Abstract
Multicopper oxidase (MCO) is an enzyme which involves in reducing the oxygen in a four electron reduction to water with concomitant one electron oxidation of reducing the substrate. We have generated the 3-D structure of MCO by homology modeling and validated on the basis of free energy while 90.4 % amino acid residues present in allowed regions of Ramachandran plot. The screening of potential hazardous aromatic compounds for MCO was performed using molecular docking. We obtained Sulfonaphthal, Thymolphthalein, Bromocresol green and Phloretin derivatives of phenol and aromatic hydrocarbon were efficient substrates for MCO. The phylogeny of MCO reveals that other bacteria restrain the homologous gene of MCO may play an important role in biodegradation of aromatic compounds. We have demonstrated the gene regulatory network of MCO with other cellular proteins which play a key role in gene regulation. These findings provide a new insight for oxidization of phenolic and aromatic compounds using biodegradation process for controlling environmental pollution.
Electronic supplementary material
The online version of this article (doi:10.1007/s11693-012-9096-9) contains supplementary material, which is available to authorized users.
Keywords: Multicopper oxidase, Homology modeling, Docking, Biodegradation, Gene regulatory network
Introduction
Multicopper oxidase (MCO) is a diverse family of metalloenzyme which widely distributed in prokaryotes and eukaryotes (Solomon et al. 1996). There are well known MCO comprise ascorbate oxidase, cytochrome C oxidase and ceruloplasmin. MCO family has grown rapidly through the addition of new enzymes include phenoxazinone synthase, bilirubin oxidase and dihydrogeodin oxidase (Freeman et al. 1993; Huang et al. 1995; Shimizu et al. 1999). MCO from fungal origin is an attractive industrial enzymes with significant applications in detoxification of toxic phenolic compounds and azo dyes (Bourbonnais et al. 1995), enzymatic bleaching of kraft pulp (Husain and Jan 2000) and delignification which help in oxidation of phenols (Youn et al. 1995).
There are several bacteria such as Pseudomonas, Proteus, Xanthomonas, Alteromonas, Aeromonas and Enterobacteriaceae highly resistant to mercury. It has been isolated from seawater and sediment samples and tested for growth in the presence of different heavy metals, pesticides, phenol, formaldehyde, formic acid, and trichloroethane for investigation of their growth (De et al. 2003). Biodegradation of phenol, 4-chlorophenol, 2,4-chlorophenol and 2,4,6 trichlorophenol using Aeromonas sp, Pseudomonas sp, Flavimonas oryzihabitans and Chryseobacterium luteola have been earlier reported (Buitron et al. 1998). Laccases are MCO that catalyzes by the oxidation of a wide range of phenols or arylamines, and their use in industrial oxidative processes (Bertrand et al. 2002). The molecular basis and mechanism of biodegradation of aromatic compounds especially phenolic compounds by Aeromonas hydrophila is not clear yet. In the present study, we have performed in silico experiment on MCO of A. hydrophila which contains the ability to bind phenolic and other aromatic compounds. It is potential enzyme which recognized for biodegradation of phenolic and aromatic compounds. However, we will require better understanding at molecular and atomic level interaction of MCO with phenolic compounds.
The 3-dimensional (3-D) structure of MCO of A. hydrophila is unknown so far. Recently there is an advancement of computational biology and bioinformatics tools for generation of 3-D structure of protein using primary amino acids sequences. Thus, homology modeling has been employed for generation of 3-D structure of unknown protein using known 3-D crystal structure. The MCO (Fet3) of yeast oxidizes the extracellular ferrous iron and later transported into the cells through the permease Ftr1. The 3-D structure of Fet3 has been derived by homology modeling (di Patti et al. 1999). Laccase is one of the well studied enzymes used for bioremediation of xenobiotics that includes phenols and anilines compounds. Protein–ligand docking has been performed using GOLD and 17 % of selected datasets showed the highest average GOLD fitness score for fungal and bacterial laccase enzyme (Suresh et al. 2008). Capability of ligninolytic fungus Trametes trogii for degradation of different xenobiotics compounds such as hexaethylbenzene, naphthalene, 1-methyl naphthalene, acenaphthylene, anthracene, fluorene and phenanthrene have been reported in vitro assay (Haglund et al. 2002).
There is recent knowledge of gene encoding protein and enzyme which interact with each other in the gene network for gene regulation. The gene networks are a complex interaction of biomolecules that includes DNA, RNA and proteins; and all these elements contribute in biological process. Systems biology uses for deeper understanding of biological system level phenomena that needs to explore the relationship between network structure and the dynamics of genes, proteins and other biomolecules (Samal and Jain 2008). The gene network analysis of gyrase subunit B encoding gene with other cellular proteins of A. hydrophila has been investigated for discovery of drug target and gene regulation (Singh et al. 2012). There are number of enzymes interacted to each other in gene networks that helps in drug discovery and development. It can further help to repress and activate the gene regulatory networks for control of cellular activity. There are proteins/enzymes serve as to activate or repress other genes which are known as transcription factors that play vital role in regulatory network. It binds to operator sites of promoter for regulation of gene expression by switching off and on system. The aim of present study was to generate 3-D structure of MCO of A. hydrophila and use for screening of phenolic and aromatic compounds. The phylogeny was constructed based on MCO of A. hydrophila protein sequences with other bacteria comprises the homology. We constructed a novel gene regulatory network of MCO of A. hydrophila.
