Abstract
Salamanders possess an extraordinary capacity for tissue and organ regeneration when compared to mammals. In our effort to characterize the unique transcriptional fingerprint emerging during the early phase of salamander limb regeneration, we identified transcriptional activation of some germline-specific genes within the Mexican axolotl (Ambystoma mexicanum) that is indicative of cellular reprogramming of differentiated cells into a germline-like state. In this work, we focus on one of these genes, the long interspersed nucleotide element-1 (LINE-1) retrotransposon, which is usually active in germ cells and silent in most of the somatic tissues in other organisms. LINE-1 was found to be dramatically upregulated during regeneration. In addition, higher genomic LINE-1 content was also detected in the limb regenerate when compared to that before amputation indicating that LINE-1 retrotransposition is indeed active during regeneration. Active LINE-1 retrotransposition has been suggested to have a potentially deleterious impact on genomic integrity. Silencing of activated LINE-1 by small RNAs has been reported to be part of the machinery aiming to maintain genomic integrity. Indeed, we were able to identify putative LINE-1-related piRNAs in the limb blastema. Transposable element-related piRNAs have been identified frequently in the germline in other organisms. Thus, we present here a scenario in which a unique germline-like state is established during axolotl limb regeneration, and the re-activation of LINE-1 may serve as a marker for cellular dedifferentiation in the early-stage of limb regeneration.
Keywords: limb, long interspaced nucleotide element-1, piRNAs, regeneration, salamander
Introduction
Among vertebrates, urodele amphibians, such as axolotls (Ambystoma mexicanum) and newts (Notophthalmus viridescens) are known for their remarkable ability to regenerate a broad variety of body tissues and organs throughout their entire lifespan. However, in mammals this capacity is highly restricted and has only been demonstrated in certain tissues or organs, such as liver and digit tip regeneration (Borgens 1982; Reginelli et al. 1995). The axolotl, a diploid salamander, has unparalleled regenerative capacity exemplified by regeneration of limb, tail, eye lens, spinal cord, portions of intestines and jaws, parts of the heart and brain, and more. Among a variety of vertebrate tissue/organ regeneration models in axolotls, limb regeneration in axolotls is the most intensively studied (Brockes & Kumar 2005). As a model for epimorphic regeneration, limb regeneration in axolotls is characterized with near perfect reconstitution of morphology and functionalities. Thus, to promote regenerative studies in mammals including humans, it is important to understand the mechanistic basis of physiologically relevant tissue/organ regeneration observed in axolotls (Tanaka 2003; Stoick-Cooper et al. 2007; Muneoka et al. 2008).
During the early stages of axolotl limb regeneration, a unique structure, the regenerating blastema arises through epithelial-mesenchymal interactions after wound healing. As the hallmark of epimorphic regeneration, the regenerating blastema has been identified as a collective pool of proliferating mesenchymal cells that are originally derived from proximal tissues of diverse types and very likely regain differential levels of multipotency via cellular dedifferentiation/reprogramming. Cellular dedifferentiation/reprogramming usually is defined as the re-acquisition of embryonic-like developmental potential (Muneoka et al. 1986; Echeverri et al. 2001). However, dedifferentiation observed during axolotl limb regeneration does not appear to be a complete reversal to the earliest pluripotent state usually exhibited in germ cells or embryonic stem cells (Kragl et al. 2009). The retention of positional information or the memory of tissue or embryonic origin of differentially reprogrammed blastemal cells has been proposed to be responsible for blastemal cell fate restriction exhibited during cellular re-differentiation in the late phase of limb regeneration. Nevertheless, lineage switching has been reported during axolotl limb regeneration. Indeed, some genes that are active during embryogenesis and remain silent postnatally have also been demonstrated to be re-activated, thus recapitulating scenarios usually occurring during embryonic limb development; for example, oocyte-type linker histone B4 is expressed during lens regeneration in newts, the Sp9 transcription factor in the basal keratinocytes of the apical blastema epithelium in axolotls and three of the four mammalian stem cell pluripotency-inducing factors, Sox-2, Klf4, and c-Myc in both lens and hind limb regeneration in newts (Maki et al., 2009, 2010;; Satoh et al. 2008b). Thus, it is conceivable that the axolotl limb blastema may possess distinctive properties that recapitulate some level of embryonic developmental potentials and might be the key for the induction of similar limb regenerative processes in mammals.
To characterize the nature of the blastema, we carried out transcriptome sequencing of innervated (NR) and denervated (DL) axolotl forelimbs on day 0, 5 and 14 post limb amputation (pa) representing unamputated, early- and medium-phase of limb regeneration, respectively (60). A group of germline-specific genes was found to be significantly upregulated in innervated limb regenerates, e.g. sperm-associated antigen 1, 7, 9, spermatid perinuclear RNA-binding protein, spermatogenesis associated, serine-rich 2, spermidine/spermine N1-acetyltransferase, spermidine/spermine N1-acetyltransferase 2, spermine synthase, Piwi-like 1, tudor domain containing 3, tudor domain containing 7, ADAM metallopeptidase domain 9 isoform 2 precursor, acrosomal vesicle protein 1 isoform A precursor and retrotransposon LINE-1 open reading frames (ORFs) 1 and 2. Here we characterize LINE-1 (long interspersed nucleotide element-1, or L1) activities during the regenerative process based on the serendipitous discovery that L1 retrotransposon is re-activated at the onset of axolotl limb regeneration.
L1 mobilization occurs via reverse transcription using an ancient mechanism shared with the group II introns of mitochondria and eubacteria (Eickbush & Malik 2002). As non-LTR (long terminal repeat) elements, L1s are abundant retrotransposons that comprise approximately 20% of mammalian genomes (Lander et al. 2001; Waterston et al. 2002; Gibbs et al. 2004). L1 retrotransposition is observed in early embryogenesis and in germ cells before the germ line becomes a distinct lineage (Muotri et al. 2005). In addition, L1 retrotransposition has been observed in certain transformed or immortalized cell lines, as well as in neuronal progenitors and embryonic stem cells, suggesting that activation of LINE-1 could be considered as a marker for cells in an undifferentiated state like that observed in stem cells, neuronal precursor cells or cancer stem cells (Moran et al. 1996; Morrish et al. 2002; Han et al. 2004). In recent years, a few studies reported that in some circumstances L1 can be expressed and/or become retrotranspositionally-active somatically. Endogenous L1 transcripts and proteins have been detected in mouse steroidogenic tissues, as well as in human somatic subsets of testicular cells (e.g. Sertoli, Leydig, testicular covering cells and epididymal columnar epithelial cells), placental syncytiotrophoblasts and vascular endothelial cells (Branciforte & Martin 1994; Trelogan & Martin 1995; Ergun et al. 2004). In rats, endogenous L1 RNA and protein were identified in cardiac tissue (Lucchinetti et al. 2006). However, all these are at significantly lower levels than in the germline tissues (Muotri et al. 2005).
Active L1 retrotransposition has been revealed to have potential deleterious impacts on the integrity of genomic DNA. For example, L1 retrotransposition has been reported to introduce potentially catastrophic alternative gene splicing patterns and gene expression profile changes in some diseases (reviewed in Chen et al. (2000); Montoya-Durango and Ramos (2000) Sinibaldi-Vallebona et al. (2006)). Therefore, silencing of activated L1 is considered to be critical as part of the negative feedback loop elicited by L1 activation aimed at maintaining essential cellular homeostasis. Among a variety of L1 silencing mechanisms, L1 piRNAs have been suggested as potential candidates contributing to the silencing process (Frost et al. 2000; Aravin et al. 2007; Kuramochi-Miyagawa et al. 2008; Xu et al. 2008; Shoji et al. 2009). Indeed, our bioinformatic analysis of small RNAs extracted from axolotl regenerating limb tissues suggested that there are putative L1-derived piRNAs in the limb regenerate. Although from other studies there are emerging novel piRNA candidates present in somatic tissues from genomic regions depleted in transposons that may have a role in the regulation of target transcripts, conventional piRNAs that are related to transposable elements (TEs) have been found much more prevalently in germ cells and are considered to be crucial for the transcriptional silencing of reactivated transposable elements and ultimately the maintenance of genomic integrity in germ cells (Siomi et al. 2011). Thus, the presence of putative L1 piRNAs further strengthens the notion that there is a germline-like state in the axolotl regenerating limb.
