Abstract
Similar morphological, physiological, and behavioral features have evolved independently in different species, a pattern known as convergence. It is known that morphological convergence can occur through changes in orthologous genes. In some cases of convergence, cis-regulatory changes generate parallel modifications in the expression patterns of orthologous genes. Our understanding of how changes in cis-regulatory regions contribute to convergence is hampered, usually, by a limited understanding of the global cis-regulatory structure of the evolving genes. Here we examine the genetic causes of a case of precise phenotypic convergence between Drosophila sechellia and Drosophila ezoana, species that diverged ∼40 Mya. Previous studies revealed that changes in multiple transcriptional enhancers of shavenbaby (svb, a transcript of the ovo locus) caused phenotypic evolution in the D. sechellia lineage. It has also been shown that the convergent phenotype of D. ezoana was likely caused by cis-regulatory evolution of svb. Here we show that the large-scale cis-regulatory architecture of svb is conserved between these Drosophila species. Furthermore, we show that the D. ezoana orthologs of the evolved D. sechellia enhancers have also evolved expression patterns that correlate precisely with the changes in the phenotype. Our results suggest that phenotypic convergence resulted from multiple noncoding changes that occurred in parallel in the D. sechellia and D. ezoana lineages.
Keywords: parallel developmental evolution, evolutionary developmental biology, enhancer function
The repeated occurrence of similar, or sometimes identical, evolutionary changes in independent lineages has occurred commonly and, at the phenotypic level, is called convergence. Phenotypic convergence is correlated often with transitions to similar environments, which provides compelling evidence that convergence resulted from response to similar patterns of natural selection. Examples include flippers and fins in cetaceans and fish, wings in birds and insects, and the eyes of mammals and octopi.
Developmental mechanisms can evolve in similar ways in independent lineages and can cause convergence, which has been called parallel developmental evolution (1). Multiple examples of parallel developmental evolution (2–13) have been reported in recent years. These data provide evidence for the contribution of similar changes in developmental mechanisms to phenotypic convergence. In particular, changes in cis-regulatory regions have contributed extensively to parallel developmental evolution. However, we do not yet have a detailed understanding of how these cis-regulatory changes contribute to convergence, partly because we currently have a limited understanding of the global cis-regulatory architecture of evolving genes.
Here we focus on a case of morphological convergence, the loss of larval dorsal and lateral cuticular extensions—michrotrichiae (hereafter called trichomes)—that occurred in evolutionary lineages that last shared a common ancestor at least 40 Mya (11). Previous studies provided preliminary evidence that similar changes in the same developmental mechanism caused this case of convergence. Given the deep evolutionary divergence between these lineages, together with the phylogenetic evidence that the loss of these trichomes is evolutionarily derived in both lineages, this appears to be a case of parallel developmental evolution. Here we provide functional evidence that supports this hypothesis.
Study System.