Materials and methods
Alignment of sequences, generation and validation of 3-D structure
The complete protein sequence of MCO of A. hydrophila was retrieved and used to search the relatedness of sequences deposited in NCBI-GenBank through BLAST (http://www.ncbi.nim.nih.gov/blast) (Altschul et al. 1997). We used BLASTP for searching of the structural similarity of sequences with protein data bank (PDB). Maximum score and E value were considered for selection of crystal structure as a template for homology modeling. The alignment was done for target protein sequences (MCO) with PDB: 1KV7 template using CLUSTAL X. Multicopper oxidase of A. hydrophila showed the structural similarity with X-ray crystal structure (PDB: 1KV7) at 1.40 Å resolution of MCO of E. coli which was used as template structure to generate 3-D structure. The crystal 3-D structure of template was taken from Protein Data Bank (http://www.rcsb.org/pdb/). The 3-D structure of MCO was generated through Modeller9v6 (Sali and Blundell 1993) and visualized using PYMOL (Delano 2002). Evaluation of generated 3-D structure of MCO was compared on the basis of Gibbs free energy of MCO with crystal structure as template. The 3-D structure of MCO was validated through PROCHECK (Laskowski et al. 1993). It generates the Ramachandran plot (RP) as we considered the amino acid residues in allowed, disallowed region and overall G-factor.
Screening of aromatic and phenolic compounds by molecular docking
Aromatic and phenolic compounds were retrieved from NCBI/PubChem compound as in SDF format and converted into 3-D structure using Open Babel. The 3-D structure of MCO and aromatic compounds were used for molecular docking experiment by AutoDock 3.0.5 (Morris et al. 1998). The docking parameters were as follows: 100 docking trials, population size 150, random starting position and conformational translation step ranges of 1.5 Ǻ, rotation step ranges 35, elitism of 1, mutation rate 0.02, cross over rate of 0.8, local search rate of 0.06 and 25 million energy evaluations. Distance dependent function of dielectric constant was used for calculation of energetic maps and all other parameters were used by default value. The selection of suitable compounds was based on minimum docked energy which indicates the highest binding affinity. Python molecule viewer was used for demonstration of amino acid in active pocket of 3-D structure of MCO with interaction of atoms level aromatic compounds includes the hydrogen bonds.
Construction and analysis of phylogenetic tree
The protein sequence of MCO of A. hydrophila was used to search the homology using BLAST and other homologous sequences were retrieved from NCBI-GenBank. All these MCO protein sequences from various bacteria were aligned using CLUSTALX (Thompson et al. 1997). It was manually checked and verified subsequently using passion correction algorithm which is implemented in MEGA4.0 for construction of phylogenetic tree by neighbor-joining method. Total 100 bootstraps values were sampled to determine the measure of support for each node on consensus tree (Tamura et al. 2007).
Protein–protein interactions
We used STRING database for generation of gene regulatory network of MCO of A. hydrophila based on known and predicted protein interactions (http://string-db.org/). The protein–protein interactions include direct (physical) and indirect (functional) associations; these are derived from four sources: (1) genomic context, (2) High-throughput experiments, (3) conserved coexpression and (4) literature knowledge. In the STRING quantitatively integrates interaction data from these sources for a large number of organisms, and transfers information between these organisms where applicable. The database currently covers 5,214,234 proteins from 1,133 organisms during protein–protein interactions in gene regulatory network.
Protein structure identifier number
The homology model of the 3-D structure of MCO of A. hydrophila subsp. hydrophila ATCC 7966 was submitted to PMDB (http://mi.caspur.it/PMDB/) which was assigned under PM0075667. It can easily accessible and use for further experiment.
Result
Structural analysis of multicopper oxidase
We identified the conserved domains of leucine zipper in MCO. Leucine zipper is an important region which contains the leucine (L) enriched amino acid that occupied every seventh position in the zipper. The identified sequence for leucine zipper LLRIRPSLIEGKGRLPDSLARL in MCO protein of A. hydrophila was located between the positions 315 and 336. MCO of A. hydrophila showed the 60 % structural similarity with MCO of E. coli (PDB: 1KV7). The complete protein sequence of MCO of A. hydrophila was aligned with 1KV7 (Supplementary Fig. S1). The asterisk showed the identical amino acids present in both protein sequences. Total five 3-D structures were generated by Modeller9v6 and free energy of 3-D structures of MCO; and template was also evaluated. The free energy of MCO was −54,848.58 kcalmol−1 which similar with crystal structure. The 3-D structure of MCO of A. hydrophila was shown (Fig. 1) which represents the restrains α-helix and β-sheets. The 3-D of MCO was validated by generating the Ramachandran plot (Supplementary Fig. S2). The favored amino acid residues in RP of 3-D structure were 90.4 %, disallowed region 0.7 % and the G-factor was 0.9.
Fig. 1.
