Abstract
Cytoplasmic dynein is the major motor protein responsible for microtubule minus-end–directed movements in most eukaryotic cells. It transports a variety of cargoes and has numerous functions during spindle assembly and chromosome segregation. It is a large complex of about 1.4 MDa composed of six different subunits, interacting with a multitude of different partners. Most biochemical studies have been performed either with the native mammalian cytoplasmic dynein complex purified from tissue or, more recently, with recombinant dynein fragments from budding yeast and Dictyostelium. Hardly any information exists about the properties of human dynein. Moreover, experiments with an entire human dynein complex prepared from recombinant subunits with a well-defined composition are lacking. Here, we reconstitute a complete cytoplasmic dynein complex using recombinant human subunits and characterize its biochemical and motile properties. Using analytical gel filtration, sedimentation-velocity ultracentrifugation, and negative-stain electron microscopy, we demonstrate that the smaller subunits of the complex have an important structural function for complex integrity. Fluorescence microscopy experiments reveal that while engaged in collective microtubule transport, the recombinant human cytoplasmic dynein complex is an active, microtubule minus-end–directed motor, as expected. However, in contrast to recombinant dynein of nonmetazoans, individual reconstituted human dynein complexes did not show robust processive motility, suggesting a more intricate mechanism of processivity regulation for the human dynein complex. In the future, the comparison of reconstituted dynein complexes from different species promises to provide molecular insight into the mechanisms regulating the various functions of these large molecular machines.
Keywords: molecular motor, protein complex
Cytoplasmic dynein (here referred to as dynein) is a motor that carries out a wide variety of tasks in the cytoplasm of most eukaryotic cells. In animal cells, it is responsible for most microtubule (MT) minus-end–directed movements, transporting various cargoes, such as vesicles, mRNA particles, and organelles (1). It is also involved in numerous mitotic processes, such as nuclear envelope breakdown and centrosome separation (2), spindle-pole focusing (3), and kinetochore activity (4). Dynein is a large multisubunit complex that interacts with several accessory proteins [such as the dynactin complex (5), the lissencephaly1 (LIS1)-nuclear distribution protein E (NudE)/NudE-like complex, and Bicaudal (6)] that modulate dynein’s properties and functions.
The mammalian dynein complex (∼1.4 MDa) consists of two identical heavy chains (∼530 kDa) (7) and several associated subunits: the intermediate chain (IC) (∼75 kDa) (8); the light intermediate chain (LIC) (50–60 kDa) (8); and three light chains, LC8 (10 kDa) (9), Roadblock (RB) (11 kDa) (10), and T-complex testis-specific protein 1 (Tctex1) (13 kDa) (11). The C-terminal two-thirds of the heavy chain form the motor domain (∼380 kDa) that converts chemical energy into mechanical work (12). The N-terminal part (the tail) associates with the accessory subunits to form the cargo-binding domain. Most of the accessory subunits have been shown to be present in the complex as dimers (9, 11, 13). The IC, as the LIC, binds directly to the heavy chain and additionally binds to all three light chains and p150, a component of the dynactin complex (14). Each of the five accessory subunits is encoded by two different genes, with IC and LIC families existing in several isoforms, giving rise to the possibility that distinct dynein holoenzymes with different subunit compositions exist in cells (15). The noncatalytic subunits are thought to link dynein to its cargoes and adaptor proteins, enabling it to carry out its diverse functions.
Electron microscopy (EM) showed that two motor domains of the mammalian dynein complex are connected to a common base by flexible linkers (7, 16). Recent crystallographic studies revealed the detailed architecture of the Dictyostelium and budding yeast motor domain (17, 18). The dynein motor domain consists of three major structural elements: a six-membered AAA ring (19), of which the first four AAA domains contain ATP-binding sites, with the AAA1 being the primary site of hydrolysis (20); a stalk protruding from AAA4 (∼15 nm long) with the MT binding domain (MTBD) at its tip (17, 18, 21); and a linker (∼60 kDa) connecting the motor domain with the tail and acting as a mechanical lever, amplifying the conformational changes originating from within the motor domain to generate movement (19, 22). Additionally, C-terminal to the AAA ring is a region (∼47 kDa) that interacts with the AAA ring and has been shown to be important for motility of Dictyostelium dynein (18, 23).
The dynein motile properties are probably best understood for the budding yeast motor. Fluorescence microscopic imaging of recombinant heavy-chain constructs labeled with fluorescent dyes showed that single yeast dynein molecules are processive motors with a velocity of 90 nm/s and run length of 1.7 μm, making predominantly 8-nm steps mostly in the minus-end direction with occasional backsteps (24). Optical trapping experiments demonstrated that single yeast dynein is able to generate stall forces of ∼7 pN (25). Remarkably, intermotor domain coordination is not required for processive stepping of budding yeast dynein (26, 27), which is fundamentally different from the hand-over-hand stepping mechanism of prototypical processive motors like kinesin-1 and myosin V. Recombinant Dictyostelium dynein differs from yeast dynein in that its velocity is higher and its processive motility has been proposed to be a consequence of motor domain coordination (23).
The motile properties of individual mammalian dynein molecules have mostly been studied with the native dynein complex that was purified from brain tissue, attached to micrometer beads and observed by differential interference contrast microscopy or optical trapping. Reported mean velocities in the range from 200 to 1,000 nm/s are considerably higher, and the average lengths of processive runs of 0.3–1 μm are slightly shorter than for the yeast motor (28–31). Mammalian dynein’s stepping behavior appears to be more variable than that of its yeast homolog. Steps of 8- to 32-nm size, stall forces of 1–8 pN, pronounced diffusional motility, and back-stepping, as well as nonprocessive motility, have all been reported (30, 32–34). Remaining controversies regarding the motile behavior of mammalian dynein are difficult to resolve, at least in part, because of the lack of a recombinant dynein complex with well-defined composition and the possibility of genetic manipulation.
Here, we have reconstituted a human cytoplasmic dynein complex from recombinant subunits and have characterized it biochemically. We show that the noncatalytic subunits are crucial for stable dynein heavy-chain dimerization. The reconstituted complex was an active minus-end–directed motor that did not, however, show any robust processivity.
Results
Reconstitution of the Human Cytoplasmic Dynein Complex.
