Abstract
Site-specific recombination plays key roles in microbe biology and is exploited extensively to manipulate the genomes of higher organisms. Cre is a well studied site-specific recombinase, responsible for establishment and maintenance of the P1 bacteriophage genome in bacteria. During recombination, Cre forms a synaptic complex between two 34-bp DNA sequences called loxP after which a pair of strand exchanges forms a Holliday junction (HJ) intermediate; HJ isomerization then allows a second pair of strand exchanges and thus formation of the final recombinant product. Despite extensive work on the Cre-loxP system, many of its mechanisms have remained unclear, mainly due to the transient nature of complexes formed and the ensemble averaging inherent to most biochemical work. Here, we address these limitations by introducing tethered fluorophore motion (TFM), a method that monitors large-scale DNA motions through reports of the diffusional freedom of a single fluorophore. We combine TFM with Förster resonance energy transfer (FRET) and simultaneously observe both large- and small-scale conformational changes within single DNA molecules. Using TFM–FRET, we observed individual recombination reactions in real time and analyzed their kinetics. Recombination was initiated predominantly by exchange of the “bottom-strands” of the DNA substrate. In productive complexes we used FRET distributions to infer rapid isomerization of the HJ intermediates and that a rate-limiting step occurs after this isomerization. We also observed two nonproductive synaptic complexes, one of which was structurally distinct from conformations in crystals. After recombination, the product synaptic complex was extremely stable and refractory to subsequent rounds of recombination.
Keywords: tethered fluorophore motion, site-specific DNA recombination, Holliday junction dynamics, alternating laser excitation, protein-DNA interactions
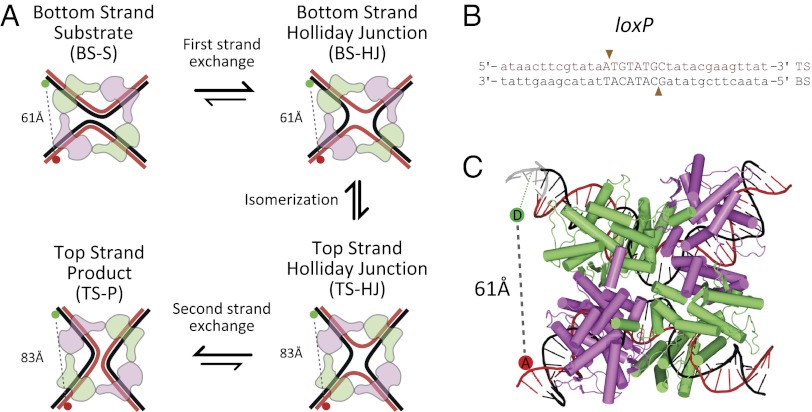
Site-specific DNA recombination is the protein-mediated cleavage, exchange, and rejoining of DNA strands between two duplexes containing a specific sequence. Tyrosine recombinases are involved in the integration and excision of viral genomes, resolution of chromosome dimers, and gene expression (1). This large family of proteins shares a general mechanism of recombination whereby a tetramer of recombinases mediates sequential pairs of strand exchange between two DNA duplexes; the first pair forms a Holliday junction (HJ) intermediate, whereas the second leads to resolution of the HJ to recombinant product (Fig. 1A). DNA cleavage requires no external energy factors, as the bond energy is stored during strand exchange as a covalent protein–DNA linkage.
Fig. 1.
Cre-mediated site-specific recombination. (A) Schematic of site-specific recombination proceeding via the BS-S complex. After DNA cleavage by the active recombinases (green) within the BS-S complex, strand exchange forms a BS-HJ. Isomerization occurs between this and the TS-HJ; after another round of cleavage and strand exchange, a TS-P synaptic complex is formed. Positions of the donor and acceptor (green and red, respectively) and interfluorophore distance measurements from crystal structure (PDB: 2HOI) are shown (SI Text). (B) Sequence of loxP site. Cre binding sites in lowercase, and asymmetric central region in uppercase; arrows indicate cleavage positions. TS and BS nomenclature as in Ghosh et al. (6) (C) Crystal structure of Cre in BS synaptic complex with DNA (PDB:2HOI, 21); DNA was extrapolated (gray) to the labeling site, and mean fluorophore positions are shown as colored circles (SI Text).
One of the best studied tyrosine recombinases is Cre, a 38 kDa protein of the bacteriophage P1 of Escherichia coli. Cre catalyzes cyclization of the viral genome and resolution of plasmid dimers during replication (2). Cre mediates recombination between 34 bp DNA sequences named loxP (Fig. 1B). Unlike other recombinases [e.g., XerCD (3) and λ Int (4)], Cre does not require accessory proteins and has been well studied genetically, biochemically, and structurally; consequently, it serves as an excellent model system for understanding the mechanisms of tyrosine recombinases. Its simplicity and compatibility with eukaryotes has also made it a versatile tool for rearrangement and manipulation of genetic elements in vivo (5).
Structural studies on Cre-loxP reaction intermediates have identified nucleoprotein complexes with remarkably similar overall structure (near-square planar) at each stage of recombination. This structure involves four Cre monomers, bound to two anti-parallel loxP sites (Fig. 1) connected through a network of protein–protein interactions; of these four monomers, only two are active at any time (Fig. 1A, green monomers). After the first pair of strand exchanges, only subtle structural changes are needed to activate the second pair of Cre monomers. It is widely believed that all steps of recombination occur within this nucleoprotein complex without major changes to its architecture (2).
