Abstract
Legumes were among the first plant species to be domesticated, and accompanied cereals in expansion of agriculture from the Fertile Crescent into diverse environments across the Mediterranean basin, Europe, Central Asia, and the Indian subcontinent. Although several recent studies have outlined the molecular basis for domestication and eco-geographic adaptation in the two main cereals from this region, wheat and barley, similar questions remain largely unexplored in their legume counterparts. Here we identify two major loci controlling differences in photoperiod response between wild and domesticated pea, and show that one of these, HIGH RESPONSE TO PHOTOPERIOD (HR), is an ortholog of EARLY FLOWERING 3 (ELF3), a gene involved in circadian clock function. We found that a significant proportion of flowering time variation in global pea germplasm is controlled by HR, with a single, widespread functional variant conferring altered circadian rhythms and the reduced photoperiod response associated with the spring habit. We also present evidence that ELF3 has a similar role in lentil, another major legume crop, with a distinct functional variant contributing to reduced photoperiod response in cultivars widely deployed in short-season environments. Our results identify the factor likely to have permitted the successful prehistoric expansion of legume cultivation to Northern Europe, and define a conserved genetic basis for major adaptive changes in flowering phenology and growth habit in an important crop group.
Keywords: crop adaptation, Pisum sativum, Lens culinaris
Many of the world’s earliest agricultural systems were based around crops from two important groups: cereals and legumes. Although molecular and genetic analyses have led to considerable progress in understanding the genetic changes underlying domestication and adaptation in several cereal crops, similar efforts in legumes are in general much less advanced. Among the legumes domesticated in the world’s oldest farming culture in the Neolithic Near East, the temperate long-day (LD) species lentil (Lens culinaris Medik.), pea (Pisum sativum L.), and chickpea (Cicer arietinum L.) all persist as crops of global economic importance. Of these crops, pea has the widest distribution, the most diverse phenology, and is the best understood genetically, and offers prospects for a detailed exploration of molecular events important in early cultivation and spread (1, 2).
P. sativum is now generally viewed as a complex species that includes a wide variety of cultivated and wild forms with pink, purple, or white flowers (1). Wild P. sativum lines are characterized by dehiscent pods and a rough, thick seed coat, and include both tall, climbing forms distributed around the Mediterranean (P. sativum var. elatius) and shorter forms restricted to the Near East (P. sativum var. humile), which intergrade in their areas of overlap. Cytogenetic differences and analyses of genetic diversity support the view that the majority of cultivated peas originated from a distinct gene pool within var. humile (1), although recent molecular studies also highlight the likely genomic contribution from other wild forms, and emphasize the importance of introgression and recombination within the complex (2, 3). Domesticated variants of P. sativum include garden or “vegetable” pea (var. sativum) and field pea (var. arvense) grown for dry seed or as a forage crop. A distinct taxon (P. sativum var. abyssinicum) found in highland regions of Ethiopia and southern Yemen is believed to represent an independent domesticate with a substantial genomic contribution from a second, related species, the yellow/orange-flowered Pisum fulvum Sibth. & Sm (1, 3).
Control of flowering time is widely acknowledged as an important feature of plant adaptation, and there has been much recent effort directed toward understanding the molecular basis for flowering time adaptation in both wild and domesticated species (4). One common feature of flowering-time adaptation is the relaxation of mechanisms that operate to delay flowering under unfavorable conditions, through loss of function in genes controlling responsiveness to photoperiod or vernalization. These changes reduce the length of the growth cycle, permitting a shift from winter to spring cropping in temperate regions, and enabling expansion to areas where the growing season is limited by short duration or water availability. Similar changes also underlie expansion of temperate crops to low latitudes where the short photoperiods and lack of vernalizing temperatures would otherwise preclude flowering. Wild Pisum in its native range displays a typical winter habit in which plants germinate in autumn, overwinter in the vegetative state, and flower in response to increasing day-length in spring (5). Anecdotal reports of experiments in controlled conditions also suggest that wild P. sativum lines generally only flower when grown under LD photoperiods (6). This obligate or near-obligate requirement for LDs suits pea to a winter cropping cycle and has been retained in certain forage cultivars (7). However, the majority of cultivated pea accessions from higher latitudes have a quantitative LD response and are grown as a spring crop (7).
In this study, we undertook an initial genetic analysis of the differences in flowering and photoperiod responsiveness between wild and domesticated pea. We identified two major loci controlling these differences, and showed that one of them likely corresponds to an ortholog of the circadian clock gene EARLY FLOWERING 3 (ELF3). We further demonstrate that variation in this gene is also associated with photoperiod-insensitive early flowering in the related crop species lentil.
Results
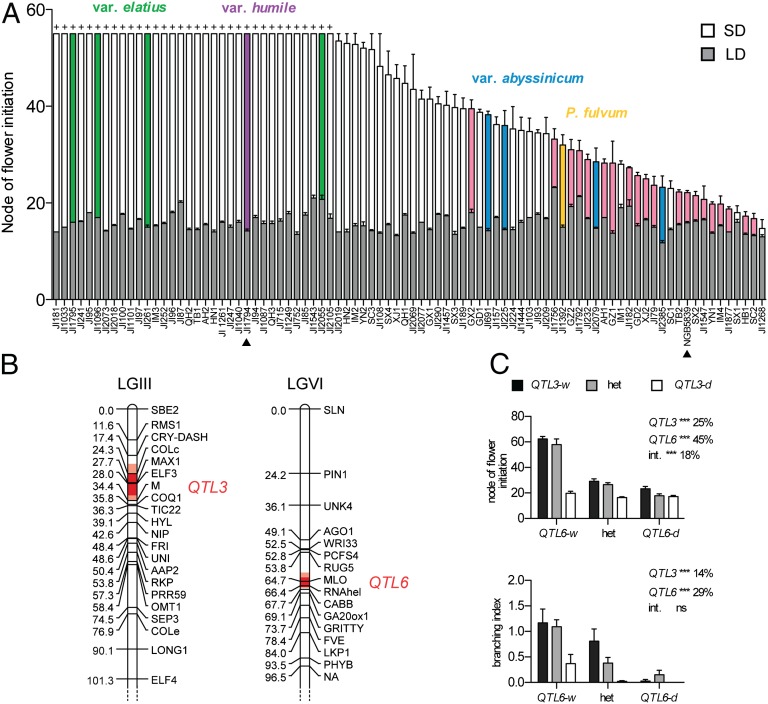
We first sought to gain an overview of natural variation for flowering and photoperiod responsiveness in pea, in a selection of lines representing a broad range of genetic diversity (3, 8) (Table S1). All lines flowered within a narrow developmental window under LD, but much wider variation was observed under short days (SD), where P. fulvum, var. abyssinicum and most domesticated lines flowered, but lines of var. elatius and humile, several land-races, and several winter cultivars did not (Fig. 1A). This finding suggests that ancestral P. sativum was an obligate LD plant, consistent with the winter habit observed for wild forms in their natural range (5, 6), and suggests that evolution of early-flowering types in which environmental constraints to flowering are genetically relaxed has been a key feature of postdomestication spread.