The 3-D structure of multicopper oxidase from A. hydrophila showing the α helix and β sheets
Screening and analysis of aromatic and phenolic compounds
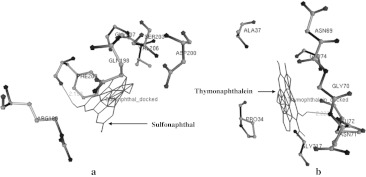
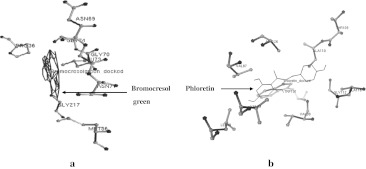
The 3-D structure of MCO of A. hydrophila was used for screening of aromatic and phenolic compounds based on binding affinity. The well-known aromatic and phenolic compounds are hazardous to environment which was targeted by MCO given in Supplementary Table S1. Total 13 well known compounds were selected for possible biodegradation by MCO enzyme and four were showed the highest binding affinity on the basis of molecular docking experiments such as Sulfonaphthal, Thymolphthalein, Bromocresol green and Phloretin. The interaction of compounds with MCO were considered on the basis of docking energy, inter molecular energy, torsional energy, root mean square deviation and internal energy (Table 1). Total 10 docking were performed on whole structure of MCO. We considered the lowest free energy of docked complex with hydrogen bonds. The docking energy of Sulfonaphthal, Thymolphthalein, Bromocresol green and Phloretin were −10.1, −11.26, −13.05 and −9.02 kcal/mol, respectively. There are several amino acids residue in 3-D structure of MCO during the interaction with phenolic compounds using molecular docking strategies (Table 2). The active pocket of amino acid residues Ser203, Gly207, Val206, Asp200, Gln198, Phe209 and Arg188 of MCO structure obtained during interaction of Sulfonaphthal, compound. It was bound with these amino acids and single hydrogen bonds (HB) was formed between UNK1:H—Phe209:O atoms with 2.153 Ǻ distance (Fig. 2a). In the other docking experiment amino acids residues Asn69, Ala37, Gly74, Gly70, Pro34, Asn71, Gly217 and Leu72 in MCO obtained during interaction of Thymolphthalein. Thymolphthalein was bound with these amino acids and single hydrogen bonds (HB) was formed between UNK1:H and Asn71:N atoms with 2.222 Ǻ distance (Fig. 2b). Bromocresol green interacted with amino acids residues include Asn69, Gly74, Gly70, Pro36, Leu73, Asn71, Gly217 and Met56 during docking experiment (Fig. 3a). There was no hydrogen bond formed but it showed the good binding affinity (−13.05 kcal/mol). In the Phloretin compound also showed the interaction with amino acids including Thr109, Ala110, Ala116, Gly112, Ala126, Val87, Trp101, Val99, Val89 and Leu49 (Fig. 3b) while no hydrogen bond was observed.
Table 1.
The interaction energy of MCO with aromatic and phenolic compounds obtained through molecular docking
| S. No. | Aromatic and phenolic compounds | Binding energy (kcalmol−1) | Docked energy (kcalmol−1) | Inter molecular energy (kcalmol−1) | Torsional energy (kcalmol−1) | Internal energy (kcalmol−1) | RMSD (Ǻ) |
|---|---|---|---|---|---|---|---|
| 1. | Sulfonaphthal | −9.57 | −10.1 | −10.5 | 0.93 | 0.41 | 34.57 |
| 2. | Bisacodyl | −8.31 | 5.95 | −10.49 | 2.18 | 16.44 | 18.62 |
| 3. | Thymolphthalein | −11.63 | −11.26 | −12.86 | 1.25 | 1.61 | 37.05 |
| 4. | 2,4 Dinitrophenol | 6.86 | 9.13 | 6.24 | 0.62 | 2.9 | 22.9 |
| 5. | Bromophenol blue | −7.87 | −10.98 | −10.98 | 3.11 | 0.0 | 14.78 |
| 6. | Bromocresol green | −6.31 | −13.05 | −12.85 | 6.54 | 0.2 | 36.98 |
| 7. | Phenolphthalein | −9.19 | −9.5 | −9.81 | 0.62 | 0.31 | 34.55 |
| 8. | Pentachlorophenol | −6.23 | −7.68 | −7.79 | 1.56 | 0.11 | 43.46 |
| 9. | Benzo[a]anthracene | −10.29 | −10.29 | −10.29 | 0.00 | 0.00 | 43.1 |
| 10. | Phenol | −5.34 | −5.22 | −5.34 | 0.00 | 0.12 | 43.23 |
| 11. | Hexachlorophene | −8.23 | −9.6 | −9.79 | 1.56 | 0.19 | 45.12 |
| 12. | Salicylic acid | 0.08 | −0.02 | −0.23 | 0.31 | 0.21 | 20.57 |
| 13. | Phloretin | −9.43 | −9.02 | −10.67 | 1.25 | 1.66 | 42.66 |
Table 2.
