Abstract
Protein-DNA interactions are the physical basis of gene expression and DNA modification. Structural models that reveal these interactions are essential for their understanding. As only a limited number of structures for protein-DNA complexes have been determined by experimental methods, computation methods provide a potential way to fill the need. We have developed the DISPLAR method to predict DNA binding sites on proteins. Predicted binding sites have been used to assist the building of structural models by docking, either by guiding the docking or by selecting near-native candidates from the docked poses. Here we applied the DISPLAR method to predict the DNA binding sites for 20 DNA-binding proteins, which have had their DNA binding sites characterized by NMR chemical shift perturbation. For two of these proteins, the structures of their complexes with DNA have also been determined. With the help of the DISPLAR predictions, we built structural models for these two complexes. Evaluations of both the 20 DNA binding sites and the structural models of the two protein-DNA complexes against experimental results demonstrate the significant promise of our model-building approach.
Keywords: protein-DNA interaction, interface prediction, interaction sites
1. INTRODUCTION
Interactions between proteins and DNA are central to life. The structures of the complexes formed between proteins and DNA are essential to understand these interactions at the atomic level. In particular, these structures reveal the mechanisms of protein-DNA recognition [1]. Experimental methods such as X-ray crystallography and NMR spectroscopy have resulted in structures for a significant number of protein-DNA complexes. However, structures of many more protein-DNA complexes are still to be determined.
In line with the unique structural and physical properties of DNA molecules, their binding sites on proteins also have distinct characteristics. DNA are highly negatively charged; correspondingly their binding sites are usually enriched with positively charged amino acids [2]. Double-stranded DNA have a regular double-helix structure, with grooves that allow protein structural elements to fill up and interact with the DNA bases. These characteristics provide the basis for computational methods to predict DNA binding sites on proteins, which in turn can be used to build structural models for protein-DNA complexes.
A number of methods have been developed to predict DNA binding sites, based on either sequences alone or the unbound structures of the proteins [2–8]. Of these our DISPLAR method [2] makes its predictions from the sequence profiles of a list of spatially neighboring residues. These predictions are of higher accuracy than the corresponding predictions of protein-protein interfaces [9, 10], perhaps reflecting the stronger characteristics of residues present in protein-DNA interfaces. Like their experimental counterparts such as hydrogen exchange and chemical shift perturbation, binding interface prediction methods can provide valuable information for characterizing the interactions between proteins and their DNA targets.
Complementary to interface prediction, structural models of protein complexes, especially those involving only proteins, have been built through docking. Docking methods have been evaluated by the CAPRI exercises (http://www.ebi.ac.uk/msd-srv/capri/). In cases where conformational changes between the unbound state and the bound state are limited (i.e., those devoid of gross conformational rearrangement and severe distortion of interfacial side chains or loops), model building by docking has been quite successful [11]. Docking of protein-DNA complexes has also been specifically investigated [12–15].
Interface prediction has shown promises in assisting the docking of protein-protein complexes in CAPRI exercises [16–18] and the docking of protein-DNA complexes [14]. The higher accuracy of predicted DNA binding sites, noted above, means that they have greater potential in providing useful constraints for building structural models of protein complexes [19].
In this study, we first assess the quality of 20 DNA binding sites predicted by DISPLAR. The assessment is based on chemical shift perturbation data collected from the literature [20–39]; a similar assessment of our protein-protein interaction site prediction was made previously [10]. Using the predicted DNA binding sites, we build structural models for two of the protein-DNA complexes, for which structures are now available from the Protein Data Bank (PDB). The study shows that the combination of interface prediction and docking produces very promising results, though with some traps along the way.
2. METHODS
2.1 Dataset Collection
The chemical shift perturbation data were collected from the literature by searching the PUBMED (www.ncbi.nlm.nih.gov/pubmed/) with keyword “chemical shift” and “DNA”, followed by manual curation. For each of the DNA-binding proteins with chemical shift perturbation data, the list of residues with significant chemical shift perturbations upon DNA binding was recorded. The criteria for what counted as significant chemical shift perturbations were somewhat arbitrary; different authors chose slightly different criteria. Because there was no easy way to choose a uniform criterion for different DNA-binding proteins, we simply followed the designations of the original authors.
The sequences of these DNA-binding proteins were aligned against each other to check for similarity. The alignment was done by the BLAST program, with the cutoff for similarity set with a threshold value of 10−3 for the expectation value. When redundancy did occur, the expectation value was always far lower than the threshold; hence redundancy was easy to recognize. When that happened, the entry with a longer sequence was retained. The retained sequences were further aligned against the training set of the DISPLAR program. Those with sequence similarity with the training set were removed.
The final list of 20 DNA-binding proteins, including names and PDB entries in the unbound state, and their residues reported as showing significant chemical shift perturbations upon DNA binding are found in Table 1. Because DISPLAR makes predictions only on surface residues, defined as those with > 10% solvent accessibility, only surface residues are listed.
Table 1.
