.00368-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Glauche I, Herberg M, Roeder I. Nanog variability and pluripotency regulation of embryonic stem cells--insights from a mathematical model analysis. PLoS One. 2010;5:e11238. doi: 10.1371/journal.pone.0011238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marks H, Kalkan T, Menafra R, et al. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TNG cells were sorted to GFP-positive and GFP-negative subpopulations. A) Gene expression between the subpopulations was analyzed after 24 hours. Sorted GFP-positive and GFP-negative cells were treated with SB, LDN, or SB+LDN. Cultures were reanalyzed for GFP expression after 3 (B) or 6 (C) days. D) Graphs depict triplicate samples.
A) BNG cells were sorted to subpopulations. They were flow analyzed to determine GFP expression. B) Expression of Nanog and T in BNG GFP-subpopulations after 24 hours. C) Nanog protein levels from subpopulations immediately after sorting. D) Mean fluorescence readings from Nanog-GFP subpopulations. E) Nanog and T expression correlate.
A, B) Comparison of LDN to Noggin and of SB431542 to SB505124 for T, Lefty and Id factors. C) Representative flow profiles after 3 days of treatment. D) Percent of GFP-low cells 3 days after treatment.
BNG ES cells were treated with SB, LDN, and PD for 3 days. Cells were analyzed by flow to determine level of Nanog-GFP expression.
A) Dox treatment of AINV-BNG Id1 cells increased Id1 expression over 20-fold. B) Protein analysis also revealed a striking increase in Id1 protein. C) Dox treatment of AINV-Smad7 increased Smad7 expression by approximately 3-fold.
A) The percentages of GFP-neg and GFP-low cells of BNG-Smad7 GFP-high sorted cells is increased with SB+LDN and SB+LDN+Smad7 treatments in serum-free media. B. Reduced colony outgrowth and increased differentiation in clonogenicity assay in serum media. C) Lower cell number in response to SB+LDN+dox after 72 hr as assessed by DNA content. D) Gene expression analysis demonstrated increased expression of Foxa2, Pax6, and Cdx2 following SB+LDN and SB+LDN+Smad7 treatments in serum-free media.