There is strong structural and biochemical evidence that Cre cleaves and exchanges DNA strands in an ordered fashion, with most studies finding recombination to be initiated with the “bottom-strands” (BSs) exchanged first (Fig. 1 A and B, black DNA) (6–8). However, many of these studies relied on mutations in either Cre or loxP, and there are conflicting reports on the order of strand exchange (9, 10).
Characterizing short-lived species using ensemble biochemical techniques and relating them directly to the progress of reactions is challenging. To overcome complications caused by ensemble averaging and observe site-specific recombination in real time, various single-molecule techniques have been used, including atomic force microscopy (11), tethered particle motion (TPM) (4, 12), and magnetic tweezers (13). In particular, the recent TPM work by Fan et al. (12) observed the overall progress of recombination reactions, measuring various association and dissociation rates as well as noting the heterogeneous behavior of complexes, but was unable to correlate these changes to nanometer-scale rearrangements within individual complexes.
To address such limitations, we introduce tethered fluorophore motion (TFM), a method that monitors large-scale DNA motions through reports of the diffusional freedom of a fluorophore attached to DNA. We use alternating-laser excitation (ALEX) (14) to combine TFM with Förster resonance energy transfer (FRET) and simultaneously observe both large- and small-scale conformational changes of single DNA molecules. Using TFM-FRET, we observed recombination within individual nucleoprotein complexes in real time and analyzed their reaction kinetics. Our results showed that recombination is initiated predominantly by exchange of the BSs rather than the “top-strands” (TSs) and that synaptic complex formation is reversible. In productive complexes, we identified a rate-limiting step that occurs after HJ isomerization. We also observed two nonproductive synaptic complexes, one of which was structurally distinct from the conformations seen in crystals. Finally, we were able to infer fast HJ dynamics in recombining complexes and demonstrate the slow dissociation of the recombined product synaptic complex.
Results
Monitoring Recombination in Real Time Within Single Cre-loxP Complexes.
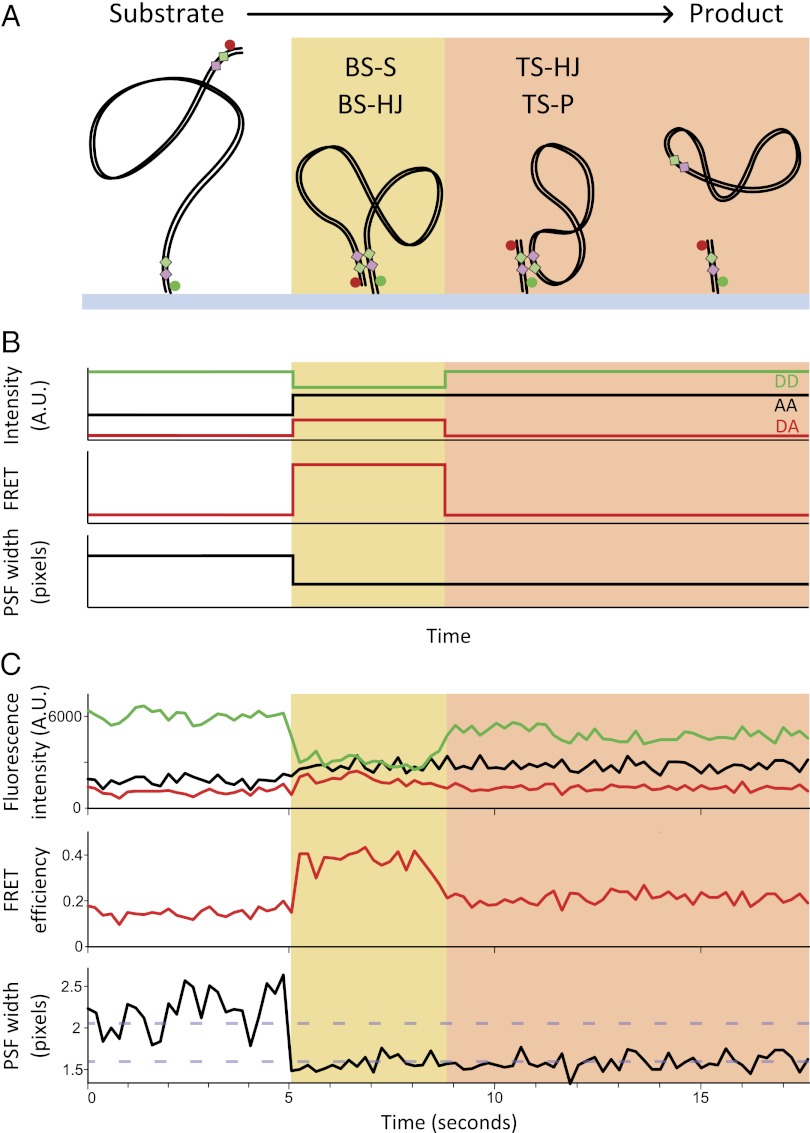
To observe recombination at the single-molecule level, we use TFM for monitoring large-scale conformational changes in DNA and combine it with the single-molecule FRET capability of ALEX for simultaneous monitoring of nanoscale changes. In our study, we prepared 1,087-bp-long DNA substrates (containing two loxP sites; SI Text and Fig. S1) labeled with a FRET donor and acceptor, tethered them to a PEG-passivated glass coverslip (Fig. 2A) using a biotin–neutravidin interaction (15), and observed them using total-internal reflection fluorescence (TIRF) microscopy. Because the fluorophores were attached at either end of the DNA substrate, the interfluorophore distance was well beyond the range of FRET.
Fig. 2.