Interface Residues Identified by NMR and by DISPLAR.
| PDB | Protein name | Ref | Interface residues by NMR and by DISPLAR |
|---|---|---|---|
| 1aa3 | C-ter RecA (270–322) | [20] | 270,290,301–304 290,301–304; 286–287,289,291,300; 285,288,292,294,297 |
| 1csp | CspB | [21] | 7–11,13,15,17,27,29–31,38 9,15,29–31; 32,37,39; 4,65 |
| 1d4u | human XPA | [22] | 44,49,68–71,78–79,84,87,108–110 68,70–71,78–79,84; 45,73,76–77,80–82; 74–75 |
| 1ewi | human RPA | [23] | 34–35,41–42,59–62,86,89,91,93 34–35,41,91; 33,36,38–39,43,88; 31,37 |
| 1h5p | sp100b SAND | [24] | 626,648–649,653–657,660,667 648–649,653–657,660,667; 608,625,641,645,647,650–652,664–665; 610–611, 617,620,628,642,663 |
| 1kft | C-ter UvrC | [25] | 28,30–31,33,35–37,39,63,65 33,35–36,39,63; 34,38,42–43,62; 41,44–46,48,51,73,76–77 |
| 1mb1 | DBD of Mbp1 | [26] | 51,55,57,60,66–67,74 66,74; 47,68,70,75; 10,37 |
| 1s7e | HNF-6 | [27] | 6–7,23,28,31–32,39,44–45,54,59–60,67,73,75,80–85,91–92,95,102–103,105, 107,123–124,140–141,146,150–152 39,102–103,105,146,150–152; 37,100–101,104,145,148–149; 98–99 |
| 1tbd | OBD of SV40 (6–124) | [28] | 24–27,76,78–79 24–27,76,78–79; 22–23,28,74,81; 19,21,96 |
| 1ub4 | MazF | [29] | Chain A: 14,17–18,28–29,42–43,54–55,57–58,61,69,86,89 Chain B: 214,217,228–229,242–243,254–255,257–258,261,269,286,289 Chain A: 29,54–55,57–58,61,69; 52–53,56,70; 50,71,74,77,79–80 Chain B: 214,217,228–229,254–255,257–258,261,269,286; 216,245,252–253, 256,271,282,291; 250,274,277,279–280 |
| 1utx | CyIR2 | [30] | Chain A: 18–19,29–30,39–40,44–45 Chain B: 17–19,29–30,39–40,44–45 Chain A: 29–30,39–40,45; 16,27–28,32,37–38,42; 4–5,7,26,36 Chain B: 17–18,29–30,39–40; 26–28,32,37–38,42; 4,36 |
| 1uw0 | Zinc finger of DL3 (4–98) | [31] | 4,7,10,12,15,17,19–20,26,28,34,36,38,40–41,43–45,49–51,59,63,66,68,70–71, 98 12,15,17,19–20,26,49–51; 11,13–14,16,21,23–24,27,47–48,52; 22 |
| 1wij | DBD of EIN3/EIL | [32] | 170,172,178–180,183,194,196,225,228,233,235,259,268 259; 255,258,260–261,266; 186,191,249,251–252,254,256,262,264–265 |
| 1wj2 | DBD of WRKY | [33] | 416–419,423,425,429,431,433,443,456 416–419,423,425,429,431,433,443; 415,420–422,424,426,428,432,435,442, 444,458; 427,440,460 |
| 1yua | C-ter of E. coli Topoisomerase I (13–120) | [34] | 22,28,31–32,35–36,41,43,45,48–50,88 88; 33–34,37,87; 90–91,93,96 |
| 2a2y | Sso10b2 (6–88) | [35] | Chain A: 17–18,21,39–40,63,75 Chain B: 17–18,21,39–40,63,70,75 Chain A: 39–40; 41–42,74; 11–13,45 Chain B: 39–40; 41–42,44,73–74; 11–13,45 |
| 2aje | C-ter TRP | [36] | 14,51,57–58,61,65,74,80 14,51,61,65,74; 12–13,15,50,55,60,64,71; 9–11,67–69,90,92,95 |
| 2fk4 | C-ter HPV (3–62) | [37] | 4,7,10–11,23,25,27–28,30–31,38,45,47,49–51,54,58,60,62 49–50,60,62; none; none |
| 2j2s | CXXC of MLL protein (1150–1201) | [38] | 1154–1155,1183–1188,1196 1154–1155,1183–1188,1196; 1153,1156,1175,1177,1182,1197; 1150–1152, 1178,1181,1198–1201 |
| 2j5o | FtsK γ domain (743–809) | [39] | 743–744,747,762,764–767,770–771,774–778,780,800 762,764–767,770–771,774–776,800; 769,773,796,798–799; 797 |
For each protein, the top row(s) are residues in the DNA binding interface identified by NMR chemical shift perturbation; the bottom row(s) are the counterparts by DISPLAR prediction. The latter are presented in three groups separated by semicolons, indicating correct, loosely correct, and incorrect predictions, respectively. In the “Protein name” column, residue numbers in parentheses in some entries indicate the protein segments used for interface prediction.
2.2 DNA Binding Site Prediction
DISPLAR is available as a web server at http://pipe.scs.fsu.edu/displar.html. It uses a trained neural network to predict DNA binding sites, with input given by the sequence profiles, obtained from running PSI-Blast, of a list of spatially neighboring residues [2]. The neighbor list was calculated from the unbound structure of the protein. Of the 20 DNA-binding proteins studied here, 16 structures were determined by NMR spectroscopy and the other 4 were determined by X-ray crystallography. In the former case we used either the first model or the model representing the average structure of an ensemble. In some cases, the N- or C-terminal was described as particularly flexible; such regions were removed. In such cases the actual segment used for prediction is listed in Table 1.