Fig. 1.
Adaptation to photoperiod in pea is controlled by two major-effect QTL. (A) Survey of photoperiod-regulated flowering in Pisum. Plants received an 8-h photoperiod of natural daylight (SD) extended with low-irradiance (10 µmol⋅m−2⋅s−1) white light (LD) from mixed fluorescent and incandescent sources. Data are mean ± SE for n = 4. Lines not flowering in SD conditions are indicated by a “+” symbol and had produced a minimum of 55 vegetative nodes before termination of the experiment 180 d after sowing. All lines are P. sativum var. sativum unless indicated. Lines carrying the hr (6C) mutation are shaded in pink, and the two lines used for subsequent genetic analysis are indicated by black arrowheads. (B) Location of QTL controlling SD flowering on linkage groups III and VI in the F2 of a domesticated (NGB5839) × wild (var. humile; JI1794) cross. The one-LOD and two-LOD confidence intervals around the peak are indicated by dark red and pale red shading, respectively. (C) Genotype means ± SE for interaction of QTL3 and QTL6 in the control of flowering and other developmental traits. Genotypes at QTL3 and QTL6 were inferred from the genotype of peak markers MAX1 (QTL3) and RNAhel (QTL6), with the wild (JI1794) and domesticated (NGB5839) alleles indicated by the suffixes -w and -d, respectively. Significance levels (***P < 0.001; ns, P > 0.05) and proportion of variance explained for the individual locus effects and their interaction (int.) were determined by two-way ANOVA and indicated to the right of each plot.
We next examined the genetic basis for changes in SD flowering under domestication in P. sativum in a cross between NGB5839, an isogenic dwarf derivative of the spring cultivar Torsdag, and the wild line JI1794, a representative accession of the northern race of P. sativum var. humile proposed as the major wild contributor to the domesticated pea gene pool (1). Like many spring cultivars, NGB5839 and its progenitor cv. Torsdag carry recessive alleles at the HIGH RESPONSE TO PHOTOPERIOD (HR) locus that confer early flowering in SD (1). We examined the association between node of flower initiation and markers for a range of flowering-related genes. As expected, we identified significant association with markers at the top of linkage group III in the region of HR, but also with markers in the middle of linkage group VI, and further genotyping of markers in these two regions defined two quantitative trait loci (QTL) (Fig. 1B and Table S2). These loci showed contrasting patterns of inheritance, with the early flowering conferred by domesticated alleles showing recessive inheritance in the case of QTL3 but dominant inheritance at QTL6 (Fig. 1C). The loci also showed significant interaction, with extreme delay of flowering under SD only seen in plants homozygous for the JI1794 allele at QTL6 and carrying at least one JI1794 allele at QTL3 (Fig. 1C). We conclude that wild alleles at both loci are required for full expression of the winter habit. QTL3 and QTL6 together explained 88% of the observed variance for node of flower initiation, as estimated by two-way ANOVA based on peak markers MAX1 and RNAhel (Fig. 1C). In addition, single marker tests failed to provide evidence for significant effects of other known flowering loci, including LF (9), members of the FT family (10), or SN (11) (Table S3). Interestingly, markers for both QTL3 and QTL6 were also associated with variation in shoot branching, which was reduced in an additive manner by homozygosity for the domesticated alleles (Fig. 1C).
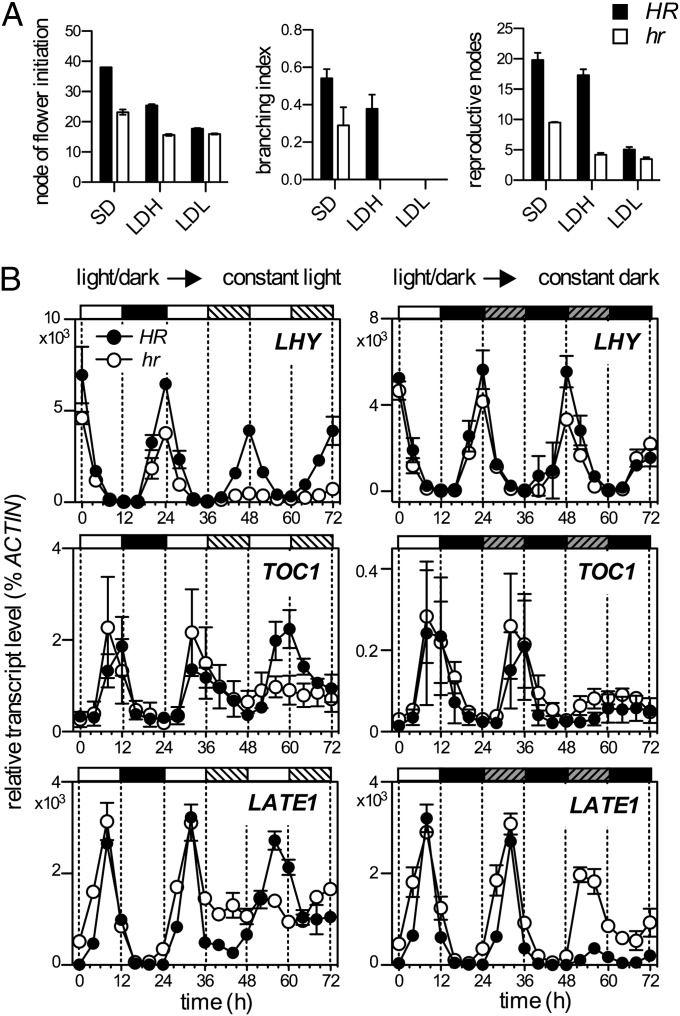