Single-molecule recombination observed in real time. (A) Schematic of BS FRET substrate during BS recombination. (B) Expected emission intensities of donor (DD, green) and acceptor under green excitation (DA, red) and acceptor under red excitation (AA, black). DD and DA are used to calculate apparent FRET efficiency (E*), and the PSF width of the acceptor is obtained from AA images. The stages of reaction (Fig. 1A) giving rise to each of these states are indicated. (C) Representative time trace of a productive recombination event.
Cre-mediated formation of a synaptic complex between the loxP sites dramatically reduced the interfluorophore distance (Fig. 2A, yellow panel). Donor and acceptor fluorophores were positioned such that formation of a Cre-loxP antiparallel synapsis [with the BSs in an active conformation for cleavage; Fig. 1A, BS synaptic (BS-S) complex] resulted in a significant FRET signal (apparent FRET efficiency, E* > 0.2). Parallel Cre-loxP synaptic complexes, which are known to assemble but not recombine (2), were not expected to show significant FRET and were not addressed in this work. Due to structural similarities of the BS-S complex and the BS-HJ (Figs. 1A and 2A), the FRET efficiency of these two states is expected to be identical. Any complexes following HJ isomerization [TS-HJ, TS product (TS-P) synaptic complex, and the product DNA], were expected to show low FRET (E* < 0.2; Fig. 2A, pink panels; SI Text). Consequently, the FRET efficiency could not distinguish directly between complexes that isomerize to a TS complex (TS-HJ or TS-P) and complexes that dissociate back to the substrate DNA.
To discriminate between the above possibilities, we developed TFM (in analogy to TPM) to probe the diffusional freedom of the surface-distal DNA end. As the 1,087 bp substrate is much longer than the persistence length of DNA (∼150 bp; ∼50 nm), the surface-distal end (and thus the attached fluorophore) is free to diffuse above the surface and around the tether point. Because this motion occurs at the millisecond timescale (16), each image of the surface-distal fluorophore represented the average position of the DNA end during the camera exposure time (100 ms). We thus expected an attenuated fluorescence emission from the fluorophore as its average position is significantly above the surface, where the excitation power is reduced due to the exponential decay of the evanescent wave generated in TIRF (17). Further, due to in-plane motion of the DNA above the surface, we expected that the surface-distal fluorophore would yield a point-spread-function (PSF) wider than the diffraction-limited PSF (∼150 nm) observed for immobilized fluorophores with a short (1–10 nm) tether. Combining the observables of TFM and FRET allowed us to directly monitor reaction intermediates during recombination.
To assess the efficiency of recombination on the surface, we incubated coverslips carrying immobilized 1,087-bp-long DNA substrate (BS FRET; Fig. S1) with Cre for 10 min. After washing the surface with SDS to disrupt protein interactions, we identified DNA molecules that had undergone recombination; such molecules were expected to be short (61 bp), be doubly labeled, and have a surface-proximal acceptor (i.e., a diffraction-limited PSF). Indeed, ∼30% of the molecules showed a PSF width corresponding to a surface-proximal fluorophore (Fig. S2), indicating that many molecules had undergone at least one pair of strand exchanges; this percentage is comparable to reactions performed in solution (Fig. S3) (18).
To monitor individual recombination reactions in real time, we acquired several 100-s movies during incubation of Cre with the surface-immobilized BS FRET substrate. We detected transient FRET signals (dwells with E* > 0.2; SI Text) from 178 of 6,548 observed DNA molecules (Fig. 2, yellow panels). The transient FRET signals were concurrent with a decrease in acceptor PSF width and were consistent with BS complex formation. The number of analyzed events was limited by the restricted observation time (due to photobleaching) and by complex photophysics of some fluorophores; however, we estimate a probability of BS complex formation per particle of P ≈ 3.8 × 10−4 s−1 (SI Text).
We then classified molecules undergoing BS complex formation according to the PSF width after loss of the transient FRET signal. The majority of molecules (76%) showed a PSF that remained narrow after the decrease in FRET, representing complexes that progressed through HJ isomerization, reaching either a TS complex (TS-HJ or TS-P) or dissociated recombinant product (Fig. 2A, pink panels). Previous studies have shown that Cre-loxP TS-HJs are efficiently resolved to recombinant product (19); we thus believe that the majority of complexes after HJ isomerization have undergone two pairs of strand exchange and formed a recombinant product, and we classify them as “productive complexes.” The remainder of molecules, where the PSF returns to that of free substrate DNA (Fig. 3B, left panel), represented BS complexes that dissociated back to the substrate (either having formed a BS-HJ intermediate or not), and we classify them as “nonproductive complexes.”
Fig. 3.
