Abstract
Aneuploidy, an abnormal number of chromosomes, is a widespread phenomenon found in unicellulars such as yeast, as well as in plants and in mammalians, especially in cancer. Aneuploidy is a genome-scale aberration that imposes a severe burden on the cell, yet under stressful conditions specific aneuploidies confer a selective advantage. This dual nature of aneuploidy raises the question of whether it can serve as a stable and sustainable evolutionary adaptation. To clarify this, we conducted a set of laboratory evolution experiments in yeast and followed the long-term dynamics of aneuploidy under diverse conditions. Here we show that chromosomal duplications are first acquired as a crude solution to stress, yet only as transient solutions that are eliminated and replaced by more efficient solutions obtained at the individual gene level. These transient dynamics of aneuploidy were repeatedly observed in our laboratory evolution experiments; chromosomal duplications gained under stress were eliminated not only when the stress was relieved, but even if it persisted. Furthermore, when stress was applied gradually rather than abruptly, alternative solutions appear to have emerged, but not aneuploidy. Our findings indicate that chromosomal duplication is a first evolutionary line of defense, that retains survivability under strong and abrupt selective pressures, yet it merely serves as a “quick fix,” whereas more refined and sustainable solutions take over. Thus, in the perspective of genome evolution trajectory, aneuploidy is a useful yet short-lived intermediate that facilitates further adaptation.
Keywords: evolutionary dynamics, environmental stress, heat tolerance, pH tolerance
Adaptation to stressful conditions often requires modified expression of certain genes that can generally be obtained by genome sequence changes. In addition, structural rearrangements of the genome, and in particular chromosomal duplications, that lead to aneuploidy, may offer a simple means to boost expression level (1, 2). Indeed, cancer cells often exercise massive genomic duplications, particularly of regions that harbor growth-promoting genes (3). Unicellular organisms, too, can duplicate chromosomes that contain genes needed at a given condition (4–9). The high prevalence of chromosomal duplications, especially under stress (10), accounts for the frequent acquisition of aneuploidies on a short evolutionary timescale (11). Chromosomal duplication may indeed offer the advantage of simultaneous elevation of a large set of genes, some of which may be beneficial under a particular selective pressure. Whole genome duplications, too, can offer selective advantage under specific conditions (12), yet genome analysis has suggested that they also survive only under specific conditions (13). Indeed, whole genome and chromosomal duplication constitute crude solutions with significant overheads on the cell that are in part associated with increased copies of DNA, RNA, and proteins (1, 2). Duplication of particular chromosomes (i.e., aneuploidy) creates, in addition, a stoichiometric imbalance between gene products (14, 15) and promotes further genome destabilizing events (16, 17). Thus, it appears that aneuploidy is concurrently advantageous and highly costly (18, 19). Therefore, it is not clear under what conditions and to what extent organisms will adopt this solution. Here we offer a resolution of the dual nature of aneuploidy, in the form of dynamic and prolonged laboratory evolution experiments in yeast. We show that aneuploidy that is rapidly gained under stress is a transient solution that is replaced by focal, refined, and sustainable solutions that require more time to evolve.
Results
Acquisition and Subsequent Elimination of Chromosome III Trisomy During Adaptation to Heat.
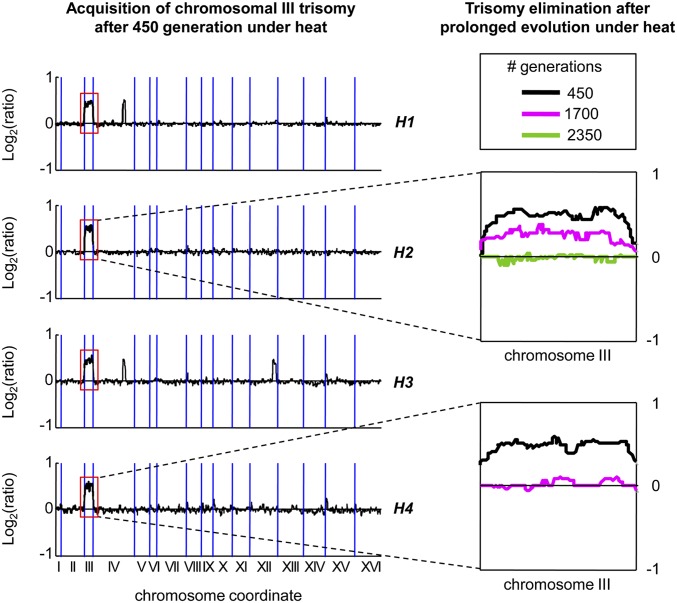
Toward evolving stress-tolerant yeast strains, we applied the well-established methodology of laboratory evolution through serial dilution (20). In this procedure, populations of yeast cells are grown under a certain condition and are diluted daily into fresh medium, still under the same condition. Our laboratory evolution experiments all together create a highly branched evolutionary scheme (Fig. 1). We started with diploid Saccharomyces cerevisiae cells that were grown in four independent repetitions in rich medium under a constant heat stress of 39 °C. After an evolution period of 450 generations, all populations were examined, and duplication of chromosome III (trisomy) was detected in all four repetitions (Fig. 2). Two additional segmental duplications occurred in two repetitions on chromosomes IV and XII. We focused on the two populations in which only chromosome III was duplicated (Fig. S1), as they provide a useful model for single acquired aneuploidy, and further evolved them under the same heat stress. Strikingly, we observed elimination of chromosome III trisomy within 2,350 generations (Fig. 2). We then turned to carefully study the dynamics of evolutionary gain and loss of aneuploidies under various growth conditions in which we vary temperature, nutrient availability, and pH.