Material and methods
Experimental animals
Axolotls (Ambystoma mexicanum) were purchased from the Ambystoma Genetic Stock Center in University of Kentucky. For isolation of RNAs and genomic DNAs, juvenile animals measuring 10–15 cm, from snout to tail tip, were used.
Genomic DNA extraction and small RNA cloning and sequencing
Axolotl genomic DNA was isolated using phenol extraction and ethanol precipitation. Contaminating RNA was digested by RNase A. For the collection of small RNAs extracted from axolotl regenerating limb blastemas, we harvested limb regenerates from early-to medium-stage during limb regeneration from different animal individuals. Pooled small RNAs (>15 and <45 nt) were gel-purified after DNase I incubation and then ligated to adaptors before reverse transcription. Reverse transcribed first-strand cDNA was polymerase chain reaction (PCR) amplified, and Solexa sequencing was carried out at University of California San Diego (UCSD).
RT–PCR and Real time quantitative PCR
Axolotl limb regenerates collected 2 or 5 days pa. Total RNAs were extracted using Trizol. aOct4 RT–PCR primer sets: aOct4 (81–106) 5′CCTCGAGGCGGGCGGGCCGGGCTTCC3′ and aOct4 (528–503) 5′CAGGTCCCGCGGGGCGCCCCCAGCCC3′; aOct4 (741–768) 5′CCAGCGCTGGCTGGTCGAGGCCGACACC3′ and aOct4 (1068–1095) 5′GCCCTGCGCGATGTAGGAGGTGGGCAGG3′. Primer P25R: GCCCAGTAGGCATTCCTTGGGTGTCCACTCGT; Primer P25F/26: GCGCTTGCATTCCCCAAAGAGGTGT; Msx2 RT–PCR primers: TAGCGCAGACCCCAAAGTCAGGGTGTCCAGC and TTCGGCAGAAACAGTACCTGTCCATTGCTGAG. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) RT–PCR primers: GACAAGGCATCTGCTCACCT and ATGTTCTGGTTGGCACCTCT. The three scrambled primers described in Fig. S3: GACTACAAAGACGATGACGACAAGGTG; ACC ATGTACCCATACGATGTTCCAGATTAC; GCATCAATGCAGAAGCTGATCTCAGAG GAGGAC. Reverse transcription was performed using Invitrogen’s Superscript II Reverse Transcriptase and the synthesis/amplification of double-stranded cDNA fragments in the following PCR was conducted in the presence of a high-fidelity DNA polymerase, Phusion DNA Polymerase from NEB. Unless specifically stated, the number of the PCR cycles in RT–PCR was 35. LINE-1 ORF1 real-time qPCR primer sets: A09: Forward: 5′GTTCCATCTTGCCCTTGA3′; Reverse: 5′TCCCGCAGGAACAATATTCG3′; D08 Forward: 5′GGGTTCCGGAGGCGG3′; Reverse: 5′GAGGTCCTTCAGAAGGGTCTCA3′. LINE-1 ORF2 real-time qPCR primer sets: 6-5: Forward: 5′TCGAGGCCGACACCCA3′; Reverse: 5′AGACCCGCAGGACCACAATA3′.
In situ hybridization
The detailed protocol used here was adapted from (Satoh et al. 2007). The RNA probes were labeled with digoxigenin (DIG). Staining was developed with 5-bromo-4-chloro-3-indolyl-phosphate (BCIP) and 4-nitroblue tetrazolium chloride (NBT) in alkaline phosphatase buffer after in situ hybridization (ISH) and anti-DIG alkaline phosphatase antibody incubation.
Consecutive limb amputation and regeneration
An axolotl was amputated first at the elbow and the lower part of the forelimb was collected for genomic DNA. After the completion of regeneration, the animal was reamputated at the wrist and the hand regenerated from the first amputation was harvested for genomic DNA extraction. When the regeneration from the second amputation was completed, the hand regenerated from the second amputation and regeneration was also collected. The LINE-1 content in the genomic DNA from these tissue samples was determined by real-time quantitative PCR (qPCR) and normalized by comparison with repetitive rRNA loci or the single copy genes TPM4 and fibroblast growth factor 8 (FGF8) giving essentially identical results. The primer set used here is the L1 ORF2 primer set: 6-5 Forward and Reverse.
Bioinformatic analysis of potential LINE-1 piRNAs
The dataset generated from the small RNA sequencing was scanned against the partial cDNA sequences of LINE-1 ORF1 (clone #7-10) and ORF2 (clone #6-5) as well as the partial genomic DNA sequence of one LINE-1 allele (Fig. S1). Putative L1 piRNAs were selected when the alignment length was ≥19 nt, the number of gap-opens was 0 and the E-value was ≤0.01, which indicated statistical significance.
Results and discussion
Characterization of retrotransposon LINE-1 transcriptional activation in regenerating limbs
In vivo, LINE-1 (L1) retrotransposition is observed in early embryogenesis and in germ cells before the germ line becomes a distinct lineage. Studies of a human L1 transgenic mouse revealed that L1 retrotransposition events are limited to germ cells and neuronal progenitors, suggesting that activation of L1 can be viewed as a marker for cells in an undifferentiated state usually observed in stem cells of different levels of potency (Muotri et al. 2005).
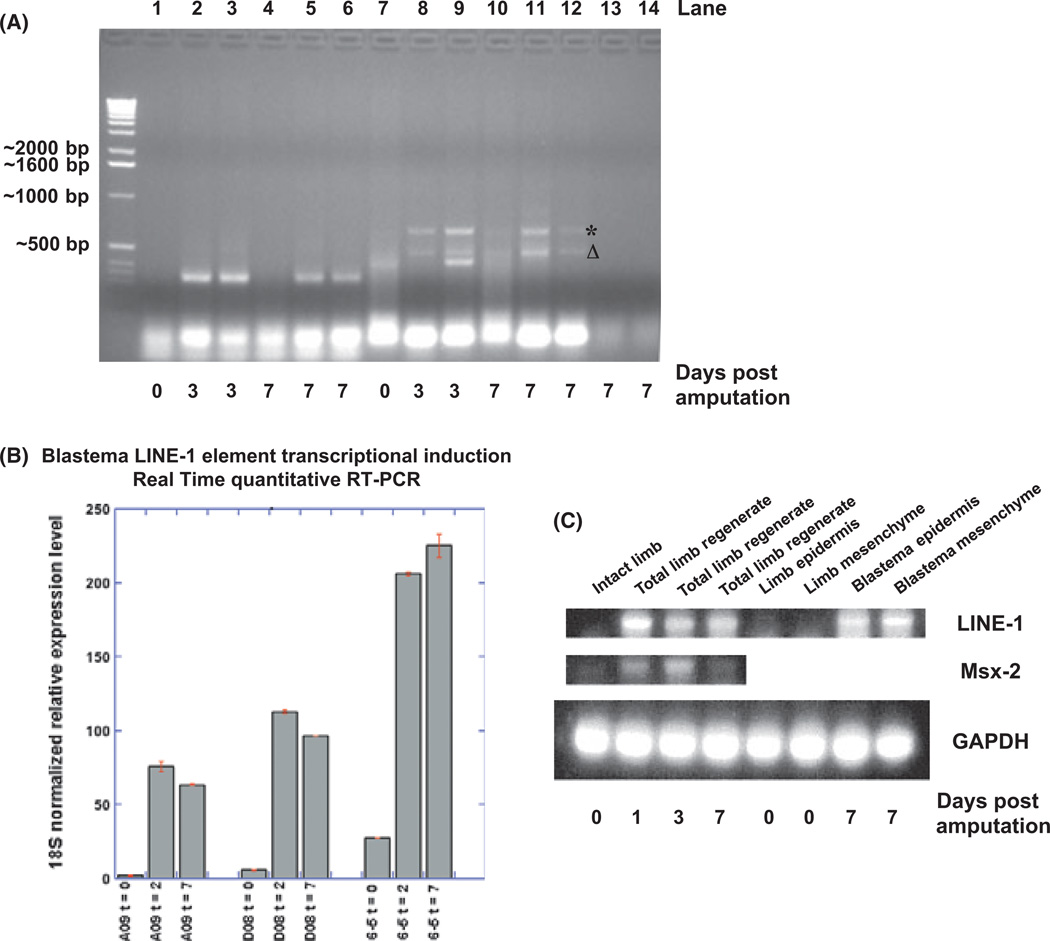
While screening for potential stem cell factors, which might be present in the limb blastema during regeneration we designed two sets of primers (aOct4 741-768/1068-1095 and aOct4 81-106/528-503) to detect the presence of the axolotl Oct4 orthologue during various timepoints during regeneration. Although none of the amplified products appeared to be the expected size, the first primer set amplified two presumably non-specific products with interesting kinetics of appearance absent in unamputated limbs and present during regeneration. These two non-specific PCR products were subsequently sequenced and found to be partial L1 ORF1 (labeled with *, called Clone #7-10) and ORF2 (labeled with Δ, called Clone #6-5) cDNA fragments, respectively. For the day 0 pa limb sample in lane 7 (day 0 pa limb tissues means the limb tissues immediately collected upon amputation), the tissue sample was collected immediately after limb amputation and frozen in liquid nitrogen. No ORF1 or ORF2 cDNA fragment was generated from this sample by RT–PCR. Lanes 13 and 14 are the controls without primers. For lanes 1–6 and 13, the RT–PCR products were produced by the other aOct4 primer set: aOct4 (81-106) and aOct4 (528-503). The nonspecific PCR products in lanes 1–6 were also sequenced but appear unlikely to be derived from L1 ORF1 or ORF2 based on sequencing. It is worth noting that the five limb regenerates collected from different individuals (lanes 8–12) all produced L1 ORF1 and ORF2 