Because NGB5839 carries recessive alleles at the HR locus, we considered that QTL3 might be largely or entirely equivalent to HR. Previous studies have characterized the effects of HR on responses to photoperiod and light quality, and in a more applied setting, on winter frost damage in field-grown plants (7, 11). To further characterize the physiological effects of HR, we generated near-isogenic HR and hr lines in which dominant HR alleles were introgressed into the NGB5839 background from the line WL1771 (Wellensiek’s Dominant) (12) through six successive backcrosses. Consistent with previous studies (11, 13), the hr allele was associated with early flowering and reduced branching in plants grown under SD and reduced sensitivity to the light quality of extended photoperiods (Fig. 2A). In HR plants, flowering was more effectively promoted by extensions with light of low red (R):far-red (FR) ratio than of high R:FR ratio, whereas in hr plants both types of photoperiod extension were equally effective (Fig. 2A). One factor that influences responses to both light quality and photoperiod is the circadian clock, and we previously noted that rhythmic expression of several circadian clock genes showed unusual light-dependent damping in NGB5839 (14). To determine whether this might be because of its hr genotype, we also examined the effect of HR on circadian rhythms of gene expression using the same near-isogenic lines. Introgression of HR to the NGB5839 background clearly restored rhythmic expression of LHY after transfer to constant light, but had little effect on LHY expression following transfer from entraining photoperiods into constant dark (Fig. 2B). HR similarly restored rhythmic expression of the clock genes TOC1 and LATE1 under constant light. Together, these results strongly implicated HR in the input of light signals to the circadian clock.
Fig. 2.
The HR locus affects photoperiod responsiveness and circadian rhythms. (A) Effect of HR on responsiveness to photoperiod and light quality. Plants received 8 h of natural daylight (SD) extended for a further 16 h with low-irradiance (10 µmol⋅m−2⋅s−1) white light of high (LDH) or low (LDL) R:FR. Data are mean ± SE for n = 8–10. (B) RT-PCR analysis of expression rhythms of clock genes in HR and hr, showing means ± SE for three biological replicates. Plants grown for 3 wk from sowing under a 12-h photoperiod (150 µmol⋅m−2⋅s−1) were transferred to constant white light (10 µmol⋅m−2⋅s−1 (Left)) or constant dark (Right).
In Arabidopsis, several genes have been shown to mediate light input to the clock, including FHY3, ELF3, and LUX (15–17), and we evaluated these genes as candidates for QTL3/HR. We also considered genes in the FRI/FRL family, based on observations that in Arabidopsis they regulate SD flowering phenotypes in a manner similar to HR, and that their regulatory target FLC influences circadian properties (e.g., refs. 18 and 19). We identified all candidate genes in Medicago truncatula (Table S4), and inferred the locations of the corresponding pea genes using the pea/Medicago comparative map (20, 21). This finding implied a location of the pea ELF3 and FRI genes on the top half of linkage group III near HR but suggested exclusion of FHY3, FRLa, FRLb, and LUX as HR candidates based on inferred positions on the bottom half of LGIII (FRLa, FHY3) or on other linkage groups (FRLb, LUX). The inferred map positions of ELF3 and FRI were confirmed by isolation and mapping of the corresponding pea orthologs (Fig. 1B). In the NGB5839 × JI1794 population, FRI was located 13 cM below the classical marker M and outside the confidence interval for QTL3, whereas ELF3 was located ∼7 cM above M, close to the previously determined position of HR (22, 23). Reanalysis identified ELF3 as the closest marker to the QTL3 peak (Table S2). Similar relative positions were obtained in a second population of 164 recombinant inbred lines derived from a cross between cultivars Térèse (hr) and Champagne (HR) (23), in which no clear recombinants were found between HR and ELF3. In addition, we observed no recombinants between HR and ELF3 in over 200 F2 plants derived from BC4 and BC5 generations of the 5839 × WL1771 cross. These data indicate a distance of less than 0.3 cM between HR and ELF3 and taken together with the physiological characteristics of HR, provided further evidence for the possibility that ELF3 might correspond to HR, or at least contribute significantly to it.