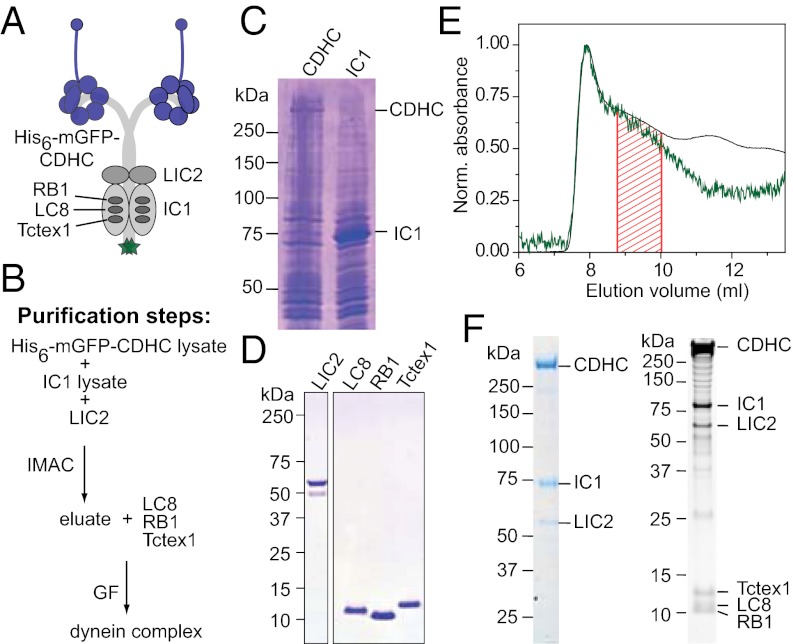
For complex assembly, we cloned an isoform of each of the six cytoplasmic dynein subunits, choosing a set of noncatalytic subunit isoforms that could easily be amplified from human brain cDNA (Fig. 1A). The subunits were expressed individually and then combined (Fig. 1B). The human cytoplasmic dynein heavy chain (CDHC) fused to an N-terminal oligo-histidine tag and a monomeric (m)GFP (His6-mGFP-CDHC; Fig. S1A, Top), and the IC1 were expressed in insect cells (Fig. 1C). The LIC2, LC8 light chain 1 (LC8), RB light chain 1 (RB1), and Tctex1 light chain 1 (Tctex1) were expressed in Escherichia coli and purified (Fig. 1D). Insect cell lysates containing overexpressed CDHC and IC1 were mixed and supplemented with the purified LIC2 (Fig. 1B). A complex of these three proteins was purified using the His6 tag present only on the heavy chain. Three purified light chains were then added, and the mixture was subjected to gel filtration where a soluble dynein complex eluted as a broad peak (region shaded in red that followed the void-volume peak of the column in Fig. 1E). Stained SDS gels revealed that the complex contained all six dynein subunits (Fig. 1F).
Fig. 1.
Reconstitution of the human cytoplasmic dynein complex. (A) Schematic of the subunit composition of the dynein complex studied here. A His6 tag, followed by mGFP (green), is fused to the N terminus of the CDHC (Fig. S1A, Top). (B) Purification scheme: lysates of cells expressing His6-mGFP-CDHC (C, lane 1; 560 kDa; referred to here as CDHC) and cells expressing IC1 (C, lane 2; 71 kDa) were mixed, supplemented with purified LIC2 (D, lane 1; 54 kDa), and subjected to immobilized metal-ion–affinity chromatography (IMAC). The eluate was supplemented with purified light chains (D, lanes 2–4; 10–13 kDa) and gel-filtered (GF). (C and D) Coomassie-stained SDS gels showing lysates of cells expressing CDHC and IC1 (C) and purified LIC2 and light chains (D), as indicated. (E) Gel-filtration profile around the position where the dynein complex elutes showing normalized absorbance values at 280 nm (black line) and 488 nm (green line). The fraction between the red lines was collected and analyzed. (F) Coomassie-stained (Left) and SYPRO Ruby–stained (Right) SDS gel showing the dynein complex after gel filtration.
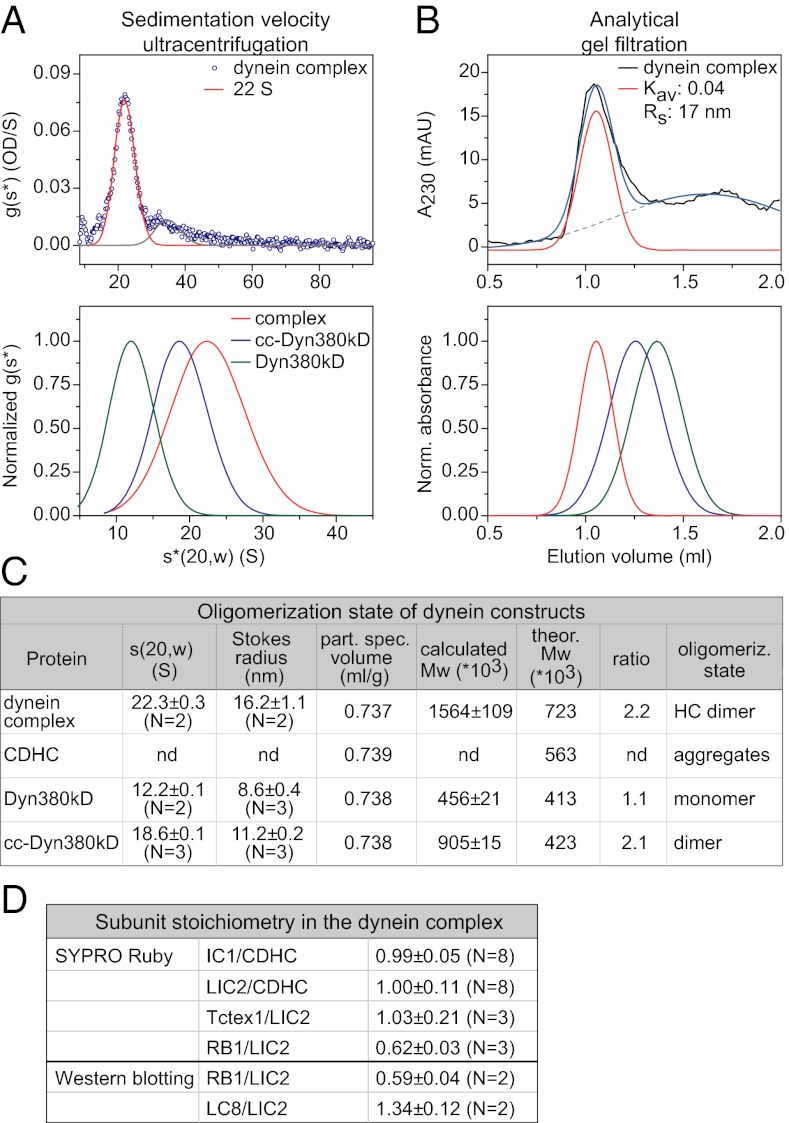
To determine the oligomerization state of the purified dynein complex, we analyzed its hydrodynamic properties. Analysis by sedimentation-velocity ultracentrifugation showed that the major population of the reconstituted complex had a sedimentation coefficient of 22S (Fig. 2A, Upper), similar to the reported values for native vertebrate dynein (∼20S; based on sucrose density gradient centrifugation analysis) (8, 15, 35). For comparison, we also purified two truncated human dynein heavy-chain constructs: the motor domain (Dyn380kD) and artificially dimerized motor domains (cc-Dyn380kD) (Fig. S1). Their sedimentation coefficients were 12S (Fig. 2A, Lower and Fig. S2A, Upper) and 19S (Fig. 2A, Lower and Fig. S2A, Lower), respectively. These results gave a first indication that a dynein complex of the correct size had formed.