amplification products when aOct4 (741–768) and aOct4 (1068–1095) were used as primers, suggesting that L1 reactivation is a ubiquitous phenomenon during axolotl limb regeneration. These initial results suggested that L1 ORF1 and ORF2 were transcriptionally activated upon limb amputation (day 3 or 7 days pa). Using real-time quantitative PCR with specifically designed primers corresponding to different regions of L1, we confirmed that there was a dramatic upregulation of L1 transcriptional activity as early as 2 days post-amputation (Fig. 1B). It is well known that even in a single organism the genomic L1 alleles are not exactly identical among themselves, and that the majority of them are incapable of retrotransposition due to cumulative mutations occurring during evolution, including substitutions, insertions and deletions. On average, a human being carries ~80–100 potentially active L1 elements in his/her diploid genome (Brouha et al. 2003) out of a total of about ~500 000 genomic loci, which are homologous to L1. Due to the polymorphic nature of L1, it is likely that for a particular set of L1 DNA primers used in RT–PCR, only a fraction of L1 transcripts will be amplified while other transcript variants bearing mutation(s) in the areas corresponding to the PCR primers will fail to produce any PCR products. Therefore, it is conceivable that the transcriptional upregulation we demonstrated by RT–PCR in Figure 1 using specific sets of L1 DNA primers only reflected the response of the subset of L1s that was recognized by those particular DNA primers. The actual L1 transcriptional activation may be much more prevalent and stronger than we have observed here. In addition, we also designed axolotl LINE-1-specific DNA primers (P25R and P25F/26) based on the partial LINE-1 ORF-2 cDNA sequence we obtained earlier to examine LINE-1 transcriptional activity in total limb regenerates as well as in regenerating epidermal or mesenchymal tissues on day 0, 3, and 7 pa for RT–PCR. To compare the intensity of LINE-1 transcriptional upregulation with other limb amputation-induced genes, we included RT –PCR of Msx2, which has been demonstrated to be transcriptionally activated during limb regeneration (Carlson et al. 1998; Koshiba et al. 1998) (Fig. 1C). As shown in Figure 1C, the transcriptional upregulation of LINE-1 upon limb regeneration was apparently more robust than that of Msx2. The kinetics of induction of LINE-1 and Msx-2 RNAs also appeared slightly different, with LINE-1 RNA coming up sooner than Msx-2 RNA, and being more sustained during regeneration. Furthermore, the RT–PCR analysis of the localization of upregulated LINE-1 transcripts revealed that LINE-1 is transcriptionally activated in both the regenerating epidermal and mesenchymal tissues.
Fig. 1.
(A) Reverse transcription–polymerase chain reaction (RT–PCR) analysis primed by the DNA primers designed specifically for axolotl Oct4. Lanes 1–6 and 13 show the RT–PCR products resulting from the aOct4 primer set: aOct4 (81-106) and aOct4 (528-503). The nonspecific PCR products in these lanes were not characterized further. Lanes 7–13 were the RT–PCR products from another aOct4 primer set: aOct4 (741-768) and aOct4 (1068-1095). Sequencing results of the nonspecific RT–PCR products primed by aOct4 (741-768) and aOct4 (1068-1095) oligonucleotides revealed that they were either partial LINE-1 ORF1 (*) or ORF2 (Δ) cDNA fragments when the templates were derived from limb regenerates of different animals on day 3 or 7 days post-amputation (pa). Lanes 13 and 14 are the controls without primers. (B) Axolotl LINE-1 cDNA Real Time quantitative (q)PCR to examine LINE-1 transcriptional activity after limb amputation. A09 and D08 are the two sets of qPCR primers for LINE-1 ORF1, while 6–5 is a pair of primers for LINE-1 ORF2. t, days after limb amputation. (C) LINE-1 RT–PCR analysis on day 0, 3 or 7 pa. RT–PCR was primed by LINE-1-specific primers (P25R and P25F/26), LINE-1 transcriptional activity was examined in the regenerating epidermal and mesenchymal tissues (lanes 4–8), respectively, in addition to the total regenerating limb tissues (lanes 1–4). The transcriptional upregulation of another limb amputation-induced gene, Msx2 RNA was also examined by RT–PCR in parallel. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) RT–PCR served as loading controls.
LINE-1 retrotransposition is active during axolotl limb regeneration
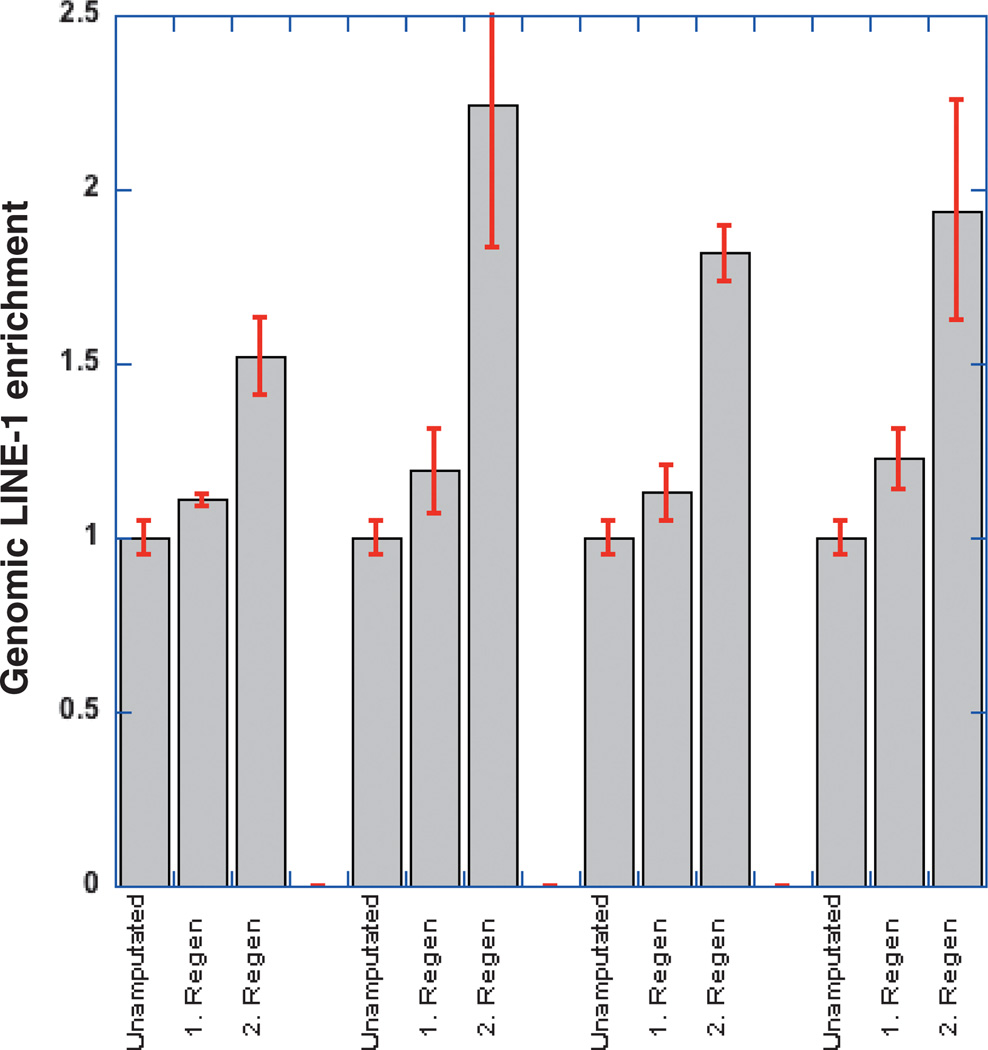
Transcriptional activation of LINE1 retrotransposons is necessary although not sufficient for active retrotransposition and mobilization of LINE1 elements. To investigate whether the LINE elements within the regenerating limb were indeed active we investigated whether these led to novel integrations. Given that in mammals the endogenous copy number is around 500 000 copies/haploid genome and the approximate number of active elements is around 80–100, we reasoned that the likely relative increase in copy number would be small if not undetectable but sequential amputations could increase the number of mobilized LINE elements exponentially if activated during regeneration. Consistent with this rationalization we observed an increase in the content of endogenous L1 elements in the genome after consecutive limb amputations (Fig. 2A). When we compared the integrated L1 element copy number in the forelimb regenerate after the first limb amputation and regeneration to the copy number in the non-regenerated region from the same limb, approximately 16% more L1 copies were found in the genome of the limb regenerate after the first regeneration. Moreover, we found an even larger increase (~70%) in L1 alleles in the genome after the second limb amputation and regeneration, suggesting that LINE-1 is actively retrotransposed during regeneration. The fold increase was normalized against various endogenous multicopy genes such as rDNA as well as single copy genes such as FGF8 and TPM4 giving identical results. These observations support the notion that LINE1 element copy number is increased during regeneration due to productive retrotransposition induced by limb amputation.