The amino acid residues and hydrogen bond formed between aromatic and phenolic compounds with multicopper oxidase
| S. No. | Protein name | Amino acids in multi-copper oxidase | Aromatic and phenolic compounds | Interaction of amino acids of MCO with compounds | Distance of hydrogen bonds (Ǻ) |
|---|---|---|---|---|---|
| 1. | MCO | Ser203, Gly207, Val206, Asp200, Gln198, Phe209, Arg188 | Sulfonaphthal | UNK1:H—Phe209:O | 2.153 |
| 2. | MCO | Asn69, Ala37, Gly74, Gly70, Pro34, Asn71, Gly217, Leu72 | Thymolphthalein | UNK1:H—Asn71:N | 2.222 |
| 3. | MCO | Asn69, Gly74, Gly70, Pro36, Leu73, Asn71, Gly217, Met56 | Bromocresol green | ND | ND |
| 4. | MCO | Thr109, Ala110, Ala116, Gly112, Ala126, Val87, Trp101, Val99, Val89, Leu49 | Phloretin | ND | ND |
ND hydrogen bond was not detected
Fig. 2.
Interaction of high affinity aromatic and phenolic compounds with MCO of A. hydrophila showing the hydrogen bond in green line. a Sulfonaphthal and b Thymolphthalein. (Color figure online)
Fig. 3.
Interaction of high affinity aromatic and phenolic compounds with MCO of A. hydrophila.a Bromocresol green and b Phloretin
Analysis of phylogeny and gene regulatory network
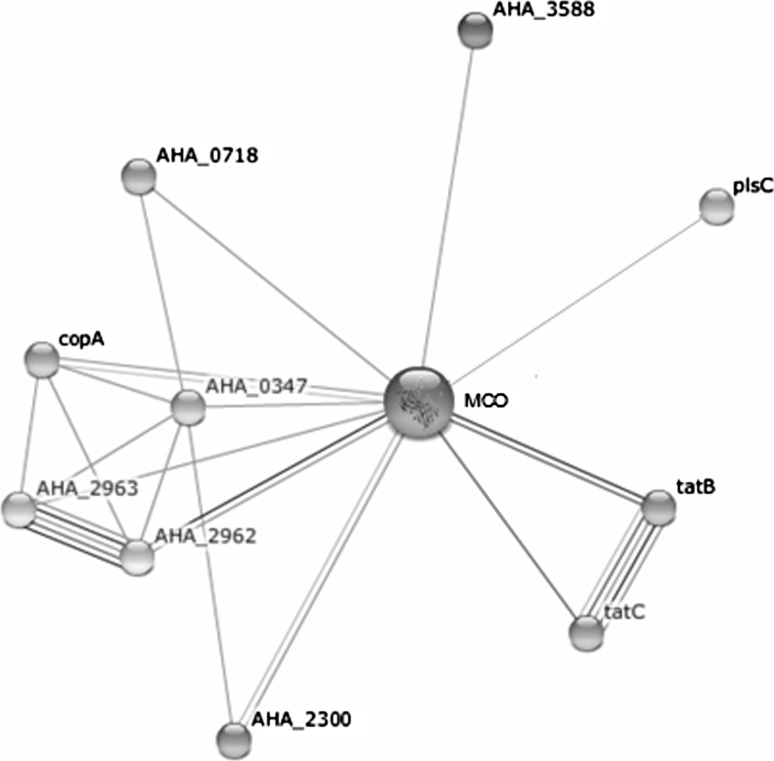
We have constructed the phylogenetic tree using MCO protein sequences of A. hydrophila and homologous proteins present in other bacteria. Total 3 major clades A, B and C were obtained in phylogeny of MCO protein sequences. The clade A shows the Salmonella species, clade B was Shigella and E. coli, while A. hydrophila present in the clade C with other bacteria (Supplementary Fig. S3). Bacteria of Clade C may play the similar role in biodegradation of aromatic and phenolic compounds because these are containing MCO enzyme. It indicates the similar compounds such as Sulfonaphthal, Thymolphthalein, Bromocresol green and Phloretin may also be degraded by functional activity of MCO. The cellular mechanism of MCO of A. hydrophila is not clear yet, thus; we have showed the interaction of MCO in cellular gene regulatory network and the interaction with other proteins/enzymes. In the MCO gene regulatory network, we found the closely related proteins such as AHA_0347, copA, AHA_2962 and tatB with >0.6 score (Table 3). We obtained the newly constructed gene regulatory network of A. hydrophila may be useful for analysis of gene regulation of MCO for detoxification. As shown in Fig. 4 the protein–protein interactions of MCO in A.hydrophila gene regulatory network.
Table 3.
Protein–protein interactions in gene regulatory network of multicopper oxidase of A. hydrophila
| Functional proteins | Score | Potential functions |
|---|---|---|
| AHA_0347 | 0.772 | Hypothetical protein (143 aa) |
| copA | 0.674 | Copper-exporting ATPase (806 aa) |
| AHA_2962 | 0.664 | Copper/silver resistance periplasmic protein (499 aa) |
| tatB | 0.601 | Twin arginine-targeting protein translocase TatB (147 aa) |
| AHA_2963 | 0.597 | Cation efflux system protein CusA (1,042 aa) |
| tatC | 0.566 | Twin arginine-targeting protein translocase TatC (251 aa) |
| plsC | 0.551 | 1-acyl-sn-glycerol-3-phosphate acyltransferase (242 aa) |
| AHA_3588 | 0.513 | Acyltransferase family protein (368 aa) |
| AHA_2300 | 0.473 | Copper-translocating P-type ATPase (799 aa) |
| AHA_0718 | 0.457 | Hypothetical protein (145 aa) |
Fig. 4.