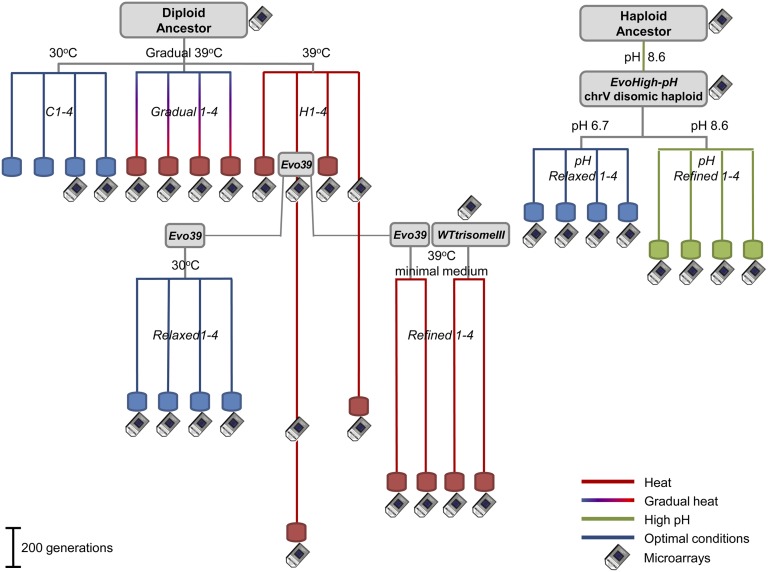
Fig. 1.
Laboratory evolution (Lab-evolution) tree describing the experimental outline. Each evolution experiment starts with an ancestor strain (gray box) that was subjected to certain growth conditions: high temperature, 39 °C (red); permissive temperature, 30 °C (blue); gradually increasing temperature, from 30 °C to 39 °C (gradient line); and high pH 8.6 (green). Parallel lines splitting from the same branch represent independent repetitions and their length is in scale with the number of generations under the specified condition. Evo39 strain was taken after 450 generations on high temperature (39 °C) as an ancestor for another two evolutionary branches (Refined 1–4 and Relaxed 1–4). Microarray icons represent points during the evolution tree in which such measurement was performed.
Fig. 2.
Aneuploidy appears and subsequently was eliminated during evolutionary adaptation to heat. Four independent repetitions (H1–4) that evolved for 450 generations in rich medium and heat (39 °C) show chromosomal duplications (black lines). Notably, duplication of chromosome III occurred in all four repetitions. H2 and H4, which carry no large-scale duplication other than chromosome III trisomy, were further evolved under the same conditions and after 1,700 generations (magenta lines) and 2,350 generations (green line), the trisomy was eliminated. All lines represent log2 intensity ratios of mRNA abundance calculated by a sliding window of heat-evolved strain over a diploid wild type, aligned according to chromosomal order where blue vertical lines differentiate between chromosomes.
Causal Link Between Chromosome III Copy Number and Heat Tolerance.
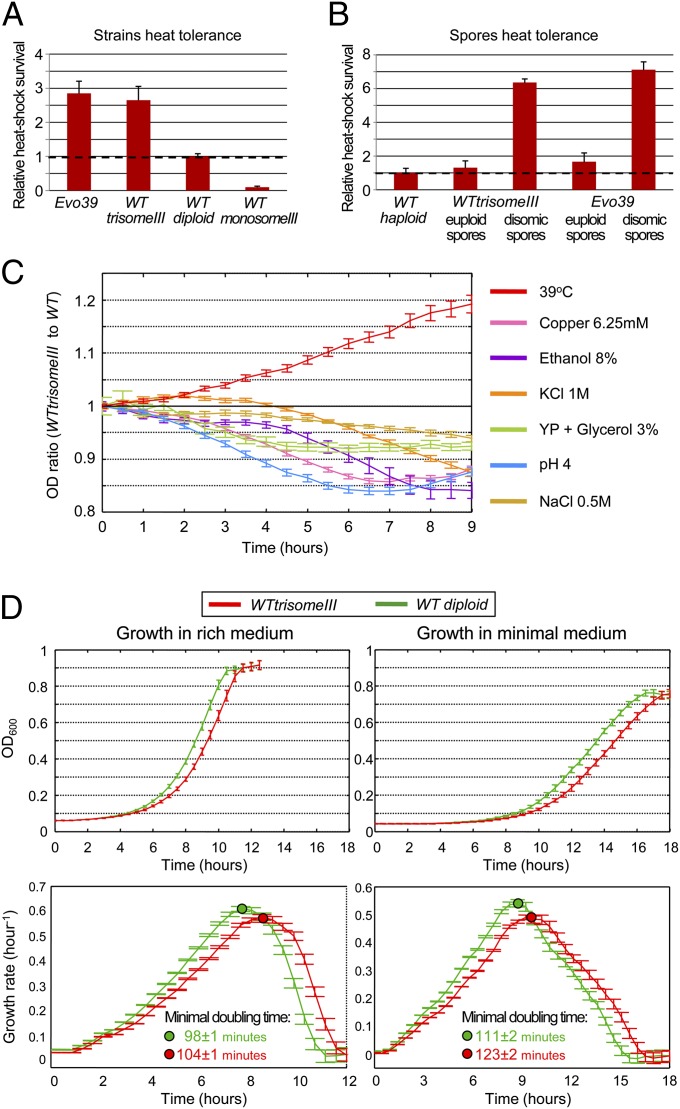
We began by determining whether the trisomy was selected for its direct contribution to heat tolerance or whether it was merely hitchhiking on another beneficial mutation and thus was lost further along the evolution under heat. To that end, strains that carry chromosome III aneuploidies (without evolution under heat) were obtained and measured for their heat tolerance (Materials and Methods). One strain is a diploid with a trisomy of chromosome III, termed here WTtrisomeIII and the other is a diploid with a monosomy of chromosome III, termed WTmonosomeIII. Interestingly, WTtrisomeIII showed elevated heat tolerance at a level that is similar to that of the trisomic strains that evolved under heat. On the other hand, WTmonosomeIII exhibited lower heat tolerance than the diploid wild type (Fig. 3A). These findings show a clear quantitative correspondence between chromosome III copy number and heat tolerance. To confirm that the extra chromosome is the predominant genetic change underlying the heat tolerance, tetrad analysis was performed both on a strain that evolved at 39 °C for 450 generations and became trisomic (H2 after 450 generations, abbreviated as evo39) and on WTtrisomeIII. These two trisomic strains were subjected to meiosis and the heat tolerance of the haploid progeny (spores) was measured. The results showed that all euploid spores (normal number of chromosomes) have heat tolerance similar to a haploid, whereas all disomic spores, which