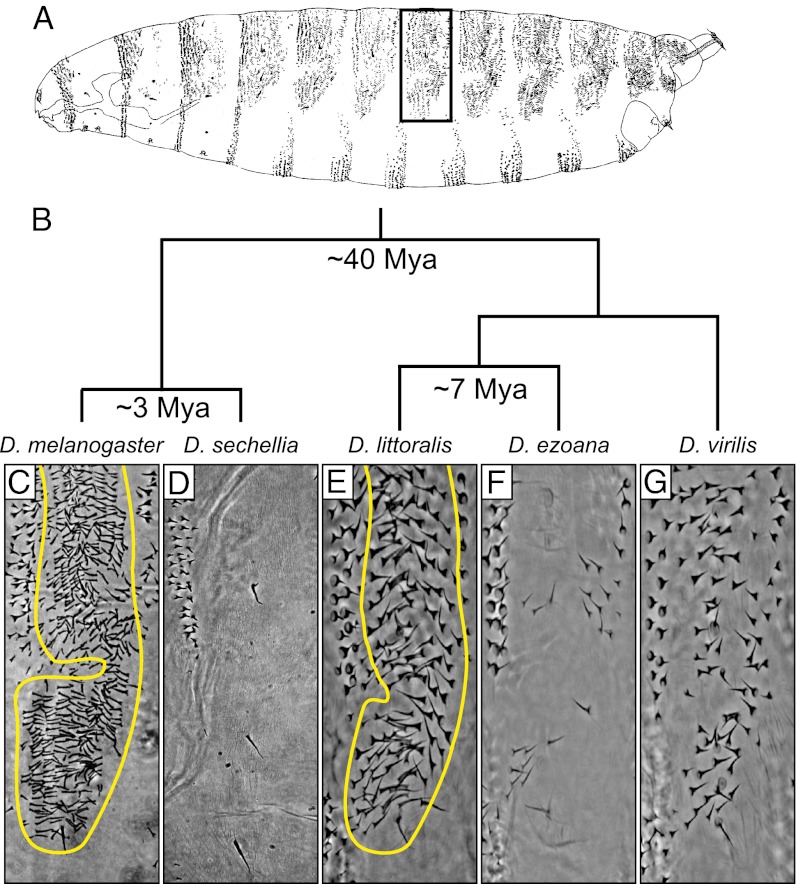
The pattern of trichomes has evolved multiple times in larvae of the genus Drosophila (14, 15), possibly in response to natural selection (16). In the Drosophila melanogaster and Drosophila virilis species groups, most species produce trichomes over much of the dorsal and lateral surface of the first-instar larva (Fig. 1A). Within the D. melanogaster species group, Drosophila sechellia has evolved first-instar larvae in which the so-called quaternary cells differentiate naked cuticle (15–18) (Fig. 1D). Likewise, several species of the D. virilis group produce first-instar larvae with different degrees of naked cuticle (11, 14). In this work we focus on the larvae of Drosophila ezoana, which differentiate quaternary cells with naked cuticle on all body segments, resembling the phenotype of D. sechellia (Fig. 1 D and F).
Fig. 1.
Convergent evolution of a naked dorso-lateral cuticle phenotype in first-instar larvae of D. sechellia and D. ezoana. (A) Drawing from the lateral perspective of a D. melanogaster first-instar larva. The dark rectangle indicates the cuticle region shown in C–G. (B) Phylogenetic relationships and estimated divergence dates between species discussed in this article. Dorso-lateral cuticle of the fourth abdominal segment for five Drosophila species. The “hairy” phenotype present in D. melanogaster (C), D. littoralis (E), and D. virilis (G) is the ancestral state in the genus Drosophila. Quaternary trichomes are outlined in D. melanogaster (C) and D. littoralis (E). Quaternary trichomes were lost independently in D. sechellia (D) and D. ezoana (F).
In Drosophila larvae, differentiation of cells with trichomes, as opposed to smooth cuticle, is controlled by the transcription factor Shavenbaby (svb) (19), whose activity is both necessary and sufficient to produce trichomes (20). The complex embryonic expression pattern of svb in D. melanogaster is determined by the activity of seven enhancers that are distributed throughout a region ∼90 kb upstream of the svb first exon (17, 18) (Fig. 2). Five of these enhancers drive expression in quaternary cells (Fig. 2). The evolution of naked cuticle in D. sechellia resulted entirely from changes in the D. sechellia orthologs of these five D. melanogaster enhancers (17, 18). All of these evolutionary changes on the D. sechellia lineage cause reduced levels of enhancer activity, leading to the absence of svb mRNA in quaternary cells.
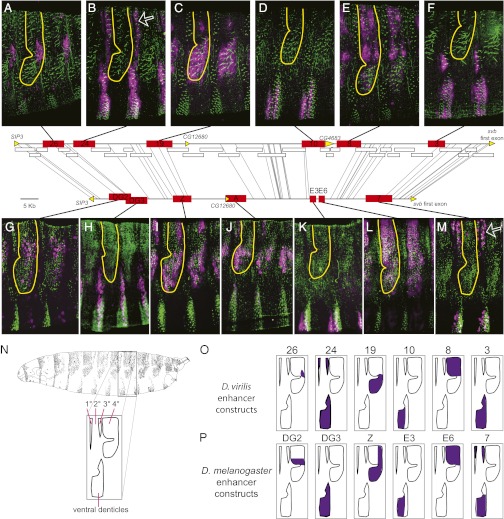
Fig. 2.