Protein–protein interactions of MCO of A. hydrophila with other cellular proteins in gene regulatory network
Discussion
There is an increasing the environmental pollution by releasing the industrial and agricultural hazardous compounds. Thus, it is transmitted in our food chain. Biodegradation can be effectively used for controlling the environmental pollutions by action of microbial enzymes (Willmott et al. 1998). Most of the non degradable aromatic and phenolic compounds are highly toxic and risk for animals health (Alcalde et al. 2002). Phenol is an important organic aromatic compound which present in waste of chemical processing industries. It is a suspected carcinogen, corrosive nature, and toxic or lethal to life. Thus, we require the degradation of these compounds for controlling of environmental contamination. Several bacteria such as Aeromonas sp, Pseudomonas sp, Flavimonas oryzihabitans and Chryseobacterium luteola used for biodegradation of phenol, 4-chlorophenol, 2,4-chlorophenol and 2,4,6 trichlorophenol compounds (Buitron et al. 1998). There are well known MCO comprise of ascorbate oxidase, laccase, cytochrome C oxidase, ceruloplasmin, phenoxazinone synthase, bilirubin oxidase and dihydrogeodin oxidase (Freeman et al. 1993; Huang et al. 1995; Shimizu et al. 1999). In this study, we have identified the MCO of A. hydrophila from NCBI- GenBank genome sequence (Seshadri et al. 2006). The size of MCO of A. hydrophila was 549 amino acids almost resembled with previously reported other bacteria Klebsiella species 601 composed of 536 amino acids (Li et al. 2008). The function of MCO has been described in copper tolerance, manganese oxidation and iron oxidation in a range of bacteria. The putative cytoplasmic membrane MCO from Legionella pneumophila have been reported earlier (Huston et al. 2008). The phytoremediation of soils contaminated with organic pollutants offers a low-cost method for removal of pollutants. An extracellular fungal enzyme laccase of Coriolus versicolor into tobacco plants was introduced into rhizosphere which was able to remove bisphenol A or pentachlorophenol (Sonoki et al. 2005).
In this study, MCO of A. hydrophila showed 60 % structural identity with MCO of E. coli. The complete protein sequence of MCO of A. hydrophila was aligned with 1KV7. The asterisk showed the identical amino acids present in both protein sequences. MCO protein sequence of A. hydrophila was used to generate 3-D structure using known crystal 3-D structure (1KV7). There are recent reports available on homology modeling for generation of 3-D structures of adenosine A2a receptor of Homo sapiens (Singh and Somvanshi 2009a), 3-oxoacyl-acyl carrier protein synthase II of Mycobacterium tuberculosis H37Rv (Singh and Somvanshi 2009b) proteins of A. hydrophila such as aerolysin (Singh and Somvanshi 2009c), hemolysin (Singh et al. 2009) and 3-oxoacyl-ACP synthase (Singh et al. 2011a). It contains α-helix which involves regularly spaced H-bonds between residues along a chain while β-sheets involves H-bonding between the backbone residues in adjacent chains. In the MCO structure the β-sheet, a single chain forms H-bonds with its neighboring chains with donor (amide) and acceptor (carbonyl) atoms pointing sideways. The stability and folding of protein are maintained in cell by α-helix and β-sheets which plays the key role in functional activity of protein. In general, many proteins do not exhibit much α-helix and β-sheets structure in solution since the entropy associated with the folding of the polypeptide chain is not compensated for sufficient amount of stabilizing interactions. We have obtained MCO enrich with α-helix and β-sheets which provide the clue for proper functioning and can show better biodegradation. The 3-D structure of MCO of A. hydrophila was used for screening of aromatic and phenolic compounds based on binding affinity.
In our study, docking energy of Sulfonphthal, Thymolphthalein, Bromocresol green and Phloretin with MCO were −10.1, −11.26, −13.05 and −9.02 kcal/mol, respectively. In the previous study, more than 4 compounds have been showed similar results include Pentachlorophenol, Benzo[a]anthracene, Hexachlorophene and Bromophenol blue were −7.68, −10.29, −9.6 and −10.98 kcal/mol. Other studied compounds have been showed comparatively lower affinity with active amino acids pocket of 3-D structure of MCO. It oxidized mono and dimethoxyphenols at 50 °C (Hirose et al. 2003). Laccase has been reported by homogeneous enzyme from Coriolus hirsutus and Coriolus zonatus. The characterization of two compounds 1-phenyl-pyrazolone class, sodium 1-phenyl-3-methyl-4-methylamino-pyrazolone-5-N(4)-methanesulfonate and 1-(3′-sulfophenyl)-3-ethylpyrazolone-5, in the reaction catalyzed by laccase. The 1-(3′-Sulfophenyl)-3- methylpyrazolone-5 was degraded about 30–40 % by laccase (Shleev et al. 2003). Laccase is a polyphenol oxidase belongs to family of blue MCOs which catalyzes the one-electron oxidation of four reducing-substrate molecules concomitant with four-electron reduction of molecular oxygen to water. Laccases oxidized a broad range of substrates preferably phenolic compounds (Piontek et al. 2002).