carry two copies of chromosome III, have a marked increase in heat tolerance (Fig. 3B). We further examined whether chromosome III trisomy confers advantage under other stress conditions by measuring the tolerance of WTtrisomeIII to a battery of other stresses. We found that duplication of chromosome III decreased the tolerance to all of the other stresses tested (Fig. 3C). Therefore, increased heat tolerance cannot be attributed to a general stress tolerance conferred by chromosome III trisomy. Together, these results establish a causal link between the relative number of chromosome III copies and heat tolerance and may explain why this chromosomal duplication was repeatedly fixated in all laboratory evolution experimental lines under heat.
Fig. 3.
An extra copy of chromosome III is beneficial under heat, yet it is maladaptive under other conditions. (A) Heat-shock tolerance rates are proportional to the copy number of chromosome III. Heat-shock survival fold change of chromosome III aneuploidic strains compared with a diploid wild type is shown (dashed line). (B) The extra copy of chromosome III is the predominant genetic trait responsible for the increased heat tolerance. Heat-shock survival fold change of spores from WTtrisomeIII and evo39, compared with a haploid wild type (dashed line), presented separately for euploid spores and for chromosome III disomic spores (P value < 8 × 10−5; for spore karyotype, see Fig. S7). (C) The growth advantage conferred by chromosome III trisomy under heat (red line) cannot be attributed to a general stress tolerance. Colored lines represent OD ratios of WTtrisomeIII over a diploid wild type during continuous growth under various stresses (Materials and Methods). (D) The cost of chromosome III trisomy at permissive temperature (30 °C) is increased on minimal medium compared with rich medium. Growth curve measurements of WTtrisomeIII (red) and of a diploid wild type (green) are shown in OD values over time during continuous growth at 30 °C (Upper), in rich medium (Left) and in minimal medium (Right). (Lower) Growth rate analyses, derived from the OD values, and the differences in minimal doubling time between WTtrisomeIII and WT. Data are presented as mean and SEM.
Cost Associated with Chromosome III Trisomy.
An additional copy of chromosome III thus clearly contributes to heat tolerance, yet its elimination later on raises the hypothesis that the substantial burden associated with chromosomal duplications (1) prevents the trisomy from serving as a sustainable solution to the stress. Therefore, a next goal was to assess the costs associated with the chromosome III trisomy and to characterize the role of these costs in the trisomy elimination. We reasoned that such costs can be measured when neutralizing the benefit, i.e., when heat is not applied. For that reason, the cost of chromosome III trisomy was measured in terms of growth defect by comparing WTtrisomeIII to a diploid wild type, at the permissive temperature (30 °C). A considerable cost of the trisomy was detected in rich medium and a further cost increase was detected in minimal medium (Fig. 3D and Fig. S2).
Optimized Adaptations Replace the Aneuploidy-Based Solution.
