Abstract
The mechanisms that underpin the varied spatial genetic structures exhibited by free-living marine microorganisms remain controversial, with most studies emphasizing a high dispersal capability that should redistribute genetic diversity in contrast to most macroorganisms whose populations often retain a genetic signature of demographic response to historic climate fluctuations. We quantified the European phylogeographic structure of the marine flagellate Oxyrrhis marina and found a marked difference in spatial genetic structure, population demography, and genetic diversity between the northwest Atlantic and Mediterranean Sea that reflects the persistent separation of these regions as well as context-dependent population responses to contrasting environments. We found similar geographic variation in the level of genetic diversity in the sister species Oxyrrhis maritima. Because the capacity for wide dispersal is not always realized, historic genetic footprints of range expansion and contraction persist in contemporary populations of marine microbes, as they do in larger species. Indeed, the well-described genetic effects of climatic variation on macroorganisms provide clear, testable hypotheses about the processes that drive genetic divergence in marine microbes and thus about the response to future environmental change.
Free-living protists (eukaryotic microbes) are key components of marine ecosystems, driving productivity and biogeochemical cycles (1), but we lack understanding of the processes that underpin the contemporary distribution of genetic diversity in such organisms. A persistent paradigm emphasizes the importance of organism size, with microorganisms (<1 mm) such as protists having huge population sizes and being widely dispersed (2, 3). The corollary is that, compared with most macroorganisms, protists have little opportunity for allopatric divergence and will not exhibit pronounced spatial genetic structure. This paradigm is thought to be typical in the marine environment where the few obvious barriers to planktonic dispersal should permit extensive gene flow, and the abundance of cryptic marine microbial species (4, 5) is hypothesized to reflect sympatric divergence (6, 7). An opposing view is that there is no clear micro/macroorganism dichotomy, and that many free-living protists have restricted distributions, low rates of dispersal, and thus exhibit parapatric or allopatric divergence (8). Resolving these dichotomous viewpoints is essential to understand the processes that drive differentiation among populations of free-living protists and, ultimately, adaptation and speciation.
An increasing number of studies therefore have evaluated the degree of population structure in free-living marine protists. A complex picture has emerged, with reports of homogeneity at regional (9, 10) and global scales (11), as well as substantial genetic divergence that occurs locally (12), regionally (9, 13–16), or globally (17–19). It is clear that marine protist populations vary considerably in the scale over which significant genetic structure occurs. However, most studies interpret patterns in terms of putative dispersal ability in response to contemporary seascape features such as the structure of ocean basins, currents, and/or fronts. This approach overlooks the legacy of historic climate change that has shaped the present-day distribution of genetic diversity in many macroscopic taxa as a consequence of geographic range shifts in response to periodic cycles of glaciation and warming throughout the Quaternary (2.59 Mya to present) (20).
Although species vary in their ability to track suitable habitat during periods of climate change, the general patterns exhibited by European marine macroorganisms (21–23) provide an insight into processes that may have affected marine protists. First, most northern populations exhibit a shallow population structure and lack genetic diversity because of recent (post-Last Glacial Maximum, 25–18 kya) expansion into formerly inhospitable glaciated areas from southern refugia, albeit with alternative genetic patterns indicating that species persisted in northern periglacial refugia (22, 24). Second, marked divergence between Atlantic and Mediterranean populations often is interpreted as vicariance associated with the oscillations in sea level that isolated the Mediterranean Sea from the Atlantic at the Strait of Gibraltar; however, this pattern is not universal, and genetic congruence between the Atlantic and Mediterranean has been reported for some pelagic species (21). Finally, contemporary Mediterranean marine fauna bear a genetic legacy resulting from an extensive period of desiccation between 5.59 and 5.3 Mya [the Messinian Salinity Crisis (25)] that extirpated most species from the region or reduced species’ distributions to small, isolated eastern refugia. Subsequent flooding of the Mediterranean basin from the Atlantic was followed by episodic changes in sea level associated with glacial cycles. Successive periods of colonization and isolation left longitudinal gradients in genetic diversity across the Mediterranean and in some cases resulted in greatest genetic diversity in the central Mediterranean as a result of bidirectional colonization and secondary contact (26).
No study has yet sought to identify the contribution of historical legacies in shaping microbial population structures, and thus patterns of colonization, spatial variation in population demography, and the occurrence of potential refugia remain undocumented for any free-living marine protist. This lack is surprising, given the evidence for complex population structures and biogeographic patterns (8, 27) in marine microbial populations, which support the more general role for divergence in geographically distinct locations, as opposed to sympatry, as hypothesized by Foissner (8). Although two studies on marine protists have dated periods of diversification to the Pleistocene (19, 28), we still do not know the extent to which contemporary marine protist populations retain a genetic legacy of historic dispersal and divergence.
The free-living marine flagellate Oxyrrhis marina (Dinoflagellata: Oxyrrhinales) is common in coastal habitats (29) and thus is a model of marine protists that have wide geographic distributions, large population sizes, and putative high dispersal capabilities. The genus Oxyrrhis comprises two sibling species (5, 30). One, O. marina, is common and widely distributed; the other, Oxyrrhis maritima, has a more limited distribution (29). By quantifying spatial genetic structure in these species across the European seascape, we tested the hypothesis that contemporary populations of free-living marine protists do not redistribute genetic diversity rapidly but rather retain the distinct genetic signatures of population responses to historic changes in climate.
Results
Sampling.
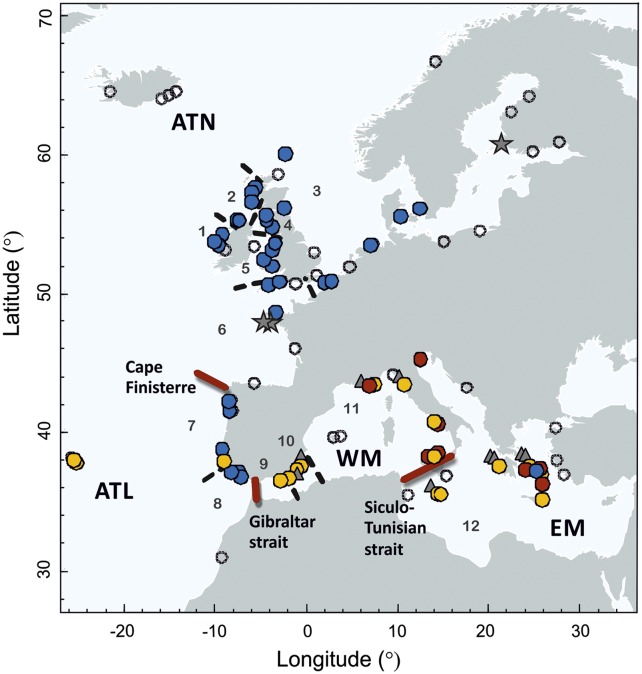
We obtained 822 environmental samples from 149 locations throughout Europe; these yielded 350 positive samples, from which we established 207 clonal isolates of Oxyrrhis from 89 locations (Fig. 1 and Table S1).