A gene encoding a MCO has been cloned from soil bacterium Klebsiella sp. 601 and its corresponding enzyme was over-expressed in E. coli. The amino acid sequence of Klebsiella sp. 601 MCO is strongly homologous to that of E. coli CueO with a similarity of 90 % and an identity of 78 %. E. coli CueO, Klebsiella sp. 601 MCO contains an extra 20 amino acids close to its C-terminus. The purified enzyme was capable of using DMP (2,6-dimethoxyphenol), ABTS (2,2′-azino-bis (3-ethylbenzthiazolinesulfonic acid)) and SGZ (syringaldazine) (Li et al. 2008). Cladosporium sphaerospermum has been isolated from soil of an aged gas manufacturing plant to degrade polycyclic aromatic hydrocarbons (PAHs). It was able to degrade benzo(a)pyrene in non-sterile soils including high molecular weight PAHs, followed by 4 weeks of incubation (Potin et al. 2004). Colorimetric assays used for laccase-catalyzed degradation of PAHs have also been developed based on studies of oxidation of 12 aromatic hydrocarbons by fungal laccases from Trametes versicolor and Myceliophthora thermophila. Moreover, Laccase catalyzed anthracene biooxidation into the orange-colored 9,10-anthrahydroquinone (Alcalde et al. 2002).
To this point, we have investigated the homology of MCO on the basis of new constructed phylogenetic tree. There are three major clades A, B and C obtained in phylogeny of MCO protein sequences. In the Clade C bacteria may play the similar role in biodegradation of aromatic and phenolic compounds that includes Sulfonaphthal, Thymolphthalein, Bromocresol green and Phloretin. Thus, there are other proteins such as hemolysin (Singh et al. 2009), aerolysin (Singh and Somvanshi 2009c), thermostable hemolysin (Singh et al. 2011b) and fab (Singh et al. 2011a) of A. hydrophila have been investigated for evolutionary relationship with other bacteria using phylogenetic analysis. In the Clade C homology of MCO which is present in other bacteria such as Erwinia, Serretia and Yersinia can also be used for biodegradation of aromatic and phenolic compounds.
In the MCO gene regulatory network, we found the closely related proteins that include AHA_0347, copA, AHA_2962 and tatB. We showed herein combinatorial influence of these newly constructed genetic networks of A. hydrophila may be useful in gene regulation of MCO for detoxification. Moreover, we have shown the protein–protein interactions with MCO in A.hydrophila gene regulatory network. We have identified two proteins such as copper-exporting ATPase (copA) and copper/silver resistance periplasmic protein (AHA_2962) helpful in biodegradation and increasing the tolerance. In our previous study, we have demonstrated the gene network of B subunit of DNA gyrase of A.hydrophila which showed the several cellular proteins interacted during gene expression (Singh et al. 2012). In the protein–protein interactions (PPI) occur when the two or several proteins/enzymes bind to each other often to carry out their biological function. There are 2,709 interactions between proteins of Saccharomycescerevisiae has been reported which facilitated the establishment of a single large network of 2,358 interactions among 1,548 proteins (Schwikowski et al. 2000). The first step needed is to define precisely what protein–protein interactions are understood as physical contacts with molecular docking between proteins that occur in cellular systems (De Las Rivas and Montanillo 2010). To elucidate the dynamics of large scale genetic regulatory networks of cells is an important goal in systems biology and synthetic biology. The system level dynamical properties of the genetic network of E.coli that regulates its metabolism and show how its design leads to biologically useful cellular properties (Samal and Jain 2008). It is a new approach for targeting any gene for regulating the cellular mechanism by knowing the expected proteins involve in gene regulatory network.
Conclusion
The present study was undertaken to generate the 3-D structure of MCO of A. hydrophila. The 3-D structure of MCO was carried out using docking and obtained four compounds such as Sulfonaphthal, Thymolphthalein, Bromocresol green and Phloretin derivatives of phenol and aromatic hydrocarbon were efficient substrates for MCO activity. The phylogeny of MCO from A. hydrophila indicates the homology present in other bacteria which may serve as a good enzyme for biodegradation that can control environmental pollutions and also prevent entry into food chains. Moreover construction of novel MCO gene regulatory network in A. hydrophila plays a key role for better understanding of the gene regulation and cellular mechanism. There is further need to evaluate these finding through in vitro experiment for biodegradation of aromatic and phenolic compounds.
Electronic supplementary material
Acknowledgments
Authors are grateful to A.K. Singh, Satya Prakash and Pritee Singh for providing the suggestions, encouragement and fruitful discussion during preparation of this manuscript. Authors are also grateful to reviewers for their helpful suggestions.
Conflict of interest
There is no competing interest.
Contributor Information
Vijai Singh, Phone: +33-169475399, FAX: +33-169474437, Email: vijaisingh15@gmail.com.