Fig. 2.
Axolotl genomic DNA quantitative polymerase chain reaction (qPCR) analysis of the change of integrated LINE-1 gene copy number in limb regenerates from consecutive limb amputations and regenerations. Primers used for genomic DNA Real Time qPCR: D08, the set of primers for LINE-1 ORF-1. Unamputated: the unamputated limb. 1.regen: the limb regenerate from the first round of limb amputation performed on the unamputated limb; 2.regen: the limb regenerate regrown from the second round of limb amputation conducted in the 1.regen. Each of the four groups of bars represents the percentage change of genomic LINE-1 content in the unamputated limb, the 1.regen and the 2.regen in four individual animals, respectively.
In situ hybridization to reveal the expression pattern of LINE-1 in the limb blastema
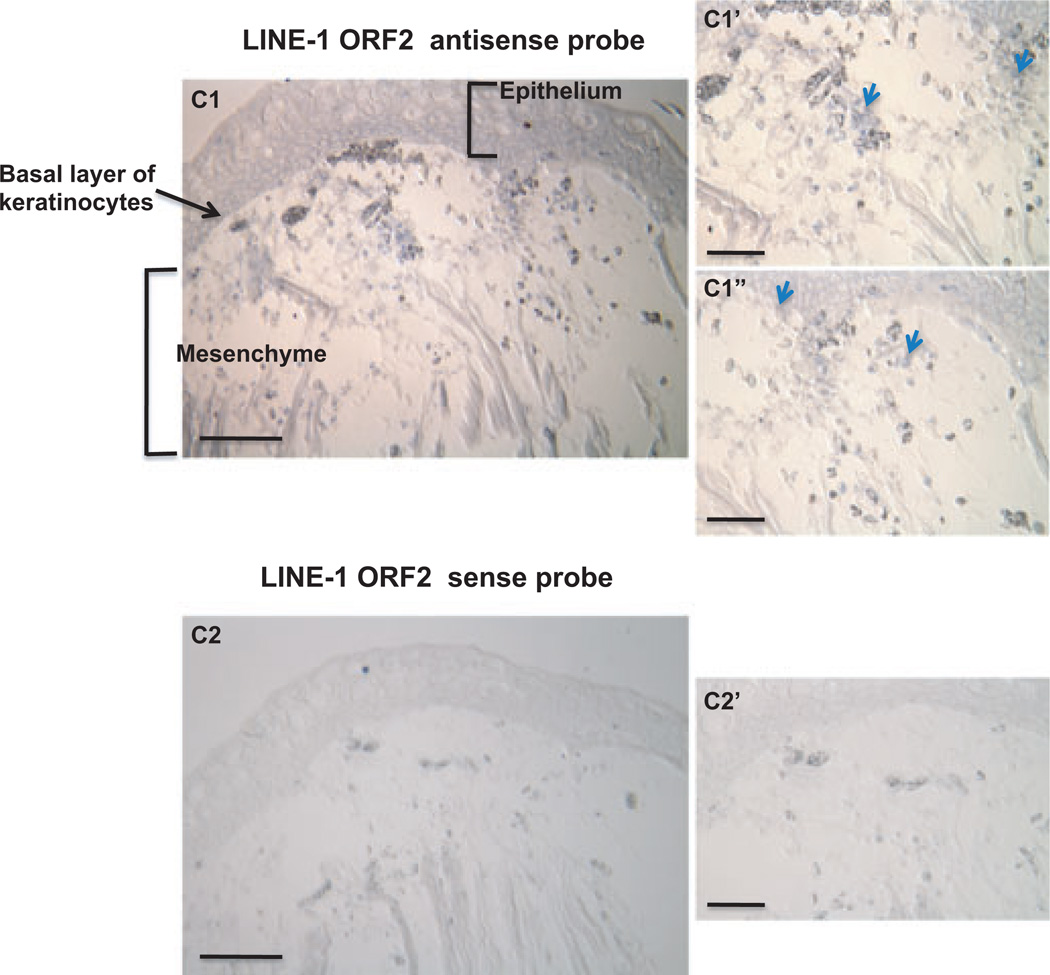
It has been suggested that the cells in the axolotl limb blastema are a heterogeneous collection of differentially dedifferentiated cells that have been reprogrammed to achieve varying levels of developmental potential (Kragl et al. 2009). Since L1 transcriptional activation is considered to be an indication of global genomic DNA derepression in most of the cases reported so far, we also examined the expression pattern of L1 in the regenerating limb blastema 5 days pa by L1 ORF2 in situ hybridization (ISH). L1 was expressed in the blastemal epidermis (labeled “Epithelium”) as well as in the mesenchyme (Fig. 3, some of the cells bearing L1 positive signals were labeled by blue arrows), the destination for the migration of the dedifferentiated stump cells towards the wound epidermis during limb regeneration. However, the signal was not evenly distributed throughout the mesenchymal area with some cells exhibiting a stronger signal than the others, reflecting the notion that the extent of global genomic DNA de-repression is likely to be heterogeneous in the blastema. In the regenerating epidermis, the basal keratinocytes and adjacent few layers of suprabasal keratinocytes seem to bear stronger ORF2 signal than other epidermal cells, which might be indicative of dedifferentiation in the epidermis. As a control, sense RNA probe for L1 ORF2 did not generate appreciable ISH positive signals in the limb blastema. Similarly, expression of the button-like transcription factor Sp9 involved in embryonic development of limb buds is re-activated in the basal layer of keratinocytes in the apical blastema epithelium, and is considered an important marker for dedifferentiation of the epidermis (Satoh et al. 2008a). Thus, the presence of L1 activity in the basal layers of keratinocytes in the regenerating limb blastema, as we demonstrated here, provides another line of evidence that the regenerating epithelium indeed undergoes some level of reprogramming during limb regeneration.
Fig. 3.
In situ hybridization of LINE-1 ORF-2 in 5 days post-amputation (pa) regenerating forelimb blastema. Sections were hybridized with either sense or anti-sense LINE-1 ORF2 RNA probe. The ORF2 sense RNA probe was used as control if the expression levels of the sense and the anti-sense transcripts were different. Staining (blue) was developed with 5-bromo-4-chloro-3-indolyl-phosphate (BCIP) and 4-nitroblue tetrazolium chloride (NBT) in alkaline phosphatase buffer after in situ hybridization (ISH) and anti-digoxygenin (DIG) alkaline phosphatase antibody incubation. Some of the cells bearing positive L1 signal were labeled by blue arrows. The scale bar for images C1 and C2 is 500 µm. The bar scale for images C1′, C1″, C2′ and C2″ is 200 µm. C1′, C1″ are the enlarged photos from different portions of the limb blastema presented in C1 similar to the relationship between C2′ and C2.