Fig. 1.
Sampling locations (n = 149) and distribution of the main clades of O. marina and O. maritima from the European seascape. For O. marina colors indicate clade/model cluster: blue, clade 1; yellow, clade 2a; red, clade 2b. For O. maritima (gray symbols) triangles indicate clade 3, and stars indicate clade 4. Open circles are locations that were sampled but did not contain Oxyrrhis. Red lines denote boundaries between the four main regions (the Siculo-Tunisian strait, the Strait of Gibraltar, and Cape Finisterre). Dashed black lines indicate the boundaries used to define the 12 populations (gray numbers) used for rarefaction analysis of haplotype diversity.
Phylogenetic Structure.
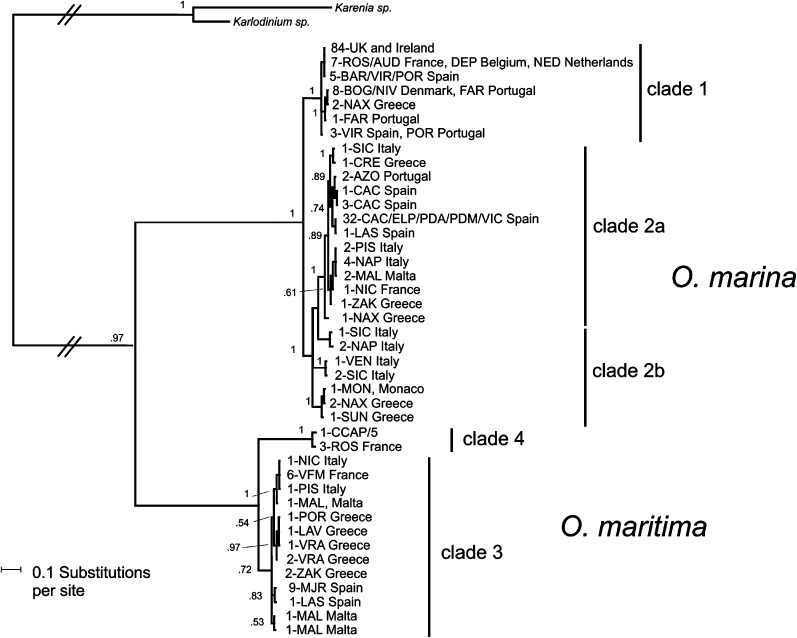
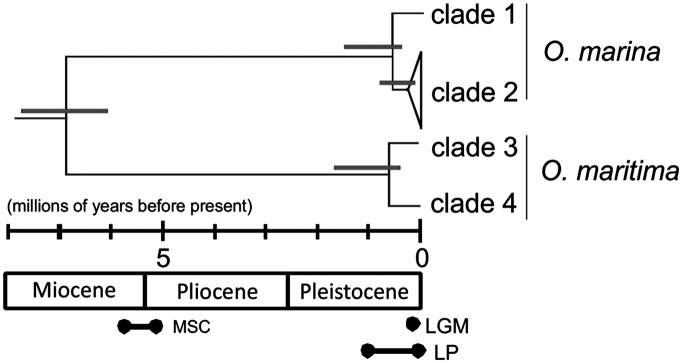
Phylogenetic analysis of the concatenated data (Fig. 2) was broadly congruent with the individual genes (Fig. S1 A–C), yielding two distinct lineages that corresponded to O. marina and O. maritima (5, 30). Each lineage comprised two well-supported clades [clades 1 and 2 for O. marina and clades 3 and 4 for O. maritima (30)] that differ in genetic diversity: clades 1 and 2 contained 7 and 20 haplotypes respectively, and clades 3 and 4 contained 13 and 2 haplotypes respectively. Based on cytochrome c oxidase I (COI) dating, O. marina and O. maritima are estimated to have diverged 5.5 Mya (3.61–7.62 95% highest posterior density, HPD) (Fig. 3). Divergence between clades within species occurred during similar times, at approximately ∼0.50 Mya (0.19–0.94 95% HPD) for O. marina and ∼0.60 Mya (0.21–1.03 95% HPD) for O. maritima. The most recent common ancestor for O. marina clade 2 was estimated to have been present at 0.26 Mya (0.03–0.57 95% HPD); comparable estimates for other clades were not possible because COI was invariable.
Fig. 2.
Majority rule consensus Bayesian phylogenetic tree of European Oxyrrhis isolates based on a concatenated alignment of three gene fragments (5.8S ITS rDNA, COI, and α-tubulin; total alignment length 1,338 bp). Node labels are posterior probabilities based on 106 permutations. Branch labels indicate the sample size (number of haplotypes) as the first number in branch label, followed by a three-letter location code and the country of origin. Clades for O. marina are synonymous with model clusters from the Structure analysis (Fig. 4).
Fig. 3.
Calibrated COI phylogenetic tree for European Oxyrrhis isolates based on a sequence substitution rate of 0.01 substitutions per million years. Errors for each dated node are upper and lower 95% posterior densities. MSC, Messinian Salinity Crisis; LP, Late Pleistocene glaciations; LGM, Last Glacial Maximum.
Spatial Genetic Structure.
O. marina was widely distributed throughout the sample region, whereas O. maritima was relatively rare and patchily distributed (Fig. 1). For O. marina, Structure analysis identified three distinct model clusters (Fig. 4 and Fig. S2) that correspond with the clades identified by phylogenetic analysis (Fig. 2). Clade 1 was equivalent to model cluster 1, whereas clade 2 was subdivided into two model clusters (2a and 2b) with cluster 2a corresponding to the most derived subclade and cluster 2b containing isolates that formed the basal subclades (Figs. 2 and 4).