For comparison, we also obtained predictions by the Patch Finder Plus (PFplus) method [40]. In this method, the DNA binding site was identified as the largest positive electrostatic patch; the electrostatic potential was calculated using the UHBD program. The predictions were done through the PFplus server (http://pfp.technion.ac.il). The DISPLAR and PFplus predictions were both assessed against the chemical shift perturbation data. For both DISPLAR and PFplus, the assessment is limited to surface residues (i.e., those with > 10% solvent accessibility).
2.3 Selection of Targets for Docking
We searched for known structures of complexes formed by the 20 proteins with their cognate DNA, to be used as targets for evaluating structural models built by docking (described below). To that end, the BLAST program was run to align the sequences of the 20 proteins against all the protein sequences in the PDB. The PDB names of the BLAST matches were then compared against all the PDB entries containing nucleic acids. A match in PDB name in the second comparison meant that a PDB entry contained a complex between a protein under study and a DNA. Structures of DNA-bound complexes were found for 7 of the 20 proteins studied. Of these, RecA C-terminal domain and CspB were bound to single-stranded DNA; HNF-6 and TRP C-terminal domain showed significant changes between the unbound structures solved by NMR spectroscopy and the bound structures solved by X-ray crystallography; and FtsK γ domain was bound to a nonspecific DNA. Hence we were left with two targets: the complexes of the CXXC domain of the mixed-lineage leukemia (MLL) protein and the origin binding domain (OBD) of SV40 with their respective DNA. The DNA-bound complex of MLL CXXC domain is in PDB entry 2kkf [41]. For SV40 OBD, two DNA-bound structures were solved, with somewhat different DNA sequences capping the cognate motif; the PDB names of the two complexes are 2itl and 2nl8, respectively [42].
2.4 Building of Structural Models by Docking
The DNA molecules used for docking with the proteins had standard B-DNA conformations, and were built using the 3D-DART program [43] from sequences; that program in turn was based on the 3DNA program [44]. The DNA sequences were taken from those in the protein-DNA complexes [41, 42]. For MLL CXXC domain, the DNA sequence of the 5′-3′ strand was CCCTGCGCAGGG. For SV40 OBD, the two DNA sequences were AGAGGCC and CGAGGCCAT. The cognate motifs are in bold.
Two docking methods were used to build models for protein-DNA complexes from the unbound protein and standard DNA structures. The HADDOCK program (version 2.1) [45] was run with default setting. The DISPLAR predicted interface residues listed in Table 1 and the cognate motifs of the DNA molecules were used to drive the docking. The predicted interface residues were designated as “active” if their solvent accessibilities were >50% and as “passive” otherwise; the DNA cognate motifs were designated as “active”. One thousand poses were generated in the rigid docking stage, with 200 of these and their “images” produced by 180° rotation selected for further refinement by semi-flexible docking to yield 200 final poses. The semi-flexible segments on the protein and DNA molecules were selected automatically in the interface of the poses.
ZDOCK 2.3 [46] was used to run rigid body docking for the two targets with 15° rotation sampling. As in our previous work [14], parameters for DNA required for running ZDOCK were taken from Fanelli and Ferrari [12]. The terminal nucleotides of the DNA molecule were blocked in ZDOCK running. After each ZDOCK run, the 2000 poses collected were ranked by using the predicted interface residues listed in Table 1 and the cognate motifs of the DNA molecules. Specifically, the percentage of these residues/nucleotides found in the interface of each pose was used as the scoring function.
The docking results were evaluated against the target structures according to L-RMSD, the RMSD between the DNA molecules in the docked structure and the target structure after the protein molecules were aligned. RMSD was calculated using the Profit program (http://www.bioinf.org.uk/software/profit/) based on Cα and P atoms on protein and DNA molecules, respectively. A docked structure with L-RMSD < 10 Å was considered near-native.
3. RESULTS AND DISCUSSION
3.1 Binding Site Predictions
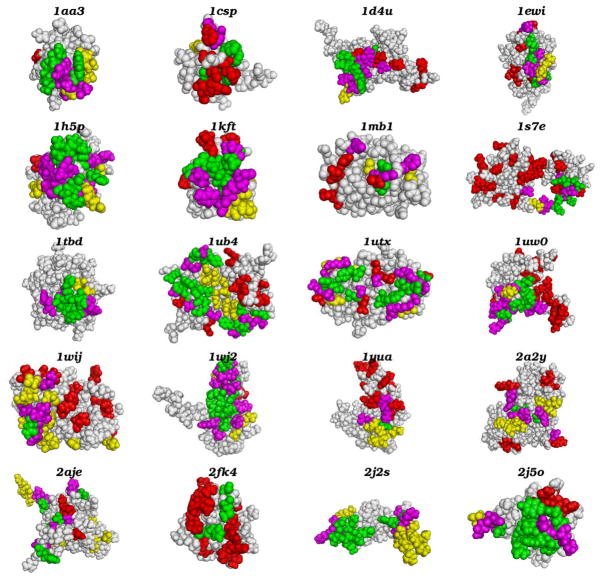
We used the DISPLAR program to predict the DNA binding sites of 20 proteins which have published chemical shift perturbation data. The predicted interface residues are listed in Table 1 and displayed in Fig. 1. Compared with the binding sites defined by chemical shift perturbation, the DISPLAR predictions overall seem quite successful. Of the predicted interface residues, 36% are identical with those identified by chemical shift perturbation; the predictions cover 45% of all the interface residues identified by chemical shift perturbation.