Fig. 2.
Oligomerization state and subunit stoichiometry of the reconstituted human dynein complex. (A, Upper) Sedimentation coefficient distribution [g(s*) plot] of the dynein complex. Gaussian functions (lines) were fitted to the data (blue circles), with the peak center at 22S for the major species (82%) of the dynein complex (red line). (A, Lower) Comparison of normalized fits to the g(s*) plots of the dynein complex (red; as above), the dynein motor domain (Dyn380kD, green; Fig. S2A, Upper), and the dynein motor domain fused to a dimerizing coiled coil (cc) sequence (cc-Dyn380kD, blue; Fig. S2A, Lower). (B, Upper) Analytical gel-filtration profile of the dynein complex. Gaussian fits (colored lines) to the data (black line) reveal a peak center at the Kav value of 0.04 (corresponding to Stokes radius, Rs, of 17 nm) for the main eluate (red line). The blue line shows the sum of the two Gaussian curves. (B, Lower) Comparison of normalized fits to the gel-filtration profiles of the dynein complex and truncated dynein constructs (color code as in A, Lower; see also Fig. S2B). (C) Summary of results from the sedimentation-velocity ultracentrifugation [s(20,w)] and analytical gel filtration (Stokes radius) and calculated partial specific volumes for the dynein complex, CDHC alone, and truncated control constructs. Mw, molecular mass. Errors are SEM. (D) Summary of the mean molar ratios of subunits in the reconstituted dynein complex. Errors are SEM. For experimental data, see Fig. S3.
To confirm this, we sought to determine the molecular mass of the reconstituted complex and the control constructs, and for this purpose, we measured their Stokes radius. Using analytical gel filtration we found that the mean Stokes radius of the reconstituted complex was 16 nm (Fig. 2 B Upper and C and Fig. S2B, Inset), which was characteristically different from the Stokes radii of the controls (9 nm for the motor domain and 11 nm for the artificially dimerized motor domains) (Fig. 2 B Lower and C and Fig. S2B). The molecular mass, as calculated from the sedimentation coefficient and the Stokes radius (SI Materials and Methods), was 1.6 MDa for the reconstituted complex (Fig. 2C). This confirms that it consists of a dimer of heavy chains and other subunits, like the native dynein complex (7, 15). In contrast, the human dynein motor domain alone was found to be monomeric, and the artificially dimerized motor domain was indeed a dimer, as expected (Fig. 2C).
Next, we determined the stoichiometry of subunits in the reconstituted human dynein complex. Based on quantitative analysis of SYPRO Ruby–stained SDS gels and Western blots, CDHC, IC1, LIC2, and Tctex1 were found to be in equimolar amounts in the purified dynein complex, whereas LC8 was present in slightly higher (1.3 molecule per CDHC) and RB1 in slightly lower amount (0.6 molecule per CDHC) (Fig. 2D and Fig. S3). In conclusion, all subunits were present in the recombinant dynein complex at roughly equimolar amounts.
Low-Resolution Structure of the Reconstituted Dynein Complex.
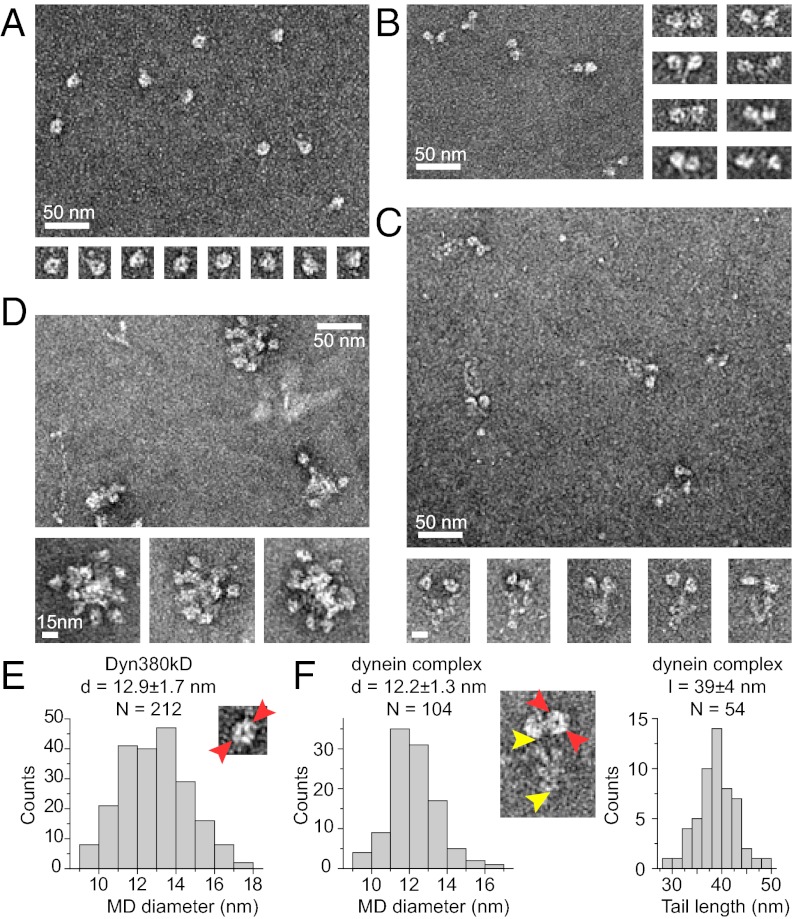
Individual dynein molecules are big enough to be observed by negative-stain EM (16). As expected, individual monomeric motor domains appeared ring-shaped (Fig. 3A) with a mean diameter of 12.9 nm (Fig. 3E), in agreement with a reported value for a recombinant dynein motor domain from Dictyostelium (18, 36). The artificially dimerized motor domains appeared, indeed, as pairs of motor domains (Fig. 3B). The reconstituted dynein complex clearly showed two identical globular motor domains connected to a tail domain (Fig. 3C). The average diameter of the motor domain was 12.2 nm, and the average length of the tail domain was 39 nm (Fig. 3F), in agreement with the dimensions of native dynein purified from tissue (7, 16). These results provide further evidence that the reconstituted human dynein complex has native properties.