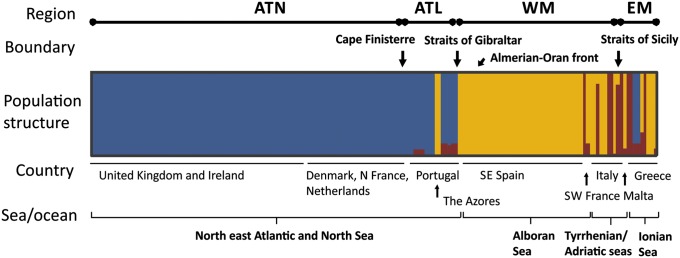
Fig. 4.
Structure plot for K = three model clusters for samples of O. marina. Each isolate of O. marina is depicted by a vertical line that is partitioned into K colored sections (colors as in Fig. 1). The length of each section is proportional to the estimated membership coefficient (Qind) of each cluster. Clusters are synonymous with clades (Fig.1): cluster 1 (blue) is equivalent to clade 1, cluster 2a (yellow) is equivalent to clade 2a, and cluster 2b (red) is equivalent to clade 2b.
These three model clusters had contrasting geographic distributions (Fig. 1), indicating the occurrence of well-defined spatial boundaries rather than widespread dispersal throughout the study region. Cluster 1 had a broad latitudinal distribution (∼38°N to 61°N) but was restricted almost exclusively to the Northeast Atlantic (only two isolates occurred in the eastern Mediterranean). Cluster 2a had a broad longitudinal distribution, with isolates from the Azores, the Iberian Atlantic coast, and the Mediterranean Sea, but a latitudinal range restricted to below 39°N in the Atlantic. Compared with the other clusters, cluster 2b had deeper genetic divergence between isolates (clade 2b, Fig. 2) and was restricted to the Mediterranean Sea but was absent from the Iberian coast (Fig. 1). O. marina isolates from European northern Atlantic (ATN) were exclusively associated with cluster 1 (average proportion of membership to cluster 1 [Q1] = 1.00; Table S2), whereas there was a western Mediterranean genetic influence (cluster 2a) in the European southern Atlantic (ATL) (Q1 = 0.76; Q2a = 0.17) as result of the isolates from the Azores and admixture with cluster 2b (Fig. 4). The eastern Mediterranean (EM) was more mixed, with roughly equal average proportions of membership in model clusters 2a and 2b (Q2a = 0.42; Q2b = 0.43; Table S2) and two isolates from model cluster 1. A qualitatively similar population structure was evident for O. maritima, with clade 3 limited to the Mediterranean Sea, whereas clade 4 was recovered only from the northern French coast and the Baltic Sea (Figs. 1 and 2).
Genetic Diversity.
Genetic diversity (concatenated sequence data) was higher in the Mediterranean Sea than in the Northeast Atlantic (Table 1). For O. marina, 21 haplotypes occurred throughout the Mediterranean [13 haplotypes in the western Mediterranean (WM) and eight haplotypes in the EM] whereas eight different haplotypes were present in the ATN and ATL (three and seven haplotypes, respectively); estimates of nucleotide diversity (π) and haplotype diversity (h) were greater in the Mediterranean than in the Atlantic (Table 1). After rarefaction analysis this gradient in genetic diversity was evident, with the greatest genetic diversity in the EM and the least diversity in the ATN (Fig. S3). Hence, model selection returned a generalized linear model (GLM) that contained longitude as the single best predictor of haplotype richness (intercept = 1.179, P = 0.0005; longitude = 0.106, P = 0.0015), accounting for 65% of the variation (R2) in diversity among groups (Fig. S3). This relationship was driven primarily by variation among Mediterranean Sea locations, because the eastern Atlantic samples lacked genetic diversity (Fig. S3). The distribution of genetic diversity for O. maritima was similar to that described for O. marina, with greatest diversity in the Mediterranean Sea (five and eight haplotypes for the WM and EM, respectively) (Table 1 and Fig. 1).
Table 1.
Regional sample sizes, estimates of genetic diversity from the concatenated gene data, and single-locus analyses of population demography for α-tubulin and 5.8S ITS rDNA for O. marina and O. maritima
| ATN | ATL | WM | EM | |
| O. marina | ||||
| n | 95 | 15 | 50 | 11 |
| S | 3 | 7 | 13 | 8 |
| π | 0.0001 | 0.013 | 0.024 | 0.065 |
| h | 0.141 | 0.876 | 0.573 | 0.946 |
| α-TUB | ||||
| Fs | ≤999*** | −8.95*** | −25.63*** | −1.97 |
| SSD | 0.000 | 0.144* | 0.006 | 0.077* |
| Rag | 0.842 | 0.371* | 0.205 | 0.158*** |
| ITS | ||||
| Fs | ≤999*** | −3.30* | −24.14*** | −0.20 |
| SSD | 0.039 | 0.106* | 0.359*** | 0.037* |
| Rag | 0.560 | 0.208* | 0.203 | 0.066* |
| O. maritima | ||||
| n | 8 | 18 | 10 | |
| S | 5 | 5 | 8 | |
| h | 0.600 | 0.667 | 0.933 | |
Demographic estimates are not completed for O. maritima because of small sample size. n, sample size; S, number of polymorphic sites; π, nucleotide diversity; h, haplotype diversity; α-TUB, α-tubulin; Fs, Fu’s index; SSD, sum of squared deviations; Rag, raggedness index; ITS, 5.8S ITS rDNA.
*P < 0.05; ***P < 0.001.
Demographic Signals.
Demographic genetic signatures were consistent between α-tubulin (α-TUB) and internal transcribed spacers 5.8S (ITS) and revealed a contrast between regions within the Atlantic and the Mediterranean Sea. O. marina from the ATN and WM likely underwent a recent population expansion, with significantly negative values of Fu’s index (Fs) (31) (with unbounded negative values in ATN resulting from the low diversity), large and nonsignificant indices of raggedness [the WM has a significant sum of squared deviations (SSD) at ITS; Table 1], and unimodal mismatch distributions (Fig. S4). Conversely, although the ATL had significantly negative values of Fs at both loci, the results for the ATL and EM are typical of more stable populations with rejected models of sudden population expansion (Table 1) and multimodal mismatch distributions (Fig. S4).