Fig. 1.
Comparison of DNA binding sites determined by DISPLAR prediction and by NMR chemical shift perturbation. The proteins are represented as van der Waals surfaces. Correct, loosely correct, and incorrect predictions are colored in green, magenta, and yellow, respectively; see Table 1 for listing of these residues. Interface residues missed by DISPLAR prediction are colored in red. All structural figures were generated using Pymol (www.pymol.org).
The predicted binding sites coincide with those identified by chemical shift perturbation data, as shown in Fig. 1. Most of the predicted residues not identical to those identified by chemical shift perturbation are located close to the latter residues. If the definition of correct predictions is extended to include four nearest spatial neighbors of any actual interface residue, the prediction accuracy increases from 36% to 74%.
As comparison, the PFplus predictions have an accuracy of 27% and cover 63% of all the interface residues identified by chemical shift perturbation. With the looser definition, the accuracy increases to 60%. DISPLAR has a significantly higher accuracy than PFplus, though with a somewhat lower coverage. The higher coverage of PFplus is the result of over-prediction: chemical shift perturbation identified 295 interface residues for the 20 DNA-binding proteins; PFplus predicted 694 residues, more than doubling the actual number. In contrast, DISPLAR predicted 368 interface residues, much closer to the actual number. Though based on different techniques, both DISPLAR and PFplus benefit from the enrichment of positively charged residues in DNA binding sites.
3.2 Structural Models by Docking
We built structural models for DNA-bound complexes of two proteins: MLL CXXC domain and SV40 OBD. The protein structures were those in the unbound state and the DNA structures were those of standard B-DNA. Two docking methods were used; both were assisted by the DISPLAR predicted interface residues of the DNA-binding proteins. We now present the results of the two docking methods separately.
3.2.1 HADDOCK models
The predicted interface residues on the protein side and the cognate motif of the DNA together were used to guide the HADDOCK process. On the DNA side, either the cognate motif on the 5′-3′ strand alone or that together with the matching nucleotides on the complementary strand was designated as “active”; the outcomes were somewhat different.
In the complex of MLL CXXC domain with its cognate DNA, the protein is mostly bound to the major groove, but the N-terminal extends to the minor groove to contact the bases which match the CTGC motif of the 5′-3′ strand [41] (Fig. 2A). With only the 5′-3′ strand designated as “active”, HADDOCK produced 10 near-native poses among the 200 final poses. Five of the 10 were found in the third largest cluster when the 200 poses were clustered with a cutoff at 7.5 Å in L_RMSD between poses. The best model, with an L_RMSD of 3.9 Å from the NMR structure of the complex, is compared with the NMR structure in Fig. 2A. When both DNA strands were designated as “active”, 9 near-native poses were produced.
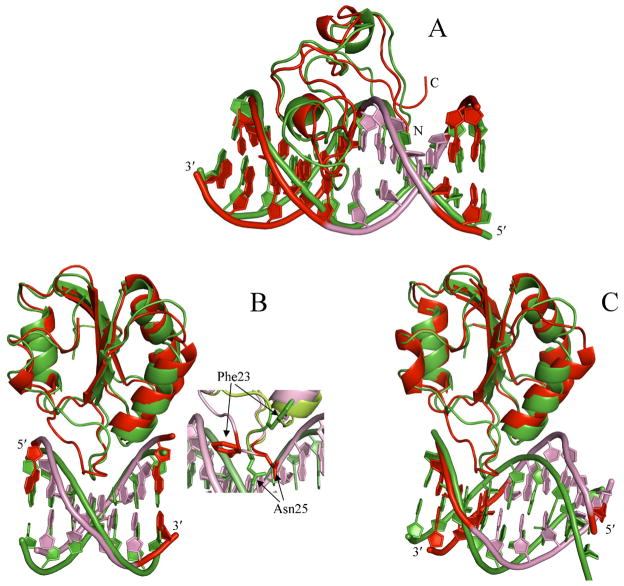
Fig. 2. Comparison of the best models from docking and the NMR or X-ray structures of the protein-DNA complexes. For each complex, the protein molecules are superimposed. Docking models are shown as green ribbons; actual structures are shown as red ribbons.
(A) Complex of MLL CXXC domain with a DNA with a 5′-3′ sequence of CCCTGCGCAGGG. The ends of this strand is labeled; the cognate motif in the 5′-3′ strand and the matching nucleotides in the complementary strand are shown in pink in the NMR structure (PDB entry 2kkf; [41]). The protein N- and C-terminals are also labeled. The best HADDOCK model, with an L_RMSD of 3.9 Å and shown here, was obtained by docking the NMR ensemble of unbound conformations in PDB entry 2j2s and the cognate DNA in a standard B-DNA conformation. The RMSD between the unbound (first model in 2j2s) and bound protein is 3.4 Å; the corresponding RMSD for the DNA is 1.0 Å.
(B) Complex of SV40 OBD with a DNA with a 5′-3′ sequence of AGAGGCC. The ends of this strand is labeled; the cognate motif in the 5′-3′ strand and the matching nucleotides in the complementary strand are shown in pink in the X-ray structure (PDB entry 2itl; [42]). The best ZDOCK model, with an L_RMSD of 3.2 Å and shown here, was obtained by docking the average structure of an NMR ensemble for the unbound protein in PDB entry 1tbd and the cognate DNA in a standard B-DNA conformation. The overall RMSD between the unbound and bound protein is 1.3 Å (the corresponding RMSD for the DNA is also 1.3 Å), but the loop that penetrates deeply into the major groove of the cognate DNA undergoes significant rearrangement, as shown by the inset. Two side chains, Phe23 and Asn25, which experience particularly large movement are presented in the inset.