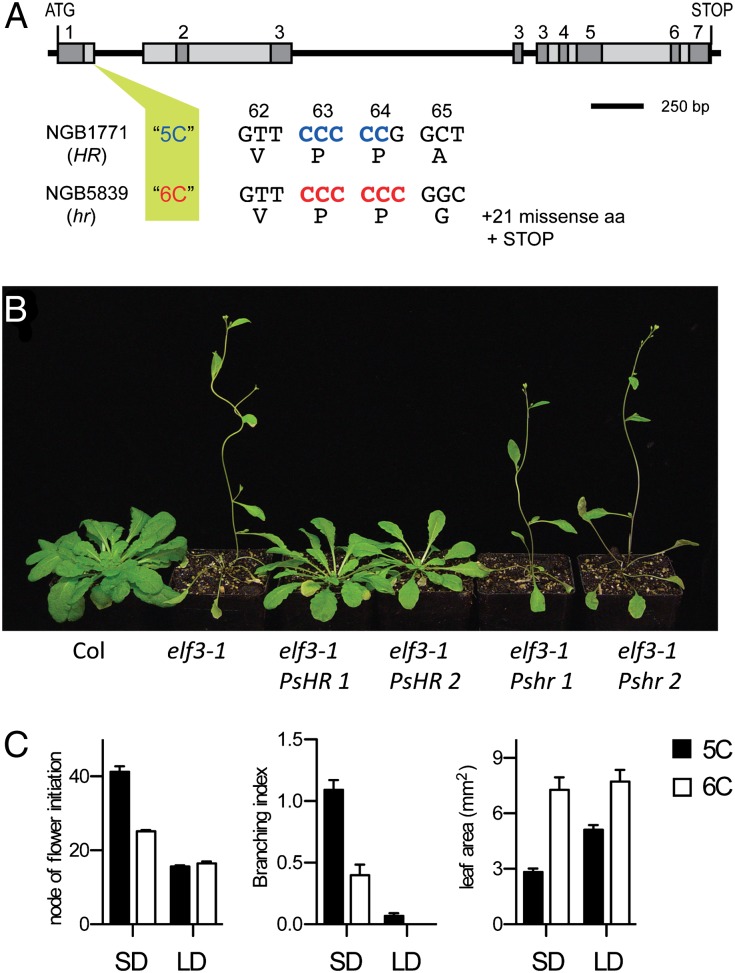
ELF3 is a highly conserved, plant-specific nuclear protein with no recognized functional domains that appears to function as a substrate adaptor enabling the physical interaction of circadian clock components and regulators (24, 25). Whereas ELF3 cDNAs from JI1794 and from HR lines WL1771 and cv. Champagne encoded full-length ELF3 proteins of 702 aa similar to other species, the corresponding cDNA from hr lines NGB5839 and cv. Térèse revealed the insertion of a single C to a 5C sequence near the end of exon 1, which introduced a frame-shift and predicted truncation of several highly conserved domains from the ELF3 protein (Fig. 3A and Fig. S1). This “6C” polymorphism was located ∼600 bp from the polymorphism used to map ELF3 in the 5839 × WL1771 population and cosegregated perfectly with this marker and with the hr early-flowering phenotype in that material. The effect of the 6C polymorphism was also tested by transgenic complementation of the Arabidopsis elf3-1 mutant, which flowers earlier than wild-type and has markedly elongated petioles. Whereas the 5C form of PsELF3 complemented elf3-1 and restored later flowering and normal petiole elongation, the 6C form had no effect on either trait (Fig. 3B). Finally, we examined the distribution of this polymorphism and its association with flowering time across the 84 lines phenotyped for photoperiod response (Fig. 1A). We found that all Pisum elatius, humile, abyssinicum, and fulvum lines and 56 of the var. sativum lines carried the 5C allele, but the 20 remaining var. sativum lines carried the 6C allele. Fig. 3C shows that in this material, the 6C mutation was associated with earlier flowering under SD but not under LD, and thus with a significant reduction in responsiveness to photoperiod, which was also reflected in other traits including branching and leaf size. We conclude that loss of ELF3 function is the likely basis for the hr/QTL3 flowering phenotype.
Fig. 3.
A mutation in ELF3 ortholog is the likely basis for the hr spring phenotype. (A) Details of the PsELF3 5C/6C polymorphism. (B) Complementation of flowering and petiole phenotypes of the Arabidopsis elf3-1 mutant by the 5C (HR) but not the 6C (hr) form of 35S::PsELF3, under 8-h SD conditions. Representative plants are shown for two independent transformants for each construct. (C) Association of the 5C/6C polymorphism with photoperiod responsiveness in a selection of P. sativum germplasm. Plants received an 8-h photoperiod of natural daylight (SD) extended with low-irradiance (10 µmol⋅m−2⋅s−1) white light (LD) from mixed fluorescent and incandescent sources. Data are mean ± SE for n = 64 (5C) and n = 20 (6C).
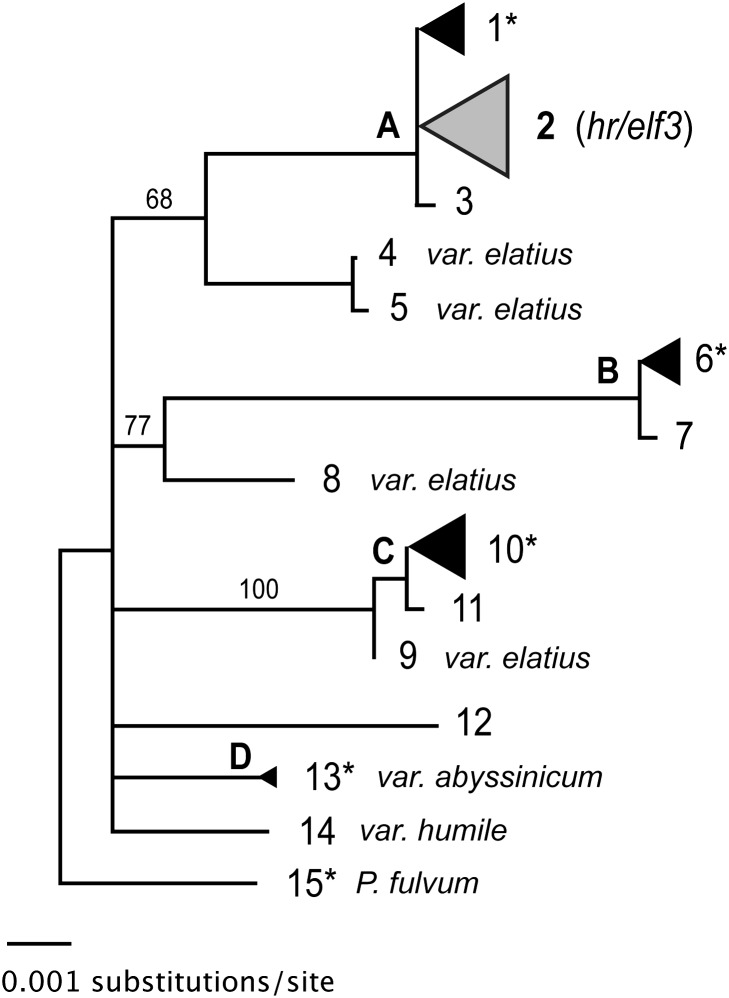
To gain insight into sequence variation at ELF3 and the origin of the hr mutation, we resequenced the entire ELF3 gene in the initial 84 accessions and in a further diversity-targeted selection of 38 accessions from the Pisum germplasm collection at the Centre des Ressources Biologiques (CRB) (26). We also surveyed the entire CRB collection for ELF3 polymorphisms using a Targeting Induced Local Lesions in Genomes (TILLING) approach (27), extending our sample size to over 450 accessions. These analyses identified over 130 sequence polymorphisms (Fig. S2), defining 15 haplotypes and four distinct haplotype groups (Fig. 4 and Table S1). Significantly, all lines carrying the 6C (hr) mutation shared the same haplotype and were associated with a single group of highly similar haplotypes (group A in Fig. 4), which contained land-races and more recently developed cultivars. Haplotype groups B and C consisted mainly of land-races and other domesticated material from Afghanistan and further east, and a single haplotype (haplotype 13) was found in all six P. sativum var. abyssinicum lines examined (Fig. 4 and Table S1). Within group A, 44 hr and 24 HR lines from diverse geographic origins were identical over the 3.8-kb genomic ELF3 sequence apart from the hr mutation, and this sequence was also shared by 250 of 251 hr lines in the CRB collection. The hr mutation and its corresponding HR haplotype are thus widespread across domesticated germplasm and represent a highly differentiated form of the ELF3 gene. Other sequence differences seem unlikely to have a major influence on flowering, in view of the fact that all 26 predicted amino acid substitutions in our ELF3 sequence dataset affected poorly conserved residues and are unlikely to be functionally significant (Fig. S2), and furthermore that there was no significant effect of haplogroup on flowering under either SD or LD (Fig. S3).