Population sizes [θ, a mutation-scaled population size (32)] were greater in the EM and declined westwards and then north into the ATN; similarly, gene flow (M, the mutation-scaled immigration rate) occurred predominantly from the EM toward the WM and ATN (Table S3) with model selection supporting the EM as the most likely source [Akaike information criterion (33) (AIC) = 2,382] over the remaining three locations (WM AIC = 4,088; ATL AIC = 2,831; ATN AIC = 3,566) (Fig. S5).
Discussion
Our systematic examination of European phylogeographic structure in a marine protist supports the hypothesis that contemporary marine microbial populations can retain prominent genetic footprints of historic divergence, as do populations of marine macrobiota (21–23, 34), and refutes the supposition that size per se is a fundamental driver of population structure (2, 3). Signals of historic vicariance and dispersal include (i) divergence between largely allopatric clades (Fig. 2), (ii) an Atlantic–Mediterranean transition at the Straits of Gibraltar (Fig. 4), (iii) longitudinal and latitudinal gradients in genetic diversity (Table 1 and Fig. S3), (iv) the prevalence of westward migration from the eastern Mediterranean, and (v) recent demographic expansion in the northern Atlantic (Table 1).
Shallow-water biota are predicted to have persisted in eastern Mediterranean refugia during the climatic extremes of the Messinian Salinity Crisis (34). The basal position of the eastern Mediterranean clade 2b (Fig. 2), combined with deeper divergence between lineages and high genetic diversity in the EM (Table 1 and Fig. S3), is evidence that coastal marine protists persisted in eastern Mediterranean refugia during the climatic extremes of the Messinian Salinity Crisis (5.59–5.3 Mya). Further support for this idea is provided by the predominantly east-to-west migration, the stable population demography in the EM, and a demographic expansion in the WM. There are fewer data for O. maritima, but this species also exhibits greatest diversity in the EM (Table 1). Moreover, for both lineages of Oxyrrhis, the absence of population subdivision at the shallow Siculo-Tunisian Strait contrasts with a persistent signal of vicariance in other marine taxa (35, 36) where previous oscillations in sea level separated the eastern and western basins. This absence of a subdivision also indicates that, like open-water protists (e.g., 37), the dispersal of coastal taxa (such as Oxyrrhis) is not limited by the present-day hydrodynamic regime (38). However, the low genetic diversity and shallow population structure in the west compared with the high diversity and a genetic mosaic in the east is more typical of sedentary benthic or demersal macroorganisms (36, 39, 40) than of dispersive pelagic taxa (35). A similar gradient in genetic diversity is likely to occur in eurytopic protists that persisted for long periods in the EM, with divergence driven by episodes of population fragmentation that accompany the repeated changes in sea-level associated with historic glacial cycles.
Only a few studies on protists have sampled the Atlantic–Mediterranean transition zone. Aurahs et al. (37) suggested that Globigerinoides ruber (Foraminifera: Polythalamea) was unaffected by a potential dispersal barrier at the Straits of Gibraltar, but their genetic marker (small subunit ribosomal DNA, SSU rDNA) lacked sufficient polymorphism to detect intraspecific subdivision. Similarly, Penna et al. (41) found no differences between Atlantic and Mediterranean isolates of Ostreopsis sp. (Dinoflagellata: Peridinea), even though they used a polymorphic locus (ITS). In contrast, Lilly et al. (42) identified different clades of Alexandrium tamarense (Dinoflagellata: Peridinea) in the Atlantic and Mediterranean. The latter two studies, however, lacked sufficient resolution to identify the location of any genetic discontinuity. The present report of a phylogeographic break at the Straits of Gibraltar in a free-living, marine protist reflects historic divergence when the Mediterranean Sea was isolated from the Atlantic, with no evidence that the Almeria–Oran front acts as a contemporary oceanographic barrier to dispersal by O. marina (Figs. 1 and 4) as it does in some species of fish (21, 43). The rare instances of clade 1 and 2a isolates inhabiting the WM and ATL, respectively, represent more recent, possibly anthropogenic, dispersals, because these samples are isolated from their main clade distributions (Fig. 1).
In addition to isolating the Mediterranean from the Atlantic, Pleistocene ice sheet expansions and contractions forced alternations in species’ ranges in the North Atlantic. Marine protists retain classic signals of recent expansion into the northeast Atlantic from southern locations (20, 22, 24): low haplotype diversity (Table 1 and Fig. S3), shallow clade structure (Fig. 2), a demographic expansion (Table 1), and directional migration (Fig. S5). Although such a population structure has not been reported previously in a marine protist, it is a common genetic footprint of macroorganisms that were extirpated from northern Atlantic habitats during the Last Glacial Maximum (25–18 kya) (22). Katz et al. (17) interpreted greater genetic diversity in Strombidum oculatum (Ciliophora: Spirotrichea) from the British Isles compared with the eastern coast of the United States (Maine) as evidence for a northeast Atlantic refugium, but their sampling in this region was limited (52–55°N) and overlooked potential southern refugia. Our extensive sampling in the northeast Atlantic provided no evidence of high diversity and divergent lineages (Table 1 and Figs. 1 and 2) that would indicate Oxyrrhis survived in periglacial refugia around the British Isles during the Last Glacial Maximum (24, 44). Rather, the distribution of haplotypes (Fig. 1) and the stable population in the ATL (Table 1) indicate that O. marina inhabited low-latitude Atlantic locations throughout the Pleistocene. Oxyrrhis is a eurytopic genus, and we expect most coastal marine protists have a similar shallow genetic structure in the North Atlantic as a consequence of a post-Last Glacial Maximum range expansion from more southern locations.