Fig. 3.
EM images of negatively stained dynein constructs. Overview of samples (large images) and galleries of single dynein particles (small images): motor domain Dyn380kD (A); artificially dimerized motor domains cc-Dyn380kD (B); dynein complex (C); and CDHC (D). Scale is the same for all single particles images in A–D. (E) Histogram of the distribution of the Dyn380kD motor domain (MD) diameter [measured as indicated by red arrowheads in the EM image (30 × 30 nm)]. (F) Histograms of the distribution of the dynein complex MD diameter (Left) and the tail domain length (Right) [measured as indicated on the EM image (60 × 80 nm) by red and yellow arrowheads, respectively]. Indicated are mean values ± SD. d, diameter; l, length.
Noncatalytic Subunits of the Dynein Complex Are Crucial for Heavy-Chain Dimerization and Stability.
The heavy chain alone (without any accessory subunits) was mostly insoluble under our standard conditions. It eluted in the void-volume peak after gel filtration (Fig. S1B, Top, and S1C), was poorly soluble (Table 1), and appeared as large aggregates in EM (Fig. 3D). Because the dynein motor domain was highly soluble and did not aggregate (Figs. S1 and S2 and Fig. 3A), this result suggests that the tail domain of the human heavy chain aggregates in the absence of the accessory subunits and that the tail domains alone are not sufficient for correct dimerization. Increasing the ionic strength of the buffer allowed the purification of a soluble heavy chain (Fig. S4A, Lower). However, sedimentation-velocity analysis showed that under these conditions, the heavy chain alone is clearly more heterogeneous than the entire dynein complex purified under the same conditions (Fig. S4 A and B). Furthermore, the most prominent soluble species at high-ionic strength had a molecular mass that is rather consistent with a monomer (Fig. S4D), in clear contrast to the entire complex that also formed a dimer of heavy chains at increased ionic strength (Fig. S4D). No conditions were found under which the heavy chain alone formed stable, soluble dimers.
Table 1.
Properties of dynein subcomplexes
| Protein | Preparative gel filtration | Solubility (mg/mL) | s(20,w) (S) | CDHC oligomerization state |
| Complex | Elutes after void volume | ∼0.2 | 21.4 | Dimer |
| CDHC + IC + LIC | Similar to the complex | ∼0.2 | 19.1 | Dimer |
| CDHC + LIC | Similar to the complex | ∼0.1 | 19.5 | Dimer |
| CDHC + IC | Elutes in void volume | ∼0.02 | ND | Mostly aggregates |
| CDHC | Elutes in void volume | ∼0.03 | ND | Aggregates |
ND, not determined
Successful dynein complex reconstitution allowed us to investigate the specific roles of the individual subunits for complex formation. Adding IC did not make the heavy chain more soluble under our standard conditions (Table 1), as demonstrated by elution of the protein in the void-volume peak after gel filtration and by the detection of mostly aggregates in EM (Fig. S5). The heavy chain in combination with LIC was partially soluble, and together with both LIC and IC, it showed similar solubility as the entire complex (Table 1). Both combinations CDHC-LIC and CDHC-IC-LIC formed mostly dimers, as demonstrated by EM (Fig. S5). However, both of these complexes did not have the entirely correct shape, as demonstrated by their reduced s values (19.5S and 19.1S, respectively) (Fig. S6B and Table 1). This was also reflected by the appearance of often less compact tails in EM images (Fig. S5). Therefore, correct complex formation requires the LIC, the IC, and the light chains, in this order of importance. These experiments identify an important structural role of the subunits for the assembly of the dynein complex.
Motile Properties of Recombinant Dynein Constructs.
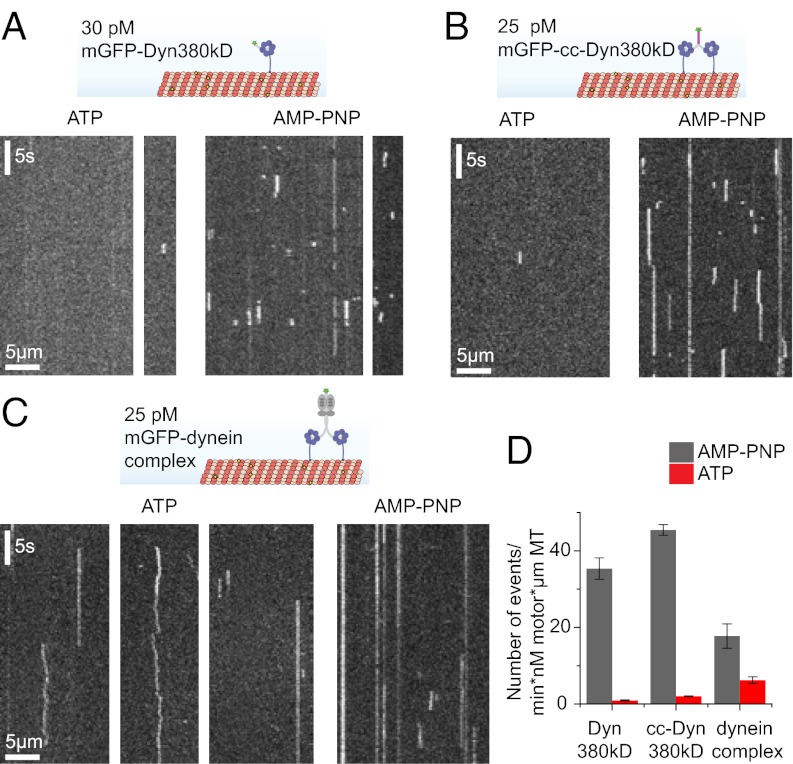
Because our dynein constructs were mGFP-tagged, we could directly observe how individual motors interact with immobilized MTs, using time-lapse total internal reflection fluorescence (TIRF) microscopy. Individual monomeric dynein motor domains showed hardly any detectable binding events in the presence of ATP and bound frequently and statically to MTs in the presence of the nonhydrolysable ATP analog adenosine 5′-(β,γ-imido)triphosphate (AMP-PNP) (Fig. 4 A and D). This is typical for a nonprocessive or weakly processive motor and is expected for truncated monomeric dynein (37). Unexpectedly, the artificially dimerized dynein motor domains exhibited similar behavior (Fig. 4 B and D). In agreement with the difference in their oligomeric state (Fig. 2C and Fig. S7A), the dimers remained bound to the MT at least three times longer than the monomeric motor domain in the presence of AMP-PNP (Fig. S7B). Their mean dwell time of 3 s in a strongly MT-bound state is, however, much shorter in comparison with that of a similar construct of processive dynein from budding yeast (38) or of processive kinesin-1 (Fig. S7C). In combination with a measured binding rate of 45 min−1⋅nM−1⋅µm−1 MT in this experiment (Fig. 4D), which is very similar to a recently measured binding rate for a dimeric dynein construct from Dictyostelium (23), this suggests that the affinity for MT binding of the human dynein motor domains in a strongly MT-binding state is rather low. These experiments show that, unlike its yeast and Dictyostelium counterparts (23, 24), the artificially dimerized recombinant human dynein is unexpectedly nonprocessive, which suggests that the intrinsic motile properties of these motors are different.
