Significance
DNA introduced into cells by infection or transfection is immediately coated by repressive histones and repressors. Viruses have evolved mechanisms to derepress their genes. What we know of these events is based on ChIP analyses that help identify the proteins involved in these processes, provided their identity is suspected and reagents are available. We report a procedure that enables identification of proteins hitherto not linked to these processes, based on the observation that HSV derepresses both viral DNA and DNAs introduced concurrently by transfection by using the transfected DNA as the surrogate for viral DNA.
Keywords: DNA binding proteins, gene derepression
Abstract
ICP0, a key herpes simplex virus regulatory protein, functions first in the nucleus and then in the cytoplasm. The duration of its nuclear sojourn in cells transfected with DNA and then infected is related to the quantity of transfected DNA. Furthermore, ICP0 transactivates both viral genes and genes encoded by the transfected DNA. The data support the hypothesis that ICP0 is retained in the nucleus until it completes the replacement of repressive chromatin with effector proteins that enable transcription of both DNA templates. To identify the effector proteins, we transfected cells with biotinylated DNA encoding a nonviral gene and then infected the cells with wild-type virus. Proteins bound to transfected biotinylated plasmid recovered from mock-treated and infected cells were identified using mass spectrometry followed by appropriate database search. The transfected DNA from mock-infected cells yielded proteins associated with repression, whereas DNA recovered from infected cells included proteins known to enable transcription and proteins that have not been previously associated with that role. To test the hypothesis that the proteins hitherto not known to associate with viral gene expression are nevertheless essential, we tested the role of the DEAD-box helicase Ddx17. We report that Ddx17 plays a critical role in the expression of early and late viral genes. Thus, biotinylated DNA recovered from transfected infected cells can function as a surrogate for viral DNA and is a rich source of proteins that play a role in viral gene expression but which have not been previously identified in that role.
Upon entry into the nucleus, the DNA of HSV-1 is immediately coated by repressive cellular proteins and bound to a dynamic nuclear body known as ND10 (1). Expression of viral genes requires sequential derepression at least at two checkpoints (2). In the first VP16, a viral tegument protein introduced into the cell during infection, recruits to the promoters of the α (immediate early) genes numerous host proteins that include the Octamer binding protein 1, Host cell factor 1, and Lysine Specific Demethylase 1 (LSD1) (1, 2). The consequence of derepression of the α genes is the synthesis of the six α proteins. One of these proteins, ICP0, plays a key role in the transition through the second checkpoint. Briefly, newly synthesized ICP0 colocalizes with ND10, recruits the ubiquitin-conjugating enzyme UbcH5a, and mediates the degradation of PML and SP100, key components of the ND10 nuclear bodies (3–5). In addition, it interacts with a repressor complex whose key components are HDAC-1/2, CoREST, LSD1, and REST. In this instance ICP0 binds to CoREST and dislodges HDAC-1/2 from the repressor complex (6–8). Last, ICP0 either recruits or enhances the recruitment of numerous host proteins to the viral replication compartment erected in the space formerly occupied by the ND10 nuclear bodies. These include cyclin D3, cdc34, CoREST, USP7, Bmal1, and the histone acetyl transferase CLOCK (9–12). The consequence of these events is that the expression of β (early) and γ (late) genes ensues and ICP0 is translocated to the cytoplasm. Nevertheless, the entire range of proteins recruited by ICP0 and its numerous partners to the viral DNA and the replication compartment is unknown.
This report describes a simple procedure to identify proteins bound to the DNA during the early stages of viral gene expression. It is based on several observations as follows:
i) Viral DNA introduced by infection or irrelevant DNA introduced by transfection colocalize at ND10 nuclear bodies on entry into the nucleus. Concomitantly, both the number and size of the ND10 bodies increase (13, 14).
ii) As noted above, newly synthesized ICP0 colocalizes with ND10 bodies. Over a period of several hours ICP0 fills the nucleus, and then it is totally translocated to the cytoplasm. In cells transfected with irrelevant DNA and then infected the duration of nuclear retention of ICP0 is related to the amount of irrelevant DNA transfected into cells (15).
iii) ICP0 transactivates the expression of both viral β and γ proteins and of genes encoded by the transfected DNA irrespective of the promoter driving the expression of these genes (16).
These observations led to two conclusions: first, ICP0 is retained in the nucleus until it has “processed” all of the DNA aggregated at the ND10 nuclear bodies. Because ICP0 does not bind DNA, its most likely function is to recruit to the ND10 bodies at which the DNA is aggregated host proteins that derepress and enable expression of the genes encoded by the DNAs (17, 18). Second, ICP0 does not discriminate between viral and irrelevant DNA deposited at the ND10 nuclear bodies. These conclusions led to the hypothesis that in uninfected cells, the transfected DNA could serve as a surrogate for viral DNA in cells that are bereft of a protein capable of transactivating viral gene expression. In infected cells the transfected DNA could serve as a surrogate for viral DNA in the process of expressing its genes.
To test this hypothesis we devised a simple procedure whereby we infected cells into which we had transfected a biotinylated plasmid encoding a nonviral gene. After a suitable time interval, the proteins were cross-linked to the DNA, then the biotinylated DNA was extracted and purified, and the bound proteins were identified. We report details of the procedure and a list of proteins captured by biotinylated DNA in mock-infected cells and in cells transfected with the biotinylated DNA and then infected with wild-type virus.
Many proteins whose functions were not known were recovered from mock-infected and infected cells. A measure of the value of this procedure is its ability to predict the putative function of a captured protein according to the environment in which it bound the biotinylated DNA. To test this, we selected among the proteins of unknown function captured by biotinylated DNA in transfected/infected cells a DEAD-box helicase designated Ddx17. We show that Ddx17 is required for optimal viral replication, in as much as depletion of Ddx17 resulted in a significant decrease in post α gene expression.
Results
Characterization of the Proteins Cross-Linked to Biotinylated DNA, Extracted, and Purified from Transfected Cells.
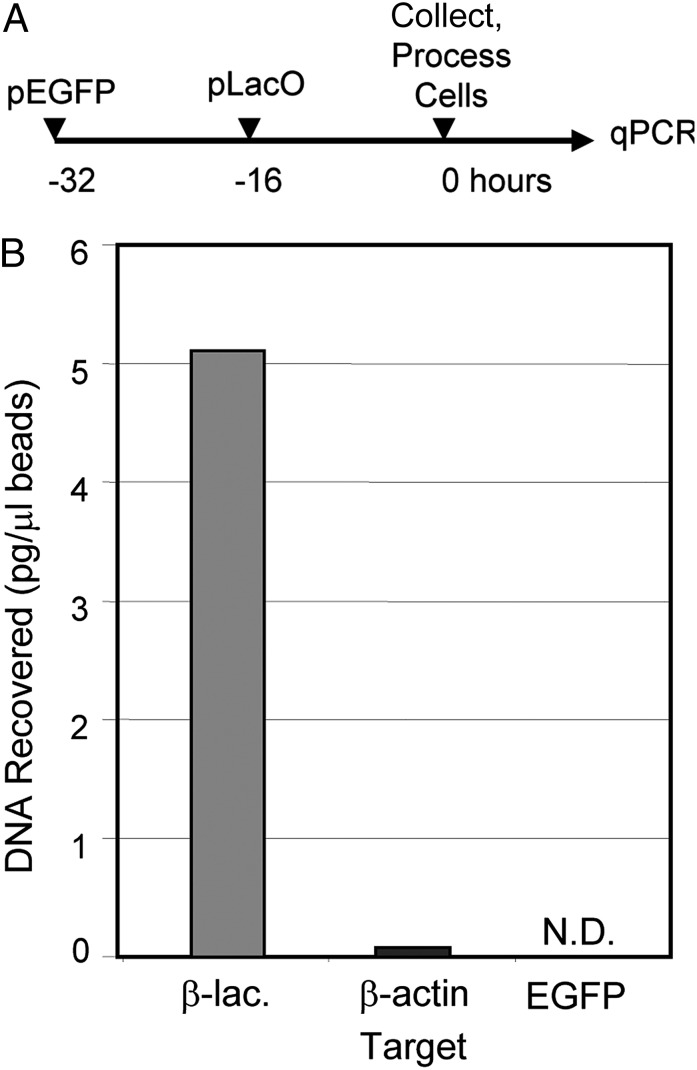
In the experiments described here (Fig. 1) replicate cultures of HeLa cells were transfected with two plasmids. Because all foreign DNAs and some cellular DNAs localize to ND10, it was possibile that nontarget DNA (i.e., any nonbiotinylated DNA) would be cross-linked to the transfected biotinylated DNA during recovery. To determine to what extent nonbiotinylated DNA localizing to ND10 would be recovered with biotinylated DNA, cells were transfected with two plasmids. Transfections were separated by 16 h to control for the possibility that the plasmids would replicate and form concatemers during or immediately after transfection. The first encoded EGFP and was not modified. The second, transfected 16 h after the first, was plasmid pLacO that was conjugated to biotin. After 16 h of incubation the cells were collected, lysed, disrupted by sonication, and incubated with streptavidin-coated beads. After an additional 16 h, the beads were collected, rinsed extensively with PBS, and subject to quantitative PCR (qPCR). Primers were selected to amplify β-lactamase, β-actin, and EGFP. The β-lactamase sequence was unique to pLacO and served as a reference to determine the amount of biotinylated DNA in the sample. EGFP and β-actin were amplified to determine the amount of control plasmid pEGFP and host DNA, respectively. The details of the procedures involved in these studies are described in Materials and Methods. The results of the experiment (Fig. 1) show that after the final step in the procedure the biotinylated DNA was enriched 68-fold relative to host chromatin. EGFP was not detected, suggesting that the plasmid encoding EGFP was not recovered to a detectable amount.