Overall, models of divergence and adaptation for coastal marine protists must incorporate the long periods of isolation through the quite different environmental histories of the Atlantic and Mediterranean basins before sympatric mechanisms of divergence (6, 7). The pattern and timing of divergence in Oxyrrhis provides a template for other European marine protists. The most parsimonious scenario is that O. marina persisted in the eastern Mediterranean throughout the Messinian Salinity Crisis, and this eastern lineage gave rise to clades 1 and 2. These clades diverged close to the start of the Late Pleistocene glacial cycles (∼0.65 Mya), thus implicating allopatric divergence associated with range contractions and expansions during glacial cycles as the key mechanism of divergence, as is likely for O. maritima (clades 3 and 4; Fig. 3) and other marine protists (19, 28). An ancestral Atlantic population of O. marina resulted in clade 1, which expanded northwards after the Last Glacial Maximum. A population that persisted in the eastern Mediterranean gave rise to clade 2b; subsequent westward expansion and divergence produced clade 2a at ∼0.26 Mya. Whether a western Mediterranean population persisted during this period and was displaced by immigration from the eastern Mediterranean cannot be determined, but colonization from the Atlantic (clade 1) has been limited (Fig. 1). Notably the estimated divergence time (∼5–7 Mya) between the two Oxyrrhis species raises the possibility that speciation was driven by the severe climatic events associated with the Messinian Salinity Crisis. Other patterns are possible, depending on life history or ecological tolerance; for example, stenotopic protists are hypothesized to have recolonized the Mediterranean Sea from Atlantic refugia after the Messinian Salinity Crisis.
The geographic location of sampling and the spatial scale have a clear impact on data interpretation. For example, sampling either the eastern Atlantic or the Mediterranean Sea could lead to alternative conclusions about inherent dispersal capability, with apparently high gene flow in the former region compared with a complex structure driven by restricted dispersal in the latter. Indeed studies of marine protists from northern Europe often indicate a lack of spatial genetic structure (9, 10), whereas studies conducted within the Mediterranean Sea tend to reveal comparatively strong population differentiation (16). Certainly, it could be inaccurate to infer dispersal potential [e.g., as emphasized to be an important goal for studies of toxic species (16)] using the pattern obtained from recently established populations that have not attained genetic equilibrium. A second issue is that most studies of marine protists collect samples separated by large distances (e.g., 19) and thus fail to differentiate between historic phylogeographic boundaries and contemporary oceanographic barriers. Nonetheless, the evidence for (Fig. 4 and ref. 42) and against (37, 41) divergence between Atlantic and Mediterranean populations in protists draws attention to the futility of considering organism size per se as the key driver of dispersal and associated evolutionary processes (2, 3). This realization should not be surprising, because the diversity of protist life histories will yield an array of population structures in response to historic and contemporary environments (45, 46). The analogy is drawn with macroorganisms, where comparisons among multiple species reveal contrasting responses to the seascape (43, 47, 48) and where no particular phylogeographic structure associates with life history characteristic or dispersal potential (21–23, 47, 49). The protist–macroorganism dichotomy per se hinders an understanding of dispersal, divergence, and adaptation in marine microbes. More useful will be further studies that target known genetic boundaries to uncover the extent of similarities within and between a range of microscopic and macroscopic taxa to identify the parallel responses to previous climatic fluctuations.
A persistent genetic signature of historic divergence contrasts with the hypothesis of high dispersal rates assumed for free-living microbial species that should rapidly erode such patterns by redistributing genetic diversity (2, 3). Although many free-living protists have broad distributions, wide dispersal potential and genetic homogeneity often are not realized (17, 46, 50). Although our data emphasize the role of vicariance, it is notable that O. marina clade 2 isolates failed to colonize high latitudes, and clade 1 is rare in the Mediterranean (Fig. 1). Because clade 2 haplotypes are broadly dispersed longitudinally, inhabiting even remote islands such as the Azores, it is likely that environmental conditions and/or intraspecific interactions rather than restricted dispersal reinforce the contemporary distribution of these divergent strains. However, there was no obvious relationship between environmental characteristics and population boundaries, and the correlation between genetic diversity and longitude is interpreted as an historical signal of east–west migration (Fig. S3).
Many microeukaryotes (e.g., rotifers and cladocerans) and some protists that inhabit lentic systems have a complex phylogeographic structure with some isolates/strains widely distributed but most populations retaining a mosaic of genetic diversity (51, 52). De Meester and coworkers’ monopolization hypothesis explains this pattern through high dispersal potential that allows the broad distribution of certain strains, but with genetic diversity maintained through stochastic colonization events that are reinforced by rapid local adaptation of the initial colonists who limit subsequent immigration (51). Whether such priority effects operate in more open marine systems has not been addressed, but the phylogeographic patterns reported here and for some other protist taxa (17) suggest that they might. The implication is a key role for local adaptation and intraspecific competition in shaping protist spatial genetic structure, in addition to interspecific interactions and historical contingency (27).
To conclude, in contrast to the expected homogeneous population structure generated by widespread contemporary dispersal, we uncover a persistent genetic footprint of historical divergence and dispersal that emphasizes the similarity between micro- and macroorganisms rather than a size-based dichotomy. Given the diversity of protists (e.g., in their evolutionary age, complexity of life cycles, and ecological requirements), future studies on a broad functional and taxonomic range of protists are likely to uncover alternative phylogeographic patterns that are distinct from the one we describe here and that indicate the diversity of microbial responses to previous changes in climate. The well-described effects of climatic variation and contemporary landscape features on populations of macroorganisms can be used to provide testable hypotheses about the processes that drive genetic divergence in marine microbes and, potentially, information about the response to future environmental challenges.
Materials and Methods
Sample Collection.
Samples of O. marina and O. maritima were collected from intertidal locations across the European seascape (Fig. 1). Isolation, culturing, and DNA extraction followed methods outlined in ref. 53. Environmental data were obtained from the World Oceanographic Database (www.nodc.noaa.gov/OC5/WOD09/pr_wod09.html).
PCR and Sequencing.
Three gene regions were studied: 358 bp of the 5.8S ITS rDNA (ITS: primers in ref. 54), 540 bp of cytochrome oxidase I (COI: primers in ref. 5), and 438 bp of α-tubulin (α-TUB: F 5′-CTG CTT GGA GCA TGG TAT TCA G-3′; R 5′-TGT CGT ACA TSG CCT CRT TGT C-3′). Sequencing in both orientations was performed from PCR products using BigDye v.3.1 chemistry and electrophoresis on an AB3130xl (Applied Biosystems). Sequence trimming and alignments were performed using DNA Star (DNA Star). Accession numbers are provided in Table S1.
Phylogenetic Analyses.