(C) Complex of SV40 OBD with a DNA with a 5′-3′ sequence of CGAGGCCAT. The ends of this strand is labeled; the presumed cognate motif, GAGGC on the 5′-3′ strand, and the matching nucleotides in the complementary strand are shown in pink in the X-ray structure (PDB entry 2nl8; [42]). Note that the interaction site as found in the X-ray structure is actually shifted downstream along the 5′-3′ strand by three base pairs, and the sequence ATGGC on the complementary strand becomes a stand-in for the cognate motif. The best ZDOCK model, with an L_RMSD of 5.6 Å, is shown here. The overall RMSD between the unbound and bound protein is 1.5 Å; the corresponding RMSD for the DNA is 1.7 Å.
For either cognate DNA sequence, SV40 OBD is bound to the major groove of the DNA (Fig. 2B and 2C). However, though the two complexes involve the same DNA binding site on the protein, its interactions with the two DNA molecules are quite different. With the AGAGGCC sequence, the protein indeed interacts with the cognate motif GAGGC on the 5′-3′ strand and the matching nucleotides on the complementary strand. In contrast, with the CGAGGCCAT sequence, the interaction site is shifted downstream along the 5′-3′ strand by three base pairs. In addition, the orientation of the DNA is flipped 180°, such that the flipped sequence ATGGC on the complementary strand becomes a stand-in for the cognate motif. For sake of specificity, we refer to the first complex as canonical and the second complex as flipped.
For the canonical complex, HADDOCK with both DNA strands designated as “active” produced 17 near-native poses in the rigid docking stage but only 1 in the final 200 poses. When only one DNA strand was designated as “active”, HADDOCK produced only 2 near-native poses in the rigid docking stage and none in the final 200 poses. The relatively poor performance of HADDOCK on this target can be attributed to conformational changes of the protein upon binding DNA. Specifically, the protein loop that deeply penetrates into the DNA major groove undergoes significant rearrangement (Fig. 2B inset); several residues, including Asn25, in the unbound structure prevent the close approach of the DNA.
HADDOCK was more successful for the flipped complex. With both DNA strands designated as “active”, 5 near-native poses were produced in the rigid docking stage and 3 poses were among the final 200. When only one DNA strand was designated as “active”, 24 near-native poses were produced in the rigid docking stage but none was among the final 200. It should be noted that we designated as “active” the presumed cognate motif, GAGGC on the 5′-3′ strand, not the flipped sequence ATGGC on the complementary strand as found in the X-ray structure.
The performance of the semi-flexible refinement is quite different for the DNA-bound complexes of MLL CXXC domain and for SV40 OBD. For MLL CXXC domain, the refinement enriched near-native poses and improved L_RMSD. In contrast, for SV40 OBD, the refinement actually reduced the number of near-native poses. The difference in performance perhaps reflects the larger conformational changes of SV40 OBD upon binding DNA; these larger conformational changes are beyond the small moves employed in the refinement stage.
Overall, for both proteins, HADDOCK generated promising results. In each case, ~20 near-native poses were produced in the rigid docking stage. Here only predicted interface residues are used to drive the docking. In many cases, experimental data (such as provided by chemical shift perturbation) may also be available; these may present valuable information for building better models. More specific constraints, such as pairwise constraints or constraints on specific atoms instead of whole residues or nucleotides, are also expected to improve docking results [15]. Here we have shown that blindly predicted information at the residue level is already providing a promising start for model building.
3.2.2 ZDOCK models
For both MLL CXXC domain and SV40 OBD, ZDOCK was able to generate dozens of near-native poses of their DNA-bound complexes among 2000 poses. We then re-ranked the 2000 poses using the DISPLAR predicted interface residues, either alone or in conjunction with the cognate motif on the 5′-3′ strand or with the matching pairs on both strands.
For MLL CXXC domain, the ranking of the near-native poses was only slightly improved by the predicted interface residues along with the cognate motif on the 5′-3′ strand (Fig. 3A) and made even worse when the predicted interface residues was either used along or in conjunction with the cognate motif basepairs on the two strands. The failure to achieve a significant improvement in re-ranking can be attributed to the fact that the 2000 ZDOCK poses were concentrated around the actual interface, with many poses differing only in the relative orientations of the two subunits. Hence the predicted interface residues did not have a strong ability to distinguish these poses. In fact, even the interface residues found from the X-ray structure of the complex or those identified by chemical shift perturbation resulted in only modest improvement in re-ranking near-native poses (Fig. 3A).
