Abstract
Sulfated molecules with diverse functions are common in biology, but sulfonation as a method to activate a metabolite for chemical catalysis is rare. Catalytic activity was characterized and crystal structures were determined for two such “activating” sulfotransferases (STs) that sulfonate β-hydroxyacyl thioester substrates. The CurM polyketide synthase (PKS) ST domain from the curacin A biosynthetic pathway of Moorea producens and the olefin synthase (OLS) ST from a hydrocarbon-producing system of Synechococcus PCC 7002 both occur as a unique acyl carrier protein (ACP), ST and thioesterase (TE) tridomain within a larger polypeptide. During pathway termination, these cyanobacterial systems introduce a terminal double bond into the β-hydroxyacyl-ACP-linked substrate by the combined action of the ST and TE. Under in vitro conditions, CurM PKS ST and OLS ST acted on β-hydroxy fatty acyl-ACP substrates; however, OLS ST was not reactive toward analogs of the natural PKS ST substrate bearing a C5-methoxy substituent. The crystal structures of CurM ST and OLS ST revealed that they are members of a distinct protein family relative to other prokaryotic and eukaryotic sulfotransferases. A common binding site for the sulfonate donor 3'-phosphoadenosine-5'-phosphosulfate was visualized in complexes with the product 3'-phosphoadenosine-5'-phosphate. Critical functions for several conserved amino acids in the active site were confirmed by site-directed mutagenesis, including a proposed glutamate catalytic base. A dynamic active-site flap unique to the “activating” ST family affects substrate selectivity and product formation, based on the activities of chimeras of the PKS and OLS STs with exchanged active-site flaps.
INTRODUCTION
Sulfotransfer is a ubiquitous and functionally diverse process across all forms of life. Although the acceptor molecule and the specific biological role of sulfonation varies widely, the sulfotransferases (STs) that catalyze transfer of sulfonate from the biological donor 3'-phosphoadenosine-5'-phosphosulfate (PAPS) to a hydroxy or amine acceptor are generally highly conserved (1–4). Sulfotransfer in the curacin A biosynthetic pathway contrasts with all of the other known sulfonation events because the natural product is transiently sulfonated, and the final structure of this potent cancer cell cytotoxin includes no sulfate group (5).
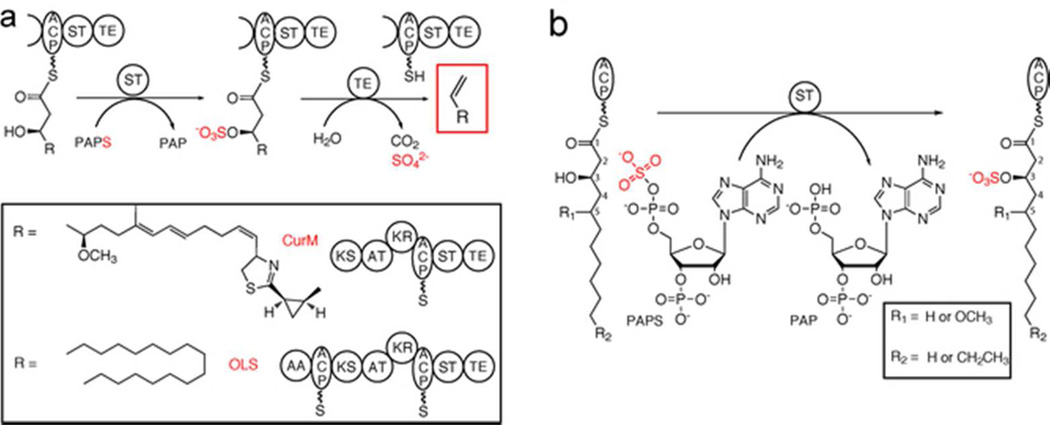
The antimitotic natural product curacin A is produced by the marine cyanobacterium Moorea producens [formerly Lyngbya majuscula (6)] in a biosynthetic pathway containing a unique ST with low sequence similarity to previously described STs. Located within the final polyketide synthase (PKS) monomodule, CurM, the ST is the penultimate domain between the acyl carrier protein (ACP) and thioesterase (TE), two common domains involved in PKS and fatty acid synthase (FAS) pathway termination (7) (Figure 1A). In previous studies, we found that CurM ST transfers sulfonate from the donor PAPS to the β-hydroxy of the pathway intermediate, which is covalently tethered to the CurM ACP (Figure 1A) (8). The unique CurM TE domain then acts specifically upon the β-sulfated substrate to hydrolyze, decarboxylate and eliminate sulfate to yield a terminal alkene product (Figure 1A) (8, 9). This mode of polyketide chain offloading to a terminal alkene was unprecedented, as typical TEs catalyze hydrolysis to the linear acyl-carboxylic acid or cyclization to a macrolactone or macrolactam product (10–14). CurM ST activates the β-hydroxy group and thus facilitates decarboxylation to yield the terminal olefin (8). To our knowledge, this work provided the first description in a metabolic pathway where sulfonation functions to activate a hydroxy group for subsequent conversion to a final product.
Figure 1.
Offloading and termination using β-hydroxy group activation and decarboxylation. a) Offloading and termination in the curacin A (CurM) and the olefin synthase (OLS) pathways. The sulfotransferase (ST) first sulfonates the β-hydroxy group from the sulfonate donor 3′-phosphoadenosine 5′-phosphosulfate (PAPS). The thioesterase (TE) then works in a β-sulfate-dependent manner to hydrolyze, decarboxylate, and eliminate sulfate to produce a terminal alkene. Other domains are acyl activating (AA), acyl carrier protein (ACP), ketosynthase (KS), acyl transferase (AT), and ketoreductase (KR). b) ST reaction in biochemical assays. Excised ACP was loaded with synthesized substrates (varying in acyl chain length, the presence of a C5-methoxy and the β-hydroxy group configuration) and reacted with PAPS. Sulfonated products were detected by HPLC.
CurM ST has low sequence similarity to all previously characterized STs (less than 25% identity). However, several additional ST domains presumed to play a role in acyl-hydroxy group activation were identified in the protein databases by virtue of a high level of amino acid sequence identity (33–52%) to the CurM ACP-ST-TE tri-domain (Supplemental Figure 1) (15). Here we designate these as “activating” STs with a common functionality to transfer a sulfonate that is subsequently removed through decarboxylative elimination. In addition to curM, where the ST is embedded within the CurM PKS module, two related genes appear in the context of uncharacterized biosynthetic gene clusters. Five additional ST genes reside within stand-alone eight-domain polypeptides that resemble monomodular PKSs or FASs. The domain organization is similar to CurM but with the addition of acyl-activating (AA) and ACP domains at the N-terminus (Figure 1A, Supplemental Figure 2). The AAs in the stand-alone systems resemble those shown to ligate a free fatty acid to ACP (16), consistent with the AA-ACP fusion. The central domains (ketosynthase (KS), acyltransferase (AT), ketoreductase (KR)), as in CurM, are presumed to catalyze a single extension of the fatty acyl chain using malonyl-CoA (AT, KS) with β-carbonyl group reduction by the KR to create a β-hydroxyacyl-ACP substrate for ST-TE mediated termination. This intriguing domain arrangement (AA-ACP-KS-AT-KR-ACP-ST-TE) (Figure 1A) suggests a pathway to convert fatty acids into terminal-alkene hydrocarbons with an odd number of carbon atoms due to the final decarboxylation step (8, 15). Indeed, the role of this biosynthetic system as an olefin synthase (OLS) was recently confirmed for the gene from Synechococcus PCC 7002 (17). Based on these findings it is evident that the unique PKS/FAS ST-TE offloading process has exciting potential applications in engineering pathways for hydrocarbon biofuel production and natural product structural diversification (18, 19).
Due to the surprising role of CurM PKS ST and OLS ST domains in hydroxy group activation for the terminal decarboxylative elimination reaction, we were motivated to obtain detailed structural information on these proteins. Previous crystal structures have primarily focused on mammalian STs, which share a common protein architecture, including a single catalytic domain, a common binding site for the sulfonate donor PAPS, and flexible loops surrounding the active site that become ordered upon PAP(S) binding (20). These structures and the PAP(S) binding mode led to the prevailing view of ST catalysis (21), in which a general base activates the acceptor substrate nucleophile for a concerted in-line transfer of sulfonate from PAPS.
Here, we report structure-function studies of activating STs from the curacin A pathway (CurM PKS ST domain) and from the Synechococcus PCC 7002 olefin synthase system (OLS ST domain, 47% sequence identity to CurM ST). Both activating STs accept substrates leading to long-chain hydrocarbon production, but the OLS ST excludes substrates bearing a C5-methoxy group (Figure 1B), which is present in the natural curacin chain elongation substrate. Crystal structures of both STs reveal the active-site residues, details of PAPS binding, and a dynamic active-site flap involved in substrate selectivity.
RESULTS
Activating STs are prototypes of a family
In addition to the eight activating STs previously identified within ACP-ST-TE tridomains (15), at least five additional ST sequences of high identity to CurM ST (31%–44%) are present in sequence databases (Supplemental Figure 2). These STs occur in the context of microbial natural product biosynthetic pathways, but unlike CurM ST, they lack a downstream decarboxylating TE. Rather, the genomic context suggests that these STs produce sulfated natural products, as has been detected for Planktothrix NIVA CYA 116 (22, 23). As we were most interested in STs with an activating functionality, the ST from the olefin synthase (OLS) in Synechococcus PCC 7002 (17) in addition to CurM ST from the curacin A PKS pathway were selected for further study.
The CurM PKS ST was demonstrated to be an activating ST (8) but no OLS ST has been characterized biochemically. Recombinant excised domains for CurM ST and OLS ST were purified as soluble, monomeric proteins and sulfotransferase activity was assayed (8). Briefly, β-hydroxyacyl substrates were loaded onto CurM ACP, and following reaction with ST, ACP products were analyzed by HPLC (Figure 1B, Supplemental Figure 3). OLS ST reacted with two OLS substrates that would produce hydrocarbons when offloaded ((R)-3-hydroxymyristoyl-ACP and (R)-3-hydroxydodecanoyl-ACP) (Table 1), as expected from previous studies (17). However to our surprise, the OLS ST had no detectable activity toward either of the curacin PKS ST substrate analogs that bear a C5-methoxy substituent ((3R/S,5R,S)-3-hydroxy-5-methoxymyristoyl-ACP) (Table 1) under the assay conditions. In contrast, CurM ST acted similarly upon the curacin PKS ST substrate mimics and the OLS substrates but with a threefold preference for the natural R-β-hydroxy enantiomer (Table 1). OLS ST displayed a slight preference for the OLS substrate with the longer (14–carbon) alkyl chain.
Table 1.
Substrate preferences of CurM ST and OLS ST
| Percent sulfonateda | ||
|---|---|---|
| CurM ST | OLS ST | |
| (R)-3-hydroxydodecanoyl-ACP | 73 ± 3 | 20.4 ± 0.3 |
| (R)-3-hydroxytetradecanoyl-ACP | 72 ± 2 | 33 ± 1 |
| (3R)-3-hydroxy-5-methoxytetradecanoyl-ACP | 74 ± 5 | 0 |
| (3S)-3-hydroxy-5-methoxytetradecanoyl-ACP | 28 ± 2 | 0 |
Raw HPLC chromatogram peak areas for the ACP-loaded substrate and sulfonated product were used to calculate the fraction of substrate sulfonated. CurM ST reactions were quenched and analyzed after 5 min and OLS ST after 3 h. Data are mean ± standard deviation from triplicate experiments.
Structures of activating STs
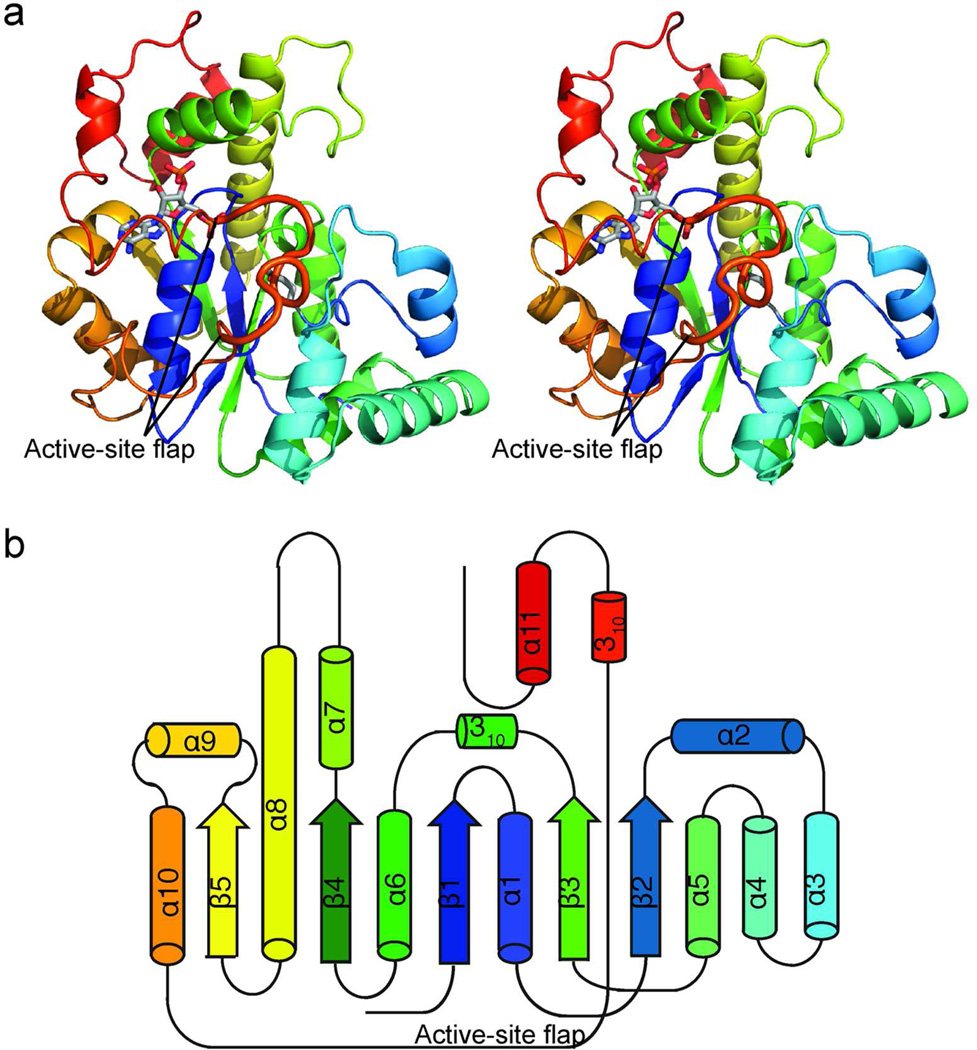
In order to more fully understand the activating STs, crystal structures were solved for CurM ST and OLS ST (Figure 2). Both enzymes were crystallized with the product PAP, which was essential for crystal growth. Crystallization of CurM ST required a double-alanine substitution at Gln259-Lys260. The structures define the limits of the ST domains as amino acids 1623–1905 of CurM and 2130–2420 of OLS. Here we use a common amino acid numbering based on the recombinant CurM ST polypeptide, as there are no gaps in the alignment of the CurM and OLS STs.
Figure 2.
Structure of activating STs. a) CurM ST polypeptide. The stereo ribbon diagram is colored as a rainbow from blue at the N-terminus to red at the C-terminus with PAP and Glu60, the proposed catalytic base, in stick (gray C). The active site flap (thick) is labeled. b) Topology diagram. CurM ST and OLS ST have an α/β core fold, like other STs, with inserted α-helices (α2–α4) and an extended loop wrapping around the core from α10–α11.
CurM and OLS STs resemble other STs in their overall architecture, as expected. However, the activating STs are much more similar to each other (RMSD = 0.79 Å for 228 Cα) than either is to the ST of next greatest structural similarity, human heparan sulfate 3-Osulfotransfease (3-OST, RMSD = 1.5 Åfor only 114 Cα atoms) (Supplemental Figure 4A,B). In the activating STs, the generic ST core is embellished with 3 additional α-helices (α2–α4) (Figure 2, Supplemental Figure 4B).
Serendipitous insight into PAPS binding
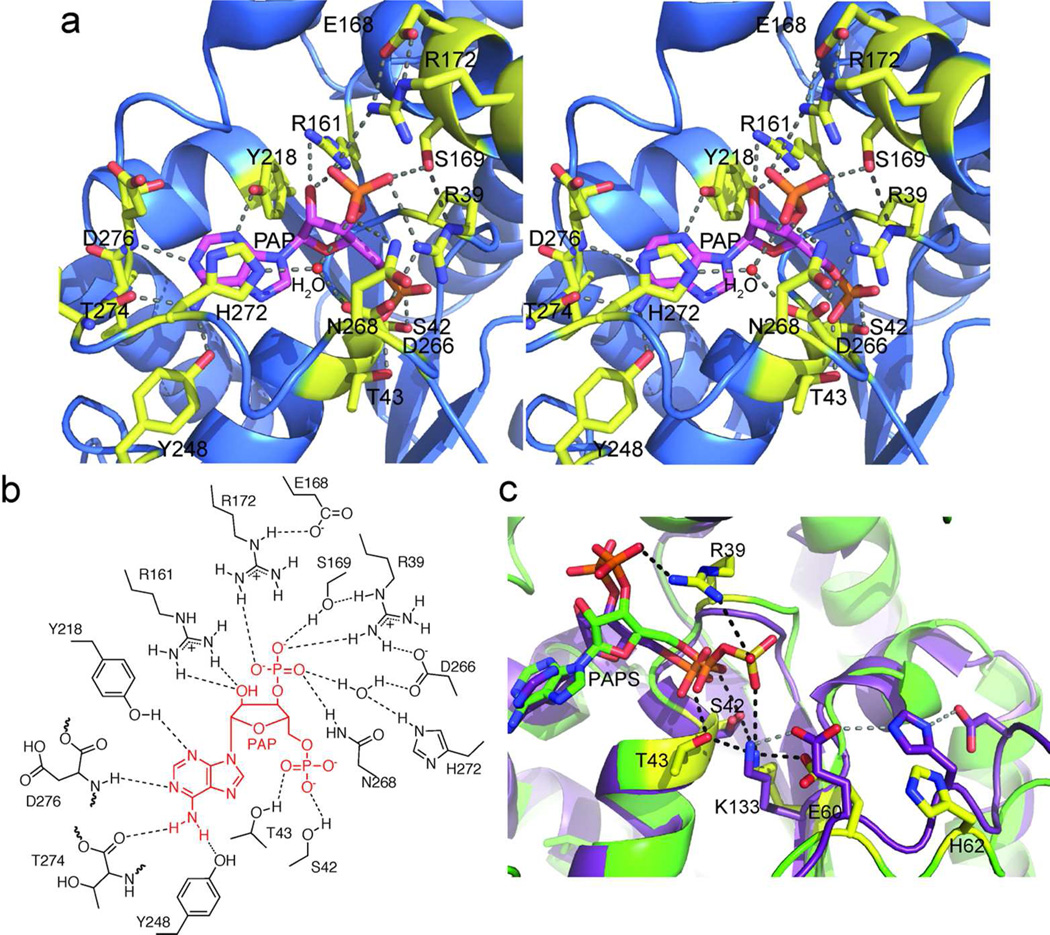
PAP binds similarly in CurM ST and OLS ST to a cleft near the C-terminal edge of a central parallel β-sheet (Figure 2A, Supplemental Figure 4). As in most other ST structures (except for the 3-OST family) (4), the adenine base is bound in the anti conformation. PAP forms an intricate network of hydrogen bonds with amino acids conserved in the activating ST sequences (Figure 3A,B, Supplemental Figure 1). Some of these interactions occur only in the activating ST family (Glu168, Tyr248, Asp266, Asn268, His272, and backbone interactions with Thr274 and Asp276) while others are common in the ST superfamily (Arg39, Gly41, Ser42, Thr43, Arg161, Ser169, Arg172, Tyr218).
Figure 3.
Active site of activating STs. a) OLS ST PAP binding site. PAP (magenta C) and surrounding amino acids (yellow C) in an intricate hydrogen-bonding network are shown in stick form in the stereo diagram. b) Schematic of PAP binding site. c) Comparison of the active sites of CurM ST and 3-OST. Key amino acids are shown in stick form in 3-OST (1T8U) (25) (purple C) and CurM ST (yellow C). Hydrogen bonds in CurM ST and the ionic interaction from Lys133 to PAP are shown in black. Hydrogen bonds in 3-OST are shown in gray. PAPS is modeled into CurM ST from the positions of PAP and ZnCl3−.
A Zn2+ ion from the CurM ST crystallization solution coordinates PAP at a position corresponding to the PAPS sulfate, and provides a serendipitous insight into PAPS binding. The Zn2+ coordinates a PAP 5' phosphate oxygen, and three Cl− ions complete the tetrahedral coordination sphere, resulting in a species with the same charge and shape as sulfate. The Zn2+ was identified by anomalous scattering, and the Cl− ions were placed at the positions of weak anomalous scattering peaks (Supplemental Figure 5). Although ZnCl3− is bulkier than SO3−, the active site accommodates ZnCl3− without perturbing the structure as the surrounding side chains are not significantly shifted relative to their positions in OLS ST (Supplemental Figure 5B). PAPS was modeled into the structure based on the position of ZnCl3− (Figure 3C, Supplemental Figure 5C). The PAPS sulfate location matches well with the PAPS complex of a distantly related mouse ST (24). Two amino acids, which are conserved in activating STs, are positioned to interact with PAPS sulfate oxygen atoms. Arg39 simultaneously recognizes the PAPS 3'-phosphate and 5'-sulfate. Lys133 forms an ionic interaction with the PAPS 5'-sulfate and hydrogen bonds with amino acids in the active site (Figure 3C).
The presence of ZnCl3− in the active site implies that Zn2+ may inhibit the ST. We tested the activity of CurM ST as a function of Zn2+ concentration, and observed a less-than twofold decrease in sulfonation over a range of 1–100 μM (Supplemental Table 1), concentrations far above estimates of free Zn2+ in the bacterial cytoplasm (25, 26). Thus, based on these data, we conclude that Zn2+ does not interfere with CurM ST activity in vivo.
Active Site
Several amino acids near the PAPS sulfate are positioned to affect catalysis (Figure 3C). The key residues Glu60 and Lys133 are invariant among the activating STs. Glu60 is well situated to serve as the catalytic base; the PAPS sulfate is in an optimal position for nucleophilic attack by the substrate β-hydroxy group following deprotonation by Glu60 (Supplemental Figure 6). In other STs, the analog of Glu60 is part of a Glu-His-Asp triad linked by hydrogen bonds (27, 28). However, the activating STs lack such a triad. His62 is too distant from Glu60 for a hydrogen bond, and no Asp is present. Lys133 anchors the active site by direct interaction with both Glu60 and the PAPS sulfate. Lys133 is further stabilized by hydrogen bonds with the Ser42 and Thr43 hydroxy groups, which in turn hydrogen bond to PAP 5' phosphate, further linking PAPS to the catalytic residues (Figure 3C). Most STs in the structure database have an “anchor” lysine like Lys133.
The importance of these and other amino acids in the active site (Arg39, Thr43, Glu60, His62, Lys133, Ser134) were probed by mutagenesis of CurM ST (Table 2). Ala and Gln substitutions at Glu60 reduced activity more than 100-fold, consistent with its proposed role as a catalytic base. A key role was confirmed for the anchor Lys133 (Figure 3), for which an alanine substitution reduced activity below detectable levels. Other perturbations of the hydrogen-bonding network through alanine substitutions at Thr43 (linking Lys133 and the PAPS 5' phosphate) and Arg39 (linking the PAPS sulfate to the 3' phosphate) led to a 100-fold reduction in activity. The mutagenesis results are consistent with a model in which Lys133 anchors key players in the active site: the PAPS sulfate, the catalytic base Glu60, and the PAPS 5' phosphate indirectly through Thr43. An Ala substitution at His62 in CurM ST reduced activity only 10-fold, in contrast to an ST with a Glu-His-Asp triad, where the analogous His is essential (27). The activity of this variant suggests His62 may interact with Glu60 during the catalytic cycle but is not necessary for catalysis.
Table 2.
Activity of CurM ST variants
| CurM ST variants | % Activitya | Location |
|---|---|---|
| Wild type | 100 | |
| R39A | 1.4 ± 1.1 | PAP binding |
| T43A | 1.1 ± 1.5 | Active site |
| E60A | 0.3 ± 1.0 | Active site |
| E60Q | 0.7 ± 0.6 | Active site |
| H62A | 8 ± 1 | Active site |
| K133A | < 0.1 | Active site |
| S134A | 52 ± 11 | Structural |
| S261A | 131 ± 19 | Flap |
| D266A | 1.9 ± 1.1 | Flap, PAP binding |
| D266N | 3.5 ± 0.9 | Flap, PAP binding |
| P267A | 58 ± 5 | Flap |
| No ST | 0 | |
| No PAPS | 0 | |
Raw HPLC chromatogram peak areas for the substrate ((R)-3-hydroxymyristoyl-ACP) and product ((R)-3-sulfomyristoyl-ACP) were used to calculate the fraction of substrate sulfonated. The activity of each variant was normalized to the wild type within each replicate. Mean ± standard deviation from triplicate experiments is shown.
Dynamic active-site flap
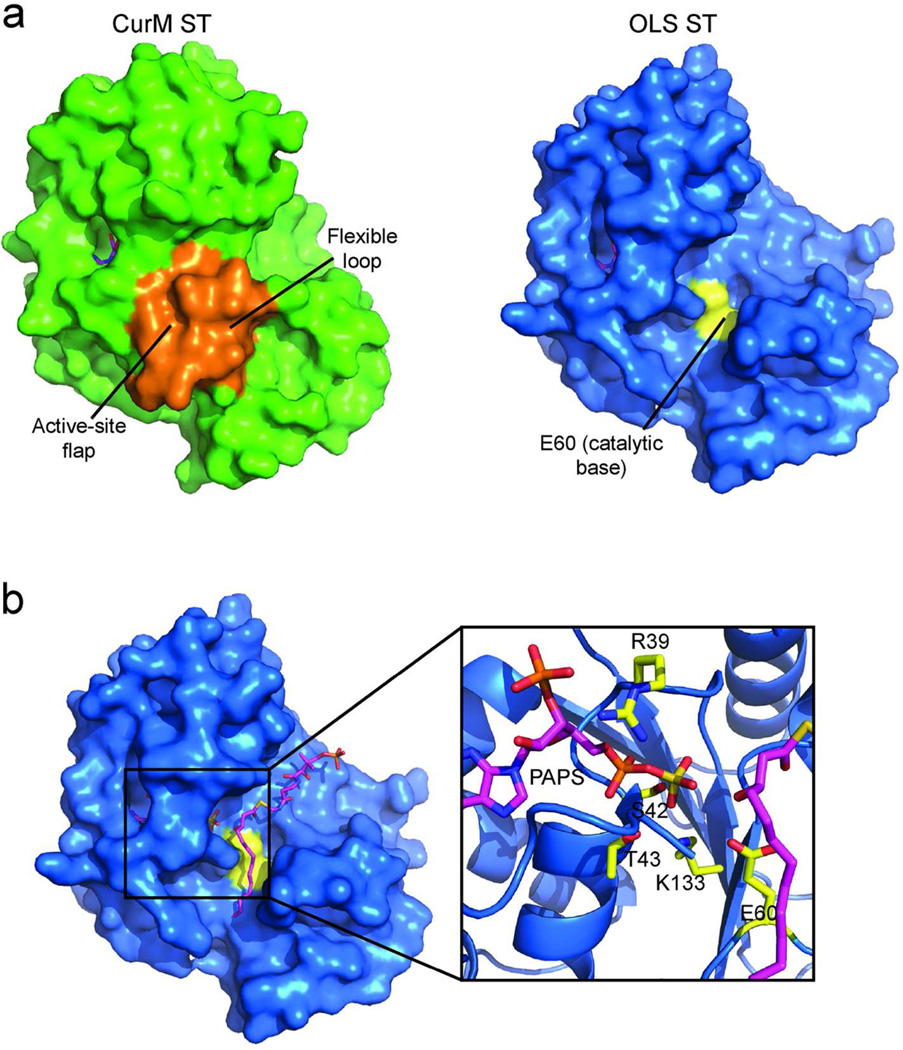
CurM ST and OLS ST possess a dynamic active-site flap (Figure 4A). A 13-residue loop between helices α10 and α11 (residues 255–267) covers the active site in CurM ST and is differently positioned (255, 264–267) and partially disordered (256–263) in OLS ST (Figure 4A, Supplemental Figure 7A). The ST superfamily exhibits substantial divergence in length and structure for this region at the entrance to the active site, in accord with the great variety of substrates that are sulfonated (4, 20). However, among the activating STs, the flap sequence is conserved, suggesting that the substrates are similar or that the flaps have a common function. Certainly the dynamic aspect of the CurM and OLS ST flaps hints that they close over the β-hydroxy substrate during catalysis. An adjacent flexible loop (residues 77–82 between α2 and α3) has somewhat different positions in CurM and OLS ST (Figure 4A), and may move in conjunction with the active-site flap. Similar order/disorder transitions have been observed in the mammalian cytosolic STs, where loops and helices surrounding the active site are progressively ordered upon the binding of PAP(S) and substrate (20).
Figure 4.
Active-site cleft. a) Surface representation of CurM ST (green) and OLS ST (blue). For both proteins PAP is shown in magenta stick and catalytic Glu60 in yellow (not visible in CurM ST). The active-site cleft in OLS ST is exposed due to disorder of the active-site flap. In CurM ST, the active-site flap and flexible loop (orange surfaces) cover the active site but their conformations are influenced by crystallization. b) Substrate modeled into OLS ST. (R)-3-hydroxymyristoylphosphopantetheine (magenta C, red O, blue N, yellow S, orange P) was modeled into the open substrate-binding cleft of OLS ST.
Two features of the CurM ST crystallization contribute to the well-ordered conformation of the active-site flap and flexible loop structure in the absence of substrate. A Zn2+ bound on the outside of the protein (not in the PAPS sulfate position) coordinates flap residues His258 and Asp266 (Supplemental Figure 5A, 7) and serves to anchor these points of the flap. Asp266 is invariant among activating STs and participates in the PAPS hydrogen bonding network in the OLS ST structure through a water-bridged hydrogen bond with the PAP 3'-phosphate and a direct hydrogen bond with Arg39 (Supplemental Figure 7A). The alanine substitutions to facilitate crystallization of CurM ST (Q259A/K260A) were at non-conserved sites in the active-site flap, and also contribute to trapping the flap in the observed conformation. Q259A/K260A mediates a crystal contact that also includes Tyr80 and Leu81 from the adjacent flexible loop (Supplemental Figure 7B). Together the crystal contact, the alanine substitutions, and the surface Zn2+ ion trapped the CurM ST flap in an ordered/closed conformation, however, this particular ordered conformation may not be relevant to ST function.
Substrate binding and specificity
We modeled substrates into the active sites in positions to interact with both the PAPS sulfate and Glu60, the proposed catalytic base. The active-site flap was ignored in the modeling because it is unknown how the flap closes over the substrate during catalysis. The structure of a ternary complex of 3-OST with PAP and a tetrasaccharide substrate [1T8U (27)] was used as a guide to place the β-hydroxy groups of the OLS ST substrate, (R)-3-hydroxymyristoyl-phosphopantethiene, and the curacin PKS ST substrate mimic, (3R)-3-hydroxy-5-methoxymyristoyl-phosphopantethiene Figure 4B). The phosphopantetheine arm and alkyl chain are well matched to the length of the substrate channel when the substrate β-hydroxy group is adjacent to Glu60 and to the PAPS sulfate.
The modeling did not provide an explanation for the OLS ST substrate selectivity, as substrates both with and without the C5-methoxy were easily modeled into both enzymes. Thus we probed the function of the active-site flap with two types of site-directed mutagenesis experiments. Single-site substitutions were made to three conserved amino acids in the active-site flap of CurM ST (Ser261, Asp266, Pro257) (Table 2). Activity with a simple alkyl substrate was reduced nearly 100-fold when Asp266 was substituted by Ala (2% of WT) or Asn (4% of WT), consistent with the Asp266 interaction with PAPS through Arg39 and an ordered water molecule (Figure 3A,B, Supplemental Figure 7A). The P267A and S261A variants had modestly reduced activity relative to the wild type for a simple alkyl substrate. We then tested the variants with the greatest activity (H62A, S134A, S261A, D266N, P267A) with all four substrates (Supplemental Figure 8A). Remarkably, alanine substitutions at conserved flap residues Ser261 and Pro267 resulted in substantial selection toward substrates lacking the C5-methoxy group, similar to OLS ST. This unexpected result suggests that the structure of the closed active-site flap affects substrate selectivity and that small changes to the flap sequence affect its structure.
The other mutagenesis experiments more directly tested the role of the active-site flap in substrate specificity. Chimeric proteins were created in which the CurM ST and OLS ST flaps (residues 254–270) were exchanged. The activity of CurM STflapOLS with C5-methoxy substrates decreased two-fold relative to wild type while the activity with the simple alkyl substrates was unchanged (Supplemental Figure 8B). This resulted in a CurM ST with activity more like that of OLS ST and demonstrates the importance of the flap in substrate selectivity. However, the flap is not the sole determinant of substrate selectivity, as OLS STflapCurM had negligible activity for all substrates (Supplemental Figure 8C).
DISCUSSION
Activating sulfotransferase family
CurM ST and OLS ST are prototypes of a family of functional-group-activating sulfotransferases in complex metabolic pathways. Both CurM ST and OLS ST catalyze β-hydroxy group sulfonation, the first step in this new mode of metabolite offloading and termination. Sulfonation activates the substrate for the unprecedented TE-catalyzed decarboxylation and sulfate elimination (8), and hence we have named the family “activating” sulfotransferases. The activating ST structure is most similar to the mammalian heparan sulfate D-glucosaminyl 3-O-sulfotransferase (3–OST) family of STs. Although the sequence identity is low (<15%), activating STs and 3-OST family members assist catalysis using a common set of amino acids. Results of site-directed mutagenesis of CurM ST are consistent with a catalytic mechanism in which Glu60 is the catalytic base and Lys133 anchors the PAPS sulfate and Glu60.
The sequence database contains several other uncharacterized gene products of high similarity to CurM ST (31%–43% identity) that occur in microbial natural product biosynthetic pathways without ACP-ST-TE tridomains (Supplemental Figure 2). These STs are not expected to activate hydroxy groups, but instead to produce sulfated natural products. The catalytic residues and active-site flap of the activating STs are conserved in these similar but “non-activating” ST sequences, indicative of recent common ancestry (Supplemental Figure 2). Natural products pathways do not include exclusively STs of the activating ST family. For example the glycopeptide ST Teg12 (29) (16% identity to CurM ST) clearly belongs to a different ST family.
OLS and PKS STs display different substrate selectivity
The assay results for OLS ST are the first biochemical studies of a domain from an olefin synthase. Consistent with genetic deletion and feeding studies (17), OLS ST acted on substrates with long alkyl chains. However, it had no detectable activity with C5-methoxy substrates in contrast to the CurM ST, which was active with all substrates tested. The exclusion of C5-methoxy substrates by OLS ST is presumably due to a feature of the closed active-site flap because the OLS ST and CurM ST surfaces “under” the active-site flap are identical in charge and structure. The flap-specificity hypothesis is consistent with the activity of the CurM STflapOLS chimera, which had decreased activity with C5-methoxy substrates but similar activity to CurM ST with the simple alkyl substrates. The active-site flap also contributes to substrate selectivity in other ST sub-families. For example, the human ST SULT2A1 active-site flap controls substrate selectivity by limiting the size of substrates that can access the active site after PAPS binding (30). Although the activating ST flap is highly diverged in sequence from the flaps of other STs such as SULT2A1, it appears to be used broadly within the ST superfamily to control substrate specificity. CurM ST had only a weak preference for the substrate mimic with the natural (R)- β-hydroxy stereochemistry over the non-natural (S) (Table 1, (8)), consistent with the idea that the ST is likely to encounter only substrates with the natural stereochemistry. The stereochemistry is predicted by sequence analysis of the upstream KR domain, which specifies an (R)-β-hydroxy product for both STs (31). Under our assay conditions, CurM ST is a much faster enzyme than OLS ST (Supplemental Figure 7B,C), even for substrates linked to the cognate OLS ACP (43% identity to CurM ACP). OLS ST may be more reactive with the natural substrate (6–8 carbons longer than the test substrates) or within the context of the full-length OLS polypeptide where substrates are linked to a fused ACP.
Distinctive structural features of activating STs
We visualized the PAPS sulfate in CurM ST from the serendipitous binding of ZnCl3 − in the sulfate position (24, 32). The PAPS sulfate is thus deduced to be less than 5 Å from the putative catalytic base, Glu60. The β-hydroxy group of the modeled substrate is in a position to be activated by Glu60 and subsequently to attack the sulfate of PAPS to complete sulfonate transfer (Figure 4B). Arg39 appears key in distinguishing PAPS from other nucleotides, as it simultaneously recognizes the 3'-phosphate and the 5'-sulfate. Additionally, four hydrogen bonds provide specificity for the adenine base (Figure 3A,B).
The structures of CurM ST and OLS ST together provide evidence of a dynamic active-site flap distinct to activating STs (Figure 4A). In the OLS ST, the flap is disordered, exposing an obvious substrate-binding cleft adjacent to the PAPS sulfate (Figure 4B). Crystallization of CurM ST captured a closed/ordered flap, as expected to follow substrate binding. However, no substrate was present, and a bound Zn2+, the two alanine substitutions, and a crystal contact contributed to the observed flap structure. The conservation of the length and amino acid sequence of the flap highlights its importance, as does the reduced activity of substitutions at invariant Asp266 in the flap. Asp266 is proposed to guide the flap into the proper closed position by interacting with PAPS, thereby ensuring that PAPS is bound before the flap closes. Other residues conserved in the flap of activating STs (Pro267, Ser261) are involved directly or indirectly in substrate recognition, as Ala variants altered the substrate specificity, an unexpected finding for amino acids conserved in the Cur and OLS enzymes. Flap chimeras resulted in a CurM ST with substrate specificity more like OLS ST but an OLS ST with negligible activity. Thus the flap is not the sole determinant of substrate selectivity, and we infer that flap-core interactions also modulate and control the substrate selectivity of the activating STs.
Functional-group activation
CurM ST and OLS ST carry out an activating sulfonation that creates a favorable leaving group to assist subsequent decarboxylation; the transient sulfate is present on an intermediate but not in the final product. The curacin A pathway also contains an “activating” halogenase, which functionalizes a carbon by chlorination for subsequent cyclopropane formation (33, 34). A similar chlorination strategy is employed to form a cyclopropane in the biosynthesis of coronamic acid (35). Activation of a hydroxy group by phosphorylation with phosphate, pyrophosphate or AMP is common in primary and secondary metabolism. The similarity of activating sulfonation and phosphorylation is striking in mevalonate-5-diphosphate decarboxylase (MDD) in the cholesterol biosynthetic pathway. MDD activates a β-hydroxy group by phosphorylation to facilitate decarboxylation and generate the terminal double bond of isopentenyl diphosphate (8, 36), chemically analogous to ST-TE catalysis. Unlike MDD, the ACP-ST-TE tridomain has the additional task of thioester hydrolysis. For the thioesterase-decarboxylase function, nature adapted the common thioesterase of PKS and NRPS systems and co-opted a sulfotransferase to facilitate the decarboxylation step. Activation by sulfonation is a rare variation on the theme of leaving-group activation by phosphorylation common to other metabolic pathways, and clearly is an effective strategy to activate substrates for combined thioester hydrolysis and decarboxylation.
Our structure-function studies define a distinct family of STs where most members have a surprising role in hydroxy group activation. This family includes ST domains from biosynthetic systems that produce natural products (polyketides, such as curacin A, and nonribosomal peptides) and hydrocarbons (OLS). The CurM ST and OLS ST structures reveal active site residues, details of PAPS binding, and a dynamic active-site flap, which was shown to influence substrate specificity. The CurM and OLS STs expand our view of nature’s remarkable synthetic toolbox and highlight promising engineering applications. The OLS ST is applicable to liquid biofuel production as it leads directly to long-chain hydrocarbons. The CurM ST, which accepts substrates containing a variety of functional groups, also holds value for use in natural product structural diversification.
METHODS
Cloning and site-directed mutagenesis
A construct encoding CurM ST (CurM residues 15981917, here referred to as 1–320) was amplified from cosmid pLM17 (7) and inserted into the pMCSG7 vector encoding an N-terminal 6xHis tag. A construct encoding OLS ST (OLSresidues 2121–2424, here referred to as 9–312) was amplified from synthetic DNA (GeneArt) and inserted into pMCSG7. Surface entropy reduction substitutions in CurM ST (E183A/E184A, K223A/K224A, Q259A/K260A, and E209A/E211A) were designed with assistance from the SERp server (37). Site directed mutagenesis of single amino acids was performed using the QuikChange protocol (Agilent). Flap chimeras were constructed using the QuikChange Lightening Multi Site Directed Mutagenesis Kit (Agilent). All constructs were verified by sequencing. CurM ACP was expressed as previously described (8).
Protein Expression, and Purification
E. coli strain BL21(DE3) was transformed with expression plasmid, grown at 37°C in 500 mL TB with 4% glycerol to an OD600 of 1.0, cooled to 20°C, induced with IPTG (final concentration 0.2 mM), and grown for an additional 18 h. Selenomethionyl (SeMet) CurM STQ259A/K260A was produced in the same E. coli strain in SelenoMet Medium (AthenaES) containing 100 µg mL−1 of seleno-DL-methionine.
All purification steps were performed at 4 °C. The cell pellet from 500 mL of cell culture was resuspended in 40 mL Buffer A (20 mM Tris pH 7.9, 500 mM NaCl, 10% glycerol) plus 20 mM imidazole and lysed by sonication, and the soluble fraction loaded onto a 5-mL HisTrap Ni NTA column (GE Healthcare). The proteins were eluted in Buffer A with a linear gradient of 20 to 650 mM imidazole. The 6xHis tag was removed by 2-h incubation with 1 mM DTT and tobacco etch virus (TEV) protease (1 mg protease per 50 mg ST) at room temperature. After overnight dialysis at 4 °C in Buffer A with 1 mM DTT, the remaining His-tagged proteins were removed by Ni-affinity chromatography, followed by size exclusion chromatography with a HiLoad 16/60 Superdex 200 column (GE Healthcare) pre-equilibrated with Buffer A. The purified proteins were concentrated to 15 mg/mL, flash frozen in liquid N2, and stored at −80 °C. SeMet CurM STQ259A/K260A was purified as the wild type with addition of 2 mM DDT to Buffer A during the size exclusion step. Of the 4 surface entropy reduction variants, 2 yielded enough soluble protein for crystallization trials (Q259A/K260A and E209A/E211A). CurM ST variants used for assay were purified as above but using a single HisTrap Ni NTA purification step followed by overnight dialysis in Buffer A with yields similar to wild type. Yields per 500 mL culture were 20 mg for CurM ST, 60 mg for OLS ST, 12 mg for CurM STQ259A/K260A, and 4 mg for SeMet CurM STQ259A/K260A.
Crystallization
Initial attempts to produce high-quality CurM ST crystals were not successful, so surface entropy reduction (SER) mutations (38) were made to promote crystallization. Double alanine substitutions were made at four sites of non-conserved, adjacent, polar amino acids; one double substitution (Q259A/K260A) resulted in crystals that diffracted to high resolution (1.6 Å. The activity of CurM STQ259A/K260A was reduced about threefold relative to wild type (Supplemental Table 1). Crystals of CurM STQ259A/K260A grew at 4 °C within 24–48 h by vapor diffusion in hanging drops from a 1:1 mix of protein stock (7 mg mL−1 ST in Buffer A, 2 mM PAP) and well solution (25–31% PEG MME 550, 2.5–12.5 mM ZnSO4, 100 mM MES pH 6.5). Crystallization was dependent upon the presence of Zn2+ and PAP. SeMet protein crystallized in similar conditions to the native CurM STQ259A/K260A with 1 mM DTT in the protein solution. OLS ST crystals grew at 20 °C within 24 h from a 1:1 mixture of protein stock (11 mg mL−1 ST in Buffer A, 2 mM PAP) and well solution (27–30% PEG 1.5K, 100 mM MMT buffer (DL-malic acid, MES, Tris) pH 6.0). Crystals of OLS ST that diffracted beyond 4.0 Åwere grown by microseeding. Crystals were harvested in loops and flash cooled in liquid N2.
Data Collection and Structure Determination
Data were collected at GM/CA beamline 23ID-D at the Advanced Photon Source (APS) at Argonne National Lab (Argonne, IL). A 2.1-Å single-wavelength anomalous diffraction dataset was collected at the wavelength of peak absorption at the selenium edge from a SeMet CurM STQ259A/K260A crystal. A 1.6-Å dataset was collected from a native CurM STQ259A/K260A crystal at E = 12.0000 keV and a 1.9-Å dataset was collected at E = 10.000 keV from a second crystal from the same drop (Table 3). A 2.1-Å dataset was collected from an OLS ST crystal. All data were processed using the HKL2000 suite (39). Using Phaser (40) in the PHENIX (41) software suite, anomalous scatterer sites were found for all eight Met residues from the one polypeptide chain in the asymmetric unit (average figure of merit (FOM) = 0.49), with two partially occupied Se sites for each of five Met side chains. Five additional anomalous scatterers were found in locations that did not correspond to Met side chains. These five anomalous scatterers were identified as Zn2+ in an anomalous difference Fourier from a dataset collected at 10.000 keV (λ = 1.23984 Å), 341 eV above the Zn K absorption edge. After density modification in RESOLVE (42) (FOM = 0.77) a 90% complete model was built by AUTOBUILD (43) and completed manually in COOT (44). Waters were added automatically in COOT and manually edited. The 1.6-Ånative dataset was used for refinement in REFMAC5 (45) from the CCP4 (46) suite using 5 TLS groups (47). Electron density was complete throughout the polypeptide chain for residues 26–308. Twenty-five residues at the N-terminus and 12 at the C-terminus were disordered. The OLS ST structure was solved by molecular replacement in Phaser (40) using CurM STQ259A/K260A as a search model (47% sequence identity). OLS ST was refined using BUSTER (48). Model quality was evaluated with MolProbity (49).
Table 3.
Crystallographic Summary
| CurM STQ259A/K260A (SeMet) |
CurM STQ259A/K260A (native) |
CurM STQ259A/K260A (native) |
OLS ST (native) |
|
|---|---|---|---|---|
| Diffraction Data | ||||
| Space group | P212121 | P212121 | P212121 | P42212 |
| X-ray source | APS 23-ID-B | APS 23-ID-B | APS 23-ID-B | APS 23-ID-D |
| a, b, c (Å) | 46.0, 67.6, 118.5 | 45.8, 67.3, 118.0 | 45.9, 67.3, 118.5 | 131.4, 131.4, 47.2 |
| a, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Wavelength (Å) | 0.97948 | 1.03320 | 1.23984 | 0.97934 |
| dmin (Å) | 2.12 (2.20-2.12)a | 1.62 (1.68-1.62) | 1.95 (2.02-1.95) | 2.15 (2.23-2.15) |
| Avg I/σI | 26.5 (14.3) | 17.5 (4.9) | 17.0 (7.8) | 24.1 (3.6) |
| Rsymmb | 0.098 (0.261) | 0.074 (0.375) | 0.085 (0.276) | 0.063 (0.634) |
| Completeness | 100 (99.9) | 98.5 (89.6) | 99.7 (99.4) | 99.8 (100.0) |
| Avg. redundancy | 14.5 (14.3) | 6.6 (5.9) | 7.2 (7.2) | 8.7 (7.5) |
| Unique reflections | 21,790 | 46,830 | 27,422 | 23,099 |
| Refinement | ||||
| Data range (Å) | 36.34-1.62 | 33.07-2.15 | ||
| No. reflections | 46,772 | 22,991 | ||
| Rwork/Rfreec | 0.158/0.182 | 0.188/0.220 | ||
| RMS deviations | ||||
| Bonds (Å) | 0.007 | .010 | ||
| Angles (°) | 1.13 | 1.06 | ||
| Avg B-factors (Å2) | ||||
| Protein | 17.1 | 61.7 | ||
| Water | 33.5 | 60.7 | ||
| Other | 22.1 | 48.2 | ||
| Ramachandran | ||||
| Allowed | 100.0% | 100.0% | ||
| Outliers | 0.0% | 0.0% | ||
| Protein Atoms | 2325 | 2211 | ||
| Water Molecules | 365 | 96 | ||
| Other Atoms | 83 | 27 | ||
Outermost shell in parentheses.
Including anomalous differences.
The Rfree data set included a random 5% of reflections.
Crystals of both STs contained one polypeptide in the asymmetric unit. OLS ST amino acid numbering within the ST domain was assigned to match with CurM ST numbering, as there are no gaps in the sequence and structure alignments. Electron density is continuous for CurM ST residues 26 to 308, and OLS ST termini are ordered in a similar range, 18 to 308. The OLS ST crystal structure has two disordered internal loops (177–184 and 255–263), which are ordered in CurM ST. Zn2+ was essential to CurM ST crystal formation. A total of five Zn positions were identified by Zn anomalous scattering (Supplemental Figure 5A). Weak anomalous scattering peaks at other positions in the tetrahedral coordination sphere of Zn2+ (Supplemental Figure 5) were interpreted as Cl− ligands.
Sequence alignment, structure alignment, and substrate modeling
The search for ST homologs was done with BLAST (50). The DALI server (51) was used to identify the closest known structures to activating STs. MUSCLE (52) was used for multiple sequence alignment. PyMOL was used to align structures and prepare figures (53). The PRODRG2 server (54) was used to generate starting models and topology files for the substrates, which were docked manually using the position of the 3-OST substrate (1T8U (27)) as a guide. Sequences were aligned for phylogenetic analysis using the MAAFT alignment algorithm followed by tree building in MrBayes (55) using the Whelan-Goldman model and gamma distribution variation. The most appropriate model was determined by adding the alignment to the TOPALi modeling program (http://www.topali.org/).
Enzyme Assay
CurM ST and OLS ST were assayed using a modification of our previous protocols (8, 9). Apo-CurM ACP was loaded with a substrate analog by 2-h incubation of 50 µM apo-ACP with 100 µM CoA-linked substrate ((3R)-3-hydroxy-5-methoxymyristoyl-CoA (8), (3S)-3-hydroxy-5-methoxymyristoyl-CoA (8), (R)-3-hydroxymyristoyl-CoA, and (R)-3-hydroxydodecanoyl-CoA), 10 µM Streptomyces verticillus Svp (56), 10 mM MgCl2, and 100 mM Tris pH 7.9 at 30 °C. Complete loading was confirmed by reverse phase HPLC using a Jupiter C4 column (250 × 2.0 mm, 5 µm, 300 Å Phenomenex) and a linear elution gradient from 30% to 90% CH3CN (0.1% CF3CO2H)/H2O (0.1% CF3CO2H) over 45 min (Supplemental Figure 3A). After exchange into Buffer A and concentration (Amicon Ultra 10 kDa concentrators, Millipore), substrate-loaded ACP was flash frozen and stored at −80 °C. It was not possible to reach saturating concentrations of ACP-linked substrates, so standard assay conditions were developed where nearly complete sulfonation of substrates was achieved with the wild type CurM ST. Reaction times were adjusted for the slower OLS ST. The reaction was initiated by the addition of ST (1 µµM) into 100 µM loaded ACP, 1.75 mM PAPS (Sigma), 100 mM Tris pH 7.9 at room temperature. After 5 min (CurM ST) or 3 h (OLS ST), the reactions were quenched with 10% formic acid. Conversion of β-hydroxy-ACP to sulfated-ACP was quantified by HPLC detection of ACP species as described above (Supplemental Figure 3B). Results were quantitated as the sulfated-ACP peak area as a fraction of the total ACP peak area (substrate + product). Under the conditions tested, CurM ST is more active than OLS ST, with 75% sulfonation of the ACP-bound substrate in 5 minutes whereas OLS ST converts approximately 50% in 3 hours.
Substrate Synthesis
The synthesis of (3R)-3-hydroxy-5-methoxymyristoyl-CoA and (3S)-3-hydroxy-5-methoxymyristoyl-CoA was previously described (8). (R)-3-hydroxymyristoyl-CoA and (R)-3-hydroxydodecanoyl-CoA were synthesized as described in Chemical Synthesis section of the Supplemental Materials.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH grants R01 DK42303 (JLS), R01 CA108874 (DHS, WHG, and JLS), and U01 TW007404 as well as the Hans W. Vahlteich Professorship (DHS). Beamline 23ID-D is supported by the NIH National Institute of General Medical Sciences (GM, Y1-GM-1104) and National Cancer Institute (CA, Y1-CO-1020) through the GM/CA at the APS, which is supported by the US Department of Energy. We appreciate the permission from the country of Curaçao and assistance of the CARMABI research station for cyanobacterial collections
Footnotes
Accession codes. Atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4GBM for CurM ST and 4GOX for OLS ST).
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Chapman E, Best MD, Hanson SR, Wong CH. Sulfotransferases: structure, mechanism, biological activity, inhibition, and synthetic utility. Angew Chem Int Ed Engl. 2004;43:3526–3548. doi: 10.1002/anie.200300631. [DOI] [PubMed] [Google Scholar]
- 2.Schelle MW, Bertozzi CR. Sulfate metabolism in mycobacteria. Chembiochem. 2006;7:1516–1524. doi: 10.1002/cbic.200600224. [DOI] [PubMed] [Google Scholar]
- 3.Bojarova P, Williams SJ. Sulfotransferases, sulfatases and formylglycinegenerating enzymes: a sulfation fascination. Curr Opin Chem Biol. 2008;12:573–581. doi: 10.1016/j.cbpa.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 4.Rath VL, Verdugo D, Hemmerich S. Sulfotransferase structural biology and inhibitor discovery. Drug Discov Today. 2004;9:1003–1011. doi: 10.1016/S1359-6446(04)03273-8. [DOI] [PubMed] [Google Scholar]
- 5.Verdier-Pinard P, Lai JY, Yoo HD, Yu J, Marquez B, Nagle DG, Nambu M, White JD, Falck JR, Gerwick WH, Day BW, Hamel E. Structureactivity analysis of the interaction of curacin, A the potent colchicine site antimitotic agent, with tubulin and effects of analogs on the growth of MCF-7 breast cancer cells. Mol Pharmacol. 1998;53:62–76. doi: 10.1124/mol.53.1.62. [DOI] [PubMed] [Google Scholar]
- 6.Engene N, Rottacker EC, Kastovsky J, Byrum T, Choi H, Ellisman MH, Komarek J, Gerwick WH. Moorea producens gen. nov., sp. nov. and Moorea bouillonii comb. nov tropical marine cyanobacteria rich in bioactive secondary metabolites. Int J Syst Evol Microbiol. 2012;62:1171–1178. doi: 10.1099/ijs.0.033761-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang Z, Sitachitta N, Rossi JV, Roberts MA, Flatt PM, Jia J, Sherman DH, Gerwick WH. Biosynthetic pathway and gene cluster analysis of curacin A an antitubulin natural product from the tropical marine cyanobacterium Lyngbya majuscula. J Nat Prod. 2004;67:1356–1367. doi: 10.1021/np0499261. [DOI] [PubMed] [Google Scholar]
- 8.Gu L, Wang B, Kulkarni A, Gehret JJ, Lloyd KR, Gerwick L, Gerwick WH, Wipf P, Hakansson K, Smith JL, Sherman DH. Polyketide decarboxylative chain termination preceded by O-sulfonation in curacin a biosynthesis. J Am Chem Soc. 2009;131:16033–16035. doi: 10.1021/ja9071578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gehret JJ, Gu L, Gerwick WH, Wipf P, Sherman DH, Smith JL. Terminal alkene formation by the thioesterase of curacin A biosynthesis: structure of a decarboxylating thioesterase. J Biol Chem. 2011;286:14445–14454. doi: 10.1074/jbc.M110.214635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giraldes JW, Akey DL, Kittendorf JD, Sherman DH, Smith JL, Fecik RA. Structural and mechanistic insights into polyketide macrolactonization from polyketide-based affinity labels. Nat Chem Biol. 2006;2:531–536. doi: 10.1038/nchembio822. [DOI] [PubMed] [Google Scholar]
- 11.Scaglione JB, Akey DL, Sullivan R, Kittendorf JD, Rath CM, Kim ES, Smith JL, Sherman DH. Biochemical and Structural Characterization of the Tautomycetin Thioesterase: Analysis of a Stereoselective Polyketide Hydrolase. Angew Chem Int Ed Engl. 2010;49:5726–5730. doi: 10.1002/anie.201000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai SC, Miercke LJ, Krucinski J, Gokhale R, Chen JC, Foster PG, Cane DE, Khosla C, Stroud RM. Crystal structure of the macrocycle-forming thioesterase domain of the erythromycin polyketide synthase: versatility from a unique substrate channel. Proc Natl Acad Sci U S A. 2001;98:14808–14813. doi: 10.1073/pnas.011399198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsai SC, Lu H, Cane DE, Khosla C, Stroud RM. Insights into channel architecture and substrate specificity from crystal structures of two macrocycleforming thioesterases of modular polyketide synthases. Biochemistry. 2002;41:12598–12606. doi: 10.1021/bi0260177. [DOI] [PubMed] [Google Scholar]
- 14.Akey DL, Kittendorf JD, Giraldes JW, Fecik RA, Sherman DH, Smith JL. Structural basis for macrolactonization by the pikromycin thioesterase. Nat Chem Biol. 2006;2:537–542. doi: 10.1038/nchembio824. [DOI] [PubMed] [Google Scholar]
- 15.Akey DL, Gehret JJ, Khare D, Smith JL. Insights from the sea: Structural biology of marine polyketide synthases. Nat Prod Rep. 2012 doi: 10.1039/c2np20016c. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leger M, Gavalda S, Guillet V, van der Rest B, Slama N, Montrozier H, Mourey L, Quemard A, Daffe M, Marrakchi H. The dual function of the Mycobacterium tuberculosis FadD32 required for mycolic acid biosynthesis. Chem Biol. 2009;16:510–519. doi: 10.1016/j.chembiol.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 17.Mendez-Perez D, Begemann MB, Pfleger BF. Modular Synthase- Encoding Gene Involved in {alpha}-Olefin Biosynthesis in Synechococcus sp. Strain PCC 7002. Appl Environ Microbiol. 2011;77:4264–4267. doi: 10.1128/AEM.00467-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goss RJ, Shankar S, Fayad AA. The generation of "unNatural" products: Synthetic biology meets synthetic chemistry. Nat Prod Rep. 2012;29:870–889. doi: 10.1039/c2np00001f. [DOI] [PubMed] [Google Scholar]
- 19.Zabala AO, Cacho RA, Tang Y. Protein engineering towards natural product synthesis and diversification. J Ind Microbiol Biotechnol. 2012;39:227–241. doi: 10.1007/s10295-011-1044-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allali-Hassani A, Pan PW, Dombrovski L, Najmanovich R, Tempel W, Dong A, Loppnau P, Martin F, Thornton J, Edwards AM, Bochkarev A, Plotnikov AN, Vedadi M, Arrowsmith CH. Structural and chemical profiling of the human cytosolic sulfotransferases. PLoS Biol. 2007;5:e97. doi: 10.1371/journal.pbio.0050097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cleland WW, Hengge AC. Enzymatic mechanisms of phosphate and sulfate transfer. Chem Rev. 2006;106:3252–3278. doi: 10.1021/cr050287o. [DOI] [PubMed] [Google Scholar]
- 22.Rounge TB, Rohrlack T, Tooming-Klunderud A, Kristensen T, Jakobsen KS. Comparison of cyanopeptolin genes in Planktothrix, Microcystis, and Anabaena strains: evidence for independent evolution within each genus. Appl Environ Microbiol. 2007;73:7322–7330. doi: 10.1128/AEM.01475-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rounge TB, Rohrlack T, Kristensen T, Jakobsen KS. Recombination and selectional forces in cyanopeptolin NRPS operons from highly similar, but geographically remote Planktothrix strains. BMC Microbiol. 2008;8:141. doi: 10.1186/1471-2180-8-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teramoto T, Sakakibara Y, Liu MC, Suiko M, Kimura M, Kakuta Y. Snapshot of a Michaelis complex in a sulfuryl transfer reaction: Crystal structure of a mouse sulfotransferase, mSULT1D1, complexed with donor substrate and accepter substrate. Biochem Biophys Res Commun. 2009;383:83–87. doi: 10.1016/j.bbrc.2009.03.146. [DOI] [PubMed] [Google Scholar]
- 25.Outten CE, O'Halloran TV. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science. 2001;292:2488–2492. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- 26.Hitomi Y, Outten CE, O'Halloran TV. Extreme zinc-binding thermodynamics of the metal sensor/regulator protein, ZntR. J Am Chem Soc. 2001;123:8614–8615. doi: 10.1021/ja016146v. [DOI] [PubMed] [Google Scholar]
- 27.Moon AF, Edavettal SC, Krahn JM, Munoz EM, Negishi M, Linhardt RJ, Liu J, Pedersen LC. Structural analysis of the sulfotransferase (3-O-sulfotransferase isoform 3) involved in the biosynthesis of an entry receptor for herpes simplex virus 1. J Biol Chem. 2004;279:45185–45193. doi: 10.1074/jbc.M405013200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moon AF, Xu Y, Woody SM, Krahn JM, Linhardt RJ, Liu J, Pedersen LC. Dissecting the substrate recognition of 3-O-sulfotransferase for the biosynthesis of anticoagulant heparin. Proc Natl Acad Sci U S A. 2012;109:5265–5270. doi: 10.1073/pnas.1117923109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bick MJ, Banik JJ, Darst SA, Brady SF. Crystal structures of the glycopeptide sulfotransferase Teg12 in a complex with the teicoplanin aglycone. Biochemistry. 2010;49:4159–4168. doi: 10.1021/bi100150v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cook I, Wang T, Falany CN, Leyh TS. A Nucleotide-Gated Molecular Pore Selects Sulfotransferase Substrates. Biochemistry. 2012 [Google Scholar]
- 31.Caffrey P. The stereochemistry of ketoreduction. Chem Biol. 2005;12:1060–1062. doi: 10.1016/j.chembiol.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 32.Pedersen LC, Petrotchenko E, Shevtsov S, Negishi M. Crystal structure of the human estrogen sulfotransferase-PAPS complex: evidence for catalytic role of Ser137 in the sulfuryl transfer reaction. J Biol Chem. 2002;277:17928–17932. doi: 10.1074/jbc.M111651200. [DOI] [PubMed] [Google Scholar]
- 33.Khare D, Wang B, Gu L, Razelun J, Sherman DH, Gerwick WH, Hakansson K, Smith JL. Conformational switch triggered by alpha-ketoglutarate in a halogenase of curacin A biosynthesis. Proc Natl Acad Sci U S A. 2010;107:14099–14104. doi: 10.1073/pnas.1006738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gu L, Wang B, Kulkarni A, Geders TW, Grindberg RV, Gerwick L, Hakansson K, Wipf P, Smith JL, Gerwick WH, Sherman DH. Metamorphic enzyme assembly in polyketide diversification. Nature. 2009;459:731–735. doi: 10.1038/nature07870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vaillancourt FH, Yeh E, Vosburg DA, O'Connor SE, Walsh CT. Cryptic chlorination by a non-haem iron enzyme during cyclopropyl amino acid biosynthesis. Nature. 2005;436:1191–1194. doi: 10.1038/nature03797. [DOI] [PubMed] [Google Scholar]
- 36.Bonanno JB, Edo C, Eswar N, Pieper U, Romanowski MJ, Ilyin V, Gerchman SE, Kycia H, Studier FW, Sali A, Burley SK. Structural genomics of enzymes involved in sterol/isoprenoid biosynthesis. Proc Natl Acad Sci U S A. 2001;98:12896–12901. doi: 10.1073/pnas.181466998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goldschmidt L, Cooper DR, Derewenda ZS, Eisenberg D. Toward rational protein crystallization: A Web server for the design of crystallizable protein variants. Protein Sci. 2007;16:1569–1576. doi: 10.1110/ps.072914007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Derewenda ZS, Vekilov PG. Entropy and surface engineering in protein crystallization. Acta Crystallogr D Biol Crystallogr. 2006;62:116–124. doi: 10.1107/S0907444905035237. [DOI] [PubMed] [Google Scholar]
- 39.Otwinowski ZaMW. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 40.Zwart PH, Afonine PV, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, McKee E, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Storoni LC, Terwilliger TC, Adams PD. Automated structure solution with the PHENIX suite. Methods Mol Biol. 2008;426:419–435. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
- 41.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 42.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Terwilliger TC. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr D Biol Crystallogr. 2003;59:38–44. doi: 10.1107/S0907444902018036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 45.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 46.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 47.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 48.Bricogne GBE, Brandl M, Flensburg C, Keller P, Paciorek W, Roversi PSA, Smart OS, Vonrhein C, Womack TO. BUSTER version 2.10.0. Cambridge, United Kingdom: Global Phasing Ltd; 2011. [Google Scholar]
- 49.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 51.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schrodinger LLC. The PyMOL Molecular Graphics System, Version 1.3r1. 2010. [Google Scholar]
- 54.Schuttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 55.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 56.Sanchez C, Du L, Edwards DJ, Toney MD, Shen B. Cloning and characterization of a phosphopantetheinyl transferase from Streptomyces verticillus ATCC15003, the producer of the hybrid peptide-polyketide antitumor drug bleomycin. Chem Biol. 2001;8:725–738. doi: 10.1016/s1074-5521(01)00047-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.