Positional and functional conservation between svb embryonic enhancers of D. melanogaster and D. virilis. Horizontal lines schematize the svb cis-regulatory region in D. virilis (Upper) and D. melanogaster (Lower). Thin lines connect identical 30 bp sequences (“anchors”) between the orthologous regions. The fact that none of these lines cross implies that the orthologous svb regions are largely collinear. White rectangles correspond to D. virilis DNA fragments tested for enhancer activity in transgenic D. virilis embryos that did not drive expression in embryonic epidermis. Yellow arrows specify coding regions. Red rectangles indicate the position of embryonic enhancers for the two species. Positional conservation is evident for six enhancer pairs. Expression patterns of D. virilis enhancers (A–F) and of D. melanogaster enhancers (G–M) are shown. Quaternary trichomes are outlined in all panels. Similar expression patterns are driven by the orthologous enhancers 26 (A) and DG2 (G), 24 (B) and DG3 (H), 19 (C) and Z (I), 10 (D) and E3 (K), 8 (E) and E6 (L), and 3 (F) and 7 (M). White arrows highlight the dorsal expression pattern encoded by different enhancers in the two species. Pictures were taken from stage 15–16 embryos. Trichomes are stained in green; lacZ reporter expression is purple. Different embryos display slightly different rotations along the dorso-ventral axis. (N) Drawing of a lateral view of the D. melanogaster first-instar larval cuticle and, below, a diagram of the major cuticular domains is shown. On the dorsal surface, the primary (1°), tertiary (3°), and quaternary (4°) cells differentiate trichomes, and the secondary (2°) cells differentiate naked cuticle (38). The ventral denticle belts are also labeled. (O) The epidermal domains in which the six D. virilis enhancers drive expression is illustrated with purple shading. (P) The epidermal domains in which the six D. melanogaster enhancers that appear to be orthologous to the six D. virilis enhancers, based on shared chromosomal synteny and sequence similarity are illustrated with purple shading. The expression patterns of the D. virilis enhancers are placed directly above the expression patterns of their putative orthologs in D. melanogaster.
Three pieces of evidence reported previously suggested, but did not prove, that cis-regulatory changes in svb caused the convergent evolution of naked cuticle in species of the D. virilis group (11). First, six genes that regulate svb expression are expressed similarly in species with divergent trichome patterns (14), suggesting that the regulatory cascades upstream of svb have been conserved in species with divergent trichome patterns. Second, interspecific crosses demonstrated that the difference in trichome patterns between two species of the D. virilis group was caused by a locus on the X chromosome—the location of svb—and that the “naked cuticle” allele is recessive to the “hairy” allele, just as svb is in the D. melanogaster species group (11). Third, analysis of svb expression in these species revealed a precise correlation between the presence of mRNA and the pattern of trichomes (11).
In this work, we performed a series of experiments revealing that the position and function of enhancers in the cis-regulatory region (what we call the functional architecture) of the svb gene has been conserved between species that diverged ∼40 Mya. We also demonstrate that orthologous enhancers from the svb cis-regulatory region have evolved independently in D. ezoana and D. sechellia and it is likely that these cis-regulatory changes have caused the precise morphological convergence between these species.
Results
Architecture of the Cis-Regulatory Region of svb in Drosophila virilis.