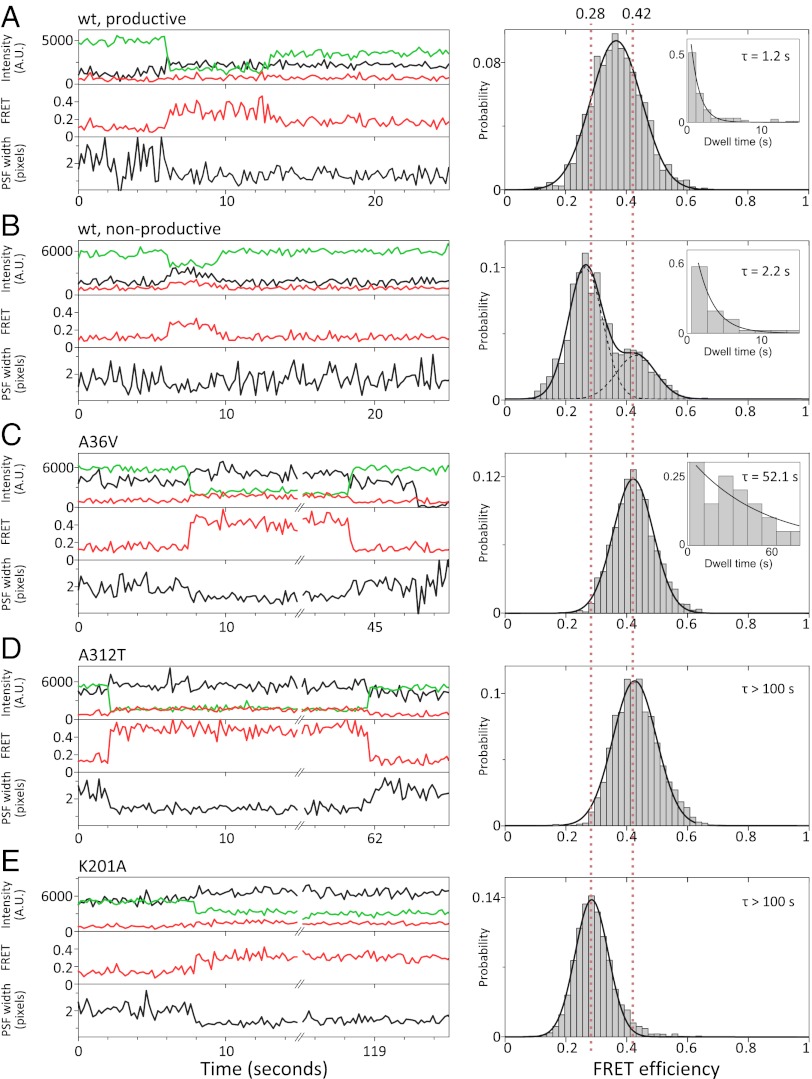
FRET efficiency histograms of protein–DNA complexes for wild-type Cre and mutants. Example time traces and population histograms for (A) wt Cre productive events (91 molecules). (B) wt Cre nonproductive synaptic complex events (48 molecules). (C) Cre A36V (27 molecules), (D) Cre A312T (25 molecules), and (E) Cre K201A (86 molecules). Histograms were plotted with E* values from each frame of an event, counts added to the histogram were normalized by the length of the event to more accurately represent the population of molecules, and histogram area was normalized to one (SI Text). Guidelines of E* = 0.42 and 0.28, corresponding to the expected BS-HJ FRET efficiency and that measured for the K201A mutant, respectively, are shown as red dotted lines. Dwell times of molecules in A, B, and C were fit to single exponentials (insets, mean lifetimes shown). D and E show very stable complexes, and dwell times could not be measured over the duration of our experiments.
BS-S Complexes Are Preferentially Assembled.
In the above experiment, we also observed events that showed PSF width changes without a concurrent FRET signal (42 of 6,548). These events can be attributed to DNA molecules recombined faster than our temporal resolution (200 ms) or to synaptic complexes that do not show a significant FRET in the BS FRET substrate (e.g., TS-S complexes; Fig. S4).
To obtain the fraction of recombination events missed due to our temporal resolution, we extrapolated the dwell-time distribution of productive FRET events (Fig. 3A, Right Inset; SI Text) and estimated the fraction missed to be ∼16% of all recombination events. As this value did not account for all events that did not exhibit FRET, we constructed a second DNA substrate (TS FRET; Fig. S1) that produced a FRET signal upon formation of TS-S (but none in the BS-S complex). Using this substrate, we observed infrequent formation of TS-S (P ≈ 4.55 × 10−5 s−1; 9 of 1,212 molecules), with ∼50% proceeding through recombination. Moreover, most recombination events did not show FRET, indicating recombination through BS-S. These results demonstrate that BS synaptic complexes are formed preferentially over TS complexes by a factor of ∼8:1.
Nonproductive Complexes.
Analysis of nonproductive complexes (i.e., transient FRET, followed by return to a broad PSF; 48 molecules) revealed two distinct populations (Fig. 3B, Right). Approximately 30% of complexes show E* = 0.43 ± 0.015 (mean of fitted Gaussian and SEM); after corrections (20), this FRET efficiency corresponds to an interfluorophore distance of 64 ± 5 Å (SI Text; Table S1). This distance agrees well with the distances in crystal structures of BS-S and BS-HJ (21), which predict a distance of ∼61 Å (Fig. 1C; SI Text). The remaining nonproductive complexes (∼70%) correspond to a population with E* of 0.26 ± 0.023 and interfluorophore distance of 75 ± 7 Å (Table S1). This distance is significantly longer than that predicted from crystal structures (21), suggesting that, in solution, Cre can form complexes with a different architecture from the structure adopted in crystals.
To link the observed intermediates with stages of the reaction, we tested three Cre mutants: A36V, shown to be partially defective in synapsis but competent in HJ resolution (21, 22); A312T, known to be proficient in synapsis and first strand exchange but inefficient in recombination, thus accumulating HJs during reactions (7); and K201A, defective in DNA cleavage but proficient in synaptic complex formation (6). We confirmed the defects of these mutants in recombination (Fig. S3) and used TFM-FRET to assess their ability to proceed through the reaction. We observed frequent synaptic complex formation with A36V (P ≈ 4.5 × 10−4 s−1, τ ≈ 52.1 s), with E* = 0.42 ± 0.018 (Fig. 3C), a value that corresponds well to the higher FRET species seen in the nonproductive complexes of wt Cre and in crystal structures (21). Despite forming synapses, the A36V mutant never completed recombination in our assays. Because A36V can resolve HJs (22), we propose that the main state formed by this mutant is likely to be the BS-S, rather than the BS-HJ, complex.