Identification of putative LINE-1 piRNAs in the axolotl limb regenerate
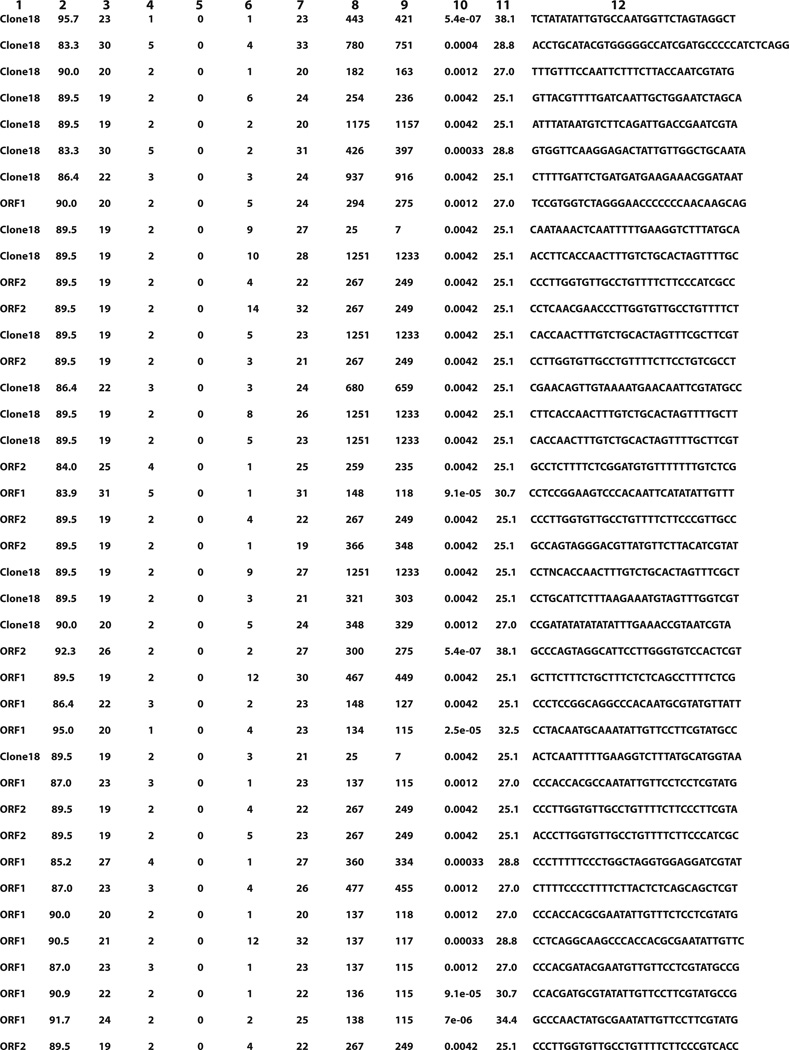
Our cDNA library sequencing of axolotl regenerating forelimbs revealed that Piwi-like 1 (PL1) and Piwi-like 2 (PL2) are also expressed/upregulated upon limb regeneration in addition to LINE-1 and other germline-specific genes. Since PL1 and PL2 are Argonaute family proteins known to process piRNA precursors to generate piRNAs, which are crucial for the silencing of activated TEs including L1 in germ cells (Juliano et al. 2000), we tried to identify potential L1 piRNA candidates from the dataset generated from our sequencing efforts of the small RNAs extracted from axolotl forelimb regenerates (Dataset S1), which might target L1 genomic DNA sequences corresponding to the partial ORF1 (Clone # 7–10) and ORF2 (Clone # 6–5) cDNA sequences as well as the protein-coding region within the genomic DNA sequence of one LINE-1 allele (Clone #18) we obtained from the axolotl genomic DNA library that we constructed (Fig. S1). Indeed, we were able to identify a group of putative axolotl L1 piRNAs (Fig. 4), which are derived from the anti-sense strand of the known L1 transcripts, which would be characteristic of the secondary piRNAs generated by the ping-pong amplification loop during piRNA biogenesis. The DNA sequence of the 5′ UTR of Clone #18 was not included in the analysis. The structure of the L1 transcript is unique in that the transcript is long, bicistronic and comprises both of the ORFs and the short (~63 bp) inter-ORF region that contains two stop codons and separates the two ORFs (Dombroski et al. 1991). Although the protein-coding region of Clone #18 contains premature stop codons, indicating that the Clone #18 may represent an inactive non-transposable L1 allele, the fact that there is a functional promoter upstream of Clone #18 ORF1 suggests that the allele is accessible to transcriptional machinery when circumstances allow.
Fig. 4.
A list of putative LINE-1 piRNA candidates that potentially target L1s. All of these putative L1 piRNAs are derived from the antisense strand of L1 transcripts. Field description for each of the 40 putative L1 piRNAs: 1, target label; 2, percent identity; 3, alignment length; 4, number of mismatches; 5, number of gap-opens; 6, one-based position of start in query; 7, one-based position of end in query; 8, one-based position of start in target; 9, one-based position of end in target; 10, E-value; 11, bit score; 12, sequence.
So far, among the intracellular small RNAs in vertebrates only siRNAs and piRNAs can be derived from the anti-sense strand of mRNA transcripts in vivo (Vagin et al. 2006; Faehnle & Joshua-Tor 2007). piRNAs usually lack conserved secondary structure motifs and are not necessarily identical to their targets (examples shown in Lee et al. (2011)). On the contrary, endo-siRNAs characteristically have well-defined structures and are faithfully identical to their targets, and also do not show an obvious nucleotide preference at their 5′ end (Czech et al. 2008; Kawamura et al. 2008; Okamura & Lai 2008). Since the L1-related anti-sense strand-derived small RNAs we present here are not identical to their targets (the partial ORF1 (Clone # 7–10) and ORF2 (Clone # 6–5) as well as the proteincoding region of Clone #18 in this case), these small RNAs may fall into the category of piRNAs. Due to L1 polymorphism, some of the L1 small RNAs we identified here that contain a few mismatching nucleotides may be completely identical to transcripts generated from other L1 alleles, and thus could act as siRNAs, but it is still possible that at least some of the L1 small RNAs are putative piRNA candidates. Nevertheless, similar to piRNAs, in mouse oocytes the endo-siRNA pathway also plays important roles in TE silencing (Murchison et al. 2007; Tam et al. 2008; Watanabe et al. 2008). The emergence of the putative LINE-1 piRNAs and/or siRNAs in the axolotl limb regenerate reflects the initiation of a negative feedback loop to suppress reactivated L1s, a phenomenon usually observed in germ cells. Moreover, considering the polymorphic nature of L1, it is likely that there are many more potential L1 piRNAs than we found here.
In order to confirm that the putative LINE-1 piRNAs are indeed derived from the genomic DNA fragments within the LINE-1 loci, we performed genomic DNA PCR in the presence of one primer (P25R) derived from one of the putative LINE-1 piRNAs listed in Figure 4, which is highly similar (with an E-value of 5.4e-07) to a fragment residing in the anti-sense strand of the partial LINE-1 ORF-2 cDNA (Clone #6–5, located between the 275th and 300th base) we identified earlier (Fig. S1) and a second primer, which was either a scrambled primer or a LINE-1-specific primer (P25F/26) in the proximity (~250 bases) upstream of the putative piRNA template in ORF-2 Clone #6–5. Indeed, we found that the PCR products primed by P25R and P25F/26 were LINE-1 homologues when we cloned the PCR fragments and sequenced them. More interestingly, the three LINE-1 clones we examined all show slight differences in their DNA sequences among themselves, although they are largely similar to each other reminiscent of the known genomic polymorphic nature of LINE-1 elements (Fig. S2). No specific PCR product was detected with the scrambled DNA primers consistent with the putative LINE-1 piRNA being derived from LINE-1 genomic loci.
