Abstract
Self-assembling peptide materials have been used extensively to mimic natural extracellular matrices (ECMs) by presenting bioactive epitopes on a synthetic matrix. Although this approach can facilitate a desired response from cells grown in the matrix, it lacks the capacity for spatial or temporal regulation of the presented signals. We describe here a photo-responsive, synthetic ECM using a supramolecular platform comprised of peptide amphiphiles (PAs) that self-assemble into cylindrical nanofibers. A photocleavable nitrobenzyl ester group was included in the peptide backbone using a novel Fmoc-amino acid that is compatible with microwave-assisted solid phase peptide synthesis. The placement of the photolabile group on the peptide backbone enabled efficient removal of the ECM-derived cell adhesion epitope RGDS from PA molecules upon exposure to light (half-life of photolysis ~ 1.9 min) without affecting the nanofiber assembly. Fibroblasts cultured on RGDS-presenting PA nanofiber substrates demonstrated increased cell spreading and more mature focal adhesions compared with unfunctionalized and control (RGES-presenting) surfaces, as determined by immunostaining and cell morphological analysis. Furthermore, we observed an arrest in fibroblast spreading on substrates containing a cleavable RGDS epitope when the culture was exposed to light; in contrast, this dynamic shift in cell response was absent when the RGDS epitope was attached to the PA molecule by a light-insensitive control linker. Light-responsive bioactive materials can contribute to the development of synthetic systems that more closely mimic the dynamic nature of native ECM.
Keywords: Supramolecular Nanofibers, Cell Morphological Analysis, Nitrobenzyl, Projected Cell Area, Photodegradable, RGDS Epitope
The extracellular matrix (ECM) forms a supporting structure around cells and plays a critical role in cell development, maturation, and repair by providing essential biochemical and biophysical cues.1 To provide cells with the necessary signals at the right time and place, the ECM is known to undergo extensive remodeling during these processes.1–2 The ability to change spatiotemporal composition of ECM allows certain amphibian species to regenerate their entire organs and limbs, where specific ECM components guide the developmental steps of regenerating cells by providing necessary instructive cues.3 This observation raises the possibility that complex regeneration in higher mammalian species also can be accomplished by providing appropriate sequence of signals in the extracellular environment. Since the discovery of short, bioactive peptide sequences in native ECM molecules,4–5 incorporation of peptide epitopes in artificial synthetic matrices has become a general strategy to impart bio-functionality to an otherwise inert material.6–9 For example, the display of the laminin-derived IKVAV epitope on a synthetic scaffold has been shown to direct the differentiation of neural progenitor cells into neurons and control the survival and maturation of differentiated neurons.9–10 One important need in the development of highly functional ECM-mimetic materials is the capacity for dynamic signaling, a process used in native ECM to deliver specific signals at distinct stages of cellular development and maturation.11 Synthetic dynamic matrices could be designed to allow for similar changes, ideally through the use of convenient external stimuli with spatial and temporal control, allowing for precise tuning of the cellular environment.12 Spatiotemporal control of signaling epitopes in dynamic scaffolds could more effectively direct cell function, for example in proliferation and differentiation of stem cells for regenerative therapies.11, 13 Additionally, dynamic matrices would allow researchers to study the effects of specific matrix modifications over time and space in vitro, reducing the inherent difficulty and complexity of studying cellular processes in vivo.
Development of dynamic matrices for regenerative medicine requires materials capable of forming robust, viscoelastic gels that are amenable to cell encapsulation and support cell viability. Self-assembling peptides have been extensively studied as bioactive gels for cell culture.14–18 These peptidic supramolecular materials have several advantages over traditional polymer hydrogels, including dense signaling capacity, the absence of toxic crosslinking agents, inherent biocompatibility and biodegradation, sheer-thinning behavior or in vivo gelation, and facile chemical modification.12 A very broad family of such materials has been developed and investigated by our group, known as peptide amphiphiles (PAs).8, 19 PAs contain peptide sequences covalently connected to hydrophobic segments such as alkyl tails.20 When the peptide component contains residues with propensity for β-sheet formation, spontaneous self-assembly of PAs in aqueous media leads to the formation of filamentous assemblies of molecules that resemble the fibrous structures in native ECM.8, 21 PA nanofibers are capable of presenting ECM derived epitopes, such as RGD and IKVAV, at a very high density,8–9 and the density of epitopes can be tuned for optimal signaling.22–23 Our group has investigated the ability of PA nanofibers to signal cells in a broad platform that includes cardiovascular disease targets,24–25 central and peripheral nervous system repair,9, 26–28 drug delivery,29–34 cancer therapeutics,32, 35 as well as bone,36–37 cartilage38, and enamel regeneration.39 Extension of the well-established signaling capacity of PAs to systems with dynamic signaling capacity would greatly expand the possibilities of self-assembling peptide materials for controllable and tunable regenerative therapies.
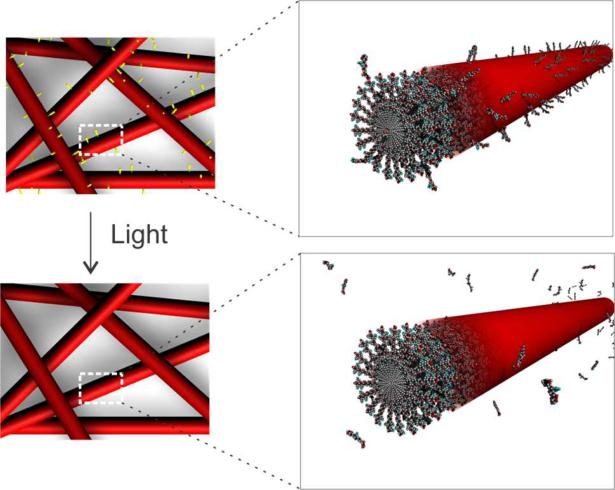
One possible feature to incorporate dynamic signaling into PA matrices is a chemical trigger that can be externally activated. The requirements for maintenance of cell viability (physiological pH and temperature, sterility, minimal shear stress, benign chemistry, among others) severely limit the possibilities for this trigger. Another prospect is the use of enzymes, a technique which has been studied in the context of initiating self-assembly by our group and others.31, 40–41 Chemically degradable bonds such as disulfides can also be used with an external stimulus (e.g., a reducing agent) to trigger a change in gel properties.42 The simplest trigger that is an orthogonal stimulus for biological processes and offers a high degree of spatial and temporal control is light, provided that the intensity and duration of exposure are sufficiently low. Recognizing this possibility, we previously employed light-mediated processes to alter the assembly state of PAs.43–44 In one case, the bioactivity of an appended epitope was shown to increase as the PA assembly shifted from spherical micelles to cylindrical nanofibers though the light-triggered reorganization of the supramolecular assembly.44 In other recent work, Shoichet used photochemical deprotection of thiols in agarose matrices for protein conjugation,45 and Anseth found 365 nm UV light, used under controlled intensity and exposure, to be a suitable stimulus for matrix or epitope removal in a PEG–acrylate based hydrogel system.13, 46 Matrices developed by Anseth have been used to promote chondrogenic differentiation of human mesenchymal stem cells through temporal changes in epitope presentation. Inspired by these previous systems, we hoped to develop a self-assembling PA that retained its capacity for supramolecular nanofiber formation with the added capacity for light-mediated dynamics in a regulated temporal and/or spatial profile (Figure 1). Such a material would extend the scope of light-responsive matrices to include self-assembled gels, which would retain all of the advantages over crosslinked polymer hydrogels noted above in addition to the added capacity of light-regulated changes. We report here the design of light-responsive PA nanofibers using a new photolabile Fmoc-amino acid compared with a similar photostable control molecule. To evaluate the light-induced dynamic epitope regulation in these self-assembled nanofibers, an integrin-binding RGDS peptide sequence was incorporated in the PA molecule and compared with a mutated RGES epitope. The effects of light-induced epitope removal were assessed by fibroblast spreading measurements and focal adhesion formation on the nanofiber matrices.