To test the hypothesis that svb enhancers evolved to generate diversity of the trichome pattern in the D. virilis group of species, we first performed a functional analysis of the svb cis-regulatory region in D. virilis. We were concerned that a search guided only by sequence conservation might generate an incomplete picture of the D. virilis svb cis-regulatory architecture (21). For example, new enhancers may have evolved in the D. virilis lineage. Therefore, we performed a comprehensive and unbiased survey of the entire cis-regulatory region of the D. virilis svb locus (a ∼132 Kb region between SIP3 and the svb first exon), by assaying 35 ∼5 Kb reporter constructs. This region corresponds to the ∼90 Kb region containing all seven svb embryonic enhancers in D. melanogaster (Fig. 2). In addition, we were concerned that some of the trans-regulatory factors that regulate svb expression might have evolved new functions between D. melanogaster and D. virilis. Therefore, instead of analyzing the constructs in D. melanogaster, we tested all constructs in transgenic D. virilis embryos. This effort uncovered six regions that drive reporter gene expression in patterns that resemble parts of the svb expression pattern (11) in epidermal cells of the embryo (Fig. 2).
Orthologous Enhancers Generate the Embryonic Expression Pattern of svb in Phylogenetically Distant Drosophila Species.
To determine whether these D. virilis enhancers represented orthologs of the D. melanogaster svb enhancers, we tested for positional conservation, functional similarity, and sequence similarity between the two species. First, we tested for positional conservation between possible orthologs by identifying conserved “anchors” of 30 bp across the locus (22) (Fig. 2). The collinear synteny of these anchors indicates that the entire cis-regulatory region of svb has not experienced rearrangements between these two species on the scale of tens of kilobases. Moreover, the locations of the conserved anchors reveal that six svb enhancers are positioned in the same relative sites within the D. melanogaster and D. virilis svb loci (Fig. 2). We did not find a D. virilis enhancer in the region orthologous to D. melanogaster enhancer A.
Second, we tested for functional similarity between the D. virilis and D. melanogaster enhancers by examining their detailed expression patterns (Fig. 2). We found that the expression patterns of all six D. virilis enhancers are similar to the patterns driven by their positional homologs (Fig. 2). Overall, putatively orthologous enhancers are active mainly in the same segmental and dorsal-ventral spatial domains. For example, the D. melanogaster putative orthologs of the three D. virilis enhancers that drive expression in cells giving rise to the ventral denticle belts also drive expression in cells giving rise to ventral denticle belts in D. melanogaster. We also detected several differences in expression between putative orthologs. First, D. melanogaster 7 drove expression in dorsal primary and tertiary cells and weakly in dorsal quaternary cells, whereas D. virilis 3 does not drive any detectable expression in dorsal cells. In contrast, D. virilis 24 drives expression in dorsal primary and tertiary cells, and D. melanogaster DG3 does not. In both species, only a single enhancer drives in primary and tertiary dorsal cells. It therefore appears that this function has shifted between enhancers during the divergence of D. melanogaster and D. virilis. We also detected apparently weaker expression in dorsal quaternary cells driven by D. virilis 19 than by D. melanogaster Z.
Third, to test whether sequence similarity supported orthology between functionally similar enhancer regions, we used D. virilis and D. melanogaster enhancer sequences as queries in reciprocal BLAST (23) searches. Despite extensive divergence, we detected significant reciprocal sequence similarity in or near each of the six enhancer pairs (Figs. S1 and S2). Thus, comparisons of position, function, and sequence suggest that six pairs of svb enhancers are true orthologs between D. melanogaster and D. virilis.
Parallel Genetic Changes in Enhancers 8 and 19 of D. ezoana and the Evolution of a Convergent Cuticular Morphology.