The A312T mutant formed stable complexes (τ > 100 s; Fig. 3D, Left) with a frequency (P ≈ 5.1 × 10−4 s−1) similar to A36V and E* = 0.42 ± 0.021 (Fig. 3D, Right). Because the E* value for A312T matches that of A36V and the existing BS-HJ crystal structure, we conclude that A312T forms mainly BS-HJs.
Finally, the K201A mutant formed stable synaptic complexes (P ≈ 7.83 × 10−4 s−1; τ > 100 s; Fig. 3E, Left), with E* = 0.28 ± 0.009 (Fig. 3E, Right), corresponding to an interfluorophore distance of 74 ± 7 Å. This value is surprising, as crystallographic studies have shown that the K201A mutant interacts with loxP sites to form a synaptic complex that is almost identical to the canonical BS conformation (21). It has previously been suggested that Lys201 plays a role in organizing the unsynapsed Cre-loxP complex into a cleavage-proficient conformation, which the K201A mutant may be unable to achieve (6, 8). Because it has been shown that crystallization conditions can alter the behavior of Cre mutants Y324F and R173K (21), we suggest that the discrepancy between our results and crystal structures could be due to crystal packing effects suppressing a structural defect of K201A. Intriguingly, the E* of K201A complexes matches that of the low-FRET nonproductive complexes observed with wt Cre (Fig. 3B, Right); although we cannot conclude the equivalence or structure of the two on the basis of a single FRET value, we suggest that both may represent the same nonrecombinogenic conformation, which could represent an out-of-plane twist of the HJ arms.
Evidence for Fast Interconversions Between HJ Isomers.
Analyzing productive complexes (Fig. 3A) that exhibit more than two frames of transient FRET (91 molecules), we observed a FRET distribution with mean E* = 0.36 ± 0.013 (Fig. 3A, Right), significantly (P value < 0.01) lower than that observed for A312T and A36V (E* = 0.42; Fig. 3 C and D, Right).
In addition, the width of the FRET distribution for productive complexes (SD = 0.085 ± 0.006; Fig. 3A, Right) is significantly greater than the width observed for A312T and A36V (SD ≈ 0.07; Fig. 3 C and D, Right; Table S2). Because FRET distributions from ensembles of single molecules are broadened due to intermolecular heterogeneity (23), we cannot rely on simple comparisons of the observed width with that expected on the basis of the statistical nature of photon emission (“shot-noise”). However, because photon counts throughout our studies are similar, it is likely that the excess FRET width in the case of productive complexes is a result of structural dynamics that cause unresolved FRET fluctuations. Specifically, because the reaction requires HJ isomerization before second strand exchange (Fig. 1A), we suggest that this broadening, along with the shift to a lower mean E* than that expected for the BS-HJ, reflects isomerization of the BS-HJ (E* ≈ 0.42) to TS-HJ (E* ≈ 0.18) occurring reversibly at timescales faster than our exposure time (100 ms).
To further explore the proposed interconversion between HJ isomers, we performed Monte Carlo simulations to infer information about the underlying process (24). Assuming a population of molecules undergoing transitions between BS-HJ and TS-HJ (SI Text), we estimate an equilibrium bias of 3:1 toward the BS-HJ and forward and backward rates in the range of 20–40 s−1 and 50–100 s−1, respectively, as these parameters best recapitulate the observed distributions (SI Text; Figs. S5 and S6). This bias may reflect the asymmetric DNA sequence of the loxP site central region, as this region affects the conformation of HJs assembled by the related recombinase XerCD (25).
Recombinant Product Synaptic Complex Is Extremely Stable.
Using the BS-FRET substrate (Fig. 2A and Fig. S1), we could not distinguish between the recombinant product complex (TS-P) and free recombinant DNA, as they have the same TFM-FRET signature (Fig. 2A, pink panels). To probe the stability of TS-P, we used a substrate with inverted loxP sites (Fig. S1); although recombination within this substrate inverts the DNA segment between loxP sites without producing a FRET signal, complex assembly and dissociation can be followed through changes in PSF width (Fig. S7A). Using this substrate, we observed that complexes were stable for longer than the 2-min acquisition time of our experiments (Fig. S7 B and C), suggesting the TS-P complex is very stable.