Fig. 4.
Single-molecule behavior of human dynein. (A–C, Upper) Schematic of the assay showing individual mGFP-labeled dyneins on fluorescent MTs that are imaged using TIRFM. (A–C, Lower) Exemplary kymographs (space–time plots) show individual monomeric Dyn380kD (A), dimeric cc-Dyn380kD (B), and dynein complex (C) molecules interacting with MTs in the presence of ATP or AMP-PNP, as indicated. (D) Total number of binding events per minute and per concentration of motor (nM) and MT length (µm). Error bars are SEM. Number N of independent experiments: dynein complex with ATP (n = 4) and AMP-PNP (n = 2), cc-Dyn380kD with ATP (n = 3) and AMP-PNP (n = 2), and Dyn380kD with ATP and AMP-PNP (both n = 2). The motor concentrations in this figure refer to monomers for Dyn380kD and dimers for cc-Dyn380kD and the dynein complex.
Surprisingly, individual mGFP–dynein complex molecules also did not show robust processive motility (Fig. 4C), although their dimeric state in this experiment was also confirmed by single-molecule fluorescence intensity analysis (Fig. S7A). The overall behavior of the dynein complex was rather heterogeneous in the presence of ATP (Fig. 4C, Left), showing both short- and long-binding events, with many molecules staying bound for many seconds. A fraction of motors showed diffusive behavior, rarely with a directional bias. In the presence of AMP-PNP, the dynein complex bound statically, as expected, with only a small fraction of diffusively moving molecules (Fig. 4C, Right). The numerous long-binding events present in both nucleotide conditions indicate that the tail domain of the dynein complex likely contributes to binding to the MT (Fig. 4C). These data suggest that recombinant human dynein is either only weakly processive, at most, or in an inactive state.
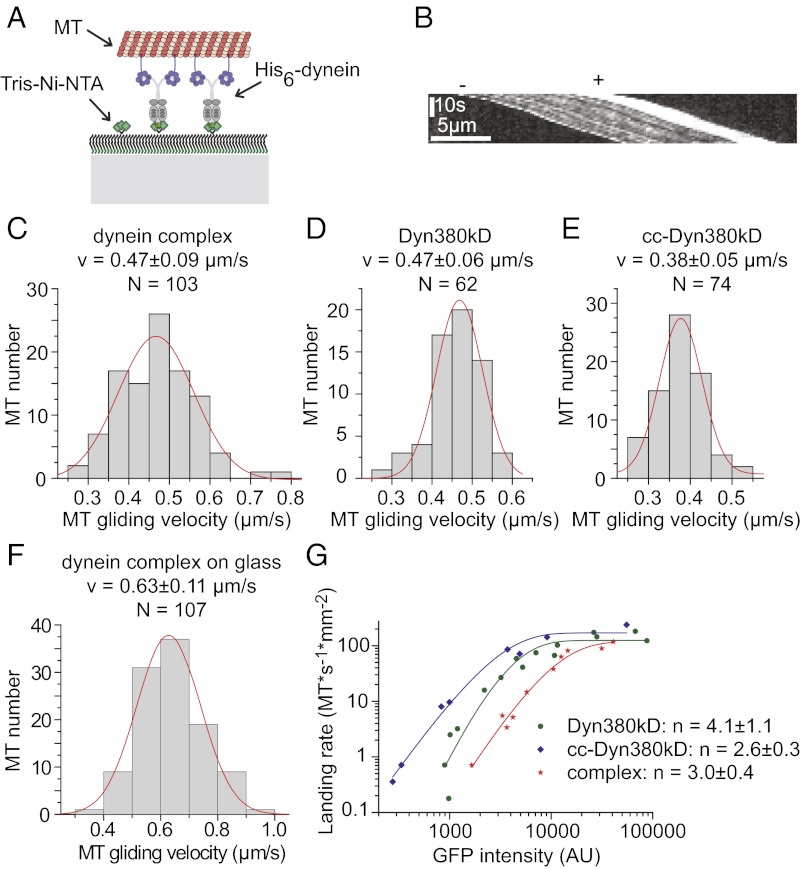
To distinguish between these possibilities, we performed MT-gliding assays. Because our constructs had an N-terminal His6 tag in addition to the mGFP tag, we could immobilize the same constructs that were used for single-molecule imaging on Tris-Ni-NTA-PEG–functionalized glass surfaces in an oriented manner (Fig. 5A). As an ensemble, the immobilized dynein complex robustly moved polarity-marked MTs with their brightly labeled plus-ends leading (Fig. 5B and Movie S1), indicative of minus-end–directed motility. MT transport-velocity distributions were Gaussian, as expected, and the mean velocity of MT transport driven by the immobilized complex was 0.5 μm/s (Fig. 5C). Similar mean velocities were observed for the truncated monomeric and artificially dimerized dynein constructs (Fig. 5 D and E and Movies S2 and S3, respectively). When the dynein complex was immobilized nonspecifically to nonfunctionalized glass, the gliding speed increased by about 35% to 0.63 μm/s (Fig. 5F and Movie S4), suggesting that the orientation of the immobilized complex might have a certain influence on dynein’s motile behavior. Taken together, the gliding velocities observed here for the various recombinant human dynein constructs are close to the reported velocities of vertebrate brain-purified dynein (0.6–1.2 μm/s) (8, 35).
Fig. 5.
Dynein motility in MT-gliding assays. (A) Schematic of the assay on Tris-Ni-NTA-PEG–functionalized glass: oligo-His–tagged motors are immobilized on a Tris-Ni-NTA–functionalized surface in an oriented manner. Fluorescently labeled MTs are imaged by TIRFM. (B) Exemplary kymograph of a polarity marked MT transported by the dynein complex. (C–E) Histograms showing MT-gliding velocity distributions of the dynein complex (C), Dyn380kD (D), and cc-Dyn380kD (E). (F) Histogram showing the MT-gliding velocity distribution of the dynein complex immobilized on nonfunctionalized glass. Gaussian fits (red lines) with mean velocity (v) ± SD as indicated. (G) Landing-rate profile for the dynein complex (red stars) immobilized on nonfunctionalized glass and Dyn380kD (green circles) and cc-Dyn380kD (blue diamonds) both immobilized on Tris-Ni-NTA–functionalized glass. The density of immobilized motor is expressed as the measured mGFP intensity of the dynein constructs. The continuous curves are fits to the data (SI Materials and Methods) yielding the minimal number n of motors required to support MT gliding.