Discussion
Studies on the biological significance of retrotransposition of LINE-1 have suggested that the L1 elements can contribute functionally as genetic modifiers in organisms. For example, in transgenic mice carrying an engineered human L1 element, activation of L1 in neuronal progenitors alters the expression of neuronal genes that can influence neuronal cell fate and result in neuronal somatic mosaicism (Muotri et al. 2005). In ES cells, L1 expression may be involved in the establishment of X inactivation (Chow et al. 2010). More interestingly, TEs have been shown to play a vital role in the maintenance of genomic integrity; for instance, the telomeric ends of linear chromosomes are maintained by successive transposition of two non-LTR retrotransposons, HeT-A and TART in Drososphila (Pardue et al. 2005). In our case, it is not clear so far whether L1 re-activation during axolotl limb regeneration plays beneficial roles during the regenerative process. However, it is likely that deregulation of L1 expression may emerge in parallel with L1 re-activation and transposition during axolotl limb regeneration, which could lead to an aberrant regeneration, and even ultimate failure after multiple rounds of regeneration. Indeed, the mobilization of L1 elements is often considered deleterious because the process often introduces harmful genomic structural rearrangements, such as deletions, duplications and inversions (reviewed in Saito & Siomi (2010)). In the fission yeast Schizosaccharomyces pombe, aberrant accumulation of complementary transcripts from centromeric heterochromatic repeats has been reported to be responsible for the impairment of centromere function (Volpe et al. 2002). In mice, loss of any of the three PIWI proteins, Miwi, Mili and Miwi2 leads to depression of TEs, spermatogenic arrest and male sterility (Kuramochi-Miyagawa et al. 2001, 2004; Aravin et al. 2007; Carmell et al. 2007). In zebrafish, the loss of Zili, a PIWI protein results in TE re-activation in the developing gonads and consequently the early failure of germline development, whereas the absence of Ziwi, another PIWI protein triggers apoptosis later in the germline, at the premeiotic stage (Houwing et al. 2007, 2008).
Although L1 transcription and/or retrotransposition have been demonstrated to occur in a variety somatic cell types (reviewed in Babushok & Kazazian (2007)), since only newly synthesized L1 alleles in primordial germ cells, germline, or the early embryo can be integrated into the germline lineage and inherited by future generations, the potential hazard directly or indirectly (e.g. L1-mediated integrations of Alu and SINE/VNTR/Alu (SVA) elements) (reviewed in Ostertag & Kazazian (2006)) induced by L1 insertion during axolotl limb regeneration is very likely to be restrained in the limb regenerate and will not be passed on to their descendants. In the future, it would be of interest to test whether a higher frequency of aberrant limb regeneration would be observed in animals that receive additional limb amputation surgeries due to the higher frequency of potentially deleterious LINE-1 integration events that occur during the process of axolotl limb regeneration. Indeed, a previous study by Dearlove & Dresden (1976) did show increasing incidence of limb abnormalities with progressive amputations. One could speculate that a higher level of L1 retrotransposition activities and the resultant increase in L1 genomic content as a consequence of progressive limb amputation might contribute to the increased occurrence of hazardous genomic integration of newly synthesized L1s, and provide a potential mechanism for the frequent occurrence of abnormal limb regeneration observed by Dearlove and Dresden.
Although we first observed the induction of LINE1 expression and retrotransposition, subsequent work showed that in addition to LINE-1, other transposable elements (TE) appear to be induced during axolotl limb regeneration. Through transcriptome Roche 454 sequencing analysis (Monaghan et al. 2009) we have observed that besides LINE1 elements a variety of other putative transposable elements appear to be expressed, although the kinetics are not always identical to those of LINE1 (Table 1, Fig. S3). Among the sequences identified, we found six retrotransposon like sequences and three retrovirus-like sequences. In addition, sequences similar to other transposable elements including DNA transposons have also been identified in the same axolotl limb regenerate transcriptome Roche 454 sequencing, which include those homologous to the Transib family similar to the RAG1 recombinase, the tigger and mariner elements, pogo as well as other retrotransposons and additional sequences that belong to the LINE1 family (Table 2, Fig. S3). These observations support the notion that regeneration does not specifically derepress LINE elements but initiates cascades present in some cells in which epigenetic silencing is reduced and allow for the general derepression of transposable elements. This environment of general derepression of transposable elements is reminiscent of the germline in most organisms, even more so due to the presence of piRNA-like sequences and the expression of germline-specific cDNAs. piRNAs might be required as in the case of the germline to moderate the amount of genetic damage due to the increase of DNA insertional and excisional events with transposon mobilization. Conclusively, these observations point towards the possible existence of a germline-like state in an undefined cell population within the blastema of the regenerating limb that might be functionally important to execute the regeneration program.
Supplementary Material
Acknowledgments
This work was supported in part by an Innovation Grant from the Salk Institute, by USPHS grants CA14195 and CA80100 from the NCI to T.H., and by the National Science Foundation through its support of the Ambystoma Genetic Stock Center at the University of Kentucky, Lexington. T.H. is a Frank and Else Schilling American Cancer Society Professor.
Footnotes
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Fig. S1. The sequences of the LINE-1 partial ORF1 (Clone # 7–10) and ORF2 (Clone # 6–5) cDNAs as well as the genomic DNA sequence of one LINE-1 allele (Clone #18). These DNA sequences were used for the screening of potential LINE-1 piRNAs (Fig. 4).
Fig. S2. Genomic DNA polymerase chain reaction (PCR) to obtain the flanking DNA sequences of a putative LINE-1 ORF-2 piRNA listed in Fig. 4. The sequences of the primers used and the 3 clones isolated from the PCR products are shown here. The alignment of these 3 ORF-2 homologous clones and the corresponding DNA fragment in LINE-1 ORF-2 cDNA (Clone #6-5, Supplemental Fig. S1) revealed the polymorphic nature of genomic LINE-1 loci.
Fig. S3. The information of Table 1 was extracted from the work by Monaghan JR, et al. (2009). Microarray and cDNA sequence analysis of transcription during nerve-dependent limb regeneration. BMC Biol 7:1. A list of differentially expressed transposon elements (TEs) other than LINE-1. These contigs have been identified by Roche 454 cDNA sequencing in innervated (NR) and denervated (DL) forelimbs after 0 (C0), 5 (NR5 or DL5), and 14 (NR14 or DL14) days post amputation. Listed in Table 2 are the sequences similar to other transposable elements that have also been identified in the same axolotl limb regenerate transcriptome Roche 454 sequencing.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- Babushok DV, Kazazian HH., Jr Progress in understanding the biology of the human mutagen LINE-1. Hum. Mutat. 2007;28:527–539. doi: 10.1002/humu.20486. [DOI] [PubMed] [Google Scholar]