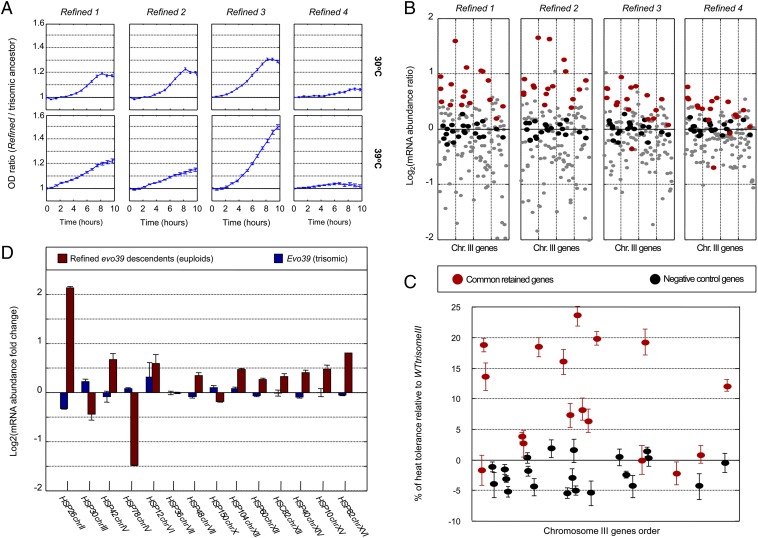
Due to the increased cost of the trisomy in minimal medium, we hypothesized that during prolonged evolution in minimal medium and heat, the trisomy would be eliminated more rapidly compared with rich medium and heat. Following this hypothesis, an additional evolution experiment was carried out. We started with two ancestral strains that carry chromosome III trisomy, namely evo39 and WTtrisomeIII. Interestingly, after 1,000 generations under heat and in minimal medium, all four repetitions (termed Refined 1–4), from both ancestral strains, eliminated the extra copy of chromosome III (Fig. S3). These evolutionarily refined strains, which eliminated the cost associated with the trisomy, exhibit improved growth both at the permissive and the high temperatures compared with their trisomic ancestors (Fig. 4A). The rapid elimination in minimal medium compared to rich medium shows that the high cost of the trisomy accelerates the evolutionary dynamics of its elimination.
Fig. 4.
The aneuploidy-based adaptation was evolutionarily eliminated and replaced by more economical solutions based on refined gene expression adaptations. (A) Descendants of chromosome III trisomic ancestors that were further evolved under heat and lost the trisomy (Refined 1–4) show improved growth under heat, yet with less cost compared with their trisomic ancestors. Each subgraph shows the OD ratios of a refined descendant over its trisomic ancestor measured during continuous growth at 30 °C (Upper) and at 39 °C (Lower). (B) Despite elimination of the trisomy in Refined 1–4, a group of genes from chromosome III retained high expression levels. Dots represent log2 ratios of mRNA abundance of chromosome III genes over a diploid wild type. Genes that retain high expression (in at least three of the four refined evolutions; for details see SI Text) are marked in red, and from the majority of genes that went back to wild-type–like expression (gray dots) a control group was selected and marked in black (used in C). These retained genes were also up-regulated in H2 and H4, which also eliminated the trisomy (χ2 P values 4 × 10−5 and 3 × 10−2). (C) The group of genes that retain high expression levels, after the elimination of the trisomy, confers increased heat tolerance when introduced into wild type. Each of the highly expressed genes (red) and the negative control genes (black) was inserted into the diploid wild type, on a centromeric plasmid, and heat-shock tolerance was compared with the heat tolerance of WTtrisomeIII (t test P value < 5 × 10−7; Materials and Methods). (D) The refined solution replacing the trisomy is characterized by changes in expression levels of most HSP genes. Log2 expression ratios over wild type are shown for all HSPs, for trisomic evo39 (blue) and its descendants that eliminated the trisomy (red). Data are presented as mean and SEM.
Focusing on these refined evolutionary resultants, it is expected that the original contribution of trisomy to heat tolerance was replaced by alternative solutions. We hypothesize that such refined solutions could take over the trisomy by conferring heat tolerance, while avoiding much of the cost associated with aneuploidy. To characterize the refined solutions, we examined the gene expression of the newly evolved refined strains from the different repetitions. A subset of 17 genes on chromosome III was found to retain elevated expression despite the elimination of the extra chromosome copy (Fig. 4B). Next, we checked whether these genes can contribute to heat tolerance when introduced to a wild-type strain, one at a time. To this end, a diploid wild-type strain was transformed with centromeric plasmids (21), each containing one of these genes. As a negative control, 22 random genes from chromosome III that did not retain elevated expression were inserted in the same manner. Reassuringly, most of the genes that retained high expression demonstrated an increased heat tolerance in the transformed wild-type cells (Table S1), with the highest contribution being as high as 23.5% of the heat tolerance of WTtrisomeIII (defined here as 100% heat tolerance) (Fig. 4C; for heat tolerance functional analysis, see SI Text). In contrast, none of the genes from the negative control set had a considerable effect on the wild-type heat tolerance (mean −2.4% ± 2.6%, Table S1). Note that the contribution of the individual genes adds up to more than 100% (defined as the tolerance of the trisomic strain). This probably reflects the fact that the trisomic level of heat tolerance includes the considerable cost of aneuploidy and may also suggest a negative epistasis between the contributions of the individual genes (22). Overall, these results indicate that a part of the solution to the heat challenge, obtained by duplicating chromosome III, was replaced during the extended evolution period under heat by solutions based on refined gene expression changes on chromosome III (and on other chromosomes, as mentioned below).