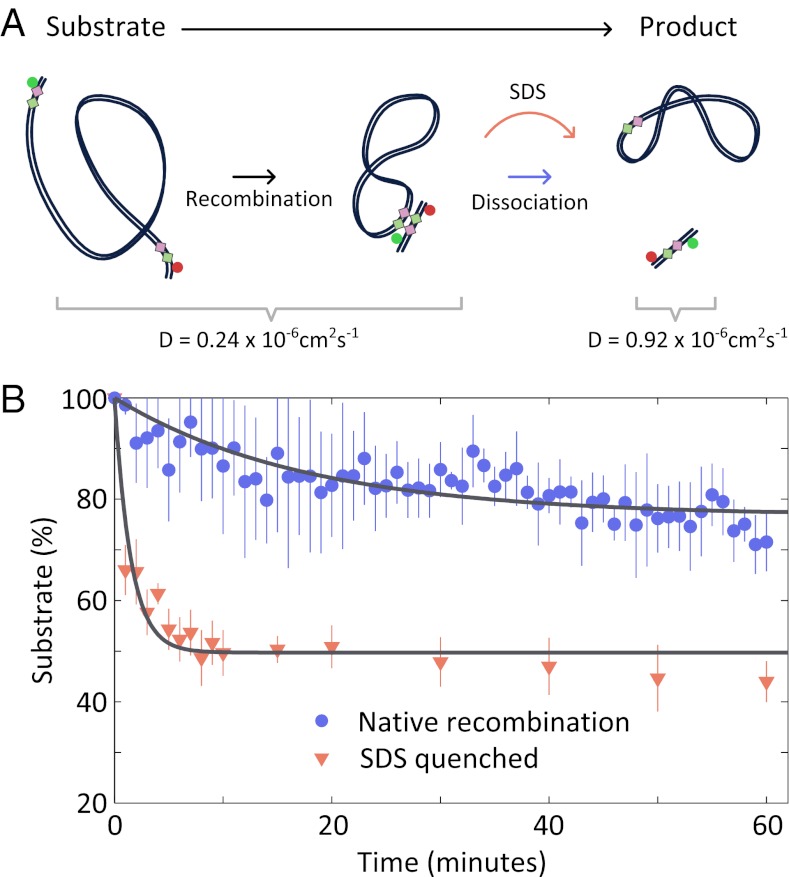
To further characterize the stability of TS-P (Fig. 1A), we followed its dissociation using a fluorescence correlation spectroscopy (FCS) assay (Fig. 4; SI Text). Using FCS, we monitored changes in translational diffusion as our BS FRET substrate was converted, via recombination, from a long DNA substrate (1,087 bp; diffusion coefficient, D ≈ 0.24 × 10−6 cm2⋅s−1) to a short DNA product (61 bp; D ≈ 0.92 × 10−6 cm2⋅s−1; SI Text), reflecting dissociation of the TS-P complex to free product DNA. To monitor TS-P dissociation after recombination, we measured the change in diffusion coefficient over time; in parallel, we measured diffusion in aliquots quenched with SDS (to disrupt protein interactions), evaluating progress of the reaction irrespective of TS-P dissociation (Fig. 4A).
Fig. 4.
Stability of product synaptic complex. (A) Schematic of the FCS assay used to investigate kinetics of TS-P dissociation. Dissociation, which can either occur naturally or be induced by addition of SDS, leads to an increase in the diffusion coefficient of the labeled species. (B) Progress of native (blue) or SDS quenched (red) recombination reactions over time.
Whereas SDS-quenched reactions displayed a rapid decrease in the fraction of substrate (similar to previous observations; ref. 18), the native reaction showed a much slower decrease, indicating that TS-P dissociation occurs at a much slower timescale (Fig. 4B). Single-exponential fits give rates of 0.01 min−1 and 0.96 min−1 for native and SDS-stopped reactions, respectively, indicating that the lifetime of the TS-P complex is ∼75 min. Our dissociation rates agree with those derived from a kinetic model of Cre recombination (18) and previous observations of highly stable complexes (26, 27).
Discussion
Here we report single-molecule fluorescence views of Cre recombination. Using our assay, TFM, in conjunction with FRET, we observe single recombination reactions in real time using wt Cre and native loxP sites. The single-molecule nature of TFM–FRET provides direct access to transient reaction intermediates and reaction paths, while uncovering molecular heterogeneity.
TFM Assay for Observing Real-Time Cre Recombination.
TFM allows observation of large-scale conformational changes of DNA, analogous to TPM (28), but without the perturbing effect of a large bead (29).The dual role of the surface-distal fluorophore (as a reporter of surface proximity and as a FRET acceptor) enables simultaneous FRET measurements that detect subtle conformational changes at the nanometer scale; as a result, the temporal coupling of global and local changes can be examined. Importantly, the FRET efficiency helps the assignment of observed intermediates to conformations seen in crystal structures. TFM is applicable to most processes involving large DNA deformations, such as looping, condensation, and changes in persistence length. On a practical note, TFM can be implemented on any single-molecule TIRF microscope without any modification.
Using TFM-FRET, we were able to identify conformational states formed during individual recombination reactions and analyze their conversion kinetics. In addition to following complete reactions, we also identified molecules that progress only partially through recombination.
Reaction Paths and a Unique Intermediate.
The direct observation of intermediates allowed us to shed light on Cre-loxP recombination. Previous work has shown a preference for recombination via BS exchange (6–8), and we confirmed that this preference arises because of an eightfold bias toward BS synaptic complex formation, rather than major differences in recombination efficiency in BS and TS complexes (BS ≈ 75%, TS ≈ 50%). We also showed that, alongside assembly of the canonical BS structure, wild-type Cre (and K201A) can form, previously unobserved, nonproductive complexes with a structure distinct from those captured in crystals. Although the role of such complexes is not clear, their existence needs to be considered in any ensemble kinetic studies of Cre recombination.
Rate-Limiting Step for Productive Complexes.
Our results establish that, once formed, synaptic complexes proceed rapidly through the reaction. For productive complexes, we measured the lifetime of the transient FRET state of E* ≈ 0.36 (representing a mixture of BS-S, BS-HJ, and TS-HJ) to be ∼1.2 s. The single-exponential decay of the dwell times in this state points to a single rate-limiting step before further progress through the reaction. Because this FRET state reflects, in part, the interconversion between HJ complexes, we conclude that the rate-limiting step is concurrent with or occurs after HJ isomerization. This rate-limiting step could be the second DNA strand exchange or a conformational change that ends HJ isomerization and/or changes the activity of the Cre monomers. We compare this to a study on λ Int (which, in contrast to Cre, requires accessory proteins) that observed that a rate-limiting step occurred before, or concomitant with, HJ formation (4).