Fig. 1.
Recovery of transfected biotinylated DNA from HeLA cells with streptavidin. HeLa cells were transfected with a plasmid encoding pEGFP that was not modified and 16 h later with a biotinylated plasmid encoding pLacO (A). The cells were collected and incubated with streptavidin-coated beads as described in the text. Beads were rinsed extensively and subject to RT-qPCR. Primers were selected to amplify β-lactamase, β-actin, and EGFP. The β-lactamase sequence was unique to pLacO and served as a reference to determine the amount of biotinylated DNA in the sample. EGFP and β-actin were used to determine the amount of pEGFP and host chromatin, respectively (B).
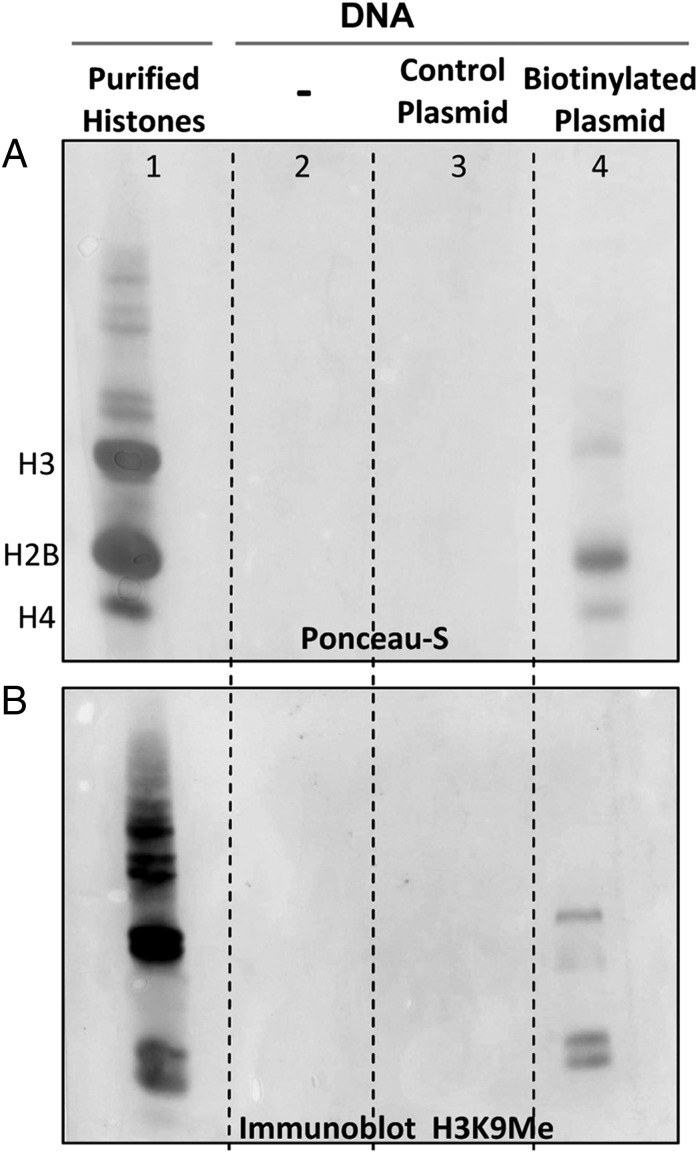
In the second series of experiments, HEp-2 cells were either mock-treated or transfected with either untreated or biotinylated plasmids encoding LacO. The samples were collected 16 h after transfection. The bound proteins were separated in an acid-urea gel and stained with ponceasu-S (Fig. 2A) or reacted with antibody directed against H3K9Me (Fig. 2B) (19). Purified histones (lane 1) were included as a reference. The results show that bound protein was detected only in samples containing biotinylated DNA (lane 4). Neither the Ponceau-S stain nor the immunoblots detected the presence of protein bands in samples recovered from mock treated cells (lane 2) or cells transfected with nonbiotinylated DNA (lane 3). The protein bands in lane 4 align with the histones in lane 1, suggesting that the canonical histones were present among the proteins bound to biotinylated DNA. The immunoblot with antibody directed against H3K9Me revealed three bands most likely reflecting the presence of various acetylated forms of the proteins.
Fig. 2.
Recovery of protein bound to biotinylated DNA. HEp-2 cells were either mock-treated or transfected with nonbiotinylated pLacO or biotinylated pLacO. Samples were collected as described in the text. Protein from samples was separated in an AU-gel, transferred to nitrocellulose, stained with Ponceau-S (A), or reacted with antibody directed against H3K9Me (B). Purified histones (lane 1) were included as a reference. Protein was present only in samples transfected with biotinylated DNA (lane 4) and form bands that aligned with the histones.
Proteome Bound to Transfected DNA Serving as a Surrogate for HSV DNA Introduced by Infection.
The procedure for the analyses of the proteins bound to biotinylated DNA was as follows. HEp-2 cells were mock-infected or infected with wild-type HSV-1(F) 16 h after transfection with a biotinylated plamsid. The cells were harvested at 2, 3, 4, 5, 6, 8, 9, or 12 h after infection and processed immediately as described in Materials and Methods. The rinsed streptavidin-bound fraction was trypsinized and subjected to liquid chromatography–tandem mass spectrometry (LC-MS/MS) as described in Materials and Methods. In total, ∼1,000 proteins were identified. The probable identification of proteins present in the biotinylated DNA fraction was based on the sequence of peptides identified by mass spectrometry analysis. Briefly, the amino acid sequence of a detected peptide was compared with a database consisting of the Homo sapiens and Human Herpesvirus 1 proteomes. Common contaminants, including keratin and epiplakin, and proteins identified by homology to fewer than two peptides were set aside. Of the remaining proteins, ∼50% were involved in processes related to chromatin regulation and nucleic acid metabolism. As expected, the most abundant proteins were the canonical histones (Table 1). The remaining proteins were sorted by scan count, a value that serves as an indicator of relative abundance within the sample. Proteins that were enriched threefold in mock-treated or HSV-1(F)–infected samples were grouped according to biological function. Two prevailing categories emerged: (i) chromatin binding and modifying proteins and (ii) proteins involved in processes related to RNA metabolism. The data presented here suggest (i) transfected DNA is bound by nucleosomes and packaged into chromatin. Supporting this notion is the observation that the canonical histones were among the most abundant proteins recovered. (ii) The chromatin is modified by host enzymes. Several proteins with known roles in modifying histone residues were identified. Included in this category were components of complexes regulating lysine methylation. (iii) Proteins involved in nucleic acid metabolism, including ATP-coordinating helicases, are involved in the response to foreign DNA.
Table 1.
Ten most abundant proteins recovered with plasmid DNA
| Accession no. | Description | Scan count, mock | Scan count, HSV(F) |
| P62805 | Histone H4 | 429.00 | 304.00 |
| Q96QV6 | Histone H2A type 1-A | 345.00 | 208.00 |
| P33778 | Histone H2B type 1-B | 299.00 | 209.00 |
| P02545 | Lamin-A/C | 177.00 | 168.00 |
| P16403 | Histone H1.2 | 165.00 | 148.00 |
| Q16695 | Histone H3.1t | 162.00 | 120.00 |
| P04908 | Histone H2A type 1-B/E | 154.00 | 83.00 |
| P62807 | Histone H2B type 1-C/E/F/G/I | 82.00 | 80.00 |
| Q96A08 | Histone H2B type 1-A | 52.00 | 60.00 |
| Q02539 | Histone H1.1 | 57.00 | 52.00 |
The data presented in Tables 2 and 3 were generated as follows. The scan count value, a measure of abundance for each protein, was used to determine its relative enrichment in mock vs. HSV-1(F)–infected samples. Proteins that were either present exclusively or enriched threefold in one sample were selected for further analysis. Tables 2 and 3 list proteins enriched or bound exclusively on biotinylated plasmids extracted from mock-infected or infected cells, respectively, and the time after infection when the protein was first recovered. In each case the proteins bound to the biotinylated DNA could be readily sorted into three categories (i.e., proteins that bound to DNA or modified DNA-bound proteins, proteins known to affect RNA metabolism, and protein that did not fit either category).
Table 2.