Phylogenetic analyses were based on a reduced haplotype alignment (a single sequence for each haplotype). Optimal substitution models, determined using Model Test v.3.06 (55), were TPM1uf (ITS and COI) and TrN+I+G (α-TUB). Bayesian analyses of individual and concatenated alignments were performed using MrBayes v.3.1.2 (56). Default model priors and random starting trees were used to initiate simulations, which were run for 5 × 106 iterations with sampling every 200th generation. Two Metropolis-coupled Markov chain Monte Carlo (MCMC) chains were run (incrementally heated, following default values), with the first 6,500 trees discarded and congruence between parallel simulations assessed using Tracer v.1.01 (57).
Estimates of the timing of divergence between major clades within Oxyrrhis were conducted using Beast v.1.6.1 (58) and COI data only, because significant rate heterogeneity was detected among ITS sequences (Table S4). The time to most recent common ancestor parameters were estimated based on a generalized substitution rate of 1% per Mya for COI (59). The MCMC simulations (four chains) were run for 1 × 107 generations, with parameters sampled every 1,000 generations and convergence between chains assessed using Tracer. Because we lack fossil data with which to calibrate our molecular clock or specific estimates of substitution rates for COI in dinoflagellates, the specific timing of divergence should be interpreted cautiously. Nonetheless, our broad inference that O. marina clades 1 and 2 diverged relatively recently (i.e., within the Pleistocene) would remain robust to a two- or threefold variation in substitution rate.
Genetic Diversity and Spatial Genetic Structure.
Samples were partitioned into 12 areas within four main geographic regions (Fig. 1). (i) the eastern Mediterranean Sea (EM) included all isolates to the East of the Strait of Sicily; (ii) the western Mediterranean Sea (WM) included isolates collected west of the Strait of Sicily and east of the Strait of Gibraltar; (iii) the European southern Atlantic (ATL) included Atlantic samples as far north as the Cape Finisterre; and (iv) the European northern Atlantic (ATN) comprised the remaining northern Atlantic samples. Basic measures of genetic diversity, number of polymorphic sites (S), nucleotide diversity (π), and haplotype diversity (h) for each region were calculated using Arlequin v.3.5.1.2 (60).
Because of the limited number of isolates of O. maritima (n = 36), analyses of population structure and diversity were performed only for O. marina. Spatial genetic structure of O. marina was quantified using Structure v.2.3.1 (61). Haplotype richness for each of the 12 areas was calculated after rarefaction resampling (n = 5) using Contrib v.1.02 (62) to standardize sample sizes. We constructed GLMs to determine which combination of potential predictor variables (location and environmental data) best explained the distribution of genetic diversity: latitude, longitude, temperature (°C), salinity (PSU), and dissolved oxygen content (mL⋅L−1). Model selection was based on minimizing corrected AIC (33) using the Dredge function within the MuMin package (63) in R v.2.12.1 (64).
Population Demographic Signals.
Historical fluctuations in population size for each geographic region were detected using Fu’s Fs (31) and mismatch analysis (65) of the ITS and α-TUB alignments (low haplotype diversity rendered COI data uninformative) using Arlequin. Although originally developed as a test of marker neutrality, Fs is sensitive to changes in population size, with a significantly negative value interpreted as a signature of population expansion; the significance of Fs was determined by randomization. A generalized least-squares approach was used to estimate parameters associated with a model of sudden population expansion (60, 65) whose validity is determined from the SSD between observed and expected mismatch distributions and also by calculating a raggedness index (66) that yields larger values for stable populations (multimodal distributions) than for expanding populations (unimodal distributions) (see Fig. S4 for mismatch distributions).
We used model selection to compare hypotheses about the potential locations of refugia and the predominant direction of dispersal (67). Each of the four geographic regions was considered as the candidate source population (Fig. S5). The maximum likelihood migration rate of each model, given the data, then was estimated using Migrate-n v.3.2.16 (32). Each model was compared with an unconstrained island migration model using the likelihood-ratio-test option to gain comparable likelihood estimates between the parameter sets of the different hypotheses. The AIC (33) was used to infer model support.
Supplementary Material
Acknowledgments
We thank the many people who provided samples (see SI Acknowledgments). This work was funded by Natural Environment Research Council Grant NE/F005237/1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database. For a list of accession numbers, see Table S1.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1214398110/-/DCSupplemental.
References
- 1.Azam F, Malfatti F. Microbial structuring of marine ecosystems. Nat Rev Microbiol. 2007;5(10):782–791. doi: 10.1038/nrmicro1747. [DOI] [PubMed] [Google Scholar]
- 2.Finlay BJ. Global dispersal of free-living microbial eukaryote species. Science. 2002;296(5570):1061–1063. doi: 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- 3.Fenchel T, Finlay B. The ubiquity of small species: Patterns of local and global diversity. Bioscience. 2004;54(8):777–784. [Google Scholar]
- 4.Šlapeta J, López-García P, Moreira D. Global dispersal and ancient cryptic species in the smallest marine eukaryotes. Mol Biol Evol. 2006;23(1):23–29. doi: 10.1093/molbev/msj001. [DOI] [PubMed] [Google Scholar]