Dharmendra Kumar Chaudhary, Email: chaudharydk12@gmail.com.
References
- Alcalde M, Bulter T, Arnold FH. Colorimetric assays for biodegradation of polycyclic aromatic hydrocarbons by fungal laccases. J Biomol Screen. 2002;7(6):547–553. doi: 10.1177/1087057102238629. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Thomas LM, Alejandro AS, Jinghui Z, Zheng Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand T, Jolivalt C, Briozzo P, Caminade E, Joly N, Madzak C, Mougin C. Crystal structure of a four-copper laccase complexed with an arylamine: insights into substrate recognition and correlation with kinetics. Biochemistry. 2002;41(23):7325–7333. doi: 10.1021/bi0201318. [DOI] [PubMed] [Google Scholar]
- Bourbonnais R, Paice MG, Reid ID, Lanthier P, Yaguchi M. Lignin oxidation by laccase isozymes from Trametes versicolor and the role of the mediator 2,29-azinobis (3-ethylbenzthiazoline-6-sulfonate) in kraft lignin depolymerization. Appl Environ Microbiol. 1995;61:1876–1880. doi: 10.1128/aem.61.5.1876-1880.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buitron G, Gonzalez A, Lopez-Marin LM. Biodegradation of phenolic compounds by an acclimated activated sludge and isolated bacteria. Water Sci Tech. 1998;37(4–5):371–378. [Google Scholar]
- Las Rivas J, Montanillo C. Protein-protein interactions essentials: key concepts to building and analyzing interactome networks. PLoS Comput Biol. 2010;6:e1000807. doi: 10.1371/journal.pcbi.1000807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De J, Ramaiah N, Mesquita A, Verlekar XN. Tolerance to various toxicants by marine bacteria highly resistant to mercury. Mar Biotechnol. 2003;5:185–193. doi: 10.1007/s10126-002-0061-6. [DOI] [PubMed] [Google Scholar]
- Delano WL (2002) The PYMOL molecular graphics system. Palo Alto CA
- Patti MC, Pascarella S, Catalucci D, Calabrese L. Homology modeling of the multicopper oxidase Fet3 gives new insights in the mechanism of iron transport in yeast. Protein Eng. 1999;12(11):895–897. doi: 10.1093/protein/12.11.895. [DOI] [PubMed] [Google Scholar]
- Freeman JC, Nayar PG, Begley TP, Villafranca JJ. Stoichiometry and spectroscopic identity of copper centers in phenoxazinone synthase—a new addition to the blue copper oxidase family. Biochemistry. 1993;32:4826–4830. doi: 10.1021/bi00069a018. [DOI] [PubMed] [Google Scholar]
- Haglund C, Levín L, Forchiassin F, López M, Viale A. Degradation of environmental pollutants by Trametes trogii. Rev Argent Microbiol. 2002;34(3):157–162. [PubMed] [Google Scholar]
- Hirose J, Nasu M, Yokoi H. Reaction of substituted phenols with thermostable laccase bound to Bacillus subtilis spores. Biotechnol Lett. 2003;25(19):1609–1612. doi: 10.1023/A:1025663931019. [DOI] [PubMed] [Google Scholar]
- Huang KX, Fujii I, Ebizuka Y, Gomi K, Sankawa U. Molecular cloning and heterologous expression of the gene encoding dihydrogeodin oxidase, a multicopper blue enzyme from Aspergillus terreus. J Biol Chem. 1995;270:21495–21502. doi: 10.1074/jbc.270.37.21495. [DOI] [PubMed] [Google Scholar]
- Husain Q, Jan U. Detoxification of phenols and aromatic amines from polluted wastewater by using phenol oxidases. J Sci Ind Res. 2000;59:286–293. [Google Scholar]
- Huston WM, Naylor J, Cianciotto NP, Jennings MP, McEwan AG. Functional analysis of the multi-copper oxidase from Legionellapneumophila. Microbes Infect. 2008;10(5):497–503. doi: 10.1016/j.micinf.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structure. J Appl Crystallogr. 1993;26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- Li Y, Yin J, Qu G, Lv L, Li Y, Yang S, Wang XG. Gene cloning, protein purification, and enzymatic properties of multicopper oxidase, from Klebsiella sp. 601. Can J Microbiol. 2008;54(9):725–733. doi: 10.1139/W08-063. [DOI] [PubMed] [Google Scholar]
- Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comp Chem. 1998;19:1639–1662. doi: 10.1002/(SICI)1096-987X(19981115)19:14<1639::AID-JCC10>3.0.CO;2-B. [DOI] [Google Scholar]
- Piontek K, Antorini M, Choinowski T. Crystal structure of a laccase from the fungus Trametes versicolor at 1.90-A resolution containing a full complement of coppers. J Biol Chem. 2002;277(40):37663–37669. doi: 10.1074/jbc.M204571200. [DOI] [PubMed] [Google Scholar]