Figure 1.
Schematic representation of epitope removal from PA nanofiber surfaces following exposure to light.
RESULTS AND DISCUSSION
Synthesis and Characterization of Light-responsive PA
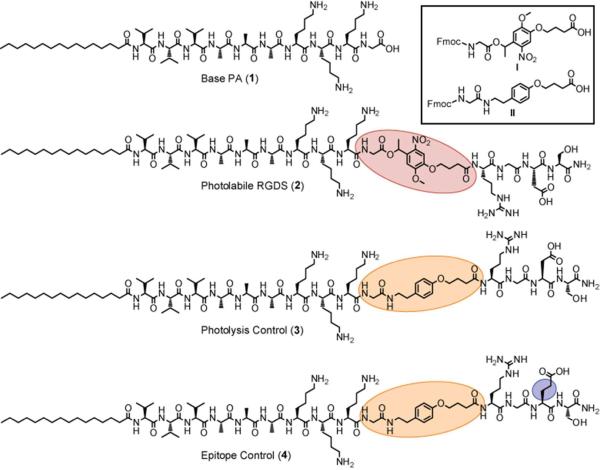
For incorporation of a photolabile group into the peptide sequence, we designed Fmoc-amino acid I (Figure 2, inset). The design employs a nitrobenzyl ester as the photocleavable component, similar to other photocleavable units in the literature.47–51 To create an Fmoc-amino acid that can be used in solid phase peptide synthesis (SPPS) like any standard amino acid, an Fmoc-protected glycine residue was also incorporated into amino acid I (see Supporting Information for synthetic details). Additionally, as a control molecule that had a similar chemical structure but lacked the capacity for cleavage under UV light, Fmoc-amino acid II was designed and synthesized.52 The strength of this approach is that the photolabile component (I) and the control (II) have similar chemical functionalities and are similar in size. Usage of I and II in comparison studies therefore allows for direct control over the variable of photostability with minimal effects on other aspects of the peptide, such as total backbone length. Fmoc-amino acids I and II were both found to be compatible with SPPS on a microwave-assisted peptide synthesizer and in manual SPPS. Couplings were carried out with HBTU and DIEA, and the amino acids were stable to cleavage with 95% TFA.
Figure 2.
Chemical structures of PAs used in this study with highlighted regions of significance. PA 1 is the base sequence. PA 2 contains a photolabile linker (red oval) and the RGDS cell-adhesion sequence; PA 3 contains a control, non-photolabile linker (orange oval) and the RGDS sequence, while PA 4 contains the same non-photolabile linker and a mutated RGES sequence (blue oval). Inset: Chemical structures of photocleavable Fmoc-amino acid I and non-photocleavable control Fmoc-amino acid II (synthetic details available in Supporting Information).
Our strategy to investigate cellular response to a dynamic change in the bioactivity of a matrix was to synthesize a group of PAs bearing the RGDS peptide sequence (Figure 2). The fibronectin-derived RGDS epitope was chosen for its well-known role in mediating cell adhesion and its consequent relevance in the design of bioactive biomaterials.6 All PAs were designed from a common base sequence of PA 1, C16V3A3K3G (where C16 = palmitic acid), a design similar to PAs commonly used in our laboratory.53 PA 2 incorporates in its sequence photolabile amino acid residue I between the base sequence and the RGDS epitope. This PA serves as the bioactive and photolabile molecule in our study. PA 3 was designed as a control for the photocleavable component and contains the non-photolabile amino acid II residue and the bioactive RGDS epitope. PA 4 is a non-photolabile control molecule for the bioactive peptide, containing the the mutated epitope RGES. All PAs were synthesized using standard SPPS conditions and purified by HPLC.
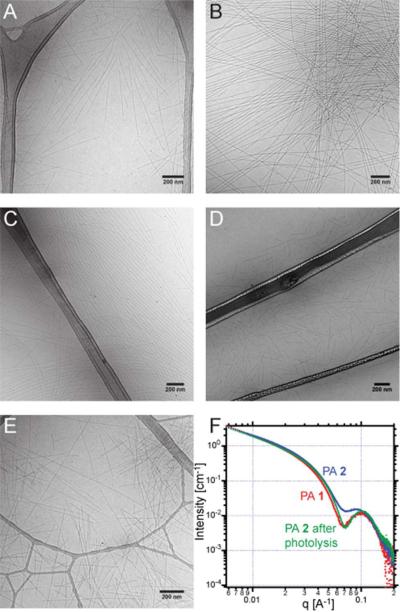
The PAs were characterized by cryogenic transmission electron microscopy (cryoTEM) and small-angle X-ray scattering (SAXS) to study their supramolecular architecture. All PAs show cylindrical nanofiber formation by cryoTEM, and SAXS data support these observations (Figure S2). Figure 3 shows the cryoTEM images for PAs 1–4, including PA 2 before and after photolysis at 365 nm in a Rayonet photoreactor (9.5 mW/cm2) for 10 min. High aspect ratio nanofibers are observed in all cases by cryoTEM, indicating that neither the addition of the photolabile or non-photolabile amino acid residues nor the irradiation have an effect on their supramolecular structure. We also used SAXS to confirm our observations by TEM and more precisely analyze fiber dimensions. The SAXS data in Figure 3F show the curves corresponding to PA 2 before and after irradiation as well as that of PA 1. PA 2 generates PA 1 upon photolytic removal of the RGDS portion of the sequence, and the scattering curve of PA 2 after irradiation is almost the same as that of an authentic sample of PA 1. We attribute the slight differences in authentic PA 1 and irradiated PA 2 to a change in the scattering length density of the solvent as it absorbs the cleaved RGDS sequence. The fitting of the SAXS data to a core-shell cylinder model yields nanofiber diameters of 8.4 nm for PA 1, 9.7 nm for PA 2 before irradiation, and 8.0 nm for PA 2 after irradiation (Table S1).
Figure 3.
CryoTEM (A–E) and SAXS (F) in H2O of PAs used in this study. CryoTEM images of PA 1 (A), PA 2 without irradiation (B), PA 2 after irradiation (C), PA 3 (D), and PA 4 (E) all show cylindrical nanofiber morphologies. SAXS curves of PAs 1, 2 and 2 after photolysis (F) indicate that the dimensions of self-assembled nanofibers of PA 2 after irradiation become that same as that observed for PA 1.