Proteins enriched in plasmid fraction recovered from mock-infected cells
| Accession no. | Official symbol | Relevant function | Scan count, mock/HSV |
| Histones and DNA-binding proteins | |||
| P16402 | HIST1H1D | Regulation of chromatin structure (20, 21) | 7/0 |
| P25685 | DNAJB1 | Unknown. Heat shock protein | 5/0 |
| Q9NZ71 | RTEL1 | Unknown. Regulates telomere length (22) | 4/0 |
| P15822 | HIVEP1 | Regulator of transcription (23) | 3/0 |
| Q5VUA4 | ZNF318 | Regulator of transcription. Interacts with HDAC2 (24, 25) | 3/0 |
| O14646 | CHD1 | Binds methylated H3K4 (26) | 2/0 |
| O15014 | ZNF609 | Protein of unknown function | 2/0 |
| P35659 | DEK | Alters chromatin structure. Transcriptional regulation (27–30) | 2/0 |
| P47902 | CDX1 | Unknown | 2/0 |
| Q6P0N0 | MIS18BP1 | Unknown | 2/0 |
| Q92522 | H1FX | Regulation of chromatin structure (20, 21, 31) | 2/0 |
| Q9Y4C1 | KDM3A | Histone demethylase. Activity demonstrated on H3K9 (32) | 2/0 |
| Q9Y618 | NCOR2 | Transcriptional corepressor. Interacts with HDACs (33, 34) | 2/0 |
| RNA metabolism | |||
| Q5TAX3 | ZCCHC11 | Unknown. Mediates RNA 3′ uridylation. mRNA stability (35) | 18/8 |
| O00148 | DDX39A | Unknown. mRNA export. Interacts with CMV UL69 (36) | 13/2 |
| Q08170 | SRSF4 | Unknown. mRNA processing. Pre-mRNA splicing | 9/0 |
| P11940 | PABPC1 | Binds Poly(A) tail. mRNA Stability (37, 38) | 4/0 |
| P61978 | HNRNPK | Unknown | 4/0 |
| Q16629 | SRSF7 | mRNA export | 4/0 |
| P07910 | HNRNPC | Unknown. Complex with remodelers SWI/SNF (39, 40) | 4/0 |
| O60812 | HNRNPCL1 | Unknown | 3/0 |
| O75494 | SRSF10 | Unknown. Inhibits splicing (41) | 3/0 |
| P09651 | HNRNPA1 | Binds viral mRNA. mRNA export (42, 43) | 3/0 |
| Q7Z2W4 | ZC3HAV1 | Antiviral protein. mRNA degradation (44, 45) | 2/0 |
| Q7Z3Z3 | PIWIL3 | Unknown. piRNA binding | 2/0 |
| Additional proteins | |||
| P21291 | CSRP1 | Unknown | 2/0 |
Table 3.
Proteins enriched in plasmid fraction recovered from HSV-1(F)–infected cells
| Accession no. | Official symbol | Relevant function | Scan count, mock/HSV | Initial detection (h after infection) |
| Histones and DNA binding proteins | ||||
| Q6FI13 | HIST2H2AA3 | Histone H2A variant (46) | 11/102 | 4 |
| Q15233 | NONO | Paraspeckle component (47) | 0/15 | 4 |
| Q9P0M6 | H2AFY | Transcription. DNA damage. Bound to latent HSV DNA (48–50) | 0/8 | 8 |
| P17947 | SPI1 | Unknown. Transcription factor (51, 52) | 0/7 | 5 |
| Q86WZ6 | ZNF227 | Unknown. Predicted transcription activator (53) | 0/6 | 3 |
| Q9Y6Y1 | CAMTA1 | Protein of unknown function | 0/5 | 3 |
| Q9UIF9 | BAZ2A | Unknown. Regulator of rRNA transcription (54) | 0/3 | 12 |
| Q92576 | PHF3 | Unknown | 0/3 | 3 |
| O94986 | CEP152 | Unknown. Required for centriole integrity (55) | 0/3 | 5 |
| Q8TEK3 | DOT1L | H3K79 methyltransferase. Maintain telomere integrity. Regulation of transcription (56–58) | 0/2 | 12 |
| RNA metabolism | ||||
| Q14865 | ARID5B | DNA binding. Histone lysine demethylase (59, 60) | 0/2 | 12 |
| P31943 | HNRNPH1 | Unknown. Splicing regulator (61) | 2/9 | 3 |
| P23246 | SFPQ | Influenza transcription. Paraspeckle formation (62, 63) | 0/9 | 5 |
| Q92841 | DDX17 | Unknown. Transcription of Influenza (64) | 0/8 | 3 |
| Q3SY52 | ZIK1 | Regulation of transcription (65) | 0/4 | 5 |
| P31942 | HNRNPH3 | Splicing. Interacts with ZNF198 (66, 67) | 0/3 | 5 |
| Q6ZSC3 | RBM43 | Protein of unknown function | 0/2 | 12 |
| Additional proteins | ||||
| Q14134 | TRIM29 | Unknown. PML domain homology (68) | 13/95 | 3 |
| 35232 | PHB | Regulation of transcription. Brg-1 interactor (69, 70) | 0/6 | 3 |
| Q99623 | PHB2 | Regulation of transcription (71) | 0/5 | 12 |
| Q13620 | CUL4B | Regulation of transcription. Ubiquitin ligase (72) | 0/3 | 12 |
Role of Ddx17 Helicase in HSV-1 Infection.
We have selected one protein, Ddx17, to test the value of the transfected biotinylated DNA as a surrogate for HSV DNA and its use in identifying proteins that bind to the DNA during infection and play a role in viral gene expression. Ddx17 is a DEAD-box helicase named for the amino acids required for ATP coordination (Asp-Glu-Ala-Asp). Its interacting partners include p300/CBP, HDAC1, RNA Pol II, etc. (73). Its activity seems to be regulated by SUMOylation. inasmuch as the mutant lacking the SUMOylation motif (K50R) was reported to stimulate transcription from an MDM2 promoter more efficiently than wild-type protein (74). We report the following results:
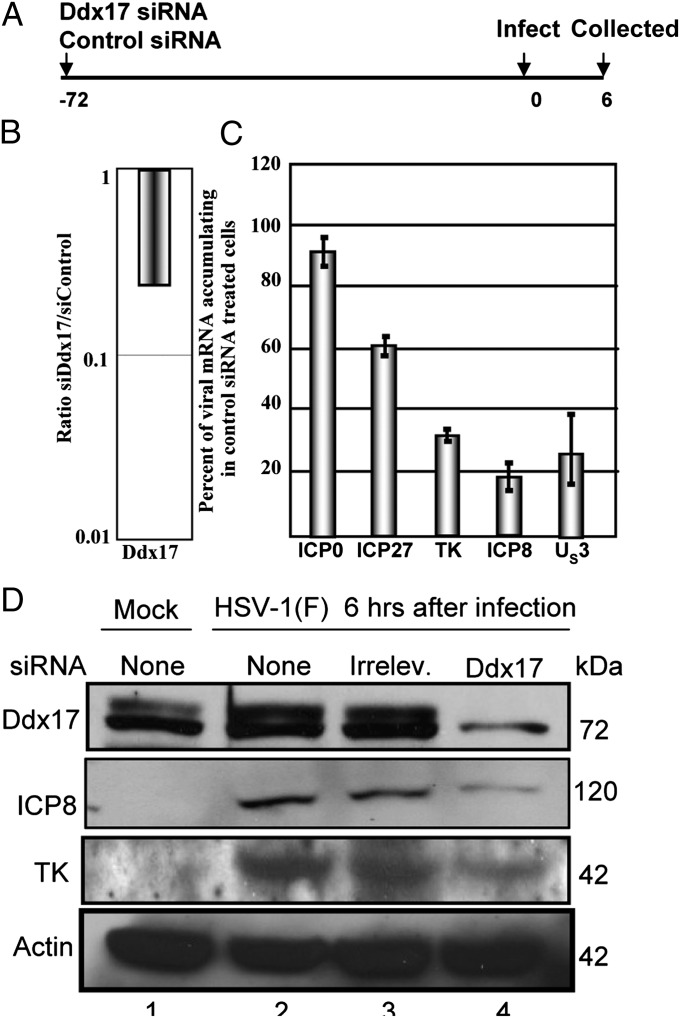
i) HEp-2 cells were transfected with 200 pmol of either control siRNA or siRNA targeting Ddx17 mRNA for knockdown. Seventy-two hours after transfection, the cells were exposed to HSV-1(F) at a ratio of 1.0 pfu per cell (Fig. 3A). After 6 h of infection the cells were collected and processed for analysis by qPCR (Fig. 3 B and C) or by immunoblotting (Fig. 3D). Fig. 3 B and C shows the results of analyses for Ddx17, ICP0, ICP27, thymidine kinase (TK), ICP8, and US3 mRNAs by RT-qPCR with probes listed in Materials and Methods. In these experiments Ddx17 mRNA was decreased by ∼80%. In these cells we observed a slight decrease in the accumulation of ICP0 mRNAs but significant decreases in mRNAs encoding ICP27, TK, ICP8, and the US3 protein kinase. The immunoblot shown in Fig. 3D is consistent with these results in that we observed a significant decrease in the accumulation of Ddx17 and ICP8 and a less pronounced but readily apparent decrease in the thymidine kinase.