HJ Dynamics.
In productive complexes we observed the presence of previously undetected dynamics of the HJ intermediates, estimating isomerization rates of 10–100 s−1. Intriguingly, these values are similar to rates observed for transitions between protein-free HJ conformers (20–100 s−1; ref. 30), suggesting that, although Cre affects the structure of the HJs formed (31), it does not substantially affect the isomerization rate. In contrast, we see no appreciable interconversion between HJ isomers using the A312T mutant, indicating that this mutant favors the BS conformation, either due to a change in isomerization rates between HJ isomers or due to an alteration in HJ cleavage proficiencies. Although from published biochemical data we do not expect that the A36V mutant will appreciably populate the HJ state, we cannot rule out that if it is able to form a HJ, it may also be undergoing some biased HJ isomerization (with the equilibrium being heavily shifted toward the BS-HJ form).
Stable Recombinant Synaptic Complexes.
After completion of recombination, we observed long-lived recombinant product synaptic complexes (TS-P) refractory to subsequent recombination. This lifetime agrees with ensemble measurements (26, 27) but is significantly longer than observed in a TPM study (12); however, we note that the use of a 200-nm bead in the TPM work generates an effective stretching force on the DNA (29) and may have increased the rate of complex dissociation. Previous ensemble biochemical data had measured similar dissociation constants (Kd) for the synaptic complexes of wt Cre and K201A (14 and 9 nM, respectively; ref 21). In our case, we observe that wt Cre complexes quickly form the TS-P complex (τ ≈ 1.2 s, Fig. 3A), whereas those formed by K201A are very stable (τ > 100 s). Unlike the ensemble assay, we were able to distinguish between BS-S and TS-P wt complexes, observing that BS-S is transient and TS-P is highly stable. Our results indicate that the previous Kd measurements for wt Cre were dominated by the TS-P complex rather than the BS-S complex (21). Our results also show that the Kd of >500 nM for the A36V synaptic complex (21) does not indicate a deficiency in complex formation but instead a ∼100-fold reduction in the complex stability, compared with wt Cre TS-P. The broadly similar association probabilities (and thus on-rates) we estimated indicate that, under our conditions, synapse formation is likely to be a diffusion-limited process. We note that, although TS-S and TS-P (i.e., TS complexes formed as either the substrate or product of the recombination reaction) could naively be assumed to be identical, our results indicate significant differences in the stability of each. This difference between complexes suggests an intriguing avenue for further study, which may shed further light on the mechanisms of regulation within Cre-loxP recombination.
Materials and Methods
Standard techniques for DNA labelling and protein purification were used and are described in detail in SI Text. Single-molecule and FCS experiments are described in SI Text. Details and procedures for data analysis and accurate FRET and distance calculation and simulations are also presented in SI Text.
Supplementary Material
Acknowledgments
We thank Gregory Van Duyne for plasmids for Cre overexpression. J.N.M.P. was supported by the UK Engineering and Physical Sciences Research Council, A.N.K. was supported by European Commission Seventh Framework Programme Grant FP7/2007-2013 HEALTH-F4-2008-201418, Biotechnology and Biological Research Council Grant BB/H01795X/1, and European Research Council Starter Grant 261227. Work in the D.J.S. laboratory was supported by Wellcome Trust Program Grant WT083469MA.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 20779.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211922109/-/DCSupplemental.
References
- 1.Grindley NDF, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu Rev Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 2.Van Duyne GD. A structural view of cre-loxp site-specific recombination. Annu Rev Biophys Biomol Struct. 2001;30:87–104. doi: 10.1146/annurev.biophys.30.1.87. [DOI] [PubMed] [Google Scholar]
- 3.Grainge I, Lesterlin C, Sherratt DJ. Activation of XerCD-dif recombination by the FtsK DNA translocase. Nucleic Acids Res. 2011;39(12):5140–5148. doi: 10.1093/nar/gkr078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mumm JP, Landy A, Gelles J. Viewing single lambda site-specific recombination events from start to finish. EMBO J. 2006;25(19):4586–4595. doi: 10.1038/sj.emboj.7601325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sauer B. Manipulation of transgenes by site-specific recombination: use of Cre recombinase. In: Wassarman PM, DePamphilis ML, editors. Guide to Techniques in Mouse Development. Vol 225. Waltham, MA: Academic Press; 1993. pp. 890–900. [DOI] [PubMed] [Google Scholar]
- 6.Ghosh K, Lau C-K, Gupta K, Van Duyne GD. Preferential synapsis of loxP sites drives ordered strand exchange in Cre-loxP site-specific recombination. Nat Chem Biol. 2005;1(5):275–282. doi: 10.1038/nchembio733. [DOI] [PubMed] [Google Scholar]
- 7.Hoess R, Wierzbicki A, Abremski K. Isolation and characterization of intermediates in site-specific recombination. Proc Natl Acad Sci USA. 1987;84(19):6840–6844. doi: 10.1073/pnas.84.19.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee L, Sadowski PD. Sequence of the loxP site determines the order of strand exchange by the Cre recombinase. J Mol Biol. 2003;326(2):397–412. doi: 10.1016/s0022-2836(02)01429-8. [DOI] [PubMed] [Google Scholar]