Genes on other chromosomes may have also evolved changes in expression level after the elimination of the extra copy of chromosome III, and it is most likely that they, too, must have contributed to the evolution of heat tolerance. A particularly interesting example are the heat-shock proteins (23) (HSPs) that are scattered on 11 different chromosomes. We analyzed all of the 14 annotated verified HSPs in the yeast genome (24) for changes in expression in the evo39 strain compared with its descendants that eliminated the trisomy. Curiously, whereas the expression levels of these heat-shock genes changed very modestly after 450 generations of evo39 strain, during which the trisomy was fixated, most of these genes showed significant up-regulation in their refined descendants, i.e., after another 1,000 generations under heat, when the trisomy was eliminated (Fig. 4D). Three of the 14 HSP genes did not show this trend, including HSP78, a gene coding for mitochondrial heat-shock protein and HSP30, which resides in the eliminated chromosome III. This indicates that the trisomy-based solution to heat does not require a concomitant up-regulation of the heat-shock genes, whereas the replacement of the trisomy is associated with an enhancement in the evolutionary expression of the heat-shock genes. This scenario may thus imply that the duplication of a certain chromosome may provide the evolutionary time window needed for the population to search for solutions not only on the duplicated chromosome but throughout the genome.
Elimination of the Extra Copy of Chromosome III at Permissive Temperature.
Another consequence that follows the above-stated properties of acquired aneuploidy would be that when the stress is relieved, i.e., when the benefit diminishes and the cost remains, aneuploidy will be selected against (25). In fact, continued evolution of evo39 under permissive temperature (30 °C) eliminates the extra copy of chromosome III after 600 generations, and with it, its tolerance to heat (Fig. S4). Reassuringly, the trisomy elimination in all four repetitions attests to its superfluous cost at permissive temperature, when it is no longer beneficial. This experiment also demonstrates the limited durability of aneuploidy-based solutions, as they are quickly eliminated when organisms evolve in the absence of the original stress.
Evolution to Gradually Applied Heat Avoids Aneuploidy-Based Solutions.
To this point, aneuploidy had been observed to evolve in response to an abrupt evolutionary challenge. We next hypothesized that an evolution experiment in which the stress is applied in a gradual manner might not select for chromosomal duplication, because under minor stress increments, the considerable cost associated with aneuploidy might surpass the benefit at each increment of the stress. To explore this hypothesis, we evolved the same ancestral strain toward tolerance to heat, but in increments of 1 °C every 50 generations (from 30 °C up to 39 °C). This evolutionary design resembles the “morbidostat” that was recently applied to evolution of drug resistance in bacteria (26). Curiously, under this regime an extra copy of chromosome III was not gained after 450 generations in any of the four repetitions (termed Gradual 1–4) of the experiment (Fig. S5A). Nonetheless, the growth of these gradual populations was better at 39 °C (and also at 30 °C) than that of the four populations (H1–4) that acquired trisomy in the same evolutionary time under constant 39 °C (Fig. S5B). This experiment indicates that gradual application of a stress alleviates the need to adopt the rapidly available yet highly costly solution of chromosomal duplication.
Acquisition and Subsequent Elimination of Chromosome V Aneuploidy During Adaptation to High pH.
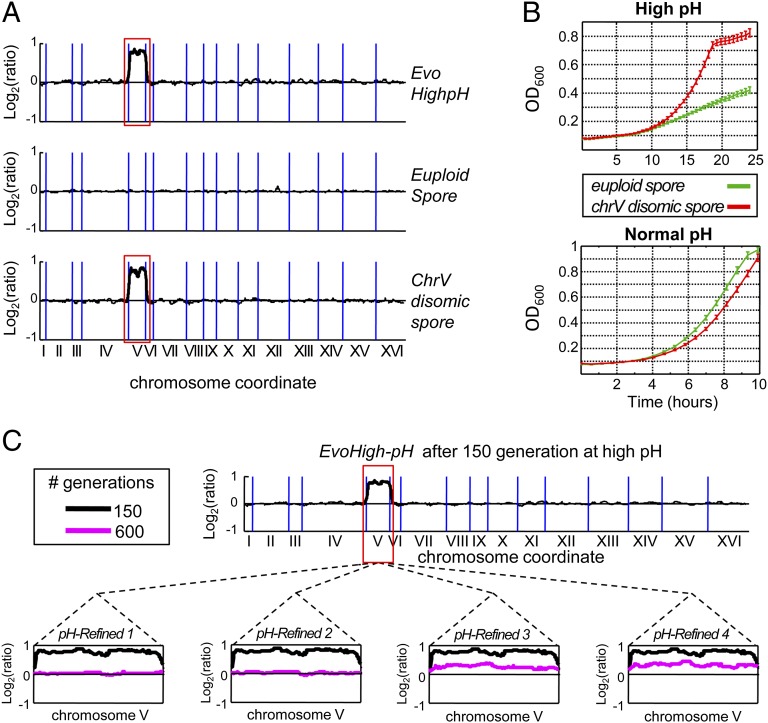
To examine the generality of the findings obtained from evolution experiments under heat, we also examined evolutionary dynamics under an additional physiological stress of high pH. For this experiment, we used an evolved strain from a previous experiment (27) in which a haploid S. cerevisiae was evolved under high pH (8.6), using a similar serial transfer protocol. We examined the evolved strain, termed here evoHigh-pH, for potential chromosomal duplications. Interestingly, in addition to seven point mutations previously detected in the genome sequence of the strains (27) (Table S2), we revealed that evoHigh-pH gained an extra copy of chromosome V (Fig. 5A). To show a direct contribution of the additional copy of chromosome V to the high pH tolerance, evoHigh-pH was crossed with its wild-type ancestor, sporulated and the meiotic products (spores) were analyzed. Among 40 spores scanned, we identified a pair of spores that carried the exact same subset of mutations (Table S2) and differed only in the copy number of chromosome V (one spore, an euploid with one copy of the chromosome, and the other, a disome with two copies; Fig. 5A). Reassuringly, the disomic spore showed better growth at high pH (Fig. 5B). However, when growth was measured at normal pH (6.7), the disomic spore exhibited a reduced growth compared with the euploid spore (Fig. 5B). These results demonstrate a causal link between the extra copy of chromosome V and high pH tolerance.