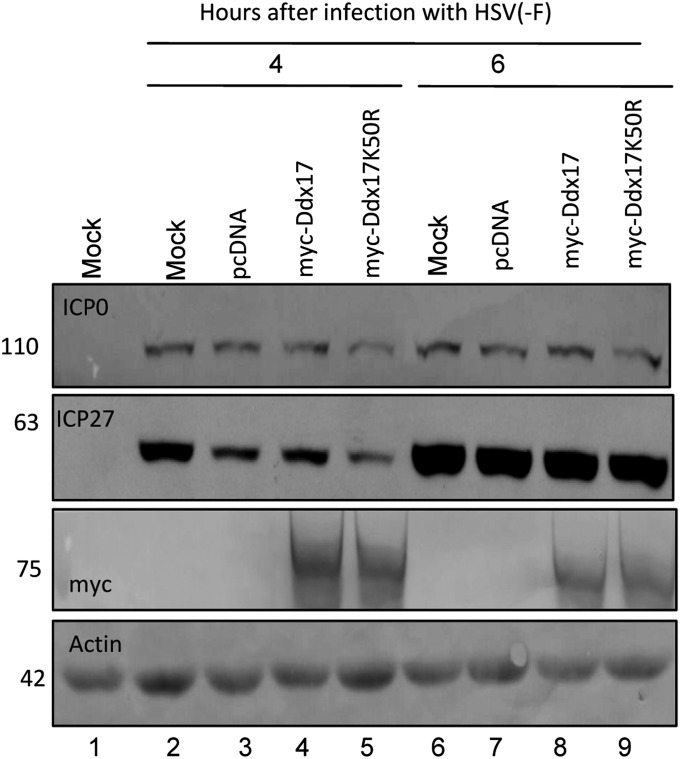
ii) In the experiment shown in Fig. 4, 293T cells were either mock-treated (lanes 1, 2, and 6) or transfected with 1.5 μg of control plasmid pcDNA (lanes 3 and 7), plasmid encoding myc-tagged Ddx17 (lanes 4 and 8), or plasmid encoding myc-tagged Ddx17 mutant K50R (lanes 5 and 9). As indicated above, the codon substitution K50R abolishes SUMOylation at residue 50. After 24 h the cells were either mock-infected (lane 1) or exposed to HSV-1(F) at a ratio of 1.0 pfu per cell. At 4 h (lanes 2–5) or 6 h (lanes 6–9) after infection the cells were harvested, solubilized, transferred to a nitrocellulose sheet, and reacted with antibodies to ICP0, ICP27, or myc-tagged Ddx17. The results show that the overexpression of wild-type or mutant myc-tagged Ddx17 had no effect on the accumulation of ICP0 or ICP27. A slight decrease in the accumulation of ICP27 and ICP0 (lane 4) was observed in cells transfected with the K50R mutant of Ddx17.
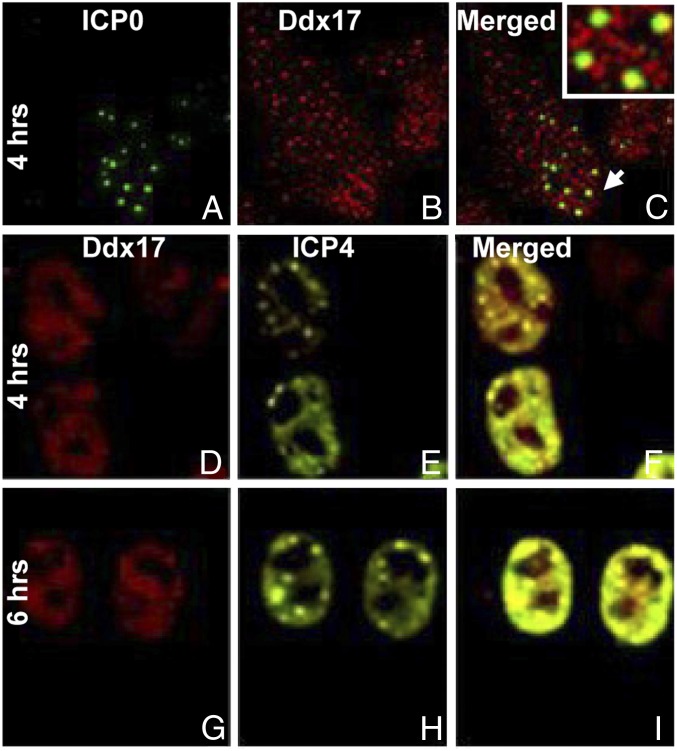
iii) In the experiment shown in Fig. 5, HEp-2 monolayers were infected with HSV-1(F) at a ratio of 10 pfu per cell. After 4 h (Fig. 5 A–F) or 6 h (Fig. 5 G–I) after infection the cells were fixed with paraformaldehyde, processed for immunofluorescence, and reacted with antibody raised against Ddx17 and either ICP0 (Fig. 5 A–C) or ICP4 (Fig. 5 D–I). The results were as follows. In both uninfected and infected cells Ddx17 aggregates in small dense bodies varying in size. ICP0 during that interval after infection localizes in ND10 structures. Some of the ND10 structures highlighted in Fig. 5C seem to be juxtaposed to Ddx17 aggregated. Viral proteins involved in viral gene expression, notably ICP4 and ICP0, ultimately form a replication compartment within the nucleus of the infected cells. In Fig. 5 E and H ICP4 is shown accumulating in dense bodies corresponding to the ND10 nuclear structures and filling parts of the nucleus. The space occupied by DdX17 coincides with the replication compartment illuminated by ICP4.
Fig. 3.
Knockdown of Ddx17 by siRNA leads to a decrease in the amount of viral mRNA and protein present at 6 h after infection. HEp-2 cells were transfected with 200 pmol of either control siRNA or siRNA targeting Ddx17 mRNA for depletion. Seventy-two hours after infection, monolayers were exposed to HSV-1(F) at a ratio of 1.0 pfu per cell. After 6 h after infection the cells were collected, and the RNA was purified and analyzed by RT-qPCR (A) or immunolotting (B). (C) The amounts of mRNAs encoding Ddx17, ICP0, ICP27, TK, ICP8, and US3 proteins. The results were normalized with respect to mRNA levels in control SI RNA-treated cells. (D) Panel depicts the amount of Ddx17, ICP8, and TK protein as determined by immunoblotting. Actin served as a loading control.
Fig. 4.
Accumulation of viral protein in cells mock-treated or transfected with control plasmid pcDNA, plasmid myc-Ddx17, or myc-Ddx17K50R. 293T cells were either mock treated (lanes 1, 2, and 6) or transfected with 1.5 μg of either control plasmid pcDNA (lanes 3 and 7), plasmid encoding myc-tagged Ddx17 (lanes 4 and 8), or plasmid encoding myc-tagged Ddx17 mutant K50R (lanes 5 and 9). Mutation K50R abolishes SUMOylation at residue 50. After 24 h, cells were either mock-treated (lane 1) or exposed to 1.0 pfu of HSV-1(F) per cell. Cells were collected at 4 h (lanes 2–5) and 6 h (6–9) after infection. Fifty micrograms of the proteins were analyzed by immunoblotting with antibodies directed against ICP0, ICP27, and myc. Actin served as a loading control.
Fig. 5.
Localization of Ddx17 in infected cells. The figure shows the localization of Ddx17 in HEp-2 cells fixed 4 h (A–F) and 6 h (G–I) after infection. The cells were also reacted with antibody directed against ICP0 (A–C) and ICP4 (D–I). As shown by the Inset in C, a portion of Ddx17 localizes to sites adjacent to and partially overlapping ICP0. The distribution of Ddx17 and ICP4 in cells fixed at 4 and 6 h after infection shows a portion of Ddx17 localized to viral transcription compartments (D–I).
We conclude from these observations that Ddx17 is intimately associated with the nuclear compartments in which viral genes are expressed.
Discussion
Two competing events take place in cells infected with HSV-1. On one hand viral DNA entering the nucleus is immediately bound by cellular proteins that repress viral genes. The entering DNA also serves as an anchor for ND10, a dynamic nuclear body whose proteins regulate transcription and, at the very least, define the host response to infection. On the other hand HSV imports a protein, VP16, that recruits several host proteins to derepress viral DNA and initiate the transcription of the α genes. An α protein, ICP0, spearheads two functions. First, it binds the UbcH5A ubiquitin-conjugating protein to degrade the ND10 proteins PML and SP100, leading to inactivation of ND10 functions (3, 4). Second, it derepresses the DNAs aggregated at the ND10 bodies to enable the expression of β and γ genes (13, 14). This report is based on the results of studies on the functions of ICP0. First, between 5 and 9 h after infection ICP0 disappears from the nucleus and accumulates in the cytoplasm (75). The duration of the nuclear sojourn is directly related to the amount of DNA delivered to the nucleus in cells transfected and then infected (15). Second, ICP0 enables the expression of viral genes encoded in the viral DNA and a reported gene encoded in the transfected plasmids (16). These results suggested that (i) ICP0 lingers in the nucleus until its “task” of derepressing genes is completed, (ii) ICP0 does not differentiate between viral and cellular genes, and (iii) the modifications of the plasmid DNA introduced into cells by transfection could serve as a surrogate for identification of viral proteins that bind to viral DNA in infected cells and enable viral gene expression. As noted in the Introduction, ICP0 does not bind DNA, but it is known to recruit proteins to the compartment in which the DNA resides (17, 18). The processing function attributed to the DNA may well be saturation of the DNA compartment with cellular proteins that modify or deplete the repressive proteins bound to the DNA on entry into the nucleus (18). The test of this proposition hinges on the demonstration that in cells that were transfected with a plasmid encoding nonviral genes and then infected, at least some proteins selectively bound to the transfected plasmids enhance the expression of viral genes.