Three of the D. virilis enhancers—8, 19, and 26—drive expression in the dorsal and lateral domains that show evolving trichome patterns in the D. virilis species group. Most of the dorso-lateral expression is driven by enhancers 8 and 19. Similarly, D. melanogaster E6 and Z, which are the orthologs of D. virilis 8 and 19, generate most, but not all, of the dorsal and lateral expression in D. melanogaster. These two enhancers lost their activity in the D. sechellia lineage, which caused the naked cuticle phenotype of this species (17, 18). To test whether these enhancers have evolved also in the D. virilis species group, we cloned the orthologous regions from two species of the D. virilis group with contrasting dorso-lateral trichome patterns (see Materials and Methods for details); Drosophila littoralis produces trichomes where enhancers 8 and 19 are active (Figs. 1E and 2), while D. ezoana displays a D. sechellia–like phenotype (Fig. 1F). As a control, we cloned the homologous regions of enhancer 3 from both species. Enhancer 3 is expressed primarily in ventral trichome-producing cells, which have not evolved between D. littoralis and D. ezoana.
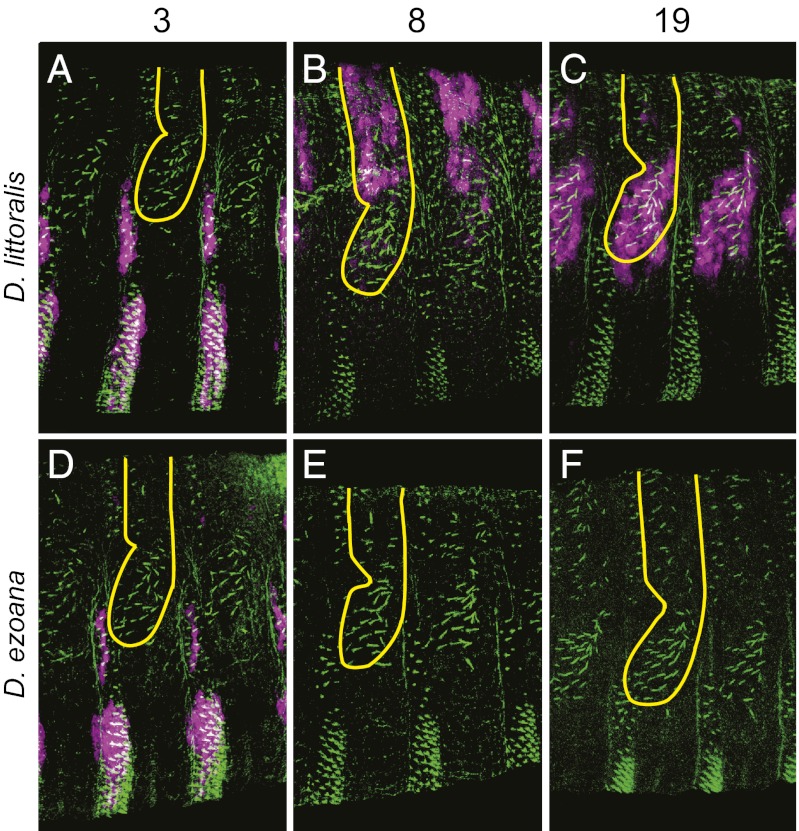
We observed that all three enhancers isolated from D. littoralis drove expression in patterns that were extremely similar to the patterns driven by the homologous D. virilis enhancers (Fig. 3). This implies that the common ancestor of D. littoralis and D. ezoana also possessed all three active enhancers. Enhancer 3 is still active in D. ezoana and drives expression in the same domains as the D. littoralis and D. virilis orthologs. In contrast, D. ezoana enhancers 8 and 19 produce no detectable expression. These results are consistent with the pattern of svb mRNA in D. ezoana embryos that was reported previously (11). Because all of these assays were performed in the common trans-regulatory landscape of D. virilis, which is competent to drive dorsal and lateral expression from the D. virilis and D. littoralis enhancers, the changes in D. ezoana enhancers 8 and 19 are likely to have caused the previously observed changes in svb expression in D. ezoana.
Fig. 3.