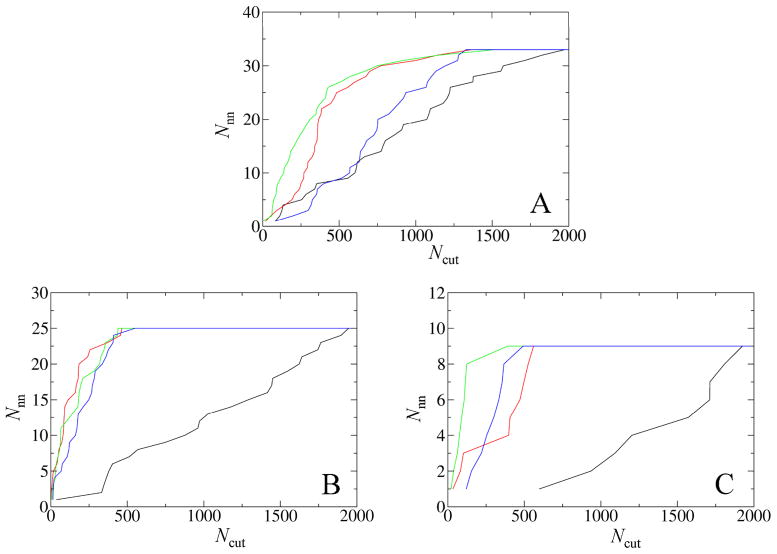
Fig. 3.
Re-ranking of near-native poses among 2000 ZDOCK poses according to DISPLAR predictions in conjunction with the cognate motif on the 5′-3′ strand. Each curve presents Nnn, the number of near-native poses, within the first Ncut of best-scored poses. The black curves are based on the original ZDOCK scores. The scoring function for each of the other three curves is the percentage of assumed interface residues/nucleotides found in the interface of each pose. The assumed interface residues are from the bound complex, the chemical shift perturbation data, and DISPLAR predictions, respectively, for the red, green, and blue curves.
(A) DNA-bound complex of Complex of MLL CXXC domain.
(B) Canonical complex of SV40 OBD.
(C) Flipped complex of SV40 OBD.
For the canonical complex of SV40 OBD, a near-native pose was re-ranked at the 1st, 2nd, and 14th places, respectively, using DISPLAR predictions alone, and in conjunction with cognate motif basepairs on the two strands and with just the 5′-3′ sequence (Fig. 3B). Overall, re-ranking in this case is quite successful.
A strength of ZDOCK is that it can tolerate clashes caused by using unbound structures. Consequently, it was able to generate much better models for this target, in which the protein loop involved in DNA binding undergoes significant rearrangement, than HADDOCK. The best ZDOCK model, with an L_RMSD of 3.2 Å, is compared in Fig. 2B with the X-ray structure of the complex.
For the flipped complex of SV40 OBD, using DISPLAR predictions along with the cognate motif on the 5′-3′ strand also significantly improved the re-ranking of near-native poses (Fig. 3C). Here again we designated as “active” the presumed cognate motif, GAGGC on the 5′-3′ strand, not the flipped sequence ATGGC on the complementary strand as found in the X-ray structure. The best ZDOCK model, with an L_RMSD of 5.6 Å, is compared in Fig. 2C with the X-ray structure of the complex.
4. CONCLUSION
We have demonstrated here that the DISPLAR method is very successful in predicting DNA binding sites on proteins. With the high prediction accuracy, the method can be used to assist the docking of protein-DNA complexes. Although protein conformational changes and distortion of DNA from the standard B-DNA structure still present obstacles, current docking methods are found to be very promising in obtaining near-native models with the help of binding site prediction. The different docking methods may performance better on some targets but worse in other targets. The complementarity of different docking methods increases the likelihood that successful structural models can be built for a given protein-DNA complex. In short, the present study suggests that binding site prediction is a useful tool for building structural models for protein-DNA complexes and for experimental design and validation.
Acknowledgments
Computations were carried out on the High-Performance Computing facility of the Florida State University. This work was supported in part by NIH grant GM058187.
References
- 1.Sarai A, Kono H. Protein-DNA recognition patterns and predictions. Annu Rev Biophys Biomol Struct. 2005;34:379–398. doi: 10.1146/annurev.biophys.34.040204.144537. [DOI] [PubMed] [Google Scholar]
- 2.Tjong H, Zhou HX. DISPLAR: an accurate method for predicting DNA-binding sites on protein surfaces. Nucleic Acids Res. 2007;35:1465–1477. doi: 10.1093/nar/gkm008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmad S, Gromiha MM, Sarai A. Analysis and prediction of DNA-binding proteins and their binding residues based on composition, sequence and structural information. Bioinformatics. 2004;20:477–486. doi: 10.1093/bioinformatics/btg432. [DOI] [PubMed] [Google Scholar]
- 4.Ahmad S, Sarai A. PSSM-based prediction of DNA binding sites in proteins. BMC Bioinformatics. 2005;6:33. doi: 10.1186/1471-2105-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang L, Brown SJ. Prediction of DNA-binding residues from sequence features. J Bioinform Comput Biol. 2006;4:1141–1158. doi: 10.1142/s0219720006002387. [DOI] [PubMed] [Google Scholar]
- 6.Hwang S, Gou Z, Kuznetsov IB. DP-Bind: a web server for sequence-based prediction of DNA-binding residues in DNA-binding proteins. Bioinformatics. 2007;23:634–636. doi: 10.1093/bioinformatics/btl672. [DOI] [PubMed] [Google Scholar]
- 7.Ofran Y, Mysore V, Rost B. Prediction of DNA-binding residues from sequence. Bioinformatics. 2007;23:347–353. doi: 10.1093/bioinformatics/btm174. [DOI] [PubMed] [Google Scholar]