Fig. 4.
Sequence diversity in the HR gene. Neighbor-joining tree representing genetic distances among haplotypes identified in a 3.8-kb region of the HR gene in 110 diverse Pisum lines (P.sativum var. sativum except where indicated). Node support (%) was obtained from 10,000 bootstrap replicates. Numbering of haplotypes corresponds to Table S1 and Fig. S2, with bold letters indicating distinct haplotype groups. Haplotypes present in more than one line are indicated by filled triangles with size proportional to the number of lines represented, with the single haplotype containing the hr mutation designated by an open triangle. Haplotypes including lines that flowered in SD conditions, despite carrying an apparently functional form of HR, are indicated by an asterisk.
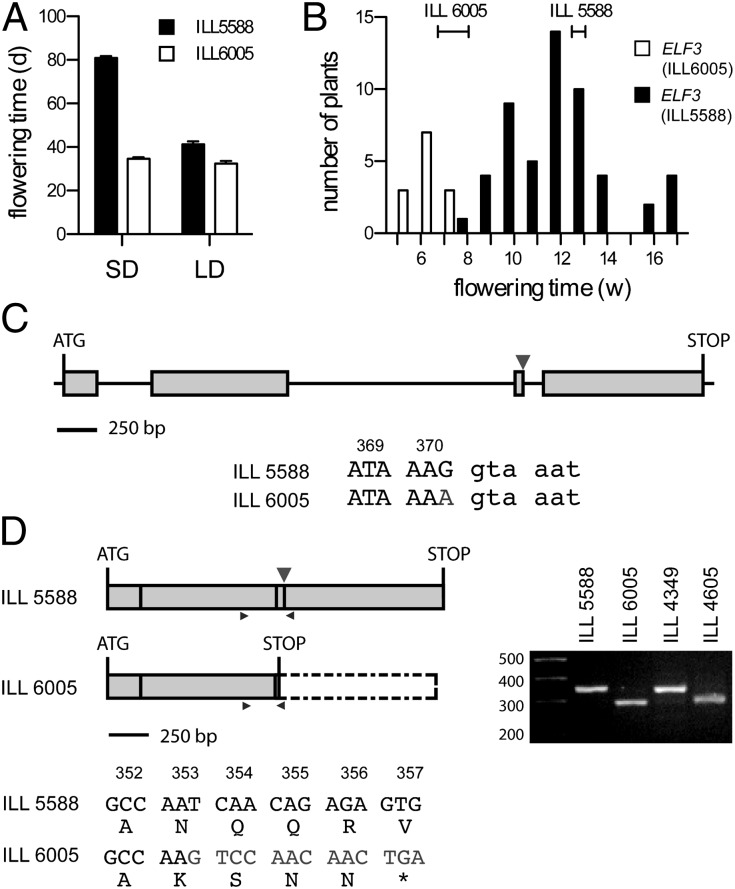
Genetic variation for flowering time and photoperiod responsiveness has been documented in other temperate LD legumes, including lentil (e.g., ref. 28) and chickpea (29). In both cases, early flowering and reduced photoperiod sensitivity has been an important adaptation to cropping in South Asia, where the season is limited by short day-lengths or terminal drought (29, 30). To examine the possibility that ELF3 orthologs might also underlie variation for phenology in other legumes, we examined the genetic control of flowering in crosses between early- and late-flowering lentil lines. We resolved a major locus controlling SD flowering (Fig. 5 A and B) in a cross between the photoperiod responsive line cv. Northfield (ILL5588) and the early-flowering line ILL6005, which is a derivative of cv. Precoz, an early-flowering line from Argentina (30). This locus likely corresponds to the SN locus previously defined in crosses with cv. Precoz (28). We assessed the genetic relationship of this locus to candidate photoperiod response genes and showed that it was tightly linked to the lentil ELF3 ortholog (Fig. 5B), effectively excluding the possibility that it is an ortholog of pea SN, which is located in pea linkage group VII in a distinct part of the legume comparative map (20, 21). Sequencing of ELF3 genomic DNA and cDNA in ILL6005 and cv. Precoz revealed a translationally silent G-to-A substitution in the last nucleotide of exon 3 (Fig. 5C) causing missplicing and skipping of exon 3. This change is predicted to cause a frame-shift in translation of exon 4 and termination after four missense amino acids. (Fig. 5D). These results suggest that this lentil locus is an ortholog of pea HR, and provides evidence that HR may play a similar role in photoperiod adaptation in lentil as in pea.
Fig. 5.
Mutation in an ELF3 ortholog is also associated with early flowering in lentil. (A) Differing photoperiod responsiveness in Lens culinaris lines ILL5588 and ILL6005. Plants received an 8-h photoperiod of natural daylight extended with 2-h (SD) or 16-h (LD) low-irradiance (10 μmol⋅m−2⋅s−1) white light from mixed fluorescent and incandescent sources. Data are mean ± SE for n = 9–10. (B) Cosegregation of flowering time under SD with a marker for LcELF3 in the F2 generation of cross ILL6005 × ILL5588. The flowering time ranges of the two parental lines are shown as horizontal bars. (C) Details of the mutation in LcELF3 genomic DNA. Early-flowering segregants carry a translationally silent G-to-A substitution in the last nucleotide of exon 3. (D) Details of splicing defect. PCR with the indicated primers (small arrowheads) revealed a 52-bp deletion in the LcELF3 mRNA from ILL6005, corresponding to skipping of exon 3. This was verified by sequencing and results in a frame-shift in translation of exon 4 and termination after four missense amino acids.