- 9.Ennifar E, Meyer JEW, Buchholz F, Stewart AF, Suck D. Crystal structure of a wild-type Cre recombinase-loxP synapse reveals a novel spacer conformation suggesting an alternative mechanism for DNA cleavage activation. Nucleic Acids Res. 2003;31(18):5449–5460. doi: 10.1093/nar/gkg732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin SS, Pulido E, Chu VC, Lechner TS, Baldwin EP. The order of strand exchanges in Cre-LoxP recombination and its basis suggested by the crystal structure of a Cre-LoxP Holliday junction complex. J Mol Biol. 2002;319(1):107–127. doi: 10.1016/S0022-2836(02)00246-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vetcher AA, et al. DNA topology and geometry in Flp and Cre recombination. J Mol Biol. 2006;357(4):1089–1104. doi: 10.1016/j.jmb.2006.01.037. [DOI] [PubMed] [Google Scholar]
- 12.Fan H-F. Real-time single-molecule tethered particle motion experiments reveal the kinetics and mechanisms of Cre-mediated site-specific recombination. Nucleic Acids Res. 2012;40(13):6208–6222. doi: 10.1093/nar/gks274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bai H, et al. Single-molecule analysis reveals the molecular bearing mechanism of DNA strand exchange by a serine recombinase. Proc Natl Acad Sci USA. 2011;108(18):7419–7424. doi: 10.1073/pnas.1018436108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kapanidis AN, et al. Fluorescence-aided molecule sorting: Analysis of structure and interactions by alternating-laser excitation of single molecules. Proc Natl Acad Sci USA. 2004;101(24):8936–8941. doi: 10.1073/pnas.0401690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kapanidis AN. Alternating-laser excitation of single molecules. In: Selvin PR, Ha T, editors. Single Molecule Techniques: A Laboratory Manual. 1st Ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2008. pp. 85–119. [Google Scholar]
- 16.Nelson PC, et al. Tethered particle motion as a diagnostic of DNA tether length. J Phys Chem B. 2006;110(34):17260–17267. doi: 10.1021/jp0630673. [DOI] [PubMed] [Google Scholar]
- 17.Axelrod D. Cell-substrate contacts illuminated by total internal reflection fluorescence. J Cell Biol. 1981;89(1):141–145. doi: 10.1083/jcb.89.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ringrose L, et al. Comparative kinetic analysis of FLP and cre recombinases: Mathematical models for DNA binding and recombination. J Mol Biol. 1998;284(2):363–384. doi: 10.1006/jmbi.1998.2149. [DOI] [PubMed] [Google Scholar]
- 19.Lee L, Sadowski PD. Directional resolution of synthetic Holliday structures by the Cre recombinase. J Biol Chem. 2001;276(33):31092–31098. doi: 10.1074/jbc.M103739200. [DOI] [PubMed] [Google Scholar]
- 20.Lee NK, et al. Accurate FRET measurements within single diffusing biomolecules using alternating-laser excitation. Biophys J. 2005;88(4):2939–2953. doi: 10.1529/biophysj.104.054114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghosh K, Guo F, Van Duyne GD. Synapsis of loxP sites by Cre recombinase. J Biol Chem. 2007;282(33):24004–24016. doi: 10.1074/jbc.M703283200. [DOI] [PubMed] [Google Scholar]
- 22.Lee L, Sadowski PD. Identification of Cre residues involved in synapsis, isomerization, and catalysis. J Biol Chem. 2003;278(38):36905–36915. doi: 10.1074/jbc.M305464200. [DOI] [PubMed] [Google Scholar]
- 23.Holden SJ, et al. Defining the limits of single-molecule FRET resolution in TIRF microscopy. Biophys J. 2010;99(9):3102–3111. doi: 10.1016/j.bpj.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santoso Y, et al. Conformational transitions in DNA polymerase I revealed by single-molecule FRET. Proc Natl Acad Sci USA. 2010;107(2):715–720. doi: 10.1073/pnas.0910909107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arciszewska LK, Baker RA, Hallet B, Sherratt DJ. Coordinated control of XerC and XerD catalytic activities during Holliday junction resolution. J Mol Biol. 2000;299(2):391–403. doi: 10.1006/jmbi.2000.3762. [DOI] [PubMed] [Google Scholar]
- 26.Hamilton DL, Abremski K. Site-specific recombination by the bacteriophage P1 lox-Cre system. Cre-mediated synapsis of two lox sites. J Mol Biol. 1984;178(2):481–486. doi: 10.1016/0022-2836(84)90154-2. [DOI] [PubMed] [Google Scholar]
- 27.Shoura MJ, et al. Measurements of DNA-loop formation via Cre-mediated recombination. Nucleic Acids Res. 2012;40(15):7452–7464. doi: 10.1093/nar/gks430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schafer DA, Gelles J, Sheetz MP, Landick R. Transcription by single molecules of RNA polymerase observed by light microscopy. Nature. 1991;352(6334):444–448. doi: 10.1038/352444a0. [DOI] [PubMed] [Google Scholar]
- 29.Segall DE, Nelson PC, Phillips R. Volume-exclusion effects in tethered-particle experiments: Bead size matters. Phys Rev Lett. 2006;96(8):088306–088310. doi: 10.1103/PhysRevLett.96.088306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKinney SA, Déclais A-C, Lilley DMJ, Ha T. Structural dynamics of individual Holliday junctions. Nat Struct Biol. 2003;10(2):93–97. doi: 10.1038/nsb883. [DOI] [PubMed] [Google Scholar]
- 31.Gopaul DN, Guo F, Van Duyne GD. Structure of the Holliday junction intermediate in Cre-loxP site-specific recombination. EMBO J. 1998;17(14):4175–4187. doi: 10.1093/emboj/17.14.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.