It should be noted that at least one partner of ICP0 relevant to its sojourn in the nucleus has been identified. ICP0 binds and recruits cyclin D3 to the replication compartment. The objective, however, appears to be the recruitment of cdk4 (76). Inhibition of cdk4 results in the retention of ICP0 in the nucleus. Overexpression of cyclin D3 reverses the effect of the cdk4 inhibitor in that ICP0 is translocated to the cytoplasm (77). Concurrently, the nuclei of infected cells show an accumulation of cdk6. These studies suggest that cdk4 is required for at least some functions of ICP0 in the nucleus, that in its absence ICP0 may use cdk6, but that the recruitment of cdk6 may require higher concentrations of cyclin D3.
Relevant to the studies reported here are the following:
i) Histones are the most abundant proteins bound to transfected DNA. This notion is supported by earlier observations that expression of reporter genes is repressed in the transfected and uninfected cells. This suggests that transfected DNA, like viral DNA, is packaged into chromatin-like structures and that both become derepressed as HSV genes are expressed. The spacing of nucleosomes on viral chromatin has been investigated. The results suggest that during latency the viral DNA is bound by regularly spaced nucleosomes, but during lytic infection the spacing becomes irregular. Nucleosomes are often displaced from active genes during transcription. This may account for the discrepancy in basic chromatin structure.
ii) Table 2 lists protein bound to transfected biotinylated DNA in mock-infected cells. At least some of these proteins would be predicted to have a repressive role on gene expression inasmuch as the gene encoded in transfected DNA is not expressed or expressed at very low levels in mock-infected cells. Examination of Table 2 shows a number of proteins that hitherto have not been associated with repression of viral genes in infected cells but nevertheless are of potential interest. Of this group, three proteins—LSD3A, NCoR2, and CHD1—are of particular interest. The lysine-specific demethylase 3A has been shown to catalyze the demethylation of di- or monomethylated H3K9 (28). This is particularly notable in light of the observation that methlyated forms of H3K9 have been detected at latent HSV promoters (78). LSD1 has also been shown to catalyze the demethylation of H3K9 (79). A series of experiments on the regulation of H3K9 levels of a reporter gene concluded that knockdown of LSD1 did not alter the recruitment of LSD3A to the promoter region but did result in an impairment of demethylation (28). The corepressor protein NCoR2, also commonly referred to as SMRT, has been described as a “hub protein” responsible for recruiting a number of chromatin-regulating factors to DNA (29). Among the proteins reported to interact with NCoR are PML, HDAC1/2, and mSIN3A (30). CHD1 is a chromatin-remodeling enzyme containing two chromodomains and an ATPase domain. Relevant to HSV, CHD1 binds methylated H3K4, a mark typically associated with transcriptional activation (22). The consequence of binding is not clear. It has been demonstrated that CHD1 is required for the deposition of Histone H3.3 into chromatin in vivo and ubiquitination of histone H2B (80, 81). CHD1 could in fact activate transcription rather than repress it.
iii) The proteins recovered from biotinylated DNA extracted from transfected and infected cells (Table 3) include the histone methyltransferase Dot1L, ARID5B, and DEAD-box helicase Ddx17. Dot1L, named as disruptor of telomeric silencing 1, is the only enzyme known to catalyze the methylation of H3K79, a mark typically associated with transcriptional activity (52). Depletion and overexpression of DotL1 causes a derepression of telomeric chromatin (82). Data support a model in which Dotl1L-mediated H3K79 methylation occludes the binding of sirtuin1 (83). Interestingly, ubiquitination of histone H2B, a modification linked to CHD1, is required for Dot1L activity (84). The AT-rich interactive domain-containing protein 5, ARID5B, has been shown to promote the recruitment of a histone methyltransferase to enable the demethylation of H3K9 (55, 56). Ddx17 is a member of the DEAD-box helicase family of proteins with known roles in regulation of transcription and mRNA metabolism and will be discussed further below. Also intriguing are the few proteins that could not be grouped into either category but were relevant to transcription, HSV-1 biology, or both. Such proteins included Trim29, a protein with a domain structure similar to PML (64) and Prohibitin 2, which has been linked to transcription (67).
iv) The premise of this study is that the proteins bound to biotinylated DNA extracted from transfected infected cells would enhance rather than repress the expression of genes encoded by the transfected DNA in transfected/infected cells. One test of this hypothesis is to define the role of an enriched protein that would not be expected to play a role in either activation or repression of chromatin. We selected Ddx17 to test this hypothesis for two reasons. Foremost, the binding of Ddx17 to the biotinylated plasmid was enriched in cells transfected with the plasmid and then infected with wild-type virus. Second, helicases, and in particular this helicase, have not been associated with enhancement of repression of HSV gene expression. The results reported here show that depletion of Ddx17 has a minimal effect on the accumulation of ICP0 transcripts but significantly impairs the expression of ICP27 and of post-α genes. Given the observation that it plays a role in accumulation of viral mRNAs, the actual finding that depletion of Ddx17 has a minimal effect on ICP0 but a very significant effect on genes representative of kinetic classes expressed later in infection does not come as a surprise. ICP0 is one of the first genes expressed in cells infected at a low ratio of pfu per cell (85) and is most likely responsible for the modification of transfected DNA that led to the binding of Ddx17 to the biotinylated DNA. At low multiplicities of infection ICP0 plays a key role in enabling the expression of β and γ genes. These observations suggest that Ddx17 plays a role in derepression of these genes by ICP0.
v) The requirement for Ddx17 in the accumulation of viral transcripts is dependent upon SUMOylation in as much as overexpression of a mutant Ddx17 lacking the consensus SUMOylation motif (mutant K50R; Fig. 4) inhibited accumulation of viral protein ICP27. SUMOylation has been linked to many cellular processes, including formation of ND10 bodies (86).
vi) It is noteworthy that depletion of Ddx17 resulted in a decrease in Influenza virus yield and a decrease in the accumulation of viral RNAs (64). Ddx17 coprecipitated with the influenza RNA-dependent RNA polymerase, suggesting it is involved directly in transcription of viral genes. The studies on influenza, an RNA virus with an entirely different mode of RNA synthesis, would not have predicted the results presented in this report.
The procedure for recovery of proteins bound to the surrogate DNA illustrated in this report is distinct and complementary to the ChiP-seq procedures. The ChIP-seq procedure defines the frequency of binding to known proteins to specific sites on viral DNA (87). The ChIP-seq assay requires prior knowledge of the proteins that may play a role in gene activation or repression and availability of reagents of high purity and specificity. In contrast, the procedures described here can easily be adapted to specific DNA sequences, and the significance of binding a specific protein can be readily determined by examining viral gene expression in cells that have been depleted or enriched for a specific cellular protein. The value of this technique is illustrated by the example cited in this report.
In the studies reported here we transfected cells with DNA driven by a nonviral promoter. This assay can easily be modified to include nonviral genes driven by viral promoters to determine whether the plasmid DNA binds virus promoter-specific proteins. This assay is also suitable for most DNA viruses because viral DNAs, as a rule, aggregate with ND10 bodies and in many instances viral gene products seem to modify ND10 proteins for efficient gene expression.
Materials and Methods
Cells, Viruses, and Plasmid.
HeLa, HEK293, and HEp-2 cells obtained from the American Type Culture Collection were propagated in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 5% FBS (vol/vol). HSV-1(F) is the prototype HSV-1 strain used by our laboratory (88). The following plasmids were used in this study: myc-Ddx17 and myc-Ddx17K50R (kindly provided by Ralf Janknecht, University of Oklahoma), pcDNA3.1(+) (Invitrogen).
Biotinylation and Recovery of Plasmid DNA.
The pSV2LacO plasmid containing 256 copies of the Escherichia coli Lac operator (kindly provided by Andrew Belmont, University of Illinois at Chicago) was modified with biotin using the Label IT Nucleic Acid Labeling Reagents (Mirus Biosciences) according to a slightly modified version of protocol suggested by the manufacturer. Briefly, 10 μg of pSV2LacO were labeled with 5 μL of LabelIT reagent at 37 °C for 1 h. Under these conditions, an approximate labeling frequency of one biotin molecule every 60 bases is expected. Unincorporated biotin was removed by filtration through a Sephadex G50 column (GE Biosciences). HEp-2 cells were grown to ∼60% confluency in 150-cm2 flasks. Transfection was carried out with the aid of Lipofectamine/PLUS reagents according to the manufacturer’s recommendation (Invitrogen): 9.75 μg of biotinylated plasmid DNAs were mixed with 50 μL of PLUS reagent (Invitrogen) in 1.2 mL of serum-free DMEM, followed by incubation at room temperature for 15 min. An equal volume (1.2 mL) of serum-free DMEM containing 40 μL of Lipofectamine (Invitrogen) was then added to the plasmid DNA mixture, followed by an additional 15-min incubation at room temperature. The transfection mixture was added to the cell monolayers along with 7.5 mL of serum-free medium. After 3 h incubation at 37 °C, the mixture was replaced with DMEM containing 5% FBS. Where indicated, transfected HEp2 cells were either mock-infected or exposed to 10 pfu of HSV-1(F) at 37 °C for 1.5 h.