- Borgens RB. Mice regrow the tips of their foretoes. Science. 1982;217:747–750. doi: 10.1126/science.7100922. [DOI] [PubMed] [Google Scholar]
- Branciforte D, Martin SL. Developmental and cell type specificity of LINE-1 expression in mouse testis: implications for transposition. Mol. Cell. Biol. 1994;14:2584–2592. doi: 10.1128/mcb.14.4.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockes JP, Kumar A. Appendage regeneration in adult vertebrates and implications for regenerative medicine. Science. 2005;310:1919–1923. doi: 10.1126/science.1115200. [DOI] [PubMed] [Google Scholar]
- Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH., Jr Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl Acad. Sci. USA. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson MR, Bryant SV, Gardiner DM. Expression of Msx-2 during development, regeneration, and wound healing in axolotl limbs. J. Exp. Zool. 1998;282:715–723. doi: 10.1002/(sici)1097-010x(19981215)282:6<715::aid-jez7>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc’his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Chen JM, Cooper DN, Ferec C, Kehrer-Sawatzki H, Patrinos GP. Genomic rearrangements in inherited disease and cancer. Semin. Cancer Biol. 2000;20:222–233. doi: 10.1016/j.semcancer.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Chow JC, Ciaudo C, Fazzari MJ, Mise N, Servant N, Glass JL, Attreed M, Avner P, Wutz A, Barillot E, Greally JM, Voinnet O, Heard E. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell. 2010;141:956–969. doi: 10.1016/j.cell.2010.04.042. [DOI] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, Hannon GJ, Brennecke J. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dearlove GE, Dresden MH. Regenerative abnormalities in Notophthalmus viridescens induced by repeated amputations. J. Exp. Zool. 1976;196:251–262. doi: 10.1002/jez.1401960212. [DOI] [PubMed] [Google Scholar]
- Dombroski BA, Mathias SL, Nanthakumar E, Scott AF, Kazazian HH., Jr Isolation of an active human transposable element. Science. 1991;254:1805–1808. doi: 10.1126/science.1662412. [DOI] [PubMed] [Google Scholar]
- Echeverri K, Clarke JD, Tanaka EM. In vivo imaging indicates muscle fiber dedifferentiation is a major contributor to the regenerating tail blastema. Dev. Biol. 2001;236:151–164. doi: 10.1006/dbio.2001.0312. [DOI] [PubMed] [Google Scholar]
- Eickbush TH, Malik HS. Origins and evolution of retrotransposons. In: Craig NL, et al., editors. Mobile DNA II. Washington, DC: ASM Press; 2002. pp. 1111–1144. [Google Scholar]
- Ergun S, Buschmann C, Heukeshoven J, Dammann K, Schnieders F, Lauke H, Chalajour F, Kilic N, Stratling WH, Schumann GG. Cell type-specific expression of LINE-1 open reading frames 1 and 2 in fetal and adult human tissues. J. Biol. Chem. 2004;279:27753–27763. doi: 10.1074/jbc.M312985200. [DOI] [PubMed] [Google Scholar]
- Faehnle CR, Joshua-Tor L. Argonautes confront new small RNAs. Curr. Opin. Chem. Biol. 2007;11:569–577. doi: 10.1016/j.cbpa.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost RJ, Hamra FK, Richardson JA, Qi X, Bassel-Duby R, Olson EN. MOV10L1 is necessary for protection of spermatocytes against retrotransposons by Piwi-interacting RNAs. Proc. Natl Acad. Sci. USA. 2000;107:11847–11852. doi: 10.1073/pnas.1007158107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, Scott G, Steffen D, Worley KC, Burch PE, Okwuonu G, Hines S, Lewis L, De-Ramo C, Delgado O, Dugan-Rocha S, Miner G, Morgan M, Hawes A, Gill R, Celera, Holt RA, Adams MD, Amanatides PG, Baden-Tillson H, Barnstead M, Chin S, Evans CA, Ferriera S, Fosler C, Glodek A, Gu Z, Jennings D, Kraft CL, Nguyen T, Pfannkoch CM, Sitter C, Sutton GG, Venter JC, Woodage T, Smith D, Lee HM, Gustafson E, Cahill P, Kana A, Doucette-Stamm L, Weinstock K, Fechtel K, Weiss RB, Dunn DM, Green ED, Blakesley RW, Bouffard GG, De Jong PJ, Osoegawa K, Zhu B, Marra M, Schein J, Bosdet I, Fjell C, Jones S, Krzywinski M, Mathewson C, Siddiqui A, Wye N, McPherson J, Zhao S, Fraser CM, Shetty J, Shatsman S, Geer K, Chen Y, Abramzon S, Nierman WC, Havlak PH, Chen R, Durbin KJ, Egan A, Ren Y, Song XZ, Li B, Liu Y, Qin X, Cawley S, Cooney AJ, D’Souza LM, Martin K, Wu JQ, Gonzalez-Garay ML, Jackson AR, Kalafus KJ, McLeod MP, Milosavljevic A, Virk D, Volkov A, Wheeler DA, Zhang Z, Bailey JA, Eichler EE, Tuzun E, Birney E, Mongin E, Ureta-Vidal A, Woodwark C, Zdobnov E, Bork P, Suyama M, Torrents D, Alexandersson M, Trask BJ, Young JM, Huang H, Wang H, Xing H, Daniels S, Gietzen D, Schmidt J, Stevens K, Vitt U, Wingrove J, Camara F, Mar Alba M, Abril JF, Guigo R, Smit A, Dubchak I, Rubin EM, Couronne O, Poliakov A, Hubner N, Ganten D, Goesele C, Hummel O, Kreitler T, Lee YA, Monti J, Schulz H, Zimdahl H, Himmelbauer H, Lehrach H, Jacob HJ, Bromberg S, Gullings-Handley J, Jensen-Seaman MI, Kwitek AE, Lazar J, Pasko D, Tonellato PJ, Twigger S, Ponting CP, Duarte JM, Rice S, Goodstadt L, Beatson SA, Emes RD, Winter EE, Webber C, Brandt P, Nyakatura G, Adetobi M, Chiaromonte F, Elnitski L, Eswara P, Hardison RC, Hou M, Kolbe D, Makova K, Miller W, Nekrutenko A, Riemer C, Schwartz S, Taylor J, Yang S, Zhang Y, Lindpaintner K, Andrews TD, Caccamo M, Clamp M, Clarke L, Curwen V, Durbin R, Eyras E, Searle SM, Cooper GM, Batzoglou S, Brudno M, Sidow A, Stone EA, Payseur BA, Bourque G, Lopez-Otin C, Puente XS, Chakrabarti K, Chatterji S, Dewey C, Pachter L, Bray N, Yap VB, Caspi A, Tesler G, Pevzner PA, Haussler D, Roskin KM, Baertsch R, Clawson H, Furey TS, Hinrichs AS, Karolchik D, Kent WJ, Rosenbloom KR, Trumbower H, Weirauch M, Cooper DN, Stenson PD, Ma B, Brent M, Arumugam M, Shteynberg D, Copley RR, Taylor MS, Riethman H, Mudunuri U, Peterson J, Guyer M, Felsenfeld A, Old S, Mockrin S, Collins F. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- Han JS, Szak ST, Boeke JD. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429:268–274. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- Houwing S, Berezikov E, Ketting RF. Zili is required for germ cell differentiation and meiosis in zebrafish. EMBO J. 2008;27:2702–2711. doi: 10.1038/emboj.2008.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, Plasterk RH, Hannon GJ, Draper BW, Ketting RF. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Juliano C, Wang J, Lin H. Uniting germline and stem cells: the function of Piwi proteins and the piRNA pathway in diverse organisms. Annu. Rev. Genet. 2000;45:447–469. doi: 10.1146/annurev-genet-110410-132541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- Koshiba K, Kuroiwa A, Yamamoto H, Tamura K, Ide H. Expression of Msx genes in regenerating and developing limbs of axolotl. J. Exp. Zool. 1998;282:703–714. doi: 10.1002/(sici)1097-010x(19981215)282:6<703::aid-jez6>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Kragl M, Knapp D, Nacu E, Khattak S, Maden M, Epperlein HH, Tanaka EM. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 2009;460:60–65. doi: 10.1038/nature08152. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, Lin H, Matsuda Y, Nakano T. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Yomogida K, Kuroiwa A, Tadokoro Y, Fujita Y, Sato M, Matsuda Y, Nakano T. Two mouse piwi-related genes: miwi and mili. Mech. Dev. 2001;108:121–133. doi: 10.1016/s0925-4773(01)00499-3. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, Hata K, Li E, Matsuda Y, Kimura T, Okabe M, Sakaki Y, Sasaki H, Nakano T. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lee EJ, Banerjee S, Zhou H, Jammalamadaka A, Arcila M, Manjunath BS, Kosik KS. Identification of piRNAs in the central nervous system. RNA. 2011;17:1090–1099. doi: 10.1261/rna.2565011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchinetti E, Feng J, Silva R, Tolstonog GV, Schaub MC, Schumann GG, Zaugg M. Inhibition of LINE-1 expression in the heart decreases ischemic damage by activation of Akt/PKB signaling. Physiol. Genomics. 2006;25:314–324. doi: 10.1152/physiolgenomics.00251.2005. [DOI] [PubMed] [Google Scholar]