To estimate the degree of nonprocessivity of the human dynein complex, we measured the MT-landing rate as a function of the relative motor density (measured directly as mGFP fluorescence that increased linearly with dynein concentration). The measured landing rates predicted (39) a minimal number of three dynein complexes or artificial dimers being required to transport a MT continuously (Fig. 5G). The number for the monomer was four (Fig. 5G), very similar to the monomeric Dictyostelium dynein motor domain (40). These results agree with our conclusions drawn from single-dynein-molecule imaging on immobilized MTs.
In summary, recombinant human dynein complexes are active motors showing robust minus-end–directed motility when acting as an ensemble, but individual complexes are not processive.
Discussion
Here, we have demonstrated that the entire human cytoplasmic dynein complex can be assembled from six recombinant proteins (Fig. 1). The reconstituted complex has a molecular mass consistent with a heavy-chain dimer (Fig. 2 A–C) and the expected shape of the native motor (Fig. 3 C and F) (7, 16). All subunits are present at roughly equimolar amounts (Fig. 2D and Fig. S3), as reported previously for most subunits in the native mammalian dynein complex (9, 11, 13). The slightly lower amount of RB1 (0.6 molecule per CDHC) might be attributable to either weaker association with the IC or to an intrinsic tendency of RB1 to form isoform heterodimers, although homodimers have also been observed previously (10).
We show that the heavy-chain–associated subunits are necessary and sufficient for correct complex formation (Fig. 3 C and D, Figs. S4–S6, and Table 1). Their structural importance in descending order is LIC, IC, and light chains. Full solubility requires both the presence of LIC and IC and correlates with stable heavy-chain dimerization, which is largely independent of the light chains. Nevertheless, the light chains are needed for complete folding of the complex. These results emphasize the essential structural role of the smaller dynein subunits in addition to mediating interactions with adaptor proteins and cargoes (1). This conclusion agrees with previous reports about the instability of a heavy chain–LIC subcomplex obtained after separation of bovine brain–purified dynein into subcomplexes using a chaotropic agent (15) and of purified recombinant full-length rat CDHC (41). The possibility to generate a recombinant complex opens the door for a systematic dissection of the functional consequences of different subunit isoforms in the complex and for the assembly of the entire dynein/dynactin complex in recombinant form in the future.
The recombinant dynein complex is an active motor showing the expected minus-end–directed motility in MT-gliding assays (Fig. 5 B, C, and F and Movies S1 and S4) (42). The measured speed of ∼0.6 µm/s was well within the range of published values for native vertebrate dynein purified from tissue (8, 35) or of human recombinant dynein heavy chain overexpressed in HEK cells and purified via an affinity tag (43). This is lower than for recombinant Dictyostelium dynein (40) and higher than for yeast dynein (24), as expected. The gliding speed was not affected by the absence or presence of the tail but, to a certain extent, by the method of immobilization (Fig. 5 C and F), suggesting that the orientation of the immobilized complex can affect aspects of its motile properties.
Although the gliding assays demonstrated that the recombinant human dynein complex has expected motile properties in this assay, single-molecule fluorescence imaging showed that it is not measurably processive (Fig. 4C), and landing assays demonstrated that surface attachment also did not “activate” the motor in the sense of making it processive (Fig. 5G). This lack of processivity was not a consequence of a possibly not correctly assembled complex but a characteristic property of the recombinant human motor domain. This was revealed by the behavior of the truncated and artificially dimerized heavy chain (Fig. 4B) that contained the entire motor domain, including the part of the linker that was also present in the corresponding yeast and Dictyostelium constructs that were shown to be processive (23, 24). Despite its largely uncoordinated stepping mechanism, recombinant yeast dynein has been demonstrated to be a processive motor even when motor domains were artificially dimerized (24), probably because the individual motor domains have a sufficiently high duty ratio (fraction of time the motor is MT-bound during its biochemical cycle) in combination with a high affinity for MT binding (26, 27, 38). Similarly, artificially dimerized Dictyostelium dynein motor domains were also observed to be processive (23). However, for this motor, direct coordination between motor domains was suggested as a reason for processivity that depended on a sequence C terminus of the motor domain that is largely absent in the yeast dynein (23). This opens the possibility that different molecular mechanisms lead to processive stepping in yeast and Dictyostelium dynein. In contrast to dynein from these two species, the artificially dimerized human dynein did not show robust processive motility (Fig. 4B), as neither did the entire recombinant human dynein complex (Fig. 4C). This could be a consequence of weak MT binding of the recombinant human dynein motor domain in combination with uncoordinated stepping.
The nonprocessivity of mGFP-labeled human dynein was surprising, because most experiments with the native vertebrate dynein complex nonspecifically adsorbed to microspheres or conjugated to quantum dots by antibodies reported processive motility of individual complexes (28, 31, 32) [although nonprocessive movement has been observed as well (34)]. At present, in contrast to the yeast or Dictyostelium dynein studies, no experiments addressing the processivity of vertebrate dynein have been reported with a dynein heavy-chain fragment or a component of the dynein complex covalently attached to a fluorescent protein or small fluorophore. A study where porcine brain dynein was observed indirectly using a fluorescently labeled interaction partner (a p150 fragment not able to bind to MTs) reported an absence of processive runs in the presence of ATP, similar to our observations with the recombinant human dynein (44). Native dynein from a transgenic mouse that was copurified together with the dynactin complex carrying the fluorescent subunit p50 displayed processive, but distinctly bidirectional movement with a minus-directed bias (33). This suggests that the dynactin complex might be required for processive motility of vertebrate dynein. A processivity-enhancing role was already reported previously (45, 46). At present, it appears that the differences in the observations regarding mammalian dynein’s processivity could be a consequence of widely varying experimental conditions (adsorption to microspheres versus genetically encoded tags, purified recombinant protein after heterologous expression versus native complex from tissue, presence versus absence of dynactin), emphasizing the need for a motor preparation with a composition and label that can be precisely controlled and varied.