At the indicated times after infection, cell monolayers were rinsed twice with PBS, scraped into PBS, and centrifuged at 500 × g. The pellet was resuspended in serum-free DMEM to which formaldehyde was added to a final concentration of 1%. To quench the cross-linking reaction after 10 min, glycine was added to a final concentration of 125 mM. The cells were then pelleted, washed twice in PBS, and resuspened in hypotonic lysis buffer containing 0.5% Triton X100 [10 mM Hepes (pH 7.5), 10 mM KCl, 3 mM MgCl2, 1 mM EDTA, 10 mM NaF, 1 mM DTT, 0.1 mM Na3VO4, and 10 mM β-glycerophosphate]. After 20 min on ice, the insoluble material was harvested by centrifugation and resuspended in low-salt binding and wash buffer [50 mM Tris·HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, and 0.25% deoxycholic acid] and sonicated on ice to disrupt the nuclear membrane with the aid of a Fisher Scientific Ultrasonic Dismembrator. Each sonication step was carried out for 10 s, and samples were kept on ice for at least 1 min between sonications. The amplitude was gradually increased every three cycles, from 20% to 50%. After sonication, the total volume of sample was increased to 750 mL with PBS. Thirty microliters of streptavadin-conjugated magnetic beads (DYNABeads, Invitrogen) resuspended in 200 μL of binding buffer were added to the sonicated nuclear extract and the mixture incubated with gentle rotation at 4 °C for 12 h. The beads were pelleted by centrifugation and rinsed four times with ice-cold PBS, followed by two washes with PBS containing 0.1% Triton X-100, and a final wash with PBS only, before suspension in a buffer appropriate for downstream application.
Analysis of Samples by LC-MS/MS.
Samples were prepared for and analyzed by LC-MS/MS as previously described (89). In brief, protein mixtures were digested with trypsin overnight at 37 °C. Peptides were then extracted twice with 70% acetonitrile, 0.1% formic acid, dried, resuspended in 6 M guanidine·HCl in 5 mM potassium phosphate and 1 mM DTT, pH 6.5, sonicated, desalted using ZipTip C18, dried, and resuspended in 0.1% formic acid in mass spectrometry-grade water containing 5% acetonitrile. Samples were subjected to LC-MS/MS analysis using an LTQ from Thermo-Fisher coupled to a Surveyor HPLC system equipped with a Micro AS auto sampler. The instrument was interfaced with an Aquasil, C18 PicoFrit capillary column (75 µm × 10 cm) from New Objective. The mobile phases consisted of 0.1% formic acid containing 5% acetonitrile (A) and 0.1% formic acid in 95% acetonitrile (B), respectively. A 120-min linear gradient was used. The ions eluted from the column were electro-sprayed at a voltage of 1.75 kV. Collected raw data were searched against a human-HSV database and the list of proteins identified.
Depletion of Ddx17 Protein.
Approximately 106 HEp-2 cells were mock-transfected or transfected with 100 pmol of either control siRNA (Dharmacon, catalog no. D-001810) or a pool of five siRNAs directed against Ddx17 (Dharmacon, catalog no. L-013450). Oligos and Dharmafect transfection reagent were purchased from Dharmacon. Unless otherwise indicated, experiments were conducted 72 h after transfection of siRNA.
RNA Extraction and cDNA Synthesis and RT-qPCR.
Total RNA was extracted at the times indicated with the aid of TRIzol reagent (Life Technologies) according to the manufacturer’s instructions. One microgram of total RNA primed with random hexamers served as template for cDNA synthesis. The reaction was performed with the SuperScriptIII First-Strand Synthesis System for RT-PCR (Invitrogen), according to the manufacturer’s instructions. After reverse transcription samples were treated with RNase to eliminate residual RNA. cDNA levels were quantified using SYBR GreenER qPCR SuperMix Universal (Invitrogen). Results were normalized using 18s rRNA sequence. Samples obtained from cells treated with control siRNA served as reference for calculating relative mRNA levels by the ΔΔCT method. For DNA enrichment experiments, streptavidin beads were used directly as template. All qPCR was performed using an Applied Biosystems StepOne real-time PCR instrument. Data were analyzed using Microsoft Excel software. Primers for amplification of 18S rRNA and β-actin were purchased from Ambion (catalog nos. 4333760F and 4333762F).
Confocal Microscopy.
Approximately 104 cells were seeded on four-well microscope slides 24 h before infection. At the indicated time points, monolayers were rinsed extensively with PBS and fixed for 10 min at room temperature with 4% paraformaldehyde. Fixed cells were permeabilized and blocked in PBS-TBH buffer, consisting of PBS plus 0.2% Triton X100, 1% BSA, and 10% horse serum. Primary antibodies [rabbit anti-Ddx17, Abcam (catalog no. 24601), mouse anti-ICP4, and mouse anti-ICP0, 1:1,000] were diluted in PBS-TBH and applied to the samples for overnight incubation at 4 °C. The slides were then rinsed several times with PBS-TBH and reacted for 2 h with fluorophore-conjugated secondary antibody (Molecular Probes) diluted 1:1,000 in PBS-TBH. After several rinses, the samples were mounted and examined with a Zeiss LSM 310 confocal microscope equipped with software provided by Zeiss.
Immunoblotting.
Cells were rinsed in PBS, collected by low-speed centrifugation, and resuspended in lysis buffer [50 mM Tris (pH 8.0), 150 mM NaCl, 0.1% SDS, 0.5% deoxycholate, 1% Nonidet P-40, and complete protease inhibitor]. Lysates were incubated on ice, sonicated, and boiled. An equal amount of protein, as determined by Bradford Reagent, was separated by denaturing polyacrylamide gel electrophoresis. The proteins were transferred to a nitrocellulose membrane, blocked in PBS plus 10% nonfat dairy milk, and incubated overnight at 4 °C with primary antibody diluted in PBS plus 1% BSA. After incubation, membranes were rinsed extensively with PBS, incubated with HRP-conjugated secondary antibody (Sigma-Aldrich), and reacted with ECL reagent (Amersham Biosciences) to visualize protein.
Acknowledgments
We thank Andrew Belmont (University of Illinois) and Ralf Janknecht (University of Oklahoma) for providing reagents. These studies were aided by National Cancer Institute Grant 5R37CA078766-12.
Footnotes
The authors declare no conflict of interest.
References
- 1.Roizman B, Knipe DM, Whitley RJ. In: Fields’ Virology, Herpes simplex viruses. 5th Ed. Knipe DM, et al., editors. New York: Lippincott-Williams and Wilkins; 2007. pp. 2501–2601. [Google Scholar]
- 2.Roizman B, Zhou G, Du T. Checkpoints in productive and latent infections with herpes simplex virus 1: Conceptualization of the issues. J Neurovirol. 2011;17(6):512–517. doi: 10.1007/s13365-011-0058-x. [DOI] [PubMed] [Google Scholar]
- 3.Boutell C, Sadis S, Everett RD. Herpes simplex virus type 1 immediate-early protein ICP0 and is isolated RING finger domain act as ubiquitin E3 ligases in vitro. J Virol. 2002;76(2):841–850. doi: 10.1128/JVI.76.2.841-850.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hagglund R, Van Sant C, Lopez P, Roizman B. Herpes simplex virus 1-infected cell protein 0 contains two E3 ubiquitin ligase sites specific for different E2 ubiquitin-conjugating enzymes. Proc Natl Acad Sci USA. 2002;99(2):631–636. doi: 10.1073/pnas.022531599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gu H, Roizman B. The degradation of promyelocytic leukemia and Sp100 proteins by herpes simplex virus 1 is mediated by the ubiquitin-conjugating enzyme UbcH5a. Proc Natl Acad Sci USA. 2003;100(15):8963–8968. doi: 10.1073/pnas.1533420100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gu H, Liang Y, Mandel G, Roizman B. Components of the REST/CoREST/histone deacetylase repressor complex are disrupted, modified, and translocated in HSV-1-infected cells. Proc Natl Acad Sci USA. 2005;102(21):7571–7576. doi: 10.1073/pnas.0502658102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gu H, Roizman B. Engagement of the lysine-specific demethylase/HDAC1/CoREST/REST complex by herpes simplex virus 1. J Virol. 2009;83(9):4376–4385. doi: 10.1128/JVI.02515-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu H, Roizman B. Herpes simplex virus-infected cell protein 0 blocks the silencing of viral DNA by dissociating histone deacetylases from the CoREST-REST complex. Proc Natl Acad Sci USA. 2007;104(43):17134–17139. doi: 10.1073/pnas.0707266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawaguchi Y, Van Sant C, Roizman B. Herpes simplex virus 1 alpha regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J Virol. 1997;71(10):7328–7336. doi: 10.1128/jvi.71.10.7328-7336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagglund R, Roizman B. Herpes simplex virus 1 mutant in which the ICP0 HUL-1 E3 ubiquitin ligase site is disrupted stabilizes cdc34 but degrades D-type cyclins and exhibits diminished neurotoxicity. J Virol. 2003;77(24):13194–13202. doi: 10.1128/JVI.77.24.13194-13202.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalamvoki M, Gu H, Roizman B. Over expression of the ubiquitin specific protease 7 resulting from transfection or mutations in the ICP0 binding site accelerates rather than depresses HSV-1 gene expression. J Virol. 