Fig. 5.
Evolution on high pH selects for transient duplication of chromosome V, which is later eliminated. (A) Haploid wild type that evolved under high pH (27) acquired chromosome V disomy (Top subgraph). We identified two spores obtained from crossing the disomic evolved strain with a haploid wild type. These spores carry the same mutation subset (Table S2) but differ in the copy number of chromosome V (Middle and Bottom subgraphs). Black lines represent log2 intensity ratios of mRNA abundance calculated by a sliding window over a haploid wild type, aligned according to chromosomal order where blue vertical lines differentiate between chromosomes. (B) An extra copy of chromosome V confers high-pH (8.6) tolerance but causes impaired growth on normal pH (6.7). Growth curves of disomic spore (red) and euploid spore (green) under high pH (Upper) and normal pH (Lower). (C) Chromosome V disomy, gained by EvoHigh-pH after 150 generations at high pH (black lines), is eliminated during further evolution under the same high pH in which it was originally gained. Four independent descendants of EvoHigh-pH (pH Refined 1–4) continued to evolve for 600 generations at high pH (8.6). All evolved populations show elimination of the disomy (magenta lines) with two populations showing complete elimination (Left two) and the other two populations showing the majority of the population's cells eliminate the disomy (Right two). All lines represent log2 intensity ratios of mRNA abundance calculated by a sliding window of the high pH evolved strain over a haploid wild type, aligned according to chromosomal order where blue vertical lines differentiate between chromosomes.
We have further extended the analogy to the heat tolerance experiments to examine potential elimination of the extra copy of chromosome V under two evolutionary tracks: one in which EvoHigh-pH continues to evolve at high pH and another in which the stress was relieved and EvoHigh-pH was switched to evolve under normal pH conditions (pH 6.7). In both tracks we detected gradual elimination of the extra copy of chromosome V from the evolving populations (Fig. 5C and Fig. S6). The fact that here too, aneuploidy is reversed even if the stress persists indicates that such large duplications do not typically serve as sustainable evolutionary solutions.
Discussion
Aneuploidy is a readily available, yet costly, evolutionary solution. Such solutions are thus expected to be followed by refinement steps that would alleviate part of the costs associated with the original solution. We predict that such refined solutions will prove to be more durable than aneuploidy-based solutions due to low reversion rate even after prolonged periods in which the stress is not applied. On the other hand, chromosomal duplications may be quickly reversed, leaving minimal imprint on the genome, a potentially desired characteristic for a genomic solution to short-term stresses. Aneuploidy, among other genomic aberrations, is common in cancer too. An interesting possibility is that during the progression of cancer, aneuploidy-based malignancies might be refined and replaced by modifying expression of specific genes. Such dynamics would be consistent with the observation that cancer driver genes are more often characterized by high mRNA levels than by high gene copy numbers (28, 29).
This study emphasizes the importance of long-term laboratory evolution experiments (30). Limiting the experiments to only a few hundred generations would have shown the chromosomal duplication but would have not revealed the massive subsequent aneuploidy elimination toward refinements. On the other hand, in order not to miss fast transient solutions, such as the current aneuploidies, one must follow long evolutionary dynamics at relatively high temporal resolution. Future full genome sequencing of the strains in this highly branched laboratory evolution tree will further sharpen our understanding of the evolutionary dynamics examined here.
Materials and Methods
Strains.