Discussion
The genetic basis for adaptive changes in flowering time has been extensively analyzed in Arabidopsis and many cereal crops, but is less well understood in other species, including legumes, which are a large and important crop group and include many of the world’s oldest domesticated species. In this study we characterized natural variation for photoperiod responsiveness in the temperate LD species pea, including a broad range of domesticated and wild material. We show that wild forms have an obligate requirement for exposure to long photoperiods to flower, which is also seen in a wide range of primitive domesticated types. In contrast, many other domesticated lines of P. sativum from diverse origins do flower in SD, indicating that the ability to flower early under restrictive photoperiods has been an important factor in the expansion of pea cultivation. In crosses between a domestic and a wild line, we distinguished major contributions from two loci corresponding to the previously studied HR locus and a novel locus, QTL6. We identified ELF3 as a strong positional candidate for HR and showed that HR influences several physiological characteristics also controlled by ELF3 in Arabidopsis, including photoperiod responsiveness, sensitivity to low R:FR (31), and light regulation of circadian gene expression rhythms (15) (Fig. 2). Furthermore, we identified sequence polymorphism in pea ELF3 that eliminated its function in transgenic Arabidopsis (Fig. 3B), was strongly associated with photoperiod responsiveness across a diverse set of pea germplasm (Fig. 3C), and was tightly linked to the HR locus. Together, these results strongly implicate ELF3 as HR, but the alternative possibility that aspects of the hr phenotype derive from other nearby genes cannot be definitively excluded.
Identification of HR as a likely ELF3 ortholog also prompted us to examine in detail the sequence diversity at the HR locus and its relationship to flowering time and photoperiod adaptation. In combined resequencing and TILLING analyses of over 500 genetically diverse pea accessions, we found evidence for only a single functional variant in the HR gene. The widespread nature of this hr mutation and the fact that it occurs within a closely related but distinct group of haplotypes suggest that the mutation originated relatively recently within an already differentiated lineage, and has undergone rapid dispersal. Archaeological evidence for the early history of pea cultivation suggests that following a relatively rapid westward spread from the inferred domestication center into Mediterranean Europe, expansion to the north occurred only after a significant lag (32, 33). We speculate that this early westward lineage may be represented by haplogroup A, and the hr mutation may have arisen within this lineage, permitting summer cropping and thereby enabling expansion to areas with colder winters. More recently, reincorporation of functional HR alleles from winter forage pea cultivars into a variety of other end-use types is an important factor in the strategy for breeding winter forms of these crops (23) and the identification of a putative causal mutation should facilitate this process.
The observation that domesticated alleles at QTL6 promote flowering under SD to an extent equivalent to the hr mutation (Fig. 1C) suggests the possibility of a second, independent route to a reduction in photoperiod response and the spring habit. The existence of HR-independent flowering variation within each of the three major HR haplogroups (Figs. 1A and Fig. S2) also indicates the likely contribution of additional genetic factors to photoperiod adaptation across the broader Pisum germplasm, which might include QTL6. In addition, although the previously characterized LF/TFL1c gene (9) did not influence SD flowering in our mapping population (Table S3), other studies of LF function and interaction indicate recessive lf alleles can confer earlier flowering in photoperiod-responsive genetic backgrounds under SD (11), and suggest LF as another candidate for some of the variation in SD flowering we observe. Interestingly, we found moderate to strong bootstrap support for association of var. elatius lines with each of the three var. sativum HR haplotype groups (Fig. 4), with haplogroup A in particular associated with var. elatius lines from Italy and Greece. This finding is consistent with the possibility that distinct lineages of var. sativum originated from different subsets of wild germplasm, and that early flowering arose independently in these lineages. The distinct form P. sativum var. abyssinicum is found in Ethiopia and Yemen and has been suggested as a probable independent domesticate with a clear genomic contribution from the second wild Pisum species, P. fulvum. We found that although var. abyssinicum lines carry a functional HR gene, they can all flower in SD, as expected from their distribution at latitudes below 10° N. Interestingly, P. fulvum itself (which also carries a predicted functional HR gene) is also able to flower in SD, which could be understood as an adaptation to a shorter effective season in its natural habitat compared with wild P. sativum forms (34). In view of the suggested relationship between P. fulvum and P. sativum var. abyssinicum, it will be interesting in future to examine whether their early-flowering phenotypes might have the same molecular basis and are related to the HR-independent early-flowering seen within var. sativum.
Evolution of early flowering under domestication has been studied in a number of other crop groups, most notably the cereals. Interestingly, these studies have identified a number of cases of convergence, with adaptation in different species conferred by changes in orthologous genes. Studies of photoperiod adaption genes in wheat and barley have identified two main genetic routes to the spring growth habit: through loss-of-function vrn2 mutations in the grass-specific ZCCT transcription factors that act as repressors of flowering, or through semidominant mutations causing deregulated expression of the flower-promoting Ppd1/PRR37 response regulator genes (35, 36). Evidence also suggests the importance of PRR37 for photoperiod response in the warm-season cereals sorghum and rice (37, 38). The convergent nature of these adaptive changes implies the potential for similar convergence in other plant groups, and we examined this possibility in another legume species, lentil, which shows significant variation for flowering time and photoperiod responsiveness (28, 39). Linkage, sequencing and expression analyses show that a locus with a large effect on SD flowering is associated with a functionally significant polymorphism in the lentil HR ortholog. The origins of the recessive early allele are obscure but it seems to have arisen relatively recently in an Argentinian macrosperma landrace, and has subsequently provided an important means of broadening the genetic base of South Asian microsperma material (30). These results implicate altered ELF3 function as a convergent route for evolution of photoperiod-hyposensitive early-flowering in two different temperate legumes in different contexts. Taken together with recent reports that ELF3 orthologs are also involved in natural variation for flowering in rice, barley, and Arabidopsis (31, 40–42), our results also suggest HR orthologs and functionally associated clock genes including ELF4 and LUX (25) as possible candidates underlying genetic variation for phenology in a wider range of legumes. More generally, our characterization of HR and QTL6 loci provides prospects for better understanding flowering-time adaptation, identifying new molecular targets in breeding for phenology and abiotic stress tolerance, and probing the early history of cultivation in this important group of crop plants.