- 5.Lowe CD, Montagnes DJS, Martin LE, Watts PC. Patterns of genetic diversity in the marine heterotrophic flagellate Oxyrrhis marina (Alveolata: Dinophyceae) Protist. 2010a;161(2):212–221. doi: 10.1016/j.protis.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 6.Bierne N, Bonhomme F, David P. Habitat preference and the marine-speciation paradox. Proc Biol Sci. 2003;270(1522):1399–1406. doi: 10.1098/rspb.2003.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koester JA, Swalwell JE, von Dassow P, Armbrust EV. Genome size differentiates co-occurring populations of the planktonic diatom Ditylum brightwellii (Bacillariophyta) BMC Evolutionary Biology. 2010;10:1. doi: 10.1186/1471-2148-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foissner W. Protist diversity and distribution: Some basic considerations. Biodiversity and Conservation. 2008;17(2):235–242. [Google Scholar]
- 9.Evans KM, Kuhn SF, Hayes PK. High levels of genetic diversity and low levels of genetic differentiation in North Sea Pseudo-Nitzschia pungens (Bacillariophyceae) Populations. Journal of Phycology. 2005;41(3):506–514. [Google Scholar]
- 10.Casteleyn G, et al. Lack of population genetic structuring in the marine planktonic diatom Pseudo-nitzschia pungens (Bacillariophyceae) in a heterogeneous area in the Southern Bight of the North Sea. Marine Biology. 2009;156(6):1149–1158. [Google Scholar]
- 11.Casteleyn G, et al. Pseudo-nitzschia pungens (Bacillariophyceae): A cosmopolitan diatom species? Harmful Algae. 2008;7(2):241–257. [Google Scholar]
- 12.Lowe CD, Montagnes DJS, Martin LE, Watts PC. High genetic diversity and fine-scale spatial structure in the marine flagellate Oxyrrhis marina (Dinophyceae) uncovered by microsatellite loci. PLoS ONE. 2010b;5(12):e15557. doi: 10.1371/journal.pone.0015557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rynearson T, Armbrust E. Genetic differentiation among populations of the planktonic marine diatom Ditylum Brightwellii (Bacillariophyceae) Journal of Phycology. 2004;40(3):34–43. [Google Scholar]
- 14.Nagai S, et al. Microsatellite markers reveal population genetic structure of the toxic dinoflagellate Alexandrium tamarense (Dinophyceae) in Japanese coastal waters. Journal of Phycology. 2007;43(1):43–54. [Google Scholar]
- 15.Nagai S, et al. Genetic structuring and transfer of marine dinoflagellate Cochlodinium polykrikoides in Japanese and Korean coastal waters revealed by microsatellites. Mol Ecol. 2009;18(11):2337–2352. doi: 10.1111/j.1365-294X.2009.04193.x. [DOI] [PubMed] [Google Scholar]
- 16.Casabianca S, et al. Population genetic structure and connectivity of the harmful dinoflagellate Alexandrium minutum in the Mediterranean Sea. Proc Biol Sci. 2012;279(1726):129–138. doi: 10.1098/rspb.2011.0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katz LA, et al. Reframing the “Everything is everywhere” debate : Evidence for high gene flow and diversity in ciliate morphospecies. Aquatic Microbial Ecology. 2005;41(1):55–65. [Google Scholar]
- 18.Darling KF, Kucera M, Wade CM. Global molecular phylogeography reveals persistent Arctic circumpolar isolation in a marine planktonic protist. Proc Natl Acad Sci USA. 2007;104(12):5002–5007. doi: 10.1073/pnas.0700520104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Casteleyn G, et al. Limits to gene flow in a cosmopolitan marine planktonic diatom. Proc Natl Acad Sci USA. 2010;107(29):12952–12957. doi: 10.1073/pnas.1001380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hewitt G. The genetic legacy of the Quaternary ice ages. Nature. 2000;405(6789):907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- 21.Patarnello T, Volckaert FA, Castilho R. Pillars of Hercules: Is the Atlantic-Mediterranean transition a phylogeographical break? Mol Ecol. 2007;16(21):4426–4444. doi: 10.1111/j.1365-294X.2007.03477.x. [DOI] [PubMed] [Google Scholar]
- 22.Maggs CA, et al. Evaluating signatures of glacial refugia for North Atlantic benthic marine taxa. Ecology. 2008;89(11) Suppl:S108–S122. doi: 10.1890/08-0257.1. [DOI] [PubMed] [Google Scholar]
- 23.Provan J, Bennett KD. Phylogeographic insights into cryptic glacial refugia. Trends Ecol Evol. 2008;23(10):564–571. doi: 10.1016/j.tree.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 24.Provan J, Wattier RA, Maggs CA. Phylogeographic analysis of the red seaweed Palmaria palmata reveals a Pleistocene marine glacial refugium in the English Channel. Mol Ecol. 2005;14(3):793–803. doi: 10.1111/j.1365-294X.2005.02447.x. [DOI] [PubMed] [Google Scholar]
- 25.Hsü KJ, et al. History of Mediterranean salinity crisis. Nature. 1977;267(5610):399–403. [Google Scholar]
- 26.Arnaud-Haond S, et al. Vicariance patterns in the Mediterranean Sea: East–west cleavage and low dispersal in the endemic seagrass Posidonia oceanica. Journal of Biogeography. 2007;34(6):963–976. [Google Scholar]
- 27.Martiny JBH, et al. Microbial biogeography: Putting microorganisms on the map. Nat Rev Microbiol. 2006;4(2):102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- 28.Darling KF, Kucera M, Pudsey CJ, Wade CM. Molecular evidence links cryptic diversification in polar planktonic protists to Quaternary climate dynamics. Proc Natl Acad Sci USA. 2004;101(20):7657–7662. doi: 10.1073/pnas.0402401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watts PC, Martin LE, Kimmance SE, Montagnes DJS, Lowe CD. The distribution of Oxyrrhis marina - a global disperser or poorly characterised endemic? Journal of Plankton Research. 2011;33(4):579–589. [Google Scholar]
- 30.Lowe CD, et al. Who is Oxyrrhis marina? Morphological and phylogenetic studies on an unusual dinoflagellate. Journal of Plankton Research. 2011;33(4):555–567. [Google Scholar]
- 31.Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133(3):693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beerli P, Felsenstein J. Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. Proc Natl Acad Sci USA. 2001;98(8):4563–4568. doi: 10.1073/pnas.081068098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akaike H. A new look at the statistical model identification. IEEE Transactions on Automatic Control. 1974;19(6):716–723. [Google Scholar]
- 34.Bianchi C, Morri C. Marine biodiversity of the Mediterranean Sea: Situation, problems and prospects for future research. Marine Pollution Bulletin. 2000;40(5):367–376. [Google Scholar]
- 35.Zardoya R, et al. Differential population structuring of two closely related fish species, the mackerel (Scomber scombrus) and the chub mackerel (Scomber japonicus), in the Mediterranean Sea. Mol Ecol. 2004;13(7):1785–1798. doi: 10.1111/j.1365-294X.2004.02198.x. [DOI] [PubMed] [Google Scholar]