- 8.Wang L, Yang MQ, Yang JY. Prediction of DNA-binding residues from protein sequence information using random forests. BMC Genomics. 2009;10(Suppl 1):S1. doi: 10.1186/1471-2164-10-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou HX, Shan Y. Prediction of protein interaction sites from sequence profiles and residue neighbor list. Proteins. 2001;44:336–343. doi: 10.1002/prot.1099. [DOI] [PubMed] [Google Scholar]
- 10.Chen H, Zhou HX. Prediction of interface residues in protein-protein complexes by a consensus neural network method: test against NMR data. Proteins. 2005;61:21–35. doi: 10.1002/prot.20514. [DOI] [PubMed] [Google Scholar]
- 11.Janin J, Wodak S. The third CAPRI assessment meeting Toronto, Canada, April 20–21, 2007. Structure. 2007;15:755–759. doi: 10.1016/j.str.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 12.Fanelli F, Ferrari S. Prediction of MEF2A-DNA interface by rigid body docking: a tool for fast estimation of protein mutational effects on DNA binding. J Struct Biol. 2006;153:278–283. doi: 10.1016/j.jsb.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 13.van Dijk M, van Dijk AD, Hsu V, Boelens R, Bonvin AM. Information-driven protein-DNA docking using HADDOCK: it is a matter of flexibility. Nucleic Acids Res. 2006;34:3317–3325. doi: 10.1093/nar/gkl412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tjong H, Qin S, Zhou HX. PI2PE: protein interface/interior prediction engine. Nucleic Acids Res. 2007;35:W357–W362. doi: 10.1093/nar/gkm231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Dijk M, Bonvin AM. Pushing the limits of what is achievable in protein-DNA docking: benchmarking HADDOCK’s performance. Nucleic Acids Res. doi: 10.1093/nar/gkq222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Dijk AD, de Vries SJ, Dominguez C, Chen H, Zhou HX, Bonvin AM. Data-driven docking: HADDOCK’s adventures in CAPRI. Proteins. 2005;60:232–238. doi: 10.1002/prot.20563. [DOI] [PubMed] [Google Scholar]
- 17.Qin S, Zhou HX. A holistic approach to protein docking. Proteins. 2007;69:743–749. doi: 10.1002/prot.21752. [DOI] [PubMed] [Google Scholar]
- 18.Qin S, Zhou HX. Selection of near-native poses in CAPRI rounds 13–19. Proteins. 2010 doi: 10.1002/prot.22772. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou HX, Qin S. Interaction-site prediction for protein complexes: a critical assessment. Bioinformatics. 2007;23:2203–2209. doi: 10.1093/bioinformatics/btm323. [DOI] [PubMed] [Google Scholar]
- 20.Aihara H, Ito Y, Kurumizaka H, Terada T, Yokoyama S, Shibata T. An interaction between a specified surface of the C-terminal domain of RecA protein and double-stranded DNA for homologous pairing. J Mol Biol. 1997;274:213–221. doi: 10.1006/jmbi.1997.1403. [DOI] [PubMed] [Google Scholar]
- 21.Zeeb M, Balbach J. Single-stranded DNA binding of the cold-shock protein CspB from Bacillus subtilis: NMR mapping and mutational characterization. Protein Sci. 2003;12:112–123. doi: 10.1110/ps.0219703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buchko GW, Daughdrill GW, de Lorimier R, BKR, Isern NG, Lingbeck JM, Taylor JS, Wold MS, Gochin M, Spicer LD, Lowry DF, Kennedy MA. Interactions of human nucleotide excision repair protein XPA with DNA and RPA70 Delta C327: chemical shift mapping and 15N NMR relaxation studies. Biochemistry. 1999;38:15116–15128. doi: 10.1021/bi991755p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daughdrill GW, Ackerman J, Isern NG, Botuyan MV, Arrowsmith C, Wold MS, Lowry DF. The weak interdomain coupling observed in the 70 kDa subunit of human replication protein A is unaffected by ssDNA binding. Nucleic Acids Res. 2001;29:3270–3276. doi: 10.1093/nar/29.15.3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bottomley MJ, Collard MW, Huggenvik JI, Liu Z, Gibson TJ, Sattler M. The SAND domain structure defines a novel DNA-binding fold in transcriptional regulation. Nat Struct Biol. 2001;8:626–633. doi: 10.1038/89675. [DOI] [PubMed] [Google Scholar]
- 25.Singh S, Folkers GE, Bonvin AMJJ, Boelens R, Wechselberger R, Niztayev A, Kaptein R. Solution structure and DNA-binding properties of the C-terminal domain of UvrC from E. coli. EMBO J. 2002;21:6257–6266. doi: 10.1093/emboj/cdf627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor IA, McIntosh PB, Pala P, Treiber MK, Howell S, Lane AN, Smerdon SJ. Characterization of the DNA-binding domains from the yeast cell-cycle transcription factors Mbp1 and Swi4. Biochemistry. 2000;39:3943–3954. doi: 10.1021/bi992212i. [DOI] [PubMed] [Google Scholar]
- 27.Sheng W, Yan H, Rausa FM, Costa RH, Liao X. Structure of the hepatocyte nuclear factor 6alpha and its interaction with DNA. J Biol Chem. 2004;279:33928–33936. doi: 10.1074/jbc.M403805200. [DOI] [PubMed] [Google Scholar]
- 28.Luo X, Sanford DG, Bullock PA, Bachovchin WW. Solution structure of the origin DNA-binding domain of SV40 T-antigen. Nat Struct Biol. 1996;3:1034–1039. doi: 10.1038/nsb1296-1034. [DOI] [PubMed] [Google Scholar]