Methods
Plant material was obtained from the Pisum germplasm collections at the John Innes Centre (Norwich, United Kingdom) and the Australian Temperate Field Crops Collection (Horsham, VIC, Australia). Lentil lines were provided by W. Erskine (International Center for Agricultural Research in the Dry Areas, Aleppo, Syria). Plants for expression experiments were grown in growth cabinets at 20 °C under 150 µmol⋅m-2⋅s−1 white light from cool-white fluorescent tubes. All other plants were grown under an 8-h photoperiod of natural daylight either with (LD) or without (SD) an 8-h extension of 10 µmol⋅m-2⋅s−1 white light from compact fluorescent tubes (LDH; R:FR = 4.8), 40 W incandescent globes (LDL; R:FR = 0.6), or a combination of both of these light sources (LD; R:FR = 1.2). The propensity to branch was quantified as the ratio of the length of lateral branches to the increase in total plant height over a specified interval. QTL analysis was performed on a population of 92 F2 plants from a cross between NGB5839 and JI1794 grown in SD, using JoinMap 4 and MapQTL 6 (Kyazma). Details of gene-based markers used are given in Table S5. Full-length pea and lentil ELF3 genes and cDNAs were isolated using PCR techniques, genome walking (GenomeWalker universal kit; Clontech), and rapid amplification of cDNA ends (SMART RACE cDNA amplification kit; Clontech) using specific primers (Table S6). Harvested tissue for expression experiments consisted of both leaflets from the uppermost fully expanded leaf. RNA extraction, reverse-transcription, and real-time PCR analysis were performed as described in ref. 14. Construct preparation and Arabidopsis transformation were also carried out as previously described (14) and several independent transformants per construct were characterized through several generations under both LD and SD conditions. Distance and parsimony-based methods were used for phylogenetic analyses in PAUP*4.0b10 (http://paup.csit.fsu.edu).
Supplementary Material
Acknowledgments
We thank N. Ellis, J. Burstin, W. Erskine, N. Weeden, I. Murfet, R. Redden, F. Sussmilch, and A. de Saint-Germain for access to materials, background information, and data; B. Potts and J. Freeman for advice on statistical and quantitative trait locus analysis; I. Cummings, T. Winterbottom, and M. Oates for technical assistance; N. Ellis, S. Abbo, and colleagues from the University of Tasmania for helpful comments on the manuscript; and I. Murfet for support in the initial stages of the lentil work. This work was supported by Australian Research Council Grant DP0878723 (to J.L.W.); New Zealand Foundation for Research Science and Technology Grant C10X0704 (to R.C.M.); an Institut National de la Recherche Agronomique Missions des Relations Internationales grant (to B.W.); and by Génoplante (project SNPEA, ANR06-GPLA019).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession nos. JN983406 and JN983407).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207943110/-/DCSupplemental.
References
- 1.Zohary D, Hopf M. Domestication of Plants in the Old World. New York: Oxford Univ Press; 2000. [Google Scholar]
- 2.Vershinin AV, Allnutt TR, Knox MR, Ambrose MJ, Ellis TH. Transposable elements reveal the impact of introgression, rather than transposition, in Pisum diversity, evolution, and domestication. Mol Biol Evol. 2003;20(12):2067–2075. doi: 10.1093/molbev/msg220. [DOI] [PubMed] [Google Scholar]
- 3.Jing R, et al. The genetic diversity and evolution of field pea (Pisum) studied by high throughput retrotransposon based insertion polymorphism (RBIP) marker analysis. BMC Evol Biol. 2010;10:44. doi: 10.1186/1471-2148-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alonso-Blanco C, et al. What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell. 2009;21(7):1877–1896. doi: 10.1105/tpc.109.068114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abbo S, Shtienberg D, Lichtenzveig J, Lev-Yadun S, Gopher A. The chickpea, summer cropping, and a new model for pulse domestication in the ancient near east. Q Rev Biol. 2003;78(4):435–448. doi: 10.1086/378927. [DOI] [PubMed] [Google Scholar]
- 6.Makasheva RK. The Pea. Rotterdam: A.A. Balkema; 1984. [Google Scholar]
- 7.Lejeune-Hénaut I, et al. Floral initiation in field-grown forage peas is delayed to a greater extent by short photoperiods, than in other types of European varieties. Euphytica. 1999;109:201–211. [Google Scholar]
- 8.Zong X, et al. Analysis of a diverse global Pisum sp. collection and comparison to a Chinese local P. sativum collection with microsatellite markers. Theor Appl Genet. 2009;118(2):193–204. doi: 10.1007/s00122-008-0887-z. [DOI] [PubMed] [Google Scholar]
- 9.Foucher F, et al. DETERMINATE and LATE FLOWERING are two TERMINAL FLOWER1/CENTRORADIALIS homologs that control two distinct phases of flowering initiation and development in pea. Plant Cell. 2003;15(11):2742–2754. doi: 10.1105/tpc.015701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hecht V, et al. The pea GIGAS gene is a FLOWERING LOCUS T homolog necessary for graft-transmissible specification of flowering but not for responsiveness to photoperiod. Plant Cell. 2011;23(1):147–161. doi: 10.1105/tpc.110.081042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murfet IC. In: CRC Handbook of Flowering. Halevy A, editor. Boca Raton: CRC Press; 1985. pp. 97–126. [Google Scholar]
- 12.Murfet IC. Flowering in Pisum. Three distinct phenotypic classes determined by the interaction of a dominant early and a dominant late gene. Heredity. 1971;26(2):243–257. [Google Scholar]
- 13.Reid JB. Flowering in peas: Effect of the gene Hr on spectral sensitivity. Crop Sci. 1982;22(2):266–268. [Google Scholar]
- 14.Liew LC, et al. DIE NEUTRALIS and LATE BLOOMER 1 contribute to regulation of the pea circadian clock. Plant Cell. 2009;21(10):3198–3211. doi: 10.1105/tpc.109.067223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McWatters HG, Bastow RM, Hall A, Millar AJ. The ELF3 zeitnehmer regulates light signalling to the circadian clock. Nature. 2000;408(6813):716–720. doi: 10.1038/35047079. [DOI] [PubMed] [Google Scholar]