Our study here has demonstrated that a recombinant human dynein complex with a well-defined composition can be assembled from recombinant subunits. The accessory subunits have an important structural role for complex integrity. Our reconstitution protocol now allows the study of all subunits in mutated form. The reconstituted human complex displays robust minus-end–directed motility when working as part of an ensemble but does not move processively when acting as an individual molecule. Thus, comparing recombinant dyneins from different species leads to the conclusion that they appear to differ in their ability to move processively, suggesting that the regulation of dynein’s processivity is more intricate than previously anticipated. A systematic exploration of the regulation of dynein’s motile properties will profit from the availability of recombinant dynein and entire dynein/dynactin complexes from different species in the future.
Materials and Methods
Proteins were expressed in E. coli or in insect cells and purified using a combination of metal ion-affinity chromatography and gel filtration. The oligomeric state of the dynein constructs and the subunit stoichiometry of the complex were determined using analytical ultracentrifugation, analytical gel filtration, negative stain electron microscopy, gel electrophoresis and western blotting. The motile properties of dynein were measured in TIRF-microscopy-based motility assays. For details, see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Imre Berger for MultiBac cells and advice; Nicholas Cade for TIRF microscopy support and single-molecule fluorescence intensity analysis; Johanna Roostalu for polarity-marked MTs; Ivo Telley for purified Kin401-mGFP; the European Molecular Biology Laboratory (EMBL) Protein Expression and Purification Facility for plasmids, Sf21 cells, and purified tobacco etch virus (TEV) protease; Jeanette Seiler for protein expressions; Claude Antony for temporary laboratory space and discussions; Iris Lüke for technical support; Hannah Armer, Raffaella Carzaniga, and Lucy Collinson [London Research Institute (LRI) EM Unit] for help with EM; the LRI Protein Analysis and Proteomics Facility for mass spectrometric analysis; and Nicholas Cade, Christian Düllberg, Franck Fourniol, Johanna Roostalu, and Martin Singleton for critical comments on the manuscript. This work was supported by the Human Frontier Science Program, Deutsche Forschungsgemeinschaft, EMBL, and Cancer Research UK.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1210573110/-/DCSupplemental.
References
- 1.Allan VJ. Cytoplasmic dynein. Biochem Soc Trans. 2011;39(5):1169–1178. doi: 10.1042/BST0391169. [DOI] [PubMed] [Google Scholar]
- 2.Gönczy P. Nuclear envelope: Torn apart at mitosis. Curr Biol. 2002;12(7):R242–R244. doi: 10.1016/s0960-9822(02)00781-9. [DOI] [PubMed] [Google Scholar]
- 3.Merdes A, Heald R, Samejima K, Earnshaw WC, Cleveland DW. Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J Cell Biol. 2000;149(4):851–862. doi: 10.1083/jcb.149.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bader JR, Vaughan KT. Dynein at the kinetochore: Timing, Interactions and Functions. Semin Cell Dev Biol. 2010;21(3):269–275. doi: 10.1016/j.semcdb.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schroer TA. Dynactin. Annu Rev Cell Dev Biol. 2004;20:759–779. doi: 10.1146/annurev.cellbio.20.012103.094623. [DOI] [PubMed] [Google Scholar]
- 6.Kardon JR, Vale RD. Regulators of the cytoplasmic dynein motor. Nat Rev Mol Cell Biol. 2009;10(12):854–865. doi: 10.1038/nrm2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vallee RB, Wall JS, Paschal BM, Shpetner HS. Microtubule-associated protein 1C from brain is a two-headed cytosolic dynein. Nature. 1988;332(6164):561–563. doi: 10.1038/332561a0. [DOI] [PubMed] [Google Scholar]
- 8.Paschal BM, Shpetner HS, Vallee RB. MAP 1C is a microtubule-activated ATPase which translocates microtubules in vitro and has dynein-like properties. J Cell Biol. 1987;105(3):1273–1282. doi: 10.1083/jcb.105.3.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.King SM, et al. Brain cytoplasmic and flagellar outer arm dyneins share a highly conserved Mr 8,000 light chain. J Biol Chem. 1996;271(32):19358–19366. doi: 10.1074/jbc.271.32.19358. [DOI] [PubMed] [Google Scholar]
- 10.Nikulina K, Patel-King RS, Takebe S, Pfister KK, King SM. The Roadblock light chains are ubiquitous components of cytoplasmic dynein that form homo- and heterodimers. Cell Motil Cytoskeleton. 2004;57(4):233–245. doi: 10.1002/cm.10172. [DOI] [PubMed] [Google Scholar]
- 11.King SM, et al. The mouse t-complex-encoded protein Tctex-1 is a light chain of brain cytoplasmic dynein. J Biol Chem. 1996;271(50):32281–32287. doi: 10.1074/jbc.271.50.32281. [DOI] [PubMed] [Google Scholar]
- 12.Nishiura M, et al. A single-headed recombinant fragment of Dictyostelium cytoplasmic dynein can drive the robust sliding of microtubules. J Biol Chem. 2004;279(22):22799–22802. doi: 10.1074/jbc.M313362200. [DOI] [PubMed] [Google Scholar]
- 13.King SM, et al. Cytoplasmic dynein contains a family of differentially expressed light chains. Biochemistry. 1998;37(43):15033–15041. doi: 10.1021/bi9810813. [DOI] [PubMed] [Google Scholar]
- 14.Vallee RB, Williams JC, Varma D, Barnhart LE. Dynein: An ancient motor protein involved in multiple modes of transport. J Neurobiol. 2004;58(2):189–200. doi: 10.1002/neu.10314. [DOI] [PubMed] [Google Scholar]
- 15.King SJ, Bonilla M, Rodgers ME, Schroer TA. Subunit organization in cytoplasmic dynein subcomplexes. Protein Sci. 2002;11(5):1239–1250. doi: 10.1110/ps.2520102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amos LA. Brain dynein crossbridges microtubules into bundles. J Cell Sci. 1989;93(Pt 1):19–28. doi: 10.1242/jcs.93.1.19. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt H, Gleave ES, Carter AP. Insights into dynein motor domain function from a 3.3-Å crystal structure. Nat Struct Mol Biol. 2012;19(5):492–497, S1. doi: 10.1038/nsmb.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kon T, et al. The 2.8 Å crystal structure of the dynein motor domain. Nature. 2012;484(7394):345–350. doi: 10.1038/nature10955. [DOI] [PubMed] [Google Scholar]