Cis-regulatory changes in evolutionarily conserved svb enhancers underlie the convergent loss of quaternary trichomes in D. ezoana. D. littoralis enhancers 3 (A), 8 (B), and 19 (C) drive expression patterns that cannot be distinguished from those produced by the D. virilis orthologous enhancers (Fig. 2). (D) The expression pattern of the D. ezoana enhancer 3 also is conserved. In contrast, D. ezoana enhancers 8 (E) and 19 (F) do not produce detectable expression. This explains, at least in part, the absence of quaternary trichomes in D. ezoana. In D. sechellia, the parallel inactivation of E6 and Z (the orthologs of D. virilis 8 and 19) likely caused the convergent loss of quaternary trichomes. The activity of all D. ezoana and D. littoralis constructs were tested in transgenic D. virilis embryos. Differences in the activity of these enhancers therefore represent differences in the enhancer sequences from each species. Colors and symbols as in Fig. 2.
svb is required for trichome development in D. melanogaster (20). We therefore tested whether svb is also required for trichome development in D. virilis by performing a knockdown of svb mRNA. Injection of two independent siRNAs in D. virilis embryos caused loss of trichomes in larvae (Figs. S3 and S4), confirming that svb is also required for trichome development in D. virilis. Taken together with previous findings of conserved expression patterns of svb regulators and genetic linkage of trichome patterns with svb in the D. virilis species group (11, 14), our current observations suggest that genetic changes in svb enhancers contributed to the evolved trichome pattern in D. ezoana. We are not able to rule out, however, additional contributions from loci closely linked to svb.
Discussion
We have found that the functional architecture of the svb cis-regulatory region was conserved between D. melanogaster and D. virilis. Similarly, in insects, some enhancers of “even-skipped” (24) and of some dorso-ventral patterning genes (25) have maintained their ancestral positions. Within single svb enhancers, DNA sequences have diverged substantially without causing major changes in enhancer function, a feature that has been observed previously (26). However, at a larger scale, the functional organization of the whole svb regulatory region has been largely conserved. This higher order conservation is reminiscent of the structural conservation of Hox gene clusters (27), suggesting that long-range molecular interactions, such as enhancer–enhancer or enhancer–promoter interactions (28), constrain the evolution of large cis-regulatory regions, such as those found in svb and in Hox genes. In other words, the spacing and order of enhancers in the cis-regulatory region of svb might be crucial for the occurrence of precise physical contacts between different regulatory elements or between regulatory elements and the core promoter.
Parallel evolution underlying convergence of trichome patterns had been inferred previously from genetic studies coupled with expression assays (11). Our current functional study supports the view that not only the same gene, svb, underlies this convergence, but that two orthologous enhancers have changed in similar ways in D. sechellia and D. ezoana.
There are multiple reasons why svb may be a favored locus of evolutionary change, and additional reasons why specific enhancers may be particularly favored. First, svb acts as a single master regulator of larval trichome development. svb function is both required for development of these trichomes and sufficient to induce expression of a large set of downstream genes that regulate and contribute to trichome development (20, 29). Thus, manipulation of the svb expression pattern can, on its own, generate diversity of trichome patterns. It is not clear that any other single gene can cause such a specific morphological change without disrupting other aspects of larval development (29). svb expression is regulated by a large number of signaling pathways and transcription factors, and manipulations of these factors can alter trichome patterns (30). However, these manipulations are likely to have pleiotropic deleterious effects, in addition to altering trichome patterns. Thus, the svb locus may be a favored target for evolutionary change underlying trichome patterns both because these changes minimize pleiotropic effects and because svb can instruct the entire module of trichome morphogenesis (31, 32).
The parallel evolution of individual enhancers of svb comes as more of a surprise. At the initiation of this work, given the extreme sequence divergence of D. melanogaster and D. virilis, it was not clear that the svb enhancer region would be both spatially and functionally conserved. The conserved svb enhancer functions between D. melanogaster and D. virilis, and the observation that orthologous enhancers seem to have evolved in similar ways to generate convergent evolution, implies that our ability to “predict” patterns of genomic evolution may improve as our understanding of genome function improves.
Although we have found some sequence similarity between D. melanogaster and D. virilis svb enhancers, the sequences align poorly. Therefore, at this stage, we cannot determine if the nