Abstract
Meiosis is a germ cell-specific cell division process through which haploid gametes are produced for sexual reproduction1. Prior to initiation of meiosis, mouse primordial germ cells (PGCs) undergo a series of epigenetic reprogramming steps2,3, including global erasure of DNA methylation on the 5-position of cytosine (5mC) at CpG4,5. Although several epigenetic regulators, such as Dnmt3l, histone methyltransferases G9a and Prdm9, have been reported to be critical for meiosis6, little is known about how the expression of meiotic genes is regulated and how their expression contributes to normal meiosis. Using a loss of function approach, here we demonstrate that the 5mC-specific dioxygenase Tet1 plays an important role in regulating meiosis in mouse oocytes. Tet1 deficiency significantly reduces female germ cell numbers and fertility. Univalent chromosomes and unresolved DNA double strand breaks are also observed in Tet1-deficient oocytes. Tet1 deficiency does not greatly affect the genome-wide demethylation that takes place in PGCs but leads to defective DNA demethylation and decreased expression of a subset of meiotic genes. Our study thus establishes a function for Tet1 in meiosis and meiotic gene activation in female germ cells.
Mouse primordial germ cells (PGCs) first appear at E7.25 on the base of the allantois and then migrate through hindgut to the genital ridge3. During migration and at the genital ridge, PGCs undergo a series of coordinated epigenetic reprogramming, including global erasure of DNA methylation4,5. Recent demonstration that the Tet family proteins are involved in DNA demethylation prompted us to evaluate the role of the Tet proteins in PGC reprogramming7–10. RT-qPCR analysis demonstrated that Tet1 is preferentially expressed in PGCs, while Tet2 is expressed in both PGCs and somatic cells, and Tet3 is mainly expressed in somatic cells during PGC development (E9.5–E13.5) (Supplementary Fig. 1). To explore a potential role of Tet1 in PGC reprogramming and/or germ cell development, we generated Tet1 gene-trap mice (Supplementary Fig. 2a–d). Homozygous mutant mice (Tet1Gt/Gt) were generated by crossing heterozygous mice. Southern blot analysis, genomic sequencing, and allele-specific PCR confirmed a single site insertion of a tandem-repeated gene-trap cassette into the first intron of Tet1 gene (Supplementary Fig. 2a–d). β-galactosidase activity analysis revealed that the transgene is almost exclusively expressed in PGCs (Supplementary Fig. 2e), which is consistent with the Tet1 expression pattern (Supplementary Fig. 1), supporting a single locus insertion. Western blot analysis demonstrated that the insertion nullified the expression of the full-length Tet1 protein (Supplementary Fig. 3a). As expected, a fusion protein between the first exon of Tet1 (aa1–621) and βGeo (1303aa) was detected in Tet1Gt/Gt ES cell. Consistent with loss of Tet1, dot-blot and mass spectrometry analyses revealed about 45% reduction of 5hmC level in E9.5 Tet1Gt/Gt embryos (Supplementary Fig. 3b–d). RT-qPCR analysis of E11.5 PGCs demonstrated that Tet1 level is less than 5% that of the wild-type level (Supplementary Fig. 3e). Consistently, Tet1 protein was also not detectable in spreads of Tet1Gt/Gt PGCs (Supplementary Fig. 3f). Furthermore, immunostaining revealed loss of the dotted 5hmC staining signal in germ cells of E14.5 Tet1Gt/Gt genital ridge (Supplementary Fig. 3g). Collectively, these data indicate that Tet1 expression is effectively abolished and 5hmC level was significantly reduced in Tet1Gt/Gt PGCs.
Analysis of early backcross generations (N1–2) revealed embryonic lethal for homozygous while heterozygous mice were born in Mendelian ratio (Supplementary Table 1). Although further backcross (N3–6) relieved the embryonic lethal phenotype, the number of viable homozygous mice is still only approximately 1/3 of the numbers expected. Since severity of embryonic lethality is affected by genetic background, we only used later than N6 generation of Tet1Gt/Gt mice for subsequent analysis. Similar to a recent report11, reduced pup numbers were observed when either homozygous male or female animals were crossed with wild-type animals, and the pup numbers are even fewer when homozygous animals were crossed (Supplementary Fig. 4a). Because male gonad was morphologically normal and no obvious defects in male germ cell development were observed (data not shown), our attention is focused on characterizing the female germ cell phenotypes.
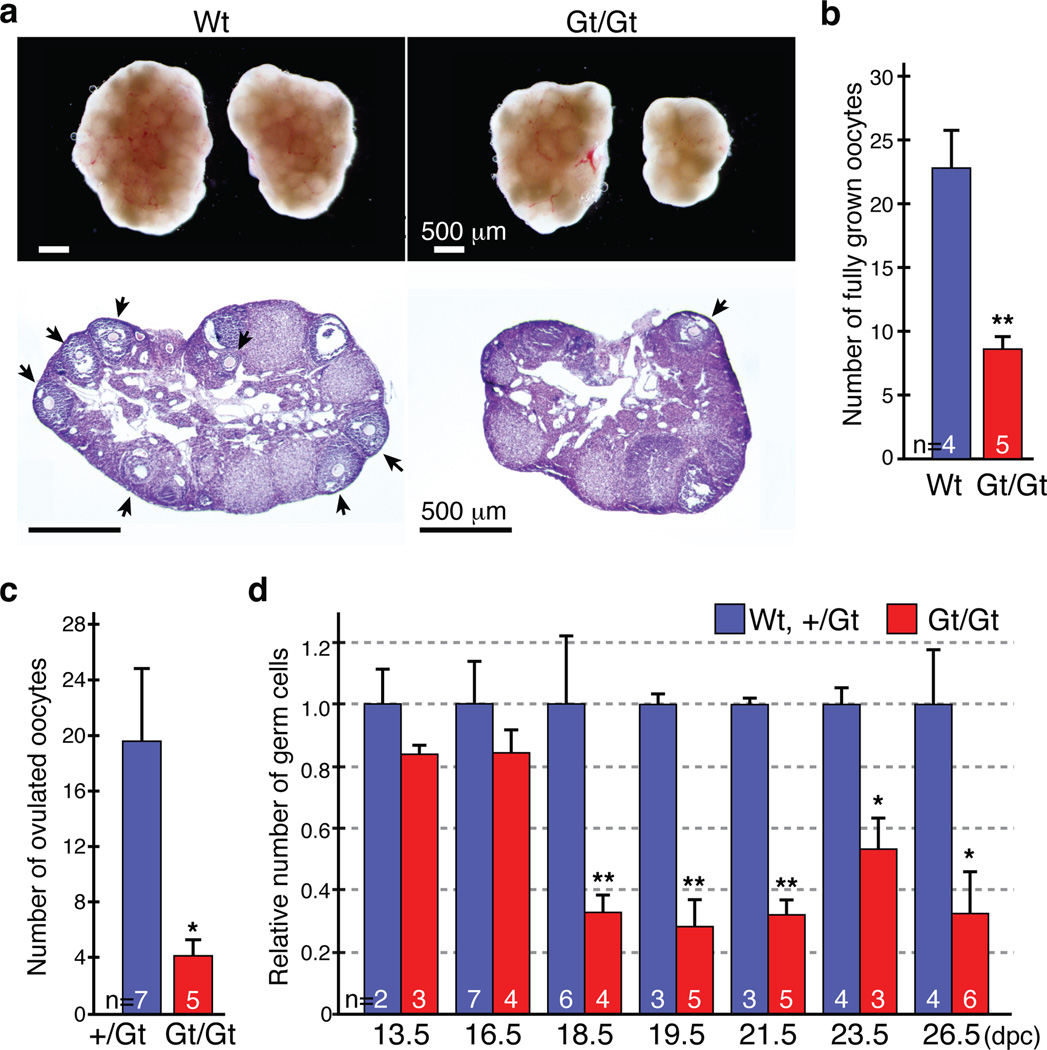
We found that the size of Tet1Gt/Gt ovary is significantly smaller, with a 30% reduction in the ovary to body weight ratio (Figs. 1a, Supplementary Fig. 4b). Interestingly, ovary agenesis-caused asymmetric ovary size is frequently observed in the Tet1Gt/Gt animals (Fig. 1a). Both fully-grown oocytes in the ovary and ovulated oocytes after hormonal stimulation are significantly reduced in the Tet1Gt/Gt animals (Fig. 1b, c). Ovary staining with germ cell-specific markers (Mvh, TRA98, and Msy2) followed by counting revealed a significantly reduction of oocyte number from E16.5–E18.5 (Fig. 1d, Supplementary Fig. 5a), concurrent with a significant increase in apoptotic oocytes (Supplementary Fig. 5b, c). These results suggest that increased apoptosis is likely one contributing factor for oocyte loss in Tet1Gt/Gt animals.
Figure 1. Subfertility of Tet1Gt/Gt mice is associated with oocyte loss in late embryonic stage.
(a) Abnormal ovaries of the Tet1Gt/Gt mice. Top panel, representative images of ovaries from 8-week-old wild-type (Wt) and Tet1 mutant (Gt/Gt) mice. Both left and right ovaries from one representative female are shown. Bottom panel, representative images of Hematoxylin and Eosin staining of adult ovary sections. Arrows indicate fully-grown oocytes.
(b) Number of fully-grown oocytes in adult ovaries. n=4 or 5. Error bars indicate SEM. **P<0.01.
(c) The average number of ovulated oocytes per female after hormonal stimulation. n=5–7. Error bars indicate SEM. *P<0.05.
(d) The relative oocyte numbers normalized to that in the wild-type (Wt) mouse. The number of oocytes in Wt mice was counted and set as 1. n=2–7. Error bars indicate SEM. *P<0.05, **P<0.01.
Since meiotic prophase takes place in the embryonic ovary and that meiotic defects can cause germ cell apoptosis12, we asked whether loss function of Tet1 might lead to a meiotic defect. Immunostaining of oocyte surface spreads with a meiotic marker SYCP3 and a centromere-marker CREST revealed that progression of meiotic prophase is severely impaired in Tet1Gt/Gt animals. About 50% of oocytes remained at the leptotene stage and no oocyte reached the pachytene stage in E16.5 Tet1Gt/Gt ovaries (Fig. 2a). Although pachytene stage oocytes were observed in E17.5 and E18.5 Tet1Gt/Gt ovaries, their percentage is significantly reduced throughout the developmental stages analyzed, indicating a developmental block rather than a developmental delay. This notion is also supported by the fact that Tet1 depletion does not affect the percentage of diplotene stage oocytes at E18.5 (Fig. 2a), and no significant difference in the body size between Tet1+/Gt and Tet1Gt/Gt embryos at E16.5 and E18.5 (Supplementary Fig. 6). The significant decrease of pachytene stage oocytes in Tet1Gt/Gt ovaries suggests that Tet1 deficiency may cause aberrant synapsis formation. Considering the increased apoptosis in E16.5–E18.5 Tet1Gt/Gt ovary (Supplemental Fig. 5b, c), severely affected germ cells might be eliminated, explaining the dramatic reduction in germ cell numbers in E18.5 Tet1Gt/Gt ovaries (Fig. 1d). Thus, meiotic defects are likely the cause of germ cell reduction in Tet1Gt/Gt ovaries.
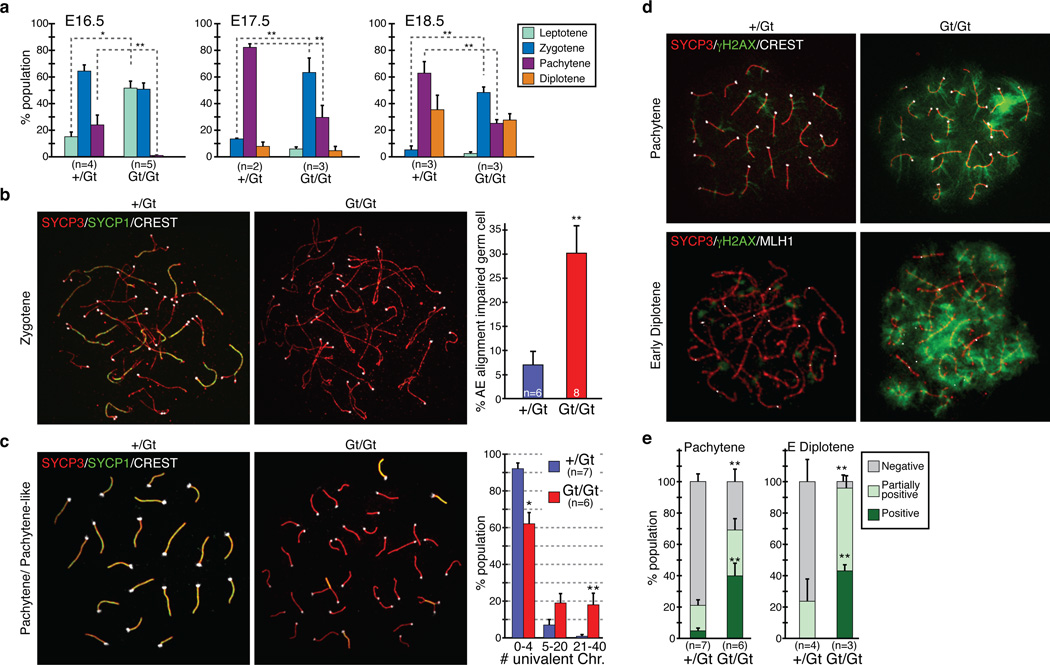
Figure 2. Meiotic defects in Tet1Gt/Gt oocytes.
(a) Distribution of E16.5, E17.5 and E18.5 oocytes in the four substages of meiotic prophase. Error bars indicate SEM. n=2–5. *P<0.05, **P<0.01, compared between +/Gt and Gt/Gt.
(b) Left, representative images of zygotene oocytes co-stained with SYCP3, SYCP1, and CREST antibodies. Right, percentage of the AE-alignment impaired oocytes at the zygotene stage. Oocytes that contain less than five SYCP1-foci are counted as AE-alignment impaired. n=6–8. Error bars indicate SEM. **P<0.01.
(c) Left, representative images of pachytene stage oocytes co-stained with SYCP3, SYCP1, and CREST antibodies. Right, distribution of oocyte types categorized by the numbers of univalent chromosomes in each pachytene stage oocyte. n=6–7. Error bars indicate SEM. *P<0.05. **P<0.01.
(d) Representative images of oocytes co-stained with antibodies against γH2AX, SYCP3, and CREST (pachytene stage), or γH2AX, SYCP3, and MLH1 (early diplotene stage).
(e) Distribution of oocyte types categorized by the staining pattern of γH2AX in each pachytene and early diplotene stage oocyte. Representative images for each group are shown in supplementary figure 9. n=3–7. Error bars indicate SEM. **P<0.01, compared to control.
In normal meiosis, the axial element (AE) of synaptonemal complex (SC) starts to form at the leptotene stage, AE-alignment and synapsis formation are initiated at the zygotene stage and completed at the pachytene stage (Supplementary Fig. 7). However, loss function of Tet1 resulted in an increase in unpaired SC at zygotene stage oocytes (Fig. 2b, Supplementary Fig. 7). Despite the presence of Sycp3-positive AE, about 30% of zygonema contained less than 4 SYCP1-positive transverse filaments (TF) in Tet1Gt/Gt oocytes (Fig. 2b) suggesting a synapsis formation defect. Defects in synapsis formation were also observed in the pachytene stage Tet1Gt/Gt oocytes. In these oocytes, the great majority of their AE failed to align and remain separated from each other as univalent chromosomes (Fig. 2c, Supplementary Fig. 7). Continuous SYCP3 in short-stretches of SC indicates that those oocytes were at a stage corresponding to pachytene. Quantification of pachytene stage oocytes revealed that 92% of them have 0–4 univalent chromosomes in wild-type oocytes, but that number dropped to 62% in the Tet1Gt/Gt oocytes, which accompanies the increase of the oocytes with 5 or more univalent chromosomes (Fig. 2c). These results indicate that loss function of Tet1 impaired synapsis formation.
Since about 62% of pachytene stage oocytes are successful for AE pairing, we asked whether they exhibited other defects. At the early phase of meiosis, DNA double strand breaks (DSBs) are introduced in the initiation of homologous recombination, which can be detected by the presence of γH2AX1. As expected, immunostaining revealed the presence of γH2AX throughout the nucleoplasm from leptotene to zygotene stages in both wild-type and Tet1Gt/Gt oocytes (Supplementary Fig. 8). However, while γH2AX is gradually decreased around pachytene stage and only few foci remained associated with fully synapsed chromosome cores in wild-type oocytes, cloud-like nuclear staining of γH2AX remained in the Tet1Gt/Gt pachytene and even early diplotene stage oocytes marked by crossover-specific marker MLH113 (Fig. 2d, Supplementary Fig. 8). Analysis of γH2AX staining pattern and quantification of the three categories (negative, partially positive, and positive) (Supplementary Fig. 9) clearly demonstrated that Tet1 depletion caused accumulation of γH2AX in pachytene and early diplotene oocytes (Fig. 2e).
The presence of γH2AX in late stage meiotic oocytes was also confirmed by co-staining of γH2AX with late stage meiosis marker Msy2 at E18.5 ovaries (Supplementary Fig. 10). Importantly, significant increase in γH2AX and cleaved Caspase3 double positive-cells were also observed (Supplementary Fig. 10b, c), suggesting that increase in apoptotic cell death is likely caused by meiotic defects. Consistent with a DSB repair defect, the DSB repair associated recombinase RAD511 remains in pachytene and diplotene oocytes (Supplementary Fig. 11). The presence of γH2AX and delayed removal of RAD51 in the chromosomes indicate that homologous recombination is impaired in Tet1Gt/Gt oocytes. Staining with the crossover mark MLH1 indicated that the MLH1 foci numbers are significantly reduced in Tet1Gt/Gt pachytene and early diplotene oocytes (Supplementary Fig. 12), further support a homologous recombination defect. Collectively, the above results support the notion that loss function of Tet1 leads to meiotic defects that include univalent chromosome formation, as well as DSB repair and homologous recombination defects.
Previous studies have shown that establishment of proper pericentric heterochromatin (PCH) structure plays an important role in meiosis14. Interestingly, immunostaining revealed specific enrichment of 5hmC at PCH of many prophase meiotic chromosomes and this enrichment is eliminated in Tet1Gt/Gt oocytes (Supplementary Fig. 13). Since prophase meiotic PCH possesses specific histone modification pattern15, we asked whether loss function of Tet1 can affect PCH structure by affecting PCH histone modification pattern. Immunostaining revealed that loss function of Tet1 neither affected PCH histone modification pattern (Supplementary Fig. 14), nor centromere clustering or HP1γ localization (Supplementary Figs. 14, 15). Collectively, these results suggest that the observed meiotic defect is unlikely due to a defect in PCH.
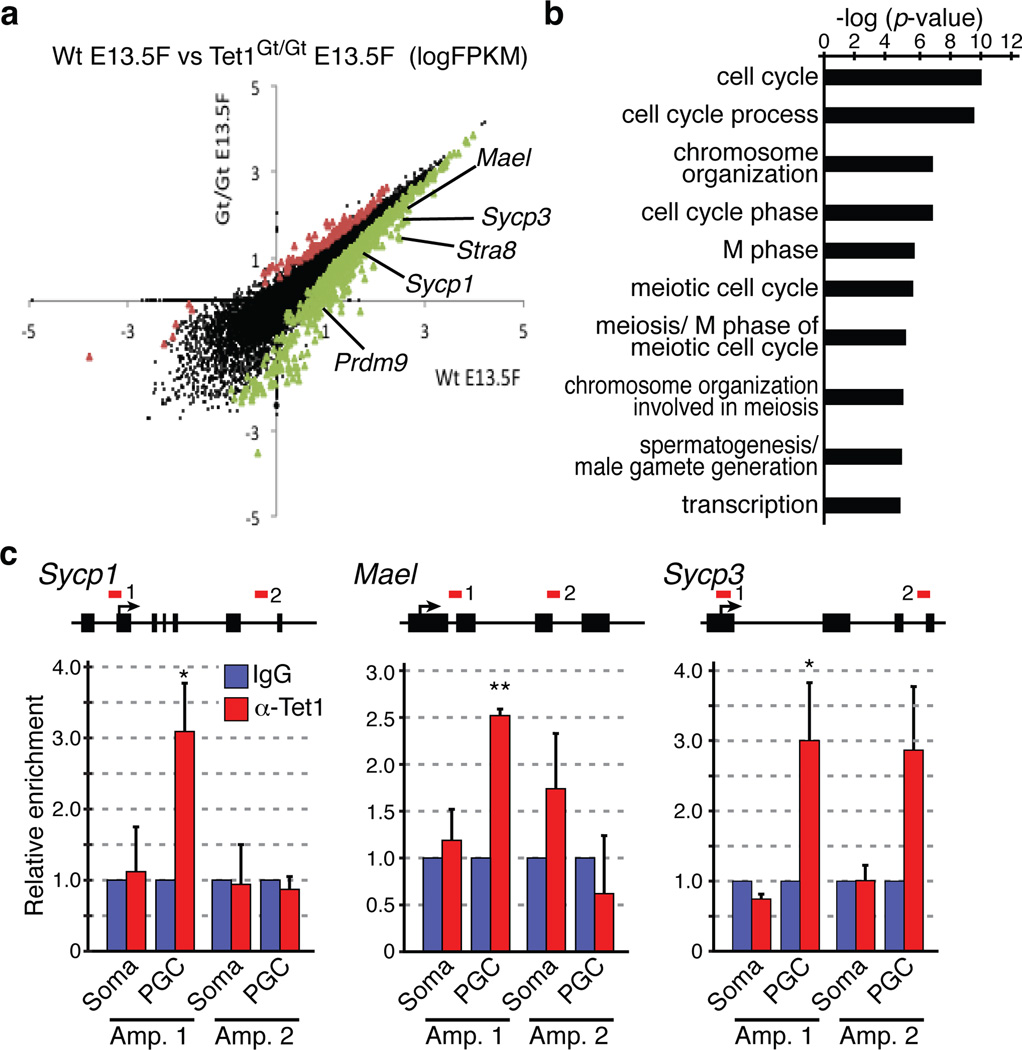
The stage- and cell type-specific Tet1 expression pattern (Supplementary Fig. 1) and the dual roles of Tet1 in transcription16,17 suggest that the meiotic defects in Tet1Gt/Gt germ cells might be due to aberrant transcription in PGCs. Thus, we purified PGCs from female E13.5 embryos and profiled their transcriptome by mRNA sequencing. We choose to focus on this time point because this is the time when female PGCs enter meiotic prophase after epigenetic reprogramming. We used a recently developed Smart-Seq method18 and generated over 20 million unique reads per sample which allowed us to identify over 13 thousands expressed transcripts in each genotype (Supplementary table 2). Hierarchical clustering and global correlation analysis indicated that the samples were clearly separated by their genotypes with Spearman correlation coefficient of 0.98/0.99 within biological replicates (Supplementary Fig. S16). Depletion of Tet1 resulted in differential expression of 1,010 genes (FDR<0.05), among which over 80% (899 genes) were down regulated (Fig. 3a, Supplementary table 3). Gene ontology (GO) analysis revealed that the most significantly enriched pathways of these down-regulated genes are related to cell cycle (p-value <9e–11) and meiosis-related processes (p-value <2e–6) (Fig. 3b, Supplemental Fig. S17a). In contrast, no significant enrichment of pathways or biological processes was identified in the up-regulated gene group. Importantly, genes known to be critical for meiosis are down-regulated in Tet1Gt/Gt PGCs (Fig. 3a, Supplementary table 3). These genes include Stra8, Prdm9, Sycp1, Mael and Sycp31,19–22. Notably, this set of meiotic genes remained down regulated even at later developmental stage (Supplemental Fig. S17b, c), consistent with the meiotic defects observed in E16.5 Tet1Gt/Gt PGCs. The effect of Tet1 on the expression of at least a subset of these genes are direct as ChIP analysis demonstrated that Tet1 occupies Sycp1, Mael, and Sycp3 gene promoters (Fig. 3c). Thus, Tet1 loss directly contributes to aberrant regulation of at least a subset of meiotic genes in PGCs.
Figure 3. Tet1 activates meiotic genes through DNA demethylation.
(a) Scatter plot comparing transcriptome of wild-type and Tet1Gt/Gt E13.5 female PGCs. There are 111 and 899 genes that are respectively up- or down-regulated (FDR<0.05). Examples of down-regulated meiotic genes include Mael, Sycp3, Stra8, Sycp1, and Prdm9 indicated.
(b) Gene ontology analysis of down-regulated genes in Tet1Gt/Gt PGCs with cutoff FDR<0.05. The most enriched biological processes based on their p-values are shown.
(c) ChIP-qPCR analysis of E13.5 wild-type female PGCs using anti-Tet1 antibody demonstrates binding of Tet1 to the promoters of Sycp1, Mael, and Sycp3. Upper panels, diagrams of Sycp1, Mael, and Sycp3 genes with the analyzed regions indicated by red lines. Bottom panels, relative enrichment of Tet1 over IgG control. n=3. Error bars indicate SEM. *P<0.05. **P<0.01.
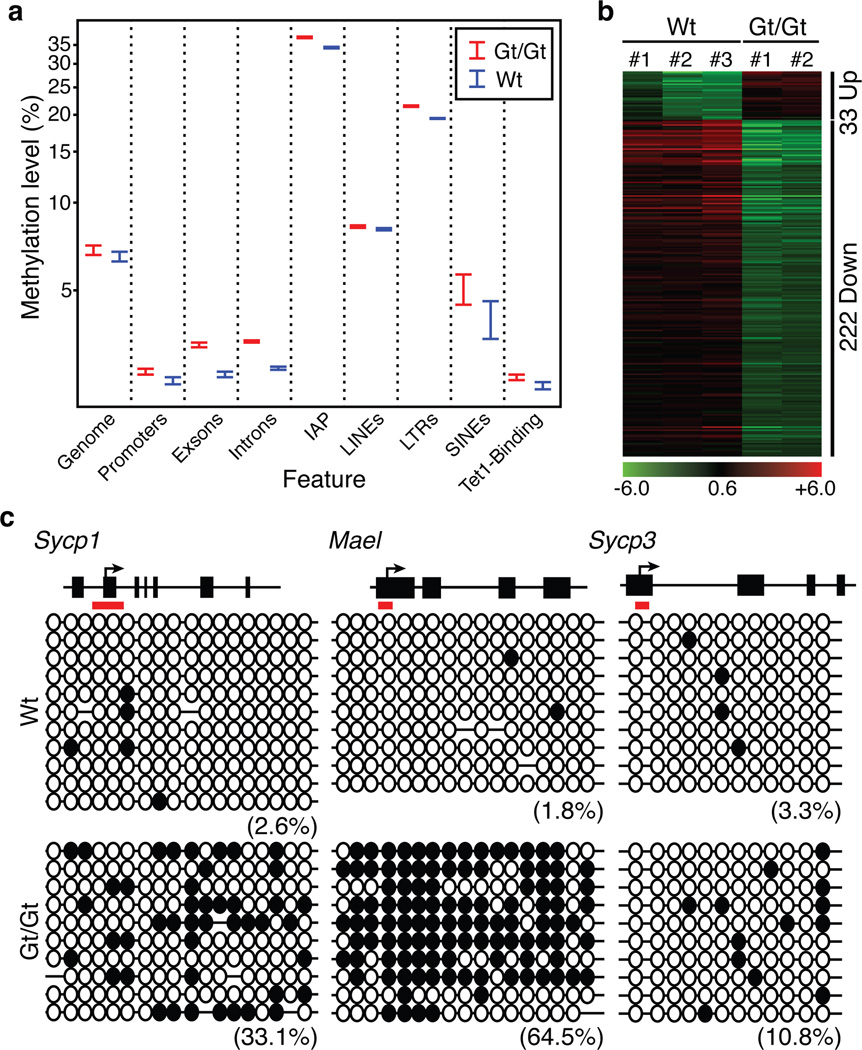
To investigate how Tet1 might be involved in activation of these meiotic genes, we performed whole-genome bisulfite sequencing (WGBS) using an ultra-low input, Tn5mC-seq method23. We generated 945 million and 302 million reads for Tet1Gt/Gt and wild-type PGCs, respectively. After removing clonal reads due to limited input cells, we obtained 14–16 million CpG sites per genotype at 1.76–2.66x genome coverage (Supplementary table 4), which is over100-fold higher than a previous effort24. To our acknowledge, this provides the most comprehensive methylation map in PGCs so far. Consistent with previous findings24, we found that PGCs are globally hypomethylated (Fig. 4a), which is verified by immunostaining (Supplemental Fig. S18). Despite no dramatic increase in global DNA methylation, DNA methylation level is generally higher in mutant PGCs, particularly in exons, introns, LTRs, and IAPs (Fig. 4a, p<0.01). With the caveat that 2X genome coverage may not allow robust identification of differentially methylated regions (DMRs) between Tet1Gt/Gt and wild-type PGCs, we nevertheless performed detailed analysis and identified 4,337 putative DMRs (Supplementary table 5, Supplemental Fig. S19) that are mostly located far away from transcriptional start sites (Supplemental Fig. S20a). These putative DMRs are associated with 5,242 genes, among which 255 exhibited altered expression (Fig. 4b, Supplementary table 6) and are enriched for cell cycle regulation as well as for reproductive and infertility processes (FDR=0.02) (Supplementary Fig. S20b, c). Furthermore, these DMRs are enriched for Tet1 binding in mouse ES cells17 (P-value <10E–100). To evaluate whether Tet1 binding affects DNA methylation in PGCs, we performed bisulfite sequencing on the three verified Tet1 target genes (Fig. 3c). Consistent with the involvement of Tet1 in DNA demethylation, the methylation level of the Sycp1, Mael, and Sycp3 promoters is increased in the Tet1Gt/Gt PGCs compared with that in the wild-type PGCs (Fig. 4c). These data indicate that Tet1-mediated demethylation of these genes is likely involved in their activation during PGC development. We note, some down-regulated genes, such as Stra8, showed no obvious change in DNA methylation indicating that they are either regulated indirectly by Tet1 or in a DNA methylation-independent manner.
Figure 4. Whole genome bisulfite analysis of the effect of Tet1 knockout on DNA methylation in PGCs.
(a) Wt E13.5 female PGCs are globally hypomethylated and depletion of Tet1 only slightly increased the global DNA methylation level. Shown are the DNA methylation levels in the entire mouse genome as well as the various genomic regions. In the last column, the DNA methylation levels at the Tet1 bound regions identified in mES cells17 were compared.
(b) Heat map of the 255 differentially expressed and DMR associated genes.
(c) Bisulfite sequencing analysis of the Sycp1, Mael, and Sycp3 gene promoters of Tet1 binding site in wild-type and Tet1Gt/Gt PGCs. Each CpG is represented by a circle. Open and filled circles represent unmethylated or methylated, respectively. Percentages of DNA methylation are indicated.
Taken together, our study provides the first evidence that Tet1 is not responsible for global demethylation in PGCs, rather it plays a specific role in meiotic gene activation at least partly through DNA demethylation. Depletion of Tet1 leads to down-regulation of meiotic genes, which causes defective meiotic prophase including accumulation of non-repaired DSBs, and formation of univalent chromosomes. The meiotic defects cause loss of oocytes and consequent decrease in fertility and small litter size. Previous studies have established that DNA methylation levels of certain meiotic genes are decreased concomitant with genomic reprogramming25. Our study has extended this observation by demonstrating that Tet1 mediates locus-specific demethylation and subsequent activation of a subset of meiotic genes, revealing a specific function of Tet1 in germ cell development.
Supplementary Material
Acknowledgements
We thank Jinzhao Wang and Wei Jiang for FACS sorting; This work was partially supported by U01DK089565 (to Y.Z.). Y.Z. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Author Contributions
Y.Z. conceived the project; S.Y., K.H., and Y.Z. designed the experiments; S.Y., K.H., R.L., L.S., and A.I. performed the experiments; S.Y., K.H., R.L., D.D., K.Z., and Y.Z. analyzed and interpreted the data; S.Y., K.H., R.L, K.Z., and Y.Z. wrote the manuscript.
RNA-seq and WGBS data have been deposited in the Gene Expression Omnibus under accession number abc, and xyz.
The authors declare of no competing interest.
References
- 1.Handel MA, Schimenti JC. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nature Reviews Genetics. 2010;11:124–136. doi: 10.1038/nrg2723. [DOI] [PubMed] [Google Scholar]
- 2.Hayashi K, Surani MA. Resetting the epigenome beyond pluripotency in the germline. Cell Stem Cell. 2009;4:493–498. doi: 10.1016/j.stem.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 3.Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet. 2008;2008:129–140. doi: 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- 4.Hajkova P, et al. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- 5.Seki Y, et al. Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev Biol. 2005;278:440–458. doi: 10.1016/j.ydbio.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 6.Kota SK, Feil R. Epigenetic Transitions in Germ Cell Development and Meiosis. Developmental cell. 2010;19:675–686. doi: 10.1016/j.devcel.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 7.He YF, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito S, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ito S, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dawlaty MM, et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011;9:166–175. doi: 10.1016/j.stem.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roeder GS, Bailis JM. The pachytene checkpoint. Trends Genet. 2000;16:395–403. doi: 10.1016/s0168-9525(00)02080-1. [DOI] [PubMed] [Google Scholar]
- 13.Edelmann W, et al. Meiotic pachytene arrest in MLH1-deficient mice. Cell. 1996;85:1125–1134. doi: 10.1016/s0092-8674(00)81312-4. [DOI] [PubMed] [Google Scholar]
- 14.Takada Y, et al. HP1{gamma} links histone methylation marks to meiotic synapsis in mice. Development. 2011;138:4207–4217. doi: 10.1242/dev.064444. [DOI] [PubMed] [Google Scholar]
- 15.Tachibana M, Nozaki M, Takeda N, Shinkai Y. Functional dynamics of H3K9 methylation during meiotic prophase progression. The EMBO Journal. 2007;26:3346–3359. doi: 10.1038/sj.emboj.7601767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams K, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu H, et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473:389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramskold D, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012 doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Vries FAT, et al. Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes & Development. 2005;19:1376–1389. doi: 10.1101/gad.329705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayashi K, Yoshida K, Matsui Y. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature. 2005;438:374–378. doi: 10.1038/nature04112. [DOI] [PubMed] [Google Scholar]
- 21.Soper SF, et al. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Developmental Cell. 2008;15:285–297. doi: 10.1016/j.devcel.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H, Höög C. Structural damage to meiotic chromosomes impairs DNA recombination and checkpoint control in mammalian oocytes. The Journal of Cell Biology. 2006;173:485–495. doi: 10.1083/jcb.200512077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adey A, Shendure J. Ultra-low-input, tagmentation-based whole-genome bisulfite sequencing. Genome Res. 2012;22:1139–1143. doi: 10.1101/gr.136242.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Popp C, et al. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maatouk DM, et al. DNA methylation is a primary mechanism for silencing postmigratory primordial germ cell genes in both germ cell and somatic cell lineages. Development. 2006;133:3411–3418. doi: 10.1242/dev.02500. [DOI] [PubMed] [Google Scholar]
- 26.Dahl JA, Collas P. A rapid micro chromatin immunoprecipitation assay (microChIP) Nature Protocols. 2008;3:1032–1045. doi: 10.1038/nprot.2008.68. [DOI] [PubMed] [Google Scholar]
References for Methods
- 27.Markoulaki S, Meissner A, Jaenisch R. Somatic cell nuclear transfer and derivation of embryonic stem cells in the mouse. Methods (San Diego, Calif.) 2008;45:101–114. doi: 10.1016/j.ymeth.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 28.Soper SFC, et al. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Developmental Cell. 2008;15:285–297. doi: 10.1016/j.devcel.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kouznetsova A, Novak I, Jessberger R, Hoog C. SYCP2 and SYCP3 are required for cohesin core integrity at diplotene but not for centromere cohesion at the first meiotic division. J Cell Sci. 2005;118:2271–2278. doi: 10.1242/jcs.02362. [DOI] [PubMed] [Google Scholar]
- 30.Diep D, et al. Library-free methylation sequencing with bisulfite padlock probes. Nat Methods. 2012;9:270–272. doi: 10.1038/nmeth.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.