- Potin O, Veignie E, Rafin C. Biodegradation of polycyclic aromatic hydrocarbons (PAHs) by Cladosporium sphaerospermum isolated from an aged PAH contaminated soil. FEMS Microbiol Ecol. 2004;51(1):71–78. doi: 10.1016/j.femsec.2004.07.013. [DOI] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modeling by satisfaction of spatial restrains. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Samal A, Jain S. The regulatory network of E. coli metabolism as a Boolean dynamical system exhibits both homeostasis and flexibility of response. BMC Syst Biol. 2008;2:21. doi: 10.1186/1752-0509-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwikowski B, Uetz P, Fields S. A network of protein-protein interactions in yeast. Nat Biotechnol. 2000;18(12):1257–1261. doi: 10.1038/82360. [DOI] [PubMed] [Google Scholar]
- Seshadri R, Joseph SW, Chopra AK, Sha J, Shaw J, Graf J, Haft D, Wu M, Ren Q, Rosovitz MJ, Madupu R, Tallon L, Kim M, Jin S, Vuong H, Stine OC, Ali A, Horneman AJ, Heidelberg JF. Genome sequence of Aeromonas hydrophila ATCC 7966T: jack of all trades. J Bacteriol. 2006;188(23):8272–8282. doi: 10.1128/JB.00621-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu A, Kwon JH, Sasaki T, Satoh T, Sakurai N, Sakurai T, Yamaguchi S, Samejima T. Myrothecium verrucaria bilirubin oxidase and its mutants for potential copper ligands. Biochemistry. 1999;38:3034–3042. doi: 10.1021/bi9819531. [DOI] [PubMed] [Google Scholar]
- Shleev SV, Khan IG, Gazaryan IG, Morozova OV, Yaropolov AI. Novel laccase redox mediators: spectral, electrochemical, and kinetic properties. Appl Biochem Biotechnol. 2003;111(3):167–184. doi: 10.1385/ABAB:111:3:167. [DOI] [PubMed] [Google Scholar]
- Singh V, Somvanshi P. Homology modeling of adenosine A2A receptor and molecular docking for exploration of appropriate potent antagonists for treatment of Parkinson’s disease. Curr Aging Sci. 2009;2(2):127–134. doi: 10.2174/1874609810902020127. [DOI] [PubMed] [Google Scholar]
- Singh V, Somvanshi P. Homology modeling of 3-oxoacyl-acyl carrier protein synthase II (KAS II) from Mycobacterium tuberculosis H37Rv and molecular docking for exploration of drugs. J Mol Model. 2009;15(5):453–460. doi: 10.1007/s00894-008-0426-5. [DOI] [PubMed] [Google Scholar]
- Singh V, Somvanshi P. Inhibition of oligomerization of aerolysin from Aeromonas hydrophila: homology modeling and docking approach for exploration of hemorrhagic septicemia. Lett Drug Des Discov. 2009;6(3):215–223. doi: 10.2174/157018009787847864. [DOI] [Google Scholar]
- Singh V, Somvanshi P, Rathore G, Kapoor D, Mishra BN. Gene cloning, expression and homology modeling of hemolysin gene from Aeromonas hydrophila. Protein Expr Purif. 2009;65:1–7. doi: 10.1016/j.pep.2008.11.015. [DOI] [PubMed] [Google Scholar]
- Singh V, Mani I, Chaudhary DK. Gene cloning and homology modeling of the 3-oxoacyl-ACP synthase from Aeromonashydrophila for drug discovery. Lett Drug Des Discov. 2011;8(7):619–625. doi: 10.2174/157018011796235293. [DOI] [Google Scholar]
- Singh V, Mani I, Chaudhary DK, Somvanshi P. Molecular detection and cloning of thermostable hemolysin gene from Aeromonas hydrophila. Mol Biol (Mosk) 2011;45(4):551–560. doi: 10.1134/S002689331104011X. [DOI] [PubMed] [Google Scholar]
- Singh V, Chaudhary DK, Mani I. Gene network analysis of Aeromonas hydrophila for novel drug target discovery. Syst Synth Biol. 2012 doi: 10.1007/s11693-012-9093-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon EI, Sundaram UM, Machonkin TE. Multicopper oxidases and oxygenases. Chem Rev. 1996;96:2563–2605. doi: 10.1021/cr950046o. [DOI] [PubMed] [Google Scholar]
- Sonoki T, Kajita S, Ikeda S, Uesugi M, Tatsumi K, Katayama Y, Iimura Y. Transgenic tobacco expressing fungal laccase promotes the detoxification of environmental pollutants. Appl Microbiol Biotechnol. 2005;67(1):138–142. doi: 10.1007/s00253-004-1770-8. [DOI] [PubMed] [Google Scholar]
- Suresh PS, Kumar A, Kumar R, Singh VP. An in silico approach to bioremediation: laccase as a case study. J Mol Graph Model. 2008;26(5):845–849. doi: 10.1016/j.jmgm.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTALX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmott N, Guthrie J, Nelson G. The biotechnology approach to colour removal from textile effluent. J Soc Dyers Colour. 1998;114:38–41. doi: 10.1111/j.1478-4408.1998.tb01943.x. [DOI] [Google Scholar]
- Youn HD, Hah YC, Kang SO. Role of laccase in lignin degradation by white-rot fungi. FEMS Microbiol Lett. 1995;132:183–188. doi: 10.1111/j.1574-6968.1995.tb07831.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.