2012;86(23):12871–12878. doi: 10.1128/JVI.01981-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalamvoki M, Roizman B. The histone acetyltransferase CLOCK is an essential component of the herpes simplex virus 1 transcriptome that includes TFIID, ICP4, ICP27, and ICP22. J Virol. 2011;85(18):9472–9477. doi: 10.1128/JVI.00876-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalamvoki M, Roizman B. Nuclear retention of ICP0 in cells exposed to HDAC inhibitor or transfected with DNA before infection with herpes simplex virus 1. Proc Natl Acad Sci USA. 2008;105(51):20488–20493. doi: 10.1073/pnas.0810879105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishov AM, Maul GG. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J Cell Biol. 1996;134(4):815–826. doi: 10.1083/jcb.134.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Everett RD, Murray J. ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J Virol. 2005;79(8):5078–5089. doi: 10.1128/JVI.79.8.5078-5089.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalamvoki M, Roizman B. Role of herpes simplex virus ICP0 in the transactivation of genes introduced by infection or transfection: a reappraisal. J Virol. 2010;84(9):4222–4228. doi: 10.1128/JVI.02585-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quinlan MP, Knipe DM. Stimulation of expression of a herpes simplex virus DNA-binding protein by two viral functions. Mol Cell Biol. 1985;5(5):957–963. doi: 10.1128/mcb.5.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cliffe AR, Knipe DM. Herpes simplex virus ICP0 promotes both histone removal and acetylation on viral DNA during lytic infection. J Virol. 2008;82(24):12030–12038. doi: 10.1128/JVI.01575-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonner WM, West MH, Stedman JD. Two-dimensional gel analysis of histones in acid extracts of nuclei, cells, and tissues. Eur J Biochem. 1980;109:17–23. doi: 10.1111/j.1432-1033.1980.tb04762.x. [DOI] [PubMed] [Google Scholar]
- 20.Happel N, Doenecke D. Histone H1 and its isoforms: Contribution to chromatin structure and function. Gene. 2009;431(1-2):1–12. doi: 10.1016/j.gene.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 21.Clausell J, Happel N, Hale TK, Doenecke D, Beato M. Histone H1 subtypes differentially modulate chromatin condensation without preventing ATP-dependent remodeling by SWI/SNF or NURF. PLoS ONE. 2009;4(10):e0007243. doi: 10.1371/journal.pone.0007243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding H, et al. Regulation of murine telomere length by Rtel: An essential gene encoding a helicase-like protein. Cell. 2004;117(7):873–886. doi: 10.1016/j.cell.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 23.Baldwin AS, Jr, LeClair KP, Singh H, Sharp PA. A large protein containing zinc finger domains binds to related sequence elements in the enhancers of the class I major histocompatibility complex and kappa immunoglobulin genes. Mol Cell Biol. 1990;10(4):1406–1414. doi: 10.1128/mcb.10.4.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishizuka M, et al. A zinc finger protein TZF is a novel corepressor of androgen receptor. Biochem Biophys Res Commun. 2005;331(4):1025–1031. doi: 10.1016/j.bbrc.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 25.Tao RH, et al. Testicular zinc finger protein recruits histone deacetylase 2 and suppresses the transactivation function and intranuclear foci formation of agonist-bound androgen receptor competitively with TIF2. Mol Cell Endocrinol. 2006;247(1-2):150–165. doi: 10.1016/j.mce.2005.12.052. [DOI] [PubMed] [Google Scholar]
- 26.Patel A, McKnight JN, Genzor P, Bowman GD. Identification of residues in chromodomain helicase DNA-binding protein 1 (Chd1) required for coupling ATP hydrolysis to nucleosome sliding. J Biol Chem. 2011;286(51):43984–43993. doi: 10.1074/jbc.M111.282970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hollenbach AD, McPherson CJ, Mientjes EJ, Iyengar R, Grosveld G. Daxx and histone deacetylase II associate with chromatin through an interaction with core histones and the chromatin-associated protein Dek. J Cell Sci. 2002;115(Pt 16):3319–3330. doi: 10.1242/jcs.115.16.3319. [DOI] [PubMed] [Google Scholar]
- 28.Waldmann T, Scholten I, Kappes F, Hu HG, Knippers R. The DEK protein—an abundant and ubiquitous constituent of mammalian chromatin. Gene. 2004;343(1):1–9. doi: 10.1016/j.gene.2004.08.029. [DOI] [PubMed] [Google Scholar]
- 29.Campillos M, García MA, Valdivieso F, Vázquez J. Transcriptional activation by AP-2alpha is modulated by the oncogene DEK. Nucleic Acids Res. 2003;31(5):1571–1575. doi: 10.1093/nar/gkg247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gamble MJ, Fisher RP. SET and PARP1 remove DEK from chromatin to permit access by the transcription machinery. Nat Struct Mol Biol. 2007;14(6):548–555. doi: 10.1038/nsmb1248. [DOI] [PubMed] [Google Scholar]
- 31.Happel N, Schulze E, Doenecke D. Characterisation of human histone H1x. Biol Chem. 2005;386(6):541–551. doi: 10.1515/BC.2005.064. [DOI] [PubMed] [Google Scholar]
- 32.Yamane K, et al. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125(3):483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 33.Guo C, et al. Regulated clearance of histone deacetylase 3 protects independent formation of nuclear receptor corepressor complexes. J Biol Chem. 2012;287(15):12111–12120. doi: 10.1074/jbc.M111.327023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Underhill C, Qutob MS, Yee SP, Torchia J. A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J Biol Chem. 2000;275(51):40463–40470. doi: 10.1074/jbc.M007864200. [DOI] [PubMed] [Google Scholar]
- 35.Heo I, et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138(4):696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 36.Lischka P, Toth Z, Thomas M, Mueller R, Stamminger T. The UL69 transactivator protein of human cytomegalovirus interacts with DEXD/H-Box RNA helicase UAP56 to promote cytoplasmic accumulation of unspliced RNA. Mol Cell Biol. 2006;26(5):1631–1643. doi: 10.1128/MCB.26.5.1631-1643.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bollig F, et al. Affinity purification of ARE-binding proteins identifies polyA-binding protein 1 as a potential substrate in MK2-induced mRNA stabilization. Biochem Biophys Res Commun. 2003;301(3):665–670. doi: 10.1016/s0006-291x(03)00015-9. [DOI] [PubMed] [Google Scholar]
- 38.Grosset C, et al. A mechanism for translationally coupled mRNA turnover: Interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell. 2000;103(1):29–40. doi: 10.1016/s0092-8674(00)00102-1. [DOI] [PubMed] [Google Scholar]
- 39.Béland M, et al. Cdx1 autoregulation is governed by a novel Cdx1-LEF1 transcription complex. Mol Cell Biol. 2004;24(11):5028–5038. doi: 10.1128/MCB.24.11.5028-5038.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahajan MC, Narlikar GJ, Boyapaty G, Kingston RE, Weissman SM. Heterogeneous nuclear ribonucleoprotein C1/C2, MeCP1, and SWI/SNF form a chromatin remodeling complex at the beta-globin locus control region. Proc Natl Acad Sci USA. 2005;102(42):15012–15017. doi: 10.1073/pnas.0507596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shin C, Feng Y, Manley JL. Dephosphorylated SRp38 acts as a splicing repressor in response to heat shock. Nature. 2004;427(6974):553–558. doi: 10.1038/nature02288. [DOI] [PubMed] [Google Scholar]
- 42.Lin JY, et al. hnRNP A1 interacts with the 5′ untranslated regions of enterovirus 71 and Sindbis virus RNA and is required for viral replication. J Virol. 2009;83(12):6106–6114. doi: 10.1128/JVI.02476-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Najera I, Krieg M, Karn J. Synergistic stimulation of HIV-1 rev-dependent export of unspliced mRNA to the cytoplasm by hnRNP A1. J Mol Biol. 1999;285(5):1951–1964. doi: 10.1006/jmbi.1998.2473. [DOI] [PubMed] [Google Scholar]
- 44.Zhu Y, et al. Zinc-finger antiviral protein inhibits HIV-1 infection by selectively targeting multiply spliced viral mRNAs for degradation. Proc Natl Acad Sci USA. 2011;108(38):15834–15839. doi: 10.1073/pnas.1101676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xuan Y, Liu L, Shen S, Deng H, Gao G. Zinc finger antiviral protein inhibits murine gammaherpesvirus 68 m2 expression and regulates viral latency in cultured cells. J Virol. 2012;86(22):12431–12434. doi: 10.1128/JVI.01514-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bonenfant D, Coulot M, Towbin H, Schindler P, van Oostrum J. Characterization of histone H2A and H2B variants and their post-translational modifications by mass spectrometry. Mol Cell Proteomics. 2006;5(3):541–552. doi: 10.1074/mcp.M500288-MCP200. [DOI] [PubMed] [Google Scholar]
- 47.Kuhnert A, et al. Proteomic identification of PSF and p54(nrb) as TopBP1-interacting proteins. J Cell Biochem. 2012;113(5):1744–1753. doi: 10.1002/jcb.24045. [DOI] [PubMed] [Google Scholar]
- 48.Bloom DC, Giordani NV, Kwiatkowski DL. Epigenetic regulation of latent HSV-1 gene expression. Biochim Biophys Acta. 2010;1799(3-4):246–256. doi: 10.1016/j.bbagrm.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Timinszky G, et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat Struct Mol Biol. 2009;16(9):923–929. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