All evolved strains in this work were based on BY4741 (MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0) or BY4743 (MATa/MATα; his3Δ1/his3Δ1; leu2Δ/leu2Δ0; met15Δ0/MET15; LYS2/lys2Δ0; ura3Δ0/ura3Δ0). WTtrisomeIII and WTmonosomeIII were obtained from the Saccharomyces Genome Deletion Consortium (31) and found to have aneuploidies of chromosome III (5) (for our verification of aneuploidy of these strains, see Figs. S8 and S9). For each of these strains, the original gene deletion was reversed by adding the deleted ORF on a centromeric plasmid (21). H1–4 are evolution strains evolved at 39 °C and in rich medium. Evo39 is an abbreviation for one of the two H1–4 strains that gained only the trisomy of chromosome III after 450 generations (H2 in Fig. 1) and was verified by comparative genomic hybridization (CGH) microarrays to carry an extra copy of chromosome III (Fig. S1). Evo30 is an evolution strain evolved in parallel to evo39 for 450 generations, but at the permissive temperature of 30 °C. Refined 1–4 are evolution descendants of chromosome III trisomic strains (1 and 2 of evo39 and 3 and 4 of WTtrisomeIII) that have been further evolved at 39 °C and in minimal medium for another 1,000 generations, with the extra copy of chromosome III eliminated, yet retaining high heat tolerance. Relaxed 1–4 are evolution descendants of the chromosome III trisomic evo39 that have been further evolved at 30 °C and in rich medium for another 600 generations, with the extra copy of chromosome III eliminated, and heat tolerance returned back, similar to a diploid wild type. Gradual 1–4 are evolution strains evolved under gradually applied heat for 450 generations and without gaining chromosome III trisomy. EvoHigh-pH is a strain obtained from the study of Romano et al. (27) that has evolved under high pH (8.6). pH Relaxed 1–4 are evolution descendants of EvoHigh-pH that have been further evolved at normal pH (6.7) for 280 generations. pH Refined 1–4 are evolution descendants of EvoHigh-pH that have been further evolved at high pH (8.6) for 600 generations.
Evolution Experiments.
All laboratory evolution experiments were carried out by serial dilution. Cells were grown until reaching stationary phase under the relevant condition and then diluted by a factor of 1:120 into fresh media (6.9 generations per dilution). This procedure was repeated daily until significant phenotype change was detected in population growth under the applied condition. In all measurements of evolved populations, we used a population sample and not selected clones (SI Text).
Media.
YPD (Yeast Extract Peptone Dextrose) [(1 L) 10 g yeast extract, 20 g peptone, 20 g dextrose, and DDW (Double distilled water)] was used as rich medium and SD (Synthetic Defined) [(1 L) 6.7 g yeast nitrogen base, 20 g dextrose, 1.5 g amino acids mixed powder, and DDW] was used as minimal medium.
Liquid Growth Measurements.
Cultures were grown at the relevant condition and OD600 measurements were taken during growth at 30-min intervals until reaching early stationary phase. Qualitative growth comparisons were performed using 96-well plates in which two strains were divided on the plate in a checkerboard manner to cancel out positional effects. For each strain, a growth curve was obtained by averaging over 48 wells.
Heat-Shock Tolerance Measurements.
To eliminate physiological adaptations (due to prior exposure to heat before the measurements) all cultures were grown for 16 generations on 30 °C. Then, cultures in midlog phase were transferred to 45 °C (t0) for 90 min (t1). At each time point, samples were taken and plated on YPD plates and incubated at 30 °C. The survival ratio was calculated by dividing the number of colonies obtained in t1 by the number of colonies obtained in t0.
Heat Tolerance of Selected Genes from Chromosome III.
To measure the heat tolerance contribution of selected genes from chromosome III when inserted as extra copies into a diploid wild type, we used centromeric plasmids (21). Heat-shock tolerance was measured for each of these strains compared with a diploid wild type with an empty plasmid. We defined 100% heat tolerance as the tolerance of WTtrisomeIII and 0% tolerance as the tolerance of diploid wild type with an empty plasmid. For each of the transformed strains, heat tolerance was calculated by dividing over the WTtrisomeIII tolerance, after subtracting the tolerance of wild type with an empty plasmid.
General Stress Tolerance.
Cultures of diploid WT and WTtrisomeIII were grown in rich medium until midlog phase and then transferred to one of the following conditions: YPD + KCl 1 M, YPD at pH = 4, YPD + ethanol 8% (vol/vol), YP + glycerol 3% (vol/vol), YPD + NaCl 0.5 M, or YPD + copper sulfate (CuSO4) 6.25 mM. OD600 measurements were taken at constant intervals.
Microarrays.
Affymetrix Yeast Genome 2.0 arrays were used to measure gene expression and to detect aneuploidies. The ability to detect large-scale genomic duplications by expression microarrays was based on previous studies that demonstrate the correspondence between chromosomal duplication and increased expression (1, 5). Agilent CGH microarrays were used to verify karyotype for selected strains (we report aneuploidy of evolved populations only when the aneuploidic part in population is >95%; see SI Text). Taking into consideration that adaptation could also be carried out at the physiological level, rather than encoded at a genetic/epigenetic level, all expression microarrays were preformed following 16 generations of growth at the normal temperature of 30 °C. This setup allows us to focus on the nonphysiological component of adaptation. Genomic DNA and RNA extractions were performed using Epicenter Yeast kits. The microarray data for detection of chromosomal duplications are shown after performing sliding window averaging with a window size of 10 genes from each side. At each point the value represents the average of the values of the current window. All Affymetrix microarray data have been deposited in the National Center for Biotechnology Information (NCBI)’s Gene Expression Omnibus and are accessible through GEO accession no. GSE40817 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE40817).
Supplementary Material
Acknowledgments
We thank Jasmin Bloch, Sharon Reikhav, and Miri Carmi for assistance with the CGH microarray. Y.P. acknowledges support from the Ben May Charitable Trust, the Minerva Foundation, and the Bikura program of the Israel Science Foundation; Y.P. is an incumbent of the Ben May Professorial Chair. M.K. acknowledges support from the Israel Science Foundation and the Israeli Ministry of Science and Technology. G.H.R. was supported by a Machiah Foundation grant and by a Safra program for bioinformatics grant.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE40817).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211150109/-/DCSupplemental.