- Maki N, Suetsugu-Maki R, Sano S, Nakamura K, Nishimura O, Tarui H, Del Rio-Tsonis K, Ohsumi K, Agata K, Tsonis PA. Oocyte-type linker histone B4 is required for transdifferentiation of somatic cells in vivo. FASEB J. 2010;24:3462–3467. doi: 10.1096/fj.10-159285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki N, Suetsugu-Maki R, Tarui H, Agata K, Del Rio-Tsonis K, Tsonis PA. Expression of stem cell pluripotency factors during regeneration in newts. Dev. Dyn. 2009;238:1613–1616. doi: 10.1002/dvdy.21959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan JR, Epp LG, Putta S, Page RB, Walker JA, Beachy CK, Zhu W, Pao GM, Verma IM, Hunter T, Bryant SV, Gardiner DM, Harkins TT, Voss SR. Microarray and cDNA sequence analysis of transcription during nerve-dependent limb regeneration. BMC Biol. 2009;7:1. doi: 10.1186/1741-7007-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya-Durango DE, Ramos KS. L1 retrotransposon and retinoblastoma: molecular linkages between epigenetics and cancer. Curr. Mol. Med. 2000;10:511–521. doi: 10.2174/156652410791608234. [DOI] [PubMed] [Google Scholar]
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- Morrish TA, Gilbert N, Myers JS, Vincent BJ, Stamato TD, Taccioli GE, Batzer MA, Moran JV. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat. Genet. 2002;31:159–165. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- Muneoka K, Fox WF, Bryant SV. Cellular contribution from dermis and cartilage to the regenerating limb blastema in axolotls. Dev. Biol. 1986;116:256–260. doi: 10.1016/0012-1606(86)90062-x. [DOI] [PubMed] [Google Scholar]
- Muneoka K, Han M, Gardiner DM. Regrowing human limbs. Sci. Am. 2008;298:56–63. doi: 10.1038/scientificamerican0408-56. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- Murchison EP, Stein P, Xuan Z, Pan H, Zhang MQ, Schultz RM, Hannon GJ. Critical roles for Dicer in the female germline. Genes Dev. 2007;21:682–693. doi: 10.1101/gad.1521307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Lai EC. Endogenous small interfering RNAs in animals. Nat. Rev. Mol. Cell Biol. 2008;9:673–678. doi: 10.1038/nrm2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostertag EM, Kazazian HH., Jr . Retrotransposition and Human Disorders. Chichester: John Wiley & Sons Ltd.; 2006. [Google Scholar]
- Pardue ML, Rashkova S, Casacuberta E, DeBaryshe PG, George JA, Traverse KL. Two retrotransposons maintain telomeres in Drosophila. Chromosome Res. 2005;13:443–453. doi: 10.1007/s10577-005-0993-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reginelli AD, Wang YQ, Sassoon D, Muneoka K. Digit tip regeneration correlates with regions of Msx1 (Hox 7) expression in fetal and newborn mice. Development. 1995;121:1065–1076. doi: 10.1242/dev.121.4.1065. [DOI] [PubMed] [Google Scholar]
- Saito K, Siomi MC. Small RNA-mediated quiescence of transposable elements in animals. Dev. Cell. 2010;19:687–697. doi: 10.1016/j.devcel.2010.10.011. [DOI] [PubMed] [Google Scholar]
- Satoh A, Gardiner DM, Bryant SV, Endo T. Nerve-induced ectopic limb blastemas in the Axolotl are equivalent to amputation-induced blastemas. Dev. Biol. 2007;312:231–244. doi: 10.1016/j.ydbio.2007.09.021. [DOI] [PubMed] [Google Scholar]
- Satoh A, Graham GM, Bryant SV, Gardiner DM. Neurotrophic regulation of epidermal dedifferentiation during wound healing and limb regeneration in the axolotl (Ambystoma mexicanum) Dev. Biol. 2008a;319:321–335. doi: 10.1016/j.ydbio.2008.04.030. [DOI] [PubMed] [Google Scholar]
- Satoh A, Graham GM, Bryant SV, Gardiner DM. Neurotrophic regulation of epidermal dedifferentiation during wound healing and limb regeneration in the axolotl (Ambystoma mexicanum) Dev. Biol. 2008b;319:321–335. doi: 10.1016/j.ydbio.2008.04.030. [DOI] [PubMed] [Google Scholar]
- Shoji M, Tanaka T, Hosokawa M, Reuter M, Stark A, Kato Y, Kondoh G, Okawa K, Chujo T, Suzuki T, Hata K, Martin SL, Noce T, Kuramochi-Miyagawa S, Nakano T, Sasaki H, Pillai RS, Nakatsuji N, Chuma S. The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev. Cell. 2009;17:775–787. doi: 10.1016/j.devcel.2009.10.012. [DOI] [PubMed] [Google Scholar]
- Sinibaldi-Vallebona P, Lavia P, Garaci E, Spadafora C. A role for endogenous reverse transcriptase in tumorigenesis and as a target in differentiating cancer therapy. Genes Chromosom. Cancer. 2006;45:1–10. doi: 10.1002/gcc.20266. [DOI] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- Stoick-Cooper CL, Moon RT, Weidinger G. Advances in signaling in vertebrate regeneration as a prelude to regenerative medicine. Genes Dev. 2007;21:1292–1315. doi: 10.1101/gad.1540507. [DOI] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, Hannon GJ. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka EM. Regeneration: if they can do it, why can’t we? Cell. 2003;113:559–562. doi: 10.1016/s0092-8674(03)00395-7. [DOI] [PubMed] [Google Scholar]
- Trelogan SA, Martin SL. Tightly regulated, developmentally specific expression of the first open reading frame from LINE-1 during mouse embryogenesis. Proc. Natl Acad. Sci. USA. 1995;92:1520–1524. doi: 10.1073/pnas.92.5.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, Surani MA, Sakaki Y, Sasaki H. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, Cawley S, Chiaromonte F, Chinwalla AT, Church DM, Clamp M, Clee C, Collins FS, Cook LL, Copley RR, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty KD, Deri J, Dermitzakis ET, Dewey C, Dickens NJ, Diekhans M, Dodge S, Dubchak I, Dunn DM, Eddy SR, Elnitski L, Emes RD, Eswara P, Eyras E, Felsenfeld A, Fewell GA, Flicek P, Foley K, Frankel WN, Fulton LA, Fulton RS, Furey TS, Gage D, Gibbs RA, Glusman G, Gnerre S, Goldman N, Goodstadt L, Grafham D, Graves TA, Green ED, Gregory S, Guigo R, Guyer M, Hardison RC, Haussler D, Hayashizaki Y, Hillier LW, Hinrichs A, Hlavina W, Holzer T, Hsu F, Hua A, Hubbard T, Hunt A, Jackson I, Jaffe DB, Johnson LS, Jones M, Jones TA, Joy A, Kamal M, Karlsson EK, Karolchik D, Kasprzyk A, Kawai J, Keibler E, Kells C, Kent WJ, Kirby A, Kolbe DL, Korf I, Kucherlapati RS, Kulbokas EJ, Kulp D, Landers T, Leger JP, Leonard S, Letunic I, Levine R, Li J, Li M, Lloyd C, Lucas S, Ma B, Maglott DR, Mardis ER, Matthews L, Mauceli E, Mayer JH, McCarthy M, McCombie WR, McLaren S, McLay K, McPherson JD, Meldrim J, Meredith B, Mesirov JP, Miller W, Miner TL, Mongin E, Montgomery KT, Morgan M, Mott R, Mullikin JC, Muzny DM, Nash WE, Nelson JO, Nhan MN, Nicol R, Ning Z, Nusbaum C, O’Connor MJ, Okazaki Y, Oliver K, Overton-Larty E, Pachter L, Parra G, Pepin KH, Peterson J, Pevzner P, Plumb R, Pohl CS, Poliakov A, Ponce TC, Ponting CP, Potter S, Quail M, Reymond A, Roe BA, Roskin KM, Rubin EM, Rust AG, Santos R, Sapojnikov V, Schultz B, Schultz J, Schwartz MS, Schwartz S, Scott C, Seaman S, Searle S, Sharpe T, Sheridan A, Shownkeen R, Sims S, Singer JB, Slater G, Smit A, Smith DR, Spencer B, Stabenau A, Stange-Thomann N, Sugnet C, Suyama M, Tesler G, Thompson J, Torrents D, Trevaskis E, Tromp J, Ucla C, Ureta-Vidal A, Vinson JP, Von Niederhausern AC, Wade CM, Wall M, Weber RJ, Weiss RB, Wendl MC, West AP, Wetterstrand K, Wheeler R, Whelan S, Wierzbowski J, Willey D, Williams S, Wilson RK, Winter E, Worley KC, Wyman D, Yang S, Yang SP, Zdobnov EM, Zody MC, Lander ES. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Xu M, You Y, Hunsicker P, Hori T, Small C, Griswold MD, Hecht NB. Mice deficient for a small cluster of Piwi-interacting RNAs implicate Piwi-interacting RNAs in transposon control. Biol. Reprod. 2008;79:51–57. doi: 10.1095/biolreprod.108.068072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.