- 16.Hazen SP, et al. LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc Natl Acad Sci USA. 2005;102(29):10387–10392. doi: 10.1073/pnas.0503029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allen T, et al. Arabidopsis FHY3 specifically gates phytochrome signaling to the circadian clock. Plant Cell. 2006;18(10):2506–2516. doi: 10.1105/tpc.105.037358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salathia N, et al. FLOWERING LOCUS C-dependent and -independent regulation of the circadian clock by the autonomous and vernalization pathways. BMC Plant Biol. 2006;6:10. doi: 10.1186/1471-2229-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michaels SD, Bezerra IC, Amasino RM. FRIGIDA-related genes are required for the winter-annual habit in Arabidopsis. Proc Natl Acad Sci USA. 2004;101(9):3281–3285. doi: 10.1073/pnas.0306778101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hecht V, et al. Conservation of Arabidopsis flowering genes in model legumes. Plant Physiol. 2005;137(4):1420–1434. doi: 10.1104/pp.104.057018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bordat A, et al. Translational genomics in legumes allowed placing in silico 5460 unigenes on the pea functional map and identified candidate genes in Pisum sativum L. G3 (Bethesda) 2011;1(2):93–103. doi: 10.1534/g3.111.000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murfet IC. Flowering in Pisum. Hr, a gene for high response to photoperiod. Heredity. 1973;31(12):157–164. [Google Scholar]
- 23.Lejeune-Hénaut I, et al. The flowering locus Hr colocalizes with a major QTL affecting winter frost tolerance in Pisum sativum L. Theor Appl Genet. 2008;116(8):1105–1116. doi: 10.1007/s00122-008-0739-x. [DOI] [PubMed] [Google Scholar]
- 24.Yu JW, et al. COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol Cell. 2008;32(5):617–630. doi: 10.1016/j.molcel.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nusinow DA, et al. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature. 2011;475(7356):398–402. doi: 10.1038/nature10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deulvot C, et al. Highly-multiplexed SNP genotyping for genetic mapping and germplasm diversity studies in pea. BMC Genomics. 2010;11:468. doi: 10.1186/1471-2164-11-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Triques K, et al. Mutation detection using ENDO1: Application to disease diagnostics in humans and TILLING and Eco-TILLING in plants. BMC Mol Biol. 2008;9:42. doi: 10.1186/1471-2199-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sarker A, Erskine W, Sharma B, Tyagi MC. Inheritance and linkage relationships of days to flower and morphological loci in lentil (Lens culinaris Medikus subsp. culinaris) J Hered. 1999;90(2):270–275. [Google Scholar]
- 29.Millan T, et al. Chickpea molecular breeding: New tools and concepts. Euphytica. 2006;147(1–2):81–103. [Google Scholar]
- 30.Erskine W, et al. A bottleneck in lentil: Widening its genetic base in south Asia. Euphytica. 1998;101(2):207–211. [Google Scholar]
- 31.Jiménez-Gómez JM, Wallace AD, Maloof JN. Network analysis identifies ELF3 as a QTL for the shade avoidance response in Arabidopsis. PLoS Genet. 2010;6(9):6. doi: 10.1371/journal.pgen.1001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Colledge S, Conolly J. The Origins and Spread of Domestic Plants in Southwest Asia and Europe. Walnut Creek, CA: Left Coast Press; 2006. [Google Scholar]
- 33.Purugganan MD, Fuller DQ. The nature of selection during plant domestication. Nature. 2009;457(7231):843–848. doi: 10.1038/nature07895. [DOI] [PubMed] [Google Scholar]
- 34.Abbo S, et al. Experimental growing of wild pea in Israel and its bearing on Near Eastern plant domestication. Ann Bot (Lond) 2011;107(8):1399–1404. doi: 10.1093/aob/mcr081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan L, et al. The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science. 2004;303(5664):1640–1644. doi: 10.1126/science.1094305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beales J, Turner A, Griffiths S, Snape JW, Laurie DA. A pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.) Theor Appl Genet. 2007;115(5):721–733. doi: 10.1007/s00122-007-0603-4. [DOI] [PubMed] [Google Scholar]
- 37.Murakami M, Matsushika A, Ashikari M, Yamashino T, Mizuno T. Circadian-associated rice pseudo response regulators (OsPRRs): Insight into the control of flowering time. Biosci Biotechnol Biochem. 2005;69(2):410–414. doi: 10.1271/bbb.69.410. [DOI] [PubMed] [Google Scholar]
- 38.Murphy RL, et al. Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum. Proc Natl Acad Sci USA. 2011;108(39):16469–16474. doi: 10.1073/pnas.1106212108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Erskine W, Ellis RH, Summerfield RJ, Roberts EH, Hussain A. Characterization of responses to temperature and photoperiod for time to flowering in a world lentil collection. Theor Appl Genet. 1990;80:193–199. doi: 10.1007/BF00224386. [DOI] [PubMed] [Google Scholar]
- 40.Zakhrabekova S, et al. Induced mutations in circadian clock regulator Mat-a facilitated short-season adaptation and range extension in cultivated barley. Proc Natl Acad Sci USA. 2012;109(11):4326–4331. doi: 10.1073/pnas.1113009109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Faure S, et al. Mutation at the circadian clock gene EARLY MATURITY 8 adapts domesticated barley (Hordeum vulgare) to short growing seasons. Proc Natl Acad Sci USA. 2012;109(21):8328–8333. doi: 10.1073/pnas.1120496109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saito H, et al. Ef7 encodes an ELF3-like protein and promotes rice flowering by negatively regulating the floral repressor gene Ghd7 under both short- and long-day conditions. Plant Cell Physiol. 2012;53(4):717–728. doi: 10.1093/pcp/pcs029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.