- 29.Li GY, Zhang Y, Chan MCY, Mal TK, Hoeflich KP, Inouye M, Ikura M. Characterization of dual substrate binding sites in the homodimeric structure of Escherichia coli mRNA interferase MazF. J Mol Biol. 2006;357:139–150. doi: 10.1016/j.jmb.2005.12.035. [DOI] [PubMed] [Google Scholar]
- 30.Rumpel S, Razeto A, Pillar CM, Vijayan V, Taylor A, Giller K, Gilmore MS, Becker S, Zweckstetter M. Structure and DNA-binding properties of the cytolysin regulator CylR2 from Enterococcus faecalis. EMBO J. 2004;23:3632–3642. doi: 10.1038/sj.emboj.7600367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kulczyk AW, Yang JC, Neuhaus D. Solution structure and DNA binding of the zinc-finger domain from DNA ligase IIIalpha. J Mol Biol. 2004;341:723–738. doi: 10.1016/j.jmb.2004.06.035. [DOI] [PubMed] [Google Scholar]
- 32.Yamasaki K, Kigawa T, Inoue M, Yamasaki T, Yabuki T, Aoki M, Seki E, Matsuda T, Tomo Y, Terada T, Shirouzu M, Tanaka A, Seki M, Shinozaki K, Yokoyama S. Solution structure of the major DNA-binding domain of Arabidopsis thaliana ethylene-insensitive3-like3. J Mol Biol. 2005;348:253–264. doi: 10.1016/j.jmb.2005.02.065. [DOI] [PubMed] [Google Scholar]
- 33.Yamasaki K, Kigawa T, Inoue M, Tateno M, Yamasaki T, Yabuki T, Aoki M, Seki E, Matsuda T, Tomo Y, Hayami N, Terada T, Shirouzu M, Tanaka A, Seki M, Shinozaki K, Yokoyama S. Solution structure of an Arabidopsis WRKY DNA binding domain. Plant Cell. 2005;17:944–956. doi: 10.1105/tpc.104.026435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu L, Zhu CX, Tse-Dinh YC, Fesik SW. Solution structure of the C-terminal single-stranded DNA-binding domain of Escherichia coli topoisomerase I. Biochemistry. 1995;34:7622–7628. doi: 10.1021/bi00023a008. [DOI] [PubMed] [Google Scholar]
- 35.Biyani K, Kahsai MA, Clark AT, Armstrong TL, Edmondson SP, Shriver JW. Solution structure, stability, and nucleic acid binding of the hyperthermophile protein Sso10b2. Biochemistry. 2005;44:14217–14230. doi: 10.1021/bi051266r. [DOI] [PubMed] [Google Scholar]
- 36.Sue SC, Hsiao HH, Chung BCP, Cheng YH, Hsueh KL, Chen CM, Ho CH, Huang TH. Solution structure of the Arabidopsis thaliana telomeric repeat-binding protein DNA binding domain: a new fold with an additional C-terminal helix. J Mol Biol. 2006;356:72–85. doi: 10.1016/j.jmb.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 37.Nomine Y, Masson M, Charbonnier S, Zanier K, Ristriani T, Deryckere F, Sibler AP, Desplancq D, Atkinson RA, Weiss E, Orfanoudakis G, Kieffer B, Trave G. Structural and functional analysis of E6 oncoprotein: insights in the molecular pathways of human papillomavirus-mediated pathogenesis. Mol Cell. 2006;21:665–678. doi: 10.1016/j.molcel.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 38.Allen MD, Grummitt CG, Hilcenko C, Min SY, Tonkin LM, Johnson CM, Freund SM, Bycroft M, Warren AJ. Solution structure of the nonmethyl-CpG-binding CXXC domain of the leukaemia-associated MLL histone methyltransferase. EMBO J. 2006;25:4503–4512. doi: 10.1038/sj.emboj.7601340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sivanathan V, Allen MD, de Bekker C, Baker R, Arciszewska LK, Freund SM, Bycroft M, Lowe J, Sherratt DJ. The FtsK gamma domain directs oriented DNA translocation by interacting with KOPS. Nat Struct Mol Biol. 2006;13:965–972. doi: 10.1038/nsmb1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shazman S, Celniker G, Haber O, Glaser F, Mandel-Gutfreund Y. Patch Finder Plus (PFplus): a web server for extracting and displaying positive electrostatic patches on protein surfaces. Nucleic Acids Res. 2007;35:W526–530. doi: 10.1093/nar/gkm401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cierpicki T, Risner LE, Grembecka J, Lukasik SM, Popovic R, Omonkowska M, Shultis DD, Zeleznik-Le NJ, Bushweller JH. Structure of the MLL CXXC domain-DNA complex and its functional role in MLL-AF9 leukemia. Nat Struct Mol Biol. 17:62–68. doi: 10.1038/nsmb.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bochkareva E, Martynowski D, Seitova A, Bochkarev A. Structure of the origin-binding domain of simian virus 40 large T antigen bound to DNA. Embo J. 2006;25:5961–5969. doi: 10.1038/sj.emboj.7601452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Dijk M, Bonvin AM. 3D-DART: a DNA structure modelling server. Nucleic Acids Res. 2009;37:W235–239. doi: 10.1093/nar/gkp287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu XJ, Olson WK. 3DNA: a software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 46.Chen R, Li L, Weng Z. ZDOCK: an initial-stage protein-docking algorithm. Proteins. 2003;52:80–87. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]