- 36.Mejri R, Arculeo M, Hassine OK, Brutto SL. Genetic architecture of the marbled goby Pomatoschistus marmoratus (Perciformes, Gobiidae) in the Mediterranean Sea. Mol Phylogenet Evol. 2011;58(2):395–403. doi: 10.1016/j.ympev.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 37.Aurahs R, Grimm GW, Hemleben V, Hemleben C, Kucera M. Geographical distribution of cryptic genetic types in the planktonic foraminifer Globigerinoides ruber. Mol Ecol. 2009;18(8):1692–1706. doi: 10.1111/j.1365-294X.2009.04136.x. [DOI] [PubMed] [Google Scholar]
- 38.Béranger K, et al. The dynamics of the Sicily Strait: A comprehensive study from observations and models. Deep Sea Research Part II Topical Studies in Oceanography. 2004;51(4-5):411–440. [Google Scholar]
- 39.Domingues VS, Santos RS, Brito A, Almada VC. Historical population dynamics and demography of the Eastern Atlantic pomacentrid Chromis limbata (Valenciennes, 1833) Mol Phylogenet Evol. 2006;40(1):139–147. doi: 10.1016/j.ympev.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 40.Pérez-Losada M, Nolte MJ, Crandall KA, Shaw PW, Holloway R. Testing hypotheses of population structuring in the Northeast Atlantic Ocean and Mediterranean Sea using the common cuttlefish Sepia officinalis. Mol Ecol. 2007;16(13):2667–2679. doi: 10.1111/j.1365-294X.2007.03333.x. [DOI] [PubMed] [Google Scholar]
- 41.Penna A, et al. A phylogeographical study of the toxic benthic dinoflagellate genus Ostreopsis Schmidt. Journal of Biogeography. 2010;37(5):830–841. [Google Scholar]
- 42.Lilly EL, Halanych KM, Anderson DM. Species boundaries and global biogeography of the Alexandrium tamarense complex (Dinophyceae) Journal of Phycology. 2007;43(6):1329–1338. [Google Scholar]
- 43.Galarza JA, et al. The influence of oceanographic fronts and early-life-history traits on connectivity among littoral fish species. Proc Natl Acad Sci USA. 2009;106(5):1473–1478. doi: 10.1073/pnas.0806804106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoarau G, Coyer JA, Veldsink JH, Stam WT, Olsen JL. Glacial refugia and recolonization pathways in the brown seaweed Fucus serratus. Mol Ecol. 2007;16(17):3606–3616. doi: 10.1111/j.1365-294X.2007.03408.x. [DOI] [PubMed] [Google Scholar]
- 45.Weisse T. Distribution and diversity of aquatic protists: An evolutionary and ecological perspective. Biodiversity and Conservation. 2008;17(2):243–259. [Google Scholar]
- 46.Caron DA, Worden AZ, Countway PD, Demir E, Heidelberg KB. Protists are microbes too: A perspective. ISME J. 2009;3(1):4–12. doi: 10.1038/ismej.2008.101. [DOI] [PubMed] [Google Scholar]
- 47.Marko PB. ‘What’s larvae got to do with it?’ Disparate patterns of post-glacial population structure in two benthic marine gastropods with identical dispersal potential. Mol Ecol. 2004;13(3):597–611. doi: 10.1046/j.1365-294x.2004.02096.x. [DOI] [PubMed] [Google Scholar]
- 48.Watts PC, Thorpe JP. Influence of contrasting larval developmental types upon the population-genetic structure of cheilostome bryozoans. Marine Biology. 2006;149(5):1093–1101. [Google Scholar]
- 49.Marko PB, et al. The ‘Expansion-Contraction’ model of Pleistocene biogeography: Rocky shores suffer a sea change? Mol Ecol. 2010;19(1):146–169. doi: 10.1111/j.1365-294x.2009.04417.x. [DOI] [PubMed] [Google Scholar]
- 50.Vanormelingen P, Verleyen E, Vyverman W. The diversity and distribution of diatoms: From cosmopolitanism to narrow endemism. Biodiversity and Conservation. 2007;17(2):393–405. [Google Scholar]
- 51.De Meester L, Gómez A, Okamura B, Schwenk K. The Monopolization Hypothesis and the dispersal–gene flow paradox in aquatic organisms. Acta Oecologica. 2002;23(3):121–135. [Google Scholar]
- 52.Logares R, Boltovskoy A, Bensch S, Laybourn-Parry J, Rengefors K. Genetic diversity patterns in five protist species occurring in lakes. Protist. 2009;160(2):301–317. doi: 10.1016/j.protis.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 53.Lowe CD, et al. Collection, isolation and culturing strategies for Oxyrrhis marina. Journal of Plankton Research. 2011;33(4):569–578. [Google Scholar]
- 54.Lowe CD, Day A, Kemp SJ, Montagnes DJS. There are high levels of functional and genetic diversity in Oxyrrhis marina. J Eukaryot Microbiol. 2005;52(3):250–257. doi: 10.1111/j.1550-7408.2005.00034.x. [DOI] [PubMed] [Google Scholar]
- 55.Posada D, Crandall KA. Bioinformatics applications note: MODELTEST : Testing the model of DNA substitution. Bioinformatics. 1998;14(9):817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 56.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 57.Rambaut A, Drummond AJ. 2007. Tracer v1.4, Available at http://beast.bio.ed.ac.uk/Tracer.
- 58.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilke T, Schultheiß R, Albrecht C. As time goes by: A simple fool’s guide to molecular clock approaches in invertebrates. American Malacological Bulletin. 2009;27(1-2):25–45. [Google Scholar]
- 60.Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- 61.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Petit RJ, El Mousadik A, Pons O. Identifying populations for conservation on the basis of genetic markers. Conservation Biology. 1998;12(4):844–855. [Google Scholar]
- 63.Barton K. 2011. MuMIn: Multi-model inference http://CRAN.R-project.org/package=MuMIn.
- 64.R Development Core Team . R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2010. [Google Scholar]
- 65.Schneider S, Excoffier L. Estimation of past demographic parameters from the distribution of pairwise differences when the mutation rates vary among sites: Application to human mitochondrial DNA. Genetics. 1999;152(3):1079–1089. doi: 10.1093/genetics/152.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harpending HC. Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Hum Biol. 1994;66(4):591–600. [PubMed] [Google Scholar]
- 67.Dépraz A, Cordellier M, Hausser J, Pfenninger M. Postglacial recolonization at a snail’s pace (Trochulus villosus): Confronting competing refugia hypotheses using model selection. Mol Ecol. 2008;17(10):2449–2462. doi: 10.1111/j.1365-294X.2008.03760.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.