Epitope Accessibility and Photolysis Kinetics
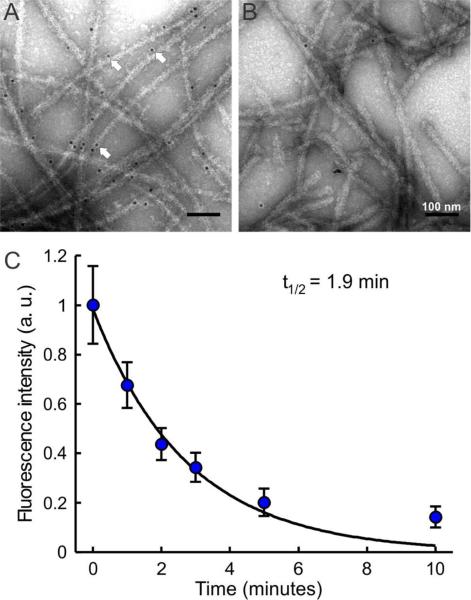
A successful design of a dynamic scaffold using photo-responsive PA molecules will not only require efficient removal of epitopes upon light treatment, but the epitopes should also be accessible to cells to serve their intended function prior to irradiation. To test the accessibility of the epitopes when presented adjacent to the photolabile segment, a biotin-containing photolabile PA was synthesized using Fmoc-amino acid I (PA 5; structure shown in Supporting Information, Figure S1). Nanofibers formed by this PA were incubated with streptavidin conjugated 10-nm gold nanoparticles (AuNPs) and observed under TEM. The AuNPs were predominantly distributed along the nanofibers (Figure 4A), indicating specific binding of streptavidin to the biotin groups displayed on the nanofiber surfaces. The binding was strongly reduced if the biotin was removed by PA irradiation before addition of the AuNPs (Figure 4B).
Figure 4.
(A and B) Conventional TEM images showing (A) streptavidin-tagged AuNPs selectively bound to the surface of biotin-containing PA 5 nanofibers (arrows point to AuNPs), and (B) streptavidin-tagged AuNPs mixed with PA 5 after irradiation showing no specific binding. (C) Kinetics of photolysis measured using PA 5 (blue data points) with fitting of the data (black line) to first order kinetics (t1/2 = 1.9 min).
To evaluate the kinetics of photolysis we used a fluorescence-based approach using the same biotin-presenting PA 5. The PA was coated on glass coverslips for this measurement to keep consistency with the methods described in subsequent cell experiments. To generate a consistent and strongly adhered PA coating, coverslips were first treated with positively charged poly-d-lysine, followed by negatively charged, non-adherent alginate, followed lastly by positively charged PA nanofibers (Figure S3). PA coated coverslips were then irradiated for specific time intervals, and at each time point the photo-irradiated coverslips were washed to remove cleaved biotin and then incubated with Alexafluor-488-tagged streptavidin. The fluorescence intensity measured from the coverslips after irradiation indicated the quantity of remaining biotin groups on the PA-coated surface. The data shows an exponential decay consistent with first order kinetics, with a photolysis half-life of 1.9 minutes (Figure 4C).
Cell Adhesion and Morphological Analysis
Since fibroblast cell response to RGD epitopes presented on material surfaces has been extensively studied,22, 54–56 we selected mouse 3T3 fibroblasts for our experiments. Adhesion and spreading of cells have been shown to be dependent on RGDS epitope density and spacing.55–57 In order to space out RGDS epitopes on the nanofiber surface, we used epitope-presenting PAs 2, 3, and 4 as binary mixtures with the base PA 1. PA mixtures were generated by lyophilizing PA solutions from hexafluoroisopropanol (HFIP), as previously reported by our group.29, 32 The mixed PA powders were then dissolved in water and coated onto glass coverslips as described above. The intermediate alginate layer in the coating not only helped to form a stable PA coating, but its poor cell attachment properties also reduced the possibility of any confounding observations that may arise from cellular attachment to underlying glass or poly-d-lysine coatings. Our group and others have found that mixtures consisting of 10% epitope-presenting peptide and 90% base peptide are optimal for cell response.22–23, 58 Our preliminary experiments also showed increased fibroblast spreading on PA coatings consisting of 10% RGDS epitope-presenting PA (PA 2) mixed with 90% base PA 1 compared with a pure PA 1 coating. The extent of enhancement was reduced when the proportion of RGDS presenting PA was increased to 20% (Figure S3). Based on these results, all further experiments were run using a 90:10 ratio of base PA (PA 1) to epitope-presenting PA.
We evaluated the cell responses to PAs using serum-free culture media in order to limit non-specific adsorption of proteins to the nanofiber surface.54 When plated on PA coated surfaces, fibroblasts showed attachment regardless of the presence or absence of RGDS epitopes. It should be noted that attachment is distinct from adhesion. The former describes any form of cell immobilization on a surface, while the latter implies cell interaction with a surface though specific receptor proteins such as integrins. The lack of epitope specificity and the fast timescale (<30 min) of this initial attachment suggests that it is driven by electrostatic interactions between the negatively charged cell surface and the positively charged nanofibers coupled with the innate property of peptide fibers to support cell attachment.59 In the presence of RGDS epitopes (PA 2 and PA 3) we observed, however, a higher degree of cell spreading after 5 h incubation time associated with a frequent polygonal cell shape (Figure 5A, Figure S3). In the absence of the epitope (PA 1) or in presence of the non-bioactive RGES epitope (PA 4), spreading was severely restricted and cells retained a circular contour. Vinculin, a protein marker for focal adhesions, was visualized by immunostaining and revealed localized expression near the periphery of polygonal cells, which provides anchorage to the substrate and facilitates the organization of actin cytoskeleton. Round fibroblasts on surfaces of PAs 1 and 4, however, rarely showed focal adhesions, and the organization of the actin cytoskeleton was also less distinct. Together these observations suggest that RGDS epitopes presented by the PA nanofibers play a crucial role in recruiting focal adhesion proteins, a process which favors cell spreading and attainment of a polygonal morphology.
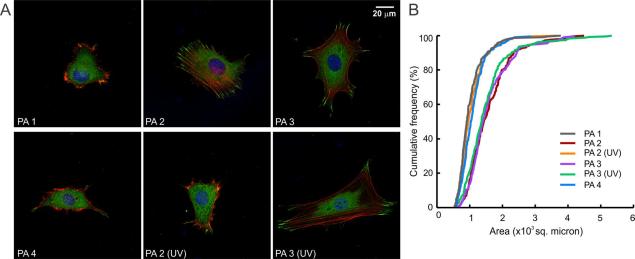
Figure 5.
Cell response to photo-responsive PA and controls. (A) Representative morphologies of 3T3 fibroblasts on various PA coated surfaces (with or without UV exposure). Staining for actin (phalloidin, red) and vinculin (green) reveals the cytoskeletal organization and focal adhesions respectively. (B) A cumulative distribution plot of projected cell surface area, where each point represents the total percentage of cells smaller than the given area. The data show an increased cell spreading in the presence of RGDS epitopes (PAs 2, 3, and 3 (UV)); this response was absent when the epitopes were not included or removed by photo-irradiation (PAs 1, 2 (UV), and 4).
To quantify our observations of cell morphology, measurements of projected cell surface area, convex hull, and circularity were performed (a minimum of 200 individual cells were analyzed from two independent experiments for each substrate; Table 1). Projected surface area was found to be ~ 1.5 fold higher on PA 2 and PA 3, which contain the RGDS epitope, than on PAs 1 and 4, which bear no epitope and contain a mutated one, respectively. A similar difference was observed when measuring convex hull, which describes the area enclosed in a polygon obtained by joining the tips of all distal cell processes. We also observed a small but statistically significant difference in both the projected cell area and the convex hull between RGES-bearing PA 4 and non-epitope-bearing PA 1. However, the absolute values of these measurements are small compared with PA 2 and PA 3, suggesting a minor effect retained by the scrambled epitope on cell spreading.60 It is also evident from these data that cell-secreted ECM does not play a major role in this timeframe, otherwise one would not expect the PA epitope to have a significant effect.
Table 1.
Summary of fibroblast morphology observed on various PA coated surfacesa
| PA 2 | PA 3 | PA 4 | PA 1 | |||
|---|---|---|---|---|---|---|
| UV exposure | No (n=205) | Yes (n=224) | No (n=202) | Yes (n=236) | No (n=217) | No (n=218) |
| Areab | 1601 ± 48*** | 1073 ± 28n.s. | 1595 ± 49*** | 1514 ± 49*** | 1115 ± 27*** | 1034 ± 30 |
| Convex hull | 3327 ± 134*** | 1855 ± 65n.s. | 3341 ± 118*** | 3142 ± 119*** | 1921 ± 56** | 1820 ± 66 |
| Circularity | 0.14 ± 0.01*** | 0.24 ± 0.01n.s. | 0.14 ± 0.01*** | 0.15 ± 0.01*** | 0.22 ± 0.01n.s. | 0.23 ± 0.01 |
Statistical correlations:
p<0.001,
p<0.01,
p>0.05;
all calculated against PA 1 group using atwo sample Kolmogorov-Smirnovtest.
Projected cell surface area
In quantifying projected cell surface area and convex hull, we observed a spectrum of cell morphologies, even on a single substrate. Therefore, to better understand the response of the entire cell population, we plotted cumulative frequency distribution against projected cell area (Figure 5B). This style of plot shows every data point (rather than only a mean and a standard deviation) and has the advantage over a traditional bar graph of illustrating trends where a wide data range exists.61 The plots for PA 2 and PA 3 overlap closely, suggesting that the difference between the photo-labile and the control linker have only minor, if any, influence on epitope presentation. Area distributions for PA 1 and PA 4 were also similar, both showing a prominent shift to the left from that of the RGDS presenting PAs, indicating that cells are less spread on these PAs. We quantified cell roundness by measuring the circularity index, where circularity = 4π(area/perimeter2) (a perfectly circular cell will have an index of 1).62 A predominant polygonal cell morphology on RGDS epitope-presenting PAs 2 and 3 matches well with the low circularity index observed (0.14 ± 0.01 for both surfaces). Cells exhibited a rounder morphology on surfaces of PAs 1 and 4, as evidenced by higher circularity indices (0.23 ± 0.01 and 0.22 ± 0.01, respectively).
To examine cell response to PA photolysis, PA coated surfaces were irradiated prior to cell plating. Fibroblasts on a surface of irradiated PA 2 were less spread and assumed a round morphology, consistent with the removal of the RGDS epitope. The cumulative distribution plot of projected surface area showed a prominent shift to the left compared with the un-irradiated PA 2 condition (p<0.001), and matched closely with RGDS deficient PA 1 (Figure 5B). Also, the average projected surface area decreased by 33%, and the convex hull was reduced by 44% when substrates were exposed to light. Furthermore, photo-irradiation of PA 3 before cell plating did not induce any changes in the cell morphology, confirming that the observed morphological changes were not due to a light-induced change in the supramolecular assembly.
Our results showed that RGDS epitopes on nanofibers facilitated the formation of focal adhesions. To investigate whether this was mediated by an enhanced expression of proteins that comprise focal adhesions, we used quantitative RT-PCR to measure the cellular expression of vinculin and β1 integrin, which are known to be recruited in mature focal adhesions.63 Analyzed at 5 h after plating when differences in both focal adhesion and cell morphology were apparent, we observed similar mRNA expression levels for both proteins regardless of surface coating or irradiation (Figure S4). This result suggests that the observed differences in focal adhesion formation in the presence of RGDS epitopes is primarily mediated by integrin binding and subsequent recruitment of focal adhesion proteins, rather than by an upregulation in protein expression.
Dynamic Cell Response
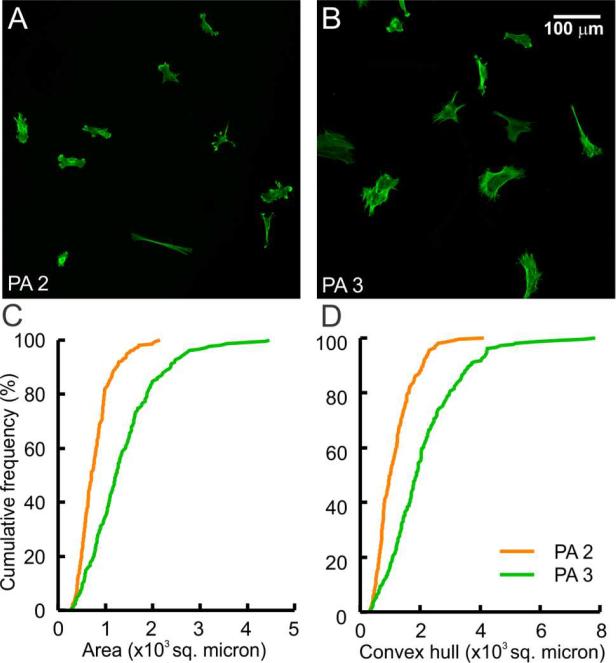
Distinct cell responses were observed on PA 2 depending on whether or not it was pre-irradiated. However, to exploit this property to mimic the dynamic nature of natural ECM, light-mediated epitope removal needs to be employed while the PA substrate is supporting living cells without causing damage to the cells. To test this possibility, we plated fibroblasts on a coating of PA 2 and allowed the cells to adhere for 1 h. Coatings with adhered cells were then exposed to 365 nm UV light for 10 min to remove the RGDS epitopes from the nanofibers. This amount of exposure to UV light is reported to be well tolerated by these cells.13 Consistent with previously reports, we observed partial retraction of the cell process soon after exposure but only minimal associated cell death. More than 90% of the cells were viable immediately after irradiation, and this proportion of live cells was maintained when observed 7 hours post-irradiation, eliminating any concerns of UV induced long-term cytotoxic effects (Figure S5). After 24 h in culture, we found the cells exhibited a limited capacity for spreading, similar to non-RGDS-bearing substrates (Figure 6A). In contrast, a much higher cell spreading was observed when the cells were seeded on the control PA 3 surface and irradiated following the same protocol (Figure 6B). As before, to better delineate the response of the entire cell population, which has heterogeneous morphologies, we plotted cumulative frequency distribution against projected cell area and convex hull (Figure 6C and D). Both measurements showed a significant leftward shift for PA 2 indicating reduced cell spreading (***p<0.001, n>195 cells), with average cell area of 783 ± 25 μm2 and 1363 ± 53 μm2 on PA 2 and PA 3, respectively. This result confirms that the photolabile residue could be used to dynamically modulate PA bioactivity, with potential to control cellular morphology and development using a light stimulus.
Figure 6.
Demonstration of cell response to a dynamic PA substrate. (A and B) Phalloidin stained images of fibroblasts on a surface of PA 2 (A) and PA 3 (B) after attachment, 10 min irradiation, and 24 h in culture. (C and D) Projected surface area (C) and convex hull (D) were found to be significantly lower on PA 2 compared with those on PA 3.
CONCLUSION
We have demonstrated that peptide amphiphile nanofiber matrices can be synthesized by incorporation of a photolabile artificial amino acid to control bioactivity. Through immunostaining and analysis of cell morphology, we showed that cells adhere through integrin-mediated processes onto surfaces displaying PAs that contain the photolabile residue and the bioactive RGDS sequence. Adhesion was dynamically modified by rapid photolytic removal of the RGDS peptide from the supramolecular nanofiber, and control PAs verified the specificity of the RGDS sequence to the observed cell response. Additionally, this method to dynamically control epitope presentation is amenable to cell culture, as cell viability was maintained after exposure to UV light. Most importantly, the incorporation of photolabile groups to generate matrices with light-mediated control over bioactivity and cell behavior is not limited to PAs or to the RGDS sequence, and several other self-assembling peptide-based materials may benefit from this approach. Moreover, peptide sequences that mediate proliferation, differentiation, or other cellular processes may also be used in combination with the photolabile linker described here. We believe that dynamic temporal control of cell-material interactions can become an important component in the design of new artificial matrices based on nanostructures for regenerative medicine research or therapies.
EXPERIMENTAL SECTION
PA Synthesis
Fmoc-amino acids I and II syntheses can be found in the Supporting Information. PAs 1–5 were synthesized at the Peptide Synthesis Core at the Institute for BioNanotechnology in Medicine. Synthesis of PAs 2–4 was performed on a CEM Liberty microwave-assisted peptide synthesizer from Rink amide MBHA resin. PA 5 was synthesized from Biotin Novatag resin. For each coupling, 5 equiv of Fmoc-protected amino acid in DMF was added with 5 equiv HBTU in DMF and 10 equiv DIEA in NMP (HBTU = O-benzotriazole-N,N,N',N'-tetramethyluronium-hexafluorophosphate; DIEA = N,N-diisopropylethylamine). Fmoc removal was accomplished using a solution of 20% piperidine in DMF and 0.1 M HoBt. Default settings for microwave power and duration were used. The palmitic acid tail was added using the same coupling conditions. The PA was cleaved from the resin in a shaker vessel using a peptide cleavage solution of 95% TFA, 2.5% TIPS and 2.5% H2O. Concentration of the cleavage solution in vacuo and precipitation of the residue into cold Et2O afforded the crude product, which was purified by preparative HPLC. MS: PA 2: (M+): calc'd: 1903.15; found: 1903.15; PA 3: (M+): calc'd: 1827.17; found: 1827.08; PA 4: (M+): calc'd: 1841.18; found: 1841.09; PA 5: (M+): calc'd: 1757.09; found: 1757.10. PA 1 was made by photolytic cleavage of PA 2 followed by purification using HPLC. MS: PA 1: (M+): calc'd: 1207.86; found: 1207.89. LCMS traces of PAs 1–4, as well as PA 2 after irradiation for 5 min in a glass capillary, can be found in the Supporting Information.
Transmission Electron Microscopy (TEM)
CryoTEM was performed on a JEOL 1230 microscope with an accelerating voltage of 100 kV. A Vitrobot Mark IV equipped with controlled humidity and temperature was used for plunge freezing samples. A small volume (5–10 μL) of PA solution at 0.25% (w/v) in water was deposited on a copper TEM grid with holey carbon support film (Electron Microscopy Sciences) and held in place with tweezers mounted to the Vitrobot. The specimen was blotted in an environment with 90–100% humidity and plunged into a liquid ethane reservoir that was cooled by liquid nitrogen. The vitrified samples were transferred in a nitrogen environment into liquid nitrogen and transferred to a Gatan 626 cryo-holder using a cryo-transfer stage. Samples were imaged using a Gatan 831 bottom-mounted camera.
Labeling with streptavidin gold nanoparticles (AuNP): A copper TEM grid with carbon support film (EMS, USA) was placed on a drop (~15 L) of PA solution (0.1% w/v in water) on a piece of clean parafilm and incubated for 10 minutes. Excess solution was wicked away with filter paper, rinsed once with milliQ water and incubated for 1 hour with streptavidin 10-nm AuNPs (Sigma; diluted 1:4 in PBS). After incubation, the samples were gently rinsed 3 times with milliQ water and were negatively stained using a 2% (w/v) uranyl acetate solution in water. Images of negatively stained samples were obtained on a Tecnai Spirit G2 microscope (FEI) operating at 120 kV.
Small Angle X-Ray Scattering (SAXS)
Measurements were performed using beam line 5ID-D, in the DuPont-Northwestern-Dow Collaborative Access team (DND-CAT) Synchrotron Research Center at the Advanced Photon Source, Argonne National Laboratory. An energy of 15 keV corresponding to a wavelength λ=0.83 Å was selected using a double-crystal monochromator. The data were collected using a CCD detector (MAR) positioned 245 cm behind the sample. The scattering intensity was recorded in the interval 0.005 < q < 0.23 Å−1. The wave vector defined as q = (4π/λ) sin(θ/2), where θ is the scattering angle. Samples lyophilized from HFIP and analyzed in 2 mm quartz capillaries at 0.5% by weight in water. The two-dimensional SAXS images were azimuthally averaged to produce one-dimensional profiles of intensity (I) vs q, using the two-dimensional data reduction program FIT2D. Scattering of a capillary containing only solvent was also collected and subtracted from the corresponding data. No attempt was made to convert the data to an absolute scale. Detailed methods for SAXS data modeling is provided in the Supporting Information.
Coverslip Coating
Sterile poly-d-lysine coated glass coverslips (12 mm diameter) were first coated with a thin layer of negatively charged biopolymer sodium alginate (0.25% in milliQ water; FMC BioPolymer) and cross-linked with 10 mM aqueous CaCl2 solution. Calcium chloride solution was pipetted out and incubated overnight with 0.05% PA solution (in milliQ water) at 37°C. After incubation, the coverslip was gently rinsed once with milliQ water, followed by one rinse with DMEM solution. The coverslip surface was never allowed to dry during the entire coating process, and it was left under DMEM until the addition of cells. For 365 nm UV irradiation of the coatings, DMEM was replaced with Dulbecco PBS (containing 1 mM CaCl2), placed inside a Rayonet photoreactor for designated amount of time, and exposed at an intensity of 9.5 mW/cm2.
Kinetics Experiments
Coverslips were coated with poly-d-lysine, alginate, and PA 5 (100%) or PA 1 as described above. After UV irradiation, coverslips were washed once with DMSO/PBS (1:1) and once with 1 mM aqueous CaCl2 and then incubated with Alexa Fluor® 488 conjugated streptavidin for one hour. The excess streptavidin was removed by washing with PBS (containing 1 mM CaCl2). Fluorescence intensities were recorded on an M5 Spectramax plate reader (Molecular Devices) using the bottom read option with λex = 488 nm and λem = 520 nm. The measurement was done at nine points on each coverslip and averaged for each timepoint (n = 3). Background fluorescence was determined from the coverslips (n = 3) coated with PA 1 and subtracted from each value. The fluorescence intensity from the 10 minute time point was statistically insignificant from the background so was not incorporated in the fit. The half-life value was extracted from the first order kinetics plot.
Cell Culture
NIH 3T3 mouse embryonic fibroblasts were maintained in Dulbecco's Modified Eagle's Medium (DMEM) with high glucose (GIBCO) supplemented with 10% fetal bovine serum (FBS, Hyclone) and 1% penicillin-streptomycin (P/S) and passaged every 3 days. To study the cell response on PA coated surfaces, cells were seeded at a low density of 25 cells/mm2 and incubated for 5 hours (37°C, 5% CO2) under serum free conditions (DMEM and 1% P/S). The lower cell seeding density was chosen to reduce the chance of cell-cell contact, so that pure cell-substrate interaction could be studied. For dynamic cell response studies, cells were incubated in serum free media for an hour to facilitate their attachment and spreading on PA surfaces, treated with UV light for 10 minutes, and then incubated with serum containing media (DMEM, 10% FBS and 1% P/S) for 24 hours.
Fluorescence Staining
Cultured cells were fixed with 4% paraformaldehyde in phosphate buffered saline (PBS) for 15 min at room temperature. The fixing solution and all solutions used in subsequent staining and washing steps were supplemented with 1 mM CaCl2 to prevent dislodgement of the PA coating (which is held on the glass surface by Ca2+ cross-linked alginate). Samples were permeabilized and blocked with 0.4% Triton X-100, 10% normal goat serum and 2% BSA in PBS, followed by overnight incubation with anti-vinculin primary antibody (mouse monoclonal, Sigma; 1:400 dilution) at 4°C. Primary antibody was detected with Alexa Fluor® conjugated secondary antibody (Life Technologies; 1 hour incubation at RT). Actin filament was visualized by staining with rhodamine or Alexa Fluor® 488 conjugated phalloidin (Life technologies; 1:100 dilution, 1 hour at RT). Cell nuclei were counterstained with DAPI (Life technologies).
Image Acquisition and Analysis
Images from fluorescently stained samples were acquired using an inverted confocal laser scanning microscope (Nikon A1R) or TissueGnostics cell imaging and analysis system, mounted to an upright microscope (Zeiss). Morphological quantifications were done on phalloidin stained images of the cells obtained at 20× objective magnification. Acquired images were thresholded using ImageJ software (NIH) and analyzed using routines written in Matlab software (MathWorks). Projected cell surface area was defined by the phalloidin positive pixels occupied by a cell. The convex hull, an index of the cell spread on the substrate, was defined as the total area obtained by joining the tips of most distal processes with straight lines. Shapes of cells were assessed from area and perimeter values by calculating the circularity index, where circularity = 4π(area/perimeter2). For accurate comparison, quantification of more than 190 randomly selected cells from 2 independent batches of culture was done for each condition.
Scanning Electron Microscopy (SEM)
Cells plated on PA coatings were fixed with 2.5% glutaraldehyde in PBS (1 hour, RT) and dehydrated in a graded series of ethanol concentration. Dehydrated samples (in 100% ethanol) were then dried using a critical point dryer (Tousimis Samdri-795) to preserve structural details. Dried samples were coated with a thin film (14 nm) of osmium metal using an osmium plasma coater (Filgen, OPC-60A) and imaged using a Hitachi S-4800 Field Emission Scanning Electron Microscope at an accelerating voltage of 5 kV.
Statistical Analysis
Normality of the distribution of cell morphology datasets was tested using the Shapiro-Wilk normality test. A two sample Kolmogorov-Smirnov test was employed to compare the distribution of morphology parameters between two PA conditions. Data in the table represent mean ± standard error of the mean.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIDCR, grant No. 2R01DE015920-06 and NIBIB, grant No. 2R01EB003806-06A2. JBM was supported by a National Institutes of Health postdoctoral fellowship (Grant No. 1F32AR061955-01). We are grateful to the following core facilities at Northwestern University: the Peptide Synthesis Core in the Institute for BioNanotechnology in Medicine (IBNAM), the Biological Imaging Facility (BIF), the Integrated Molecular Structure Education and Research Center (IMSERC), the Cell Imaging Facility (CIF), and Keck Biophysics for instrument use. We are grateful to Mark Seniw for the illustrations. We acknowledge Dr. Steven Weigand and the DuPont-Northwestern-Dow Collaborative Access Team (DND-CAT) Synchrotron Research Center at the Advanced Photon Source (APS) at Argonne National Lab for assistance with SAXS measurements. Use of the APS was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357.
Footnotes
Supporting Information Available. SAXS traces and fitting parameters, SEM images, and PCR data, including experimental procedures for each, as well as synthetic details and characterization for new amino acids. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Rozario T, DeSimone DW. The Extracellular Matrix in Development and Morphogenesis: A Dynamic View. Dev. Biol. 2010;341:126–140. doi: 10.1016/j.ydbio.2009.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larsen M, Artym VV, Green JA, Yamada KM. The Matrix Reorganized: Extracellular Matrix Remodeling and Integrin Signaling. Curr. Opin. Cell Biol. 2006;18:463–471. doi: 10.1016/j.ceb.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 3.Calve S, Odelberg SJ, Simon HG. A Transitional Extracellular Matrix Instructs Cell Behavior During Muscle Regeneration. Dev. Biol. 2010;344:259–271. doi: 10.1016/j.ydbio.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pierschbacher MD, Ruoslahti E. Cell Attachment Activity of Fibronectin Can Be Duplicated by Small Synthetic Fragments of the Molecule. Nature. 1984;309:30–33. doi: 10.1038/309030a0. [DOI] [PubMed] [Google Scholar]
- 5.Tashiro K, Sephel GC, Weeks B, Sasaki M, Martin GR, Kleinman HK, Yamada Y. A Synthetic Peptide Containing the Ikvav Sequence from the a-Chain of Laminin Mediates Cell Attachment, Migration, and Neurite Outgrowth. J. Biol. Chem. 1989;264:16174–16182. [PubMed] [Google Scholar]
- 6.Hersel U, Dahmen C, Kessler H. Rgd Modified Polymers: Biomaterials for Stimulated Cell Adhesion and Beyond. Biomaterials. 2003;24:4385–4415. doi: 10.1016/s0142-9612(03)00343-0. [DOI] [PubMed] [Google Scholar]
- 7.Shin H, Jo S, Mikos AG. Biomimetic Materials for Tissue Engineering. Biomaterials. 2003;24:4353–4364. doi: 10.1016/s0142-9612(03)00339-9. [DOI] [PubMed] [Google Scholar]