References
- 1.Torres EM, et al. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317(5840):916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 2.Pavelka N, et al. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature. 2010;468(7321):321–325. doi: 10.1038/nature09529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nat Rev Genet. 2012;13(3):189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- 4.Gresham D, et al. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet. 2008;4(12):e1000303. doi: 10.1371/journal.pgen.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hughes TR, et al. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat Genet. 2000;25(3):333–337. doi: 10.1038/77116. [DOI] [PubMed] [Google Scholar]
- 6.Rancati G, et al. Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell. 2008;135(5):879–893. doi: 10.1016/j.cell.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Selmecki A, Gerami-Nejad M, Paulson C, Forche A, Berman J. An isochromosome confers drug resistance in vivo by amplification of two genes, ERG11 and TAC1. Mol Microbiol. 2008;68(3):624–641. doi: 10.1111/j.1365-2958.2008.06176.x. [DOI] [PubMed] [Google Scholar]
- 8.Dunham MJ, et al. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2002;99(25):16144–16149. doi: 10.1073/pnas.242624799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poláková S, et al. Formation of new chromosomes as a virulence mechanism in yeast Candida glabrata. Proc Natl Acad Sci USA. 2009;106(8):2688–2693. doi: 10.1073/pnas.0809793106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen G, Bradford WD, Seidel CW, Li R. Hsp90 stress potentiates rapid cellular adaptation through induction of aneuploidy. Nature. 2012;482(7384):246–250. doi: 10.1038/nature10795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chester M, et al. Extensive chromosomal variation in a recently formed natural allopolyploid species, Tragopogon miscellus (Asteraceae) Proc Natl Acad Sci USA. 2012;109(4):1176–1181. doi: 10.1073/pnas.1112041109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fawcett JA, Maere S, Van de Peer Y. Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proc Natl Acad Sci USA. 2009;106(14):5737–5742. doi: 10.1073/pnas.0900906106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van de Peer Y, Maere S, Meyer A. The evolutionary significance of ancient genome duplications. Nat Rev Genet. 2009;10(10):725–732. doi: 10.1038/nrg2600. [DOI] [PubMed] [Google Scholar]
- 14.Anders KR, et al. A strategy for constructing aneuploid yeast strains by transient nondisjunction of a target chromosome. BMC Genet. 2009;10:36. doi: 10.1186/1471-2156-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torres EM, Williams BR, Amon A. Aneuploidy: Cells losing their balance. Genetics. 2008;179(2):737–746. doi: 10.1534/genetics.108.090878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sheltzer JM, et al. Aneuploidy drives genomic instability in yeast. Science. 2011;333(6045):1026–1030. doi: 10.1126/science.1206412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Torres EM, et al. Identification of aneuploidy-tolerating mutations. Cell. 2010;143(1):71–83. doi: 10.1016/j.cell.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pavelka N, Rancati G, Li R. Dr Jekyll and Mr Hyde: Role of aneuploidy in cellular adaptation and cancer. Curr Opin Cell Biol. 2010;22(6):809–815. doi: 10.1016/j.ceb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheltzer JM, Amon A. The aneuploidy paradox: Costs and benefits of an incorrect karyotype. Trends Genet. 2011;27(11):446–453. doi: 10.1016/j.tig.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elena SF, Lenski RE. Evolution experiments with microorganisms: The dynamics and genetic bases of adaptation. Nat Rev Genet. 2003;4(6):457–469. doi: 10.1038/nrg1088. [DOI] [PubMed] [Google Scholar]
- 21.Ho CH, et al. A molecular barcoded yeast ORF library enables mode-of-action analysis of bioactive compounds. Nat Biotechnol. 2009;27(4):369–377. doi: 10.1038/nbt.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khan AI, Dinh DM, Schneider D, Lenski RE, Cooper TF. Negative epistasis between beneficial mutations in an evolving bacterial population. Science. 2011;332(6034):1193–1196. doi: 10.1126/science.1203801. [DOI] [PubMed] [Google Scholar]
- 23.Parsell DA, Lindquist S. The function of heat-shock proteins in stress tolerance: Degradation and reactivation of damaged proteins. Annu Rev Genet. 1993;27:437–496. doi: 10.1146/annurev.ge.27.120193.002253. [DOI] [PubMed] [Google Scholar]
- 24.Saccharomyces Genome Database. Available at www.yeastgenome.org.
- 25.Selmecki A, Forche A, Berman J. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science. 2006;313(5785):367–370. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toprak E, et al. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat Genet. 2012;44(1):101–105. doi: 10.1038/ng.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romano GH, et al. Different sets of QTLs influence fitness variation in yeast. Mol Syst Biol. 2010;6:346. doi: 10.1038/msb.2010.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akavia UD, et al. An integrated approach to uncover drivers of cancer. Cell. 2010;143(6):1005–1017. doi: 10.1016/j.cell.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pe’er D, Hacohen N. Principles and strategies for developing network models in cancer. Cell. 2011;144(6):864–873. doi: 10.1016/j.cell.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barrick JE, et al. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461(7268):1243–1247. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- 31.Winzeler EA, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285(5429):901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.