- 50.Gamble MJ, Frizzell KM, Yang C, Krishnakumar R, Kraus WL. The histone variant macroH2A1 marks repressed autosomal chromatin, but protects a subset of its target genes from silencing. Genes Dev. 2010;24(1):21–32. doi: 10.1101/gad.1876110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goswami R, Kaplan MH. Gcn5 Is required for PU.1-dependent IL-9 induction in Th9 cells. J Immunol. 2012;189(6):3026–3033. doi: 10.4049/jimmunol.1201496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hagman J, Lukin K. Transcription factors drive B cell development. Curr Opin Immunol. 2006;18(2):127–134. doi: 10.1016/j.coi.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 53.Bouché N, Scharlat A, Snedden W, Bouchez D, Fromm H. A novel family of calmodulin-binding transcription activators in multicellular organisms. J Biol Chem. 2002;277(24):21851–21861. doi: 10.1074/jbc.M200268200. [DOI] [PubMed] [Google Scholar]
- 54.Németh A, Strohner R, Grummt I, Längst G. The chromatin remodeling complex NoRC and TTF-I cooperate in the regulation of the mammalian rRNA genes in vivo. Nucleic Acids Res. 2004;32(14):4091–4099. doi: 10.1093/nar/gkh732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cizmecioglu O, et al. Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J Cell Biol. 2010;191(4):731–739. doi: 10.1083/jcb.201007107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Steger DJ, et al. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol. 2008;28(8):2825–2839. doi: 10.1128/MCB.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shah S, Henriksen MA. A novel disrupter of telomere silencing 1-like (DOT1L) interaction is required for signal transducer and activator of transcription 1 (STAT1)-activated gene expression. J Biol Chem. 2011;286(48):41195–41204. doi: 10.1074/jbc.M111.284190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nguyen AT, Zhang Y. The diverse functions of Dot1 and H3K79 methylation. Genes Dev. 2011;25(13):1345–1358. doi: 10.1101/gad.2057811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baba A, et al. PKA-dependent regulation of the histone lysine demethylase complex PHF2-ARID5B. Nat Cell Biol. 2011;13(6):668–675. doi: 10.1038/ncb2228. [DOI] [PubMed] [Google Scholar]
- 60.Zhu L, et al. Dynamics of the Mrf-2 DNA-binding domain free and in complex with DNA. Biochemistry. 2001;40(31):9142–9150. doi: 10.1021/bi010476a. [DOI] [PubMed] [Google Scholar]
- 61.Lefave CV, et al. Splicing factor hnRNPH drives an oncogenic splicing switch in gliomas. EMBO J. 2011;30(19):4084–4097. doi: 10.1038/emboj.2011.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Landeras-Bueno S, Jorba N, Pérez-Cidoncha M, Ortín J. The splicing factor proline-glutamine rich (SFPQ/PSF) is involved in influenza virus transcription. PLoS Pathog. 2011;7(11):e1002397. doi: 10.1371/journal.ppat.1002397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Souquere S, Beauclair G, Harper F, Fox A, Pierron G. Highly ordered spatial organization of the structural long noncoding NEAT1 RNAs within paraspeckle nuclear bodies. Mol Biol Cell. 2010;21(22):4020–4027. doi: 10.1091/mbc.E10-08-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bortz E, et al. Host- and strain-specific regulation of influenza virus polymerase activity by interacting cellular proteins. MBio. 2011 doi: 10.1128/mBio.00151-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Denisenko ON, O’Neill B, Ostrowski J, Van Seuningen I, Bomsztyk K. Zik1, a transcriptional repressor that interacts with the heterogeneous nuclear ribonucleoprotein particle K protein. J Biol Chem. 1996;271(44):27701–27706. doi: 10.1074/jbc.271.44.27701. [DOI] [PubMed] [Google Scholar]
- 66.Honoré B. The hnRNP 2H9 gene, which is involved in the splicing reaction, is a multiply spliced gene. Biochim Biophys Acta. 2000;1492(1):108–119. doi: 10.1016/s0167-4781(00)00092-0. [DOI] [PubMed] [Google Scholar]
- 67.Kasyapa CS, Kunapuli P, Cowell JK. Mass spectroscopy identifies the splicing-associated proteins, PSF, hnRNP H3, hnRNP A2/B1, and TLS/FUS as interacting partners of the ZNF198 protein associated with rearrangement in myeloproliferative disease. Exp Cell Res. 2005;309(1):78–85. doi: 10.1016/j.yexcr.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 68.Yuan Z, et al. The ATDC (TRIM29) protein binds p53 and antagonizes p53-mediated functions. Mol Cell Biol. 2010;30(12):3004–3015. doi: 10.1128/MCB.01023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Joshi B, et al. Differential regulation of human YY1 and caspase 7 promoters by prohibitin through E2F1 and p53 binding sites. Biochem J. 2007;401(1):155–166. doi: 10.1042/BJ20060364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dai Y, Ngo D, Jacob J, Forman LW, Faller DV. Prohibitin and the SWI/SNF ATPase subunit BRG1 are required for effective androgen antagonist-mediated transcriptional repression of androgen receptor-regulated genes. Carcinogenesis. 2008;29(9):1725–1733. doi: 10.1093/carcin/bgn117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee SJ, et al. PHB2 interacts with RNF2 and represses CP2c-stimulated transcription. Mol Cell Biochem. 2008;319(1-2):69–77. doi: 10.1007/s11010-008-9878-2. [DOI] [PubMed] [Google Scholar]
- 72.Abbas T, et al. CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol Cell. 2010;40(1):9–21. doi: 10.1016/j.molcel.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wilson BJ, et al. The p68 and p72 DEAD box RNA helicases interact with HDAC1 and repress transcription in a promoter-specific manner. BMC Mol Biol. 2004;5:11. doi: 10.1186/1471-2199-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mooney SM, Grande JP, Salisbury JL, Janknecht R. Sumoylation of p68 and p72 RNA helicases affects protein stability and transactivation potential. Biochemistry. 2010;49(1):1–10. doi: 10.1021/bi901263m. [DOI] [PubMed] [Google Scholar]
- 75.Lopez P, Van Sant C, Roizman B. Requirements for the nuclear-cytoplasmic translocation of infected-cell protein 0 of herpes simplex virus 1. J Virol. 2001;75(8):3832–3840. doi: 10.1128/JVI.75.8.3832-3840.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Van Sant C, Lopez P, Advani SJ, Roizman B. Role of cyclin D3 in the biology of herpes simplex virus 1 ICPO. J Virol. 2001;75(4):1888–1898. doi: 10.1128/JVI.75.4.1888-1898.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kalamvoki M, Roizman B. Interwoven roles of cyclin D3 and cdk4 recruited by ICP0 and ICP4 in the expression of herpes simplex virus genes. J Virol. 2010;84(19):9709–9717. doi: 10.1128/JVI.01050-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cliffe AR, Garber DA, Knipe DM. Transcription of the herpes simplex virus latency-associated transcript promotes the formation of facultative heterochromatin on lytic promoters. J Virol. 2009;83(16):8182–8190. doi: 10.1128/JVI.00712-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nicholson TB, Chen T. LSD1 demethylates histone and non-histone proteins. Epigenetics. 2009;4(3):129–132. doi: 10.4161/epi.4.3.8443. [DOI] [PubMed] [Google Scholar]
- 80.Konev AY, et al. CHD1 motor protein is required for deposition of histone variant H3.3 into chromatin in vivo. Science. 2007;317(5841):1087–1090. doi: 10.1126/science.1145339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee JS, et al. Codependency of H2B monoubiquitination and nucleosome reassembly on Chd1. Genes Dev. 2012;26(9):914–919. doi: 10.1101/gad.186841.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ng HH, et al. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 2002;16(12):1518–1527. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang D, Li S, Cruz P, Kone BC. Sirtuin 1 functionally and physically interacts with disruptor of telomeric silencing-1 to regulate alpha-ENaC transcription in collecting duct. J Biol Chem. 2009;284(31):20917–20926. doi: 10.1074/jbc.M109.020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature. 2008;453(7196):812–816. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kalamvoki M, Roizman B. Circadian CLOCK histone acetyl transferase localizes at ND10 nuclear bodies and enables herpes simplex virus gene expression. Proc Natl Acad Sci USA. 2010;107(41):17721–17726. doi: 10.1073/pnas.1012991107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chung I, Leonhardt H, Rippe K. De novo assembly of a PML nuclear subcompartment occurs through multiple pathways and induces telomere elongation. J Cell Sci. 2011;124(Pt 21):3603–3618. doi: 10.1242/jcs.084681. [DOI] [PubMed] [Google Scholar]
- 87.Robertson G, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4(8):651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 88.Ejercito PM, Kieff ED, Roizman B. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol. 1968;2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 89.Dunmire JJ, et al. Novel serum proteomic signatures in a non-human primate model of retinal injury. Mol Vis. 2011;17:779–791. [PMC free article] [PubMed] [Google Scholar]