- 19.Roberts AJ, et al. AAA+ Ring and linker swing mechanism in the dynein motor. Cell. 2009;136(3):485–495. doi: 10.1016/j.cell.2008.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kon T, Nishiura M, Ohkura R, Toyoshima YY, Sutoh K. Distinct functions of nucleotide-binding/hydrolysis sites in the four AAA modules of cytoplasmic dynein. Biochemistry. 2004;43(35):11266–11274. doi: 10.1021/bi048985a. [DOI] [PubMed] [Google Scholar]
- 21.Gee MA, Heuser JE, Vallee RB. An extended microtubule-binding structure within the dynein motor domain. Nature. 1997;390(6660):636–639. doi: 10.1038/37663. [DOI] [PubMed] [Google Scholar]
- 22.Kon T, Mogami T, Ohkura R, Nishiura M, Sutoh K. ATP hydrolysis cycle-dependent tail motions in cytoplasmic dynein. Nat Struct Mol Biol. 2005;12(6):513–519. doi: 10.1038/nsmb930. [DOI] [PubMed] [Google Scholar]
- 23.Numata N, Shima T, Ohkura R, Kon T, Sutoh K. C-sequence of the Dictyostelium cytoplasmic dynein participates in processivity modulation. FEBS Lett. 2011;585(8):1185–1190. doi: 10.1016/j.febslet.2011.03.036. [DOI] [PubMed] [Google Scholar]
- 24.Reck-Peterson SL, et al. Single-molecule analysis of dynein processivity and stepping behavior. Cell. 2006;126(2):335–348. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gennerich A, Carter AP, Reck-Peterson SL, Vale RD. Force-induced bidirectional stepping of cytoplasmic dynein. Cell. 2007;131(5):952–965. doi: 10.1016/j.cell.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeWitt MA, Chang AY, Combs PA, Yildiz A. Cytoplasmic dynein moves through uncoordinated stepping of the AAA+ ring domains. Science. 2012;335(6065):221–225. doi: 10.1126/science.1215804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiu W, et al. Dynein achieves processive motion using both stochastic and coordinated stepping. Nat Struct Mol Biol. 2012;19(2):193–200. doi: 10.1038/nsmb.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, Khan S, Sheetz MP. Single cytoplasmic dynein molecule movements: Characterization and comparison with kinesin. Biophys J. 1995;69(5):2011–2023. doi: 10.1016/S0006-3495(95)80071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mallik R, Petrov D, Lex SA, King SJ, Gross SP. Building complexity: An in vitro study of cytoplasmic dynein with in vivo implications. Curr Biol. 2005;15(23):2075–2085. doi: 10.1016/j.cub.2005.10.039. [DOI] [PubMed] [Google Scholar]
- 30.Toba S, Watanabe TM, Yamaguchi-Okimoto L, Toyoshima YY, Higuchi H. Overlapping hand-over-hand mechanism of single molecular motility of cytoplasmic dynein. Proc Natl Acad Sci USA. 2006;103(15):5741–5745. doi: 10.1073/pnas.0508511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ori-McKenney KM, Xu J, Gross SP, Vallee RB. A cytoplasmic dynein tail mutation impairs motor processivity. Nat Cell Biol. 2010;12(12):1228–1234. doi: 10.1038/ncb2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mallik R, Carter BC, Lex SA, King SJ, Gross SP. Cytoplasmic dynein functions as a gear in response to load. Nature. 2004;427(6975):649–652. doi: 10.1038/nature02293. [DOI] [PubMed] [Google Scholar]
- 33.Ross JL, Wallace K, Shuman H, Goldman YE, Holzbaur EL. Processive bidirectional motion of dynein-dynactin complexes in vitro. Nat Cell Biol. 2006;8(6):562–570. doi: 10.1038/ncb1421. [DOI] [PubMed] [Google Scholar]
- 34.Walter WJ, Koonce MP, Brenner B, Steffen W. Two independent switches regulate cytoplasmic dynein’s processivity and directionality. Proc Natl Acad Sci USA. 2012;109(14):5289–5293. doi: 10.1073/pnas.1116315109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schroer TA, Sheetz MP. Two activators of microtubule-based vesicle transport. J Cell Biol. 1991;115(5):1309–1318. doi: 10.1083/jcb.115.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Samsó M, Koonce MP. 25 Angstrom resolution structure of a cytoplasmic dynein motor reveals a seven-member planar ring. J Mol Biol. 2004;340(5):1059–1072. doi: 10.1016/j.jmb.2004.05.063. [DOI] [PubMed] [Google Scholar]
- 37.Imamula K, Kon T, Ohkura R, Sutoh K. The coordination of cyclic microtubule association/dissociation and tail swing of cytoplasmic dynein. Proc Natl Acad Sci USA. 2007;104(41):16134–16139. doi: 10.1073/pnas.0702370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang J, Roberts AJ, Leschziner AE, Reck-Peterson SL. Lis1 acts as a “clutch” between the ATPase and microtubule-binding domains of the dynein motor. Cell. 2012;150(5):975–986. doi: 10.1016/j.cell.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hancock WO, Howard J. Processivity of the motor protein kinesin requires two heads. J Cell Biol. 1998;140(6):1395–1405. doi: 10.1083/jcb.140.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shima T, Imamula K, Kon T, Ohkura R, Sutoh K. Head-head coordination is required for the processive motion of cytoplasmic dynein, an AAA+ molecular motor. J Struct Biol. 2006;156(1):182–189. doi: 10.1016/j.jsb.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 41.Mazumdar M, Mikami A, Gee MA, Vallee RB. In vitro motility from recombinant dynein heavy chain. Proc Natl Acad Sci USA. 1996;93(13):6552–6556. doi: 10.1073/pnas.93.13.6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paschal BM, Vallee RB. Retrograde transport by the microtubule-associated protein MAP 1C. Nature. 1987;330(6144):181–183. doi: 10.1038/330181a0. [DOI] [PubMed] [Google Scholar]
- 43.Ichikawa M, Watanabe Y, Murayama T, Toyoshima YY. Recombinant human cytoplasmic dynein heavy chain 1 and 2: Observation of dynein-2 motor activity in vitro. FEBS Lett. 2011;585(15):2419–2423. doi: 10.1016/j.febslet.2011.06.026. [DOI] [PubMed] [Google Scholar]
- 44.Miura M, Matsubara A, Kobayashi T, Edamatsu M, Toyoshima YY. Nucleotide-dependent behavior of single molecules of cytoplasmic dynein on microtubules in vitro. FEBS Lett. 2010;584(11):2351–2355. doi: 10.1016/j.febslet.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 45.Kardon JR, Reck-Peterson SL, Vale RD. Regulation of the processivity and intracellular localization of Saccharomyces cerevisiae dynein by dynactin. Proc Natl Acad Sci USA. 2009;106(14):5669–5674. doi: 10.1073/pnas.0900976106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.King SJ, Schroer TA. Dynactin increases the processivity of the cytoplasmic dynein motor. Nat Cell Biol. 2000;2(1):20–24. doi: 10.1038/71338. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.