- 8.Hartgerink JD, Beniash E, Stupp SI. Self-Assembly and Mineralization of Peptide-Amphiphile Nanofibers. Science. 2001;294:1684–1688. doi: 10.1126/science.1063187. [DOI] [PubMed] [Google Scholar]
- 9.Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, Stupp SI. Selective Differentiation of Neural Progenitor Cells by High-Epitope Density Nanofibers. Science. 2004;303:1352–1355. doi: 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]
- 10.Sur S, Pashuck ET, Guler MO, Ito M, Stupp SI, Launey T. A Hybrid Nanofiber Matrix to Control the Survival and Maturation of Brain Neurons. Biomaterials. 2012;33:545–555. doi: 10.1016/j.biomaterials.2011.09.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daley WP, Peters SB, Larsen M. Extracellular Matrix Dynamics in Development and Regenerative Medicine. J. Cell Sci. 2008;121:255–264. doi: 10.1242/jcs.006064. [DOI] [PubMed] [Google Scholar]
- 12.Matson JB, Stupp SI. Self-Assembling Peptide Scaffolds for Regenerative Medicine. Chem. Commun. 2012;48:26–33. doi: 10.1039/c1cc15551b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Photodegradable Hydrogels for Dynamic Tuning of Physical and Chemical Properties. Science. 2009;324:5963. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matson JB, Zha RH, Stupp SI. Peptide Self-Assembly for Crafting Functional Biological Materials. Curr. Opin. Solid State Mater. Sci. 2011;15:225–235. doi: 10.1016/j.cossms.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hauser CAE, Zhang SG. Designer Self-Assembling Peptide Nanofiber Biological Materials. Chem. Soc. Rev. 2010;39:2780–2790. doi: 10.1039/b921448h. [DOI] [PubMed] [Google Scholar]
- 16.Nagarkar RP, Hule RA, Pochan DJ, Schneider JP. Domain Swapping in Materials Design. Biopolymers. 2010;94:141–155. doi: 10.1002/bip.21332. [DOI] [PubMed] [Google Scholar]
- 17.Webber MJ, Kessler JA, Stupp SI. Emerging Peptide Nanomedicine to Regenerate Tissues and Organs. J. Intern.Med. 2010;267:71–88. doi: 10.1111/j.1365-2796.2009.02184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collier JH, Rudra JS, Gasiorowski JZ, Jung JP. Multi-Component Extracellular Matrices Based on Peptide Self-Assembly. Chem. Soc. Rev. 2010;39:3413–3424. doi: 10.1039/b914337h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu YC, Berndt P, Tirrell M, Fields GB. Self-Assembling Amphiphiles for Construction of Protein Molecular Architecture. J. Am. Chem. Soc. 1996;118:12515–12520. [Google Scholar]
- 20.Hartgerink JD, Beniash E, Stupp SI. Peptide-Amphiphile Nanofibers: A Versatile Scaffold for the Preparation of Self-Assembling Materials. Proc. Natl. Acad. Sci. U.S.A. 2002;99:5133–5138. doi: 10.1073/pnas.072699999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Velichko YS, Stupp SI, de la Cruz MO. Molecular Simulation Study of Peptide Amphiphile Self-Assembly. J. Phys. Chem. B. 2008;112:2326–2334. doi: 10.1021/jp074420n. [DOI] [PubMed] [Google Scholar]
- 22.Storrie H, Guler MO, Abu-Amara SN, Volberg T, Rao M, Geiger B, Stupp SI. Supramolecular Crafting of Cell Adhesion. Biomaterials. 2007;28:4608–4618. doi: 10.1016/j.biomaterials.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 23.Webber MJ, Tongers J, Renault MA, Roncalli JG, Losordo DW, Stupp SI. Development of Bioactive Peptide Amphiphiles for Therapeutic Cell Delivery. Acta Biomater. 2010;6:3–11. doi: 10.1016/j.actbio.2009.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Webber MJ, Han XQ, Murthy SNP, Rajangam K, Stupp SI, Lomasney JW. Capturing the Stem Cell Paracrine Effect Using Heparin-Presenting Nanofibres to Treat Cardiovascular Diseases. J. Tissue Eng. Regen. Med. 2010;4:600–610. doi: 10.1002/term.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Webber MJ, Tongers J, Newcomb CJ, Marquardt K-T, Bauersachs J, Losordo DW, Stupp SI. Supramolecular Nanostructures That Mimic Vegf as a Strategy for Ischemic Tissue Repair. Proc. Natl. Acad. Sci. U.S.A. 2011;108:13438–13443. doi: 10.1073/pnas.1016546108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tysseling-Mattiace VM, Sahni V, Niece KL, Birch D, Czeisler C, Fehlings MG, Stupp SI, Kessler JA. Self-Assembling Nanofibers Inhibit Glial Scar Formation and Promote Axon Elongation after Spinal Cord Injury. J. Neurosci. 2008;28:3814–3823. doi: 10.1523/JNEUROSCI.0143-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Angeloni NL, Bond CW, Tang Y, Harrington DA, Zhang SM, Stupp SI, McKenna KE, Podlasek CA. Regeneration of the Cavernous Nerve by Sonic Hedgehog Using Aligned Peptide Amphiphile Nanofibers. Biomaterials. 2011;32:1091–1101. doi: 10.1016/j.biomaterials.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bond CW, Angeloni NL, Harrington DA, Stupp SI, McKenna KE, Podlasek CA. Peptide Amphiphile Nanofiber Delivery of Sonic Hedgehog Protein to Reduce Smooth Muscle Apoptosis in the Penis after Cavernous Nerve Resection. J. Sex. Med. 2011;8:78–89. doi: 10.1111/j.1743-6109.2010.02001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matson JB, Stupp SI. Drug Release from Hydrazone-Containing Peptide Amphiphiles. Chem. Commun. 2011;47:7962–7964. doi: 10.1039/c1cc12570b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matson JB, Newcomb CJ, Bitton R, Stupp SI. Nanostructure-Templated Control of Drug Release from Peptide Amphiphile Nanofiber Gels. Soft Matter. 2012;8:3586–3595. doi: 10.1039/C2SM07420F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Webber MJ, Newcomb CJ, Bitton R, Stupp SI. Switching of Self-Assembly in a Peptide Nanostructure with a Specific Enzyme. Soft Matter. 2011;7:9665–9672. doi: 10.1039/c1sm05610g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soukasene S, Toft DJ, Moyer TJ, Lu H, Lee H-K, Standley SM, Cryns VL, Stupp SI. Antitumor Activity of Peptide Amphiphile Nanofiber-Encapsulated Camptothecin. ACS Nano. 2011;5:9113–9121. doi: 10.1021/nn203343z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matson JB, Webber MJ, Tamboli VK, Weber B, Stupp SI. A Peptide-Based Material for Therapeutic Carbon Monoxide Delivery. Soft Matter. 2012;8:2689–2692. doi: 10.1039/C2SM25785H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Webber MJ, Matson JB, Tamboli VK, Stupp SI. Controlled Release of Dexamethasone from Peptide Nanofiber Gels to Modulate Inflammatory Response. Biomaterials. 2012;33:6823–6832. doi: 10.1016/j.biomaterials.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Standley SM, Toft DJ, Cheng H, Soukasene S, Chen J, Raja SM, Band V, Band H, Cryns VL, Stupp SI. Induction of Cancer Cell Death by Self-Assembling Nanostructures Incorporating a Cytotoxic Peptide. Cancer Res. 2010;70:3020–3026. doi: 10.1158/0008-5472.CAN-09-3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mata A, Geng YB, Henrikson KJ, Aparicio C, Stock SR, Satcher RL, Stupp SI. Bone Regeneration Mediated by Biomimetic Mineralization of a Nanofiber Matrix. Biomaterials. 2010;31:6004–6012. doi: 10.1016/j.biomaterials.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spoerke ED, Anthony SG, Stupp SI. Enzyme Directed Templating of Artificial Bone Mineral. Adv. Mater. 2009;21:425–430. doi: 10.1002/adma.200802242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shah RN, Shah NA, Lim MMD, Hsieh C, Nuber G, Stupp SI. Supramolecular Design of Self-Assembling Nanofibers for Cartilage Regeneration. Proc. Natl. Acad. Sci. U.S.A. 2010;107:3293–3298. doi: 10.1073/pnas.0906501107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang Z, Newcomb CJ, Bringas P, Jr., Stupp SI, Snead ML. Biological Synthesis of Tooth Enamel Instructed by an Artificial Matrix. Biomaterials. 2010;31:9202–11. doi: 10.1016/j.biomaterials.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang ZM, Gu HW, Fu DG, Gao P, Lam JK, Xu B. Enzymatic Formation of Supramolecular Hydrogels. Adv. Mater. 2004;16:1440–1444. [Google Scholar]
- 41.Williams RJ, Smith AM, Collins R, Hodson N, Das AK, Ulijn RV. Enzyme-Assisted Self-Assembly under Thermodynamic Control. Nat. Nanotechnol. 2009;4:19–24. doi: 10.1038/nnano.2008.378. [DOI] [PubMed] [Google Scholar]
- 42.Madsen J, Armes SP, Bertal K, Lomas H, MacNeil S, Lewis AL. Biocompatible Wound Dressings Based on Chemically Degradable Triblock Copolymer Hydrogels. Biomacromolecules. 2008;9:2265–2275. doi: 10.1021/bm8005006. [DOI] [PubMed] [Google Scholar]
- 43.Muraoka T, Cui H, Stupp SI. Quadruple Helix Formation of a Photoresponsive Peptide Amphiphile and Its Light-Triggered Dissociation into Single Fibers. J. Am. Chem. Soc. 2008;130:2946–2947. doi: 10.1021/ja711213s. [DOI] [PubMed] [Google Scholar]
- 44.Muraoka T, Koh CY, Cui HG, Stupp SI. Light-Triggered Bioactivity in Three Dimensions. Angew. Chem. Int. Ed. 2009;48:5946–5949. doi: 10.1002/anie.200901524. [DOI] [PubMed] [Google Scholar]
- 45.Luo Y, Shoichet MS. A Photolabile Hydrogel for Guided Three-Dimensional Cell Growth and Migration. Nat. Mater. 2004;3:249–253. doi: 10.1038/nmat1092. [DOI] [PubMed] [Google Scholar]
- 46.DeForest CA, Anseth KS. Cytocompatible Click-Based Hydrogels with Dynamically Tunable Properties through Orthogonal Photoconjugation and Photocleavage Reactions. Nat. Chem. 2011;3:925–931. doi: 10.1038/nchem.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson JA, Lewis DR, Az D, Finn MG, Koberstein JT, Turro NJ. Synthesis of Degradable Model Networks Via Atrp and Click Chemistry. J. Am. Chem. Soc. 2006;128:6564–6565. doi: 10.1021/ja0612910. [DOI] [PubMed] [Google Scholar]
- 48.Holmes CP. Model Studies for New O-Nitrobenzyl Photolabile Linkers: Substituent Effects on the Rates of Photochemical Cleavage. J. Org. Chem. 1997;62:2370–2380. doi: 10.1021/jo961602x. [DOI] [PubMed] [Google Scholar]
- 49.Ramos R, Manning B, Avino A, Gargallo R, Eritja R. Photocleavage of Peptides and Oligodeoxynucleotides Carrying 2-Nitrobenzyl Groups. Helv. Chim. Acta. 2009;92:613–622. [Google Scholar]
- 50.Loewik DWPM, Meijer JT, Minten IJ, Van Kalkeren H, Heckenmuller L, Schulten I, Sliepen K, Smittenaar P, Van Hest JCM. Controlled Disassembly of Peptide Amphiphile Fibres. J. Pept. Sci. 2008;14:127–133. doi: 10.1002/psc.969. [DOI] [PubMed] [Google Scholar]
- 51.Fomina N, McFearin C, Sermsakdi M, Edigin O, Almutairi A. Uv and near-Ir Triggered Release from Polymeric Nanoparticles. J. Am. Chem. Soc. 2010;132:9540–9542. doi: 10.1021/ja102595j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fmoc-Amino Acid I Was Also Synthesized without the Nitro Group but Was Found to Be Unstable in Tfa, Necessitating the Redesign of the Fmoc-Amino Acid to Linker Stable II
- 53.Capito RM, Azevedo HS, Velichko YS, Mata A, Stupp SI. Self-Assembly of Large and Small Molecules into Hierarchically Ordered Sacs and Membranes. Science. 2008;319:1812–1816. doi: 10.1126/science.1154586. [DOI] [PubMed] [Google Scholar]
- 54.Grafahrend D, Heffels K-H, Beer MV, Gasteier P, Moeller M, Boehm G, Dalton PD, Groll J. Degradable Polyester Scaffolds with Controlled Surface Chemistry Combining Minimal Protein Adsorption with Specific Bioactivation. Nat. Mater. 2011;10:67–73. doi: 10.1038/nmat2904. [DOI] [PubMed] [Google Scholar]
- 55.Massia SP, Hubbell JA. An Rgd Spacing of 440 Nm Is Sufficient for Integrin Alpha-V-Beta-3-Mediated Fibroblast Spreading and 140 Nm for Focal Contact and Stress Fiber Formation. J. Cell Biol. 1991;114:1089–1100. doi: 10.1083/jcb.114.5.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schvartzman M, Palma M, Sable J, Abramson J, Hu X, Sheetz MP, Wind SJ. Nanolithographic Control of the Spatial Organization of Cellular Adhesion Receptors at the Single-Molecule Level. Nano Lett. 2011;11:1306–1312. doi: 10.1021/nl104378f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gasiorowski JZ, Collier JH. Directed Intermixing in Multicomponent Self-Assembling Biomaterials. Biomacromolecules. 2011;12:3549–3558. doi: 10.1021/bm200763y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jung JP, Nagaraj AK, Fox EK, Rudra JS, Devgun JM, Collier JH. Co-Assembling Peptides as Defined Matrices for Endothelial Cells. Biomaterials. 2009;30:2400–2410. doi: 10.1016/j.biomaterials.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang S, Holmes TC, DiPersio CM, Hynes RO, Su X, Rich A. Self-Complementary Oligopeptide Matrices Support Mammalian Cell Attachment. Biomaterials. 1995;16:1385–93. doi: 10.1016/0142-9612(95)96874-y. [DOI] [PubMed] [Google Scholar]
- 60.Dsouza SE, Ginsberg MH, Plow EF. Arginyl-Glycyl-Aspartic Acid (Rgd)- a Cell-Adhesion Motif. Trends. Biochem. Sci. 1991;16:246–250. doi: 10.1016/0968-0004(91)90096-e. [DOI] [PubMed] [Google Scholar]
- 61.Cingolani LA, Thalhammer A, Yu LMY, Catalano M, Ramos T, Colicos MA, Goda Y. Activity-Dependent Regulation of Synaptic Ampa Receptor Composition and Abundance by Beta 3 Integrins. Neuron. 2008;58:749–762. doi: 10.1016/j.neuron.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sung KE, Yang N, Pehlke C, Keely PJ, Eliceiri KW, Friedl A, Beebe DJ. Transition to Invasion in Breast Cancer: A Microfluidic in vitro Model Enables Examination of Spatial and Temporal Effects. Integr. Biol. 2011;3:439–450. doi: 10.1039/c0ib00063a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Puklin-Faucher E, Sheetz MP. The Mechanical Integrin Cycle. J. Cell Sci. 2009;122:179–186. doi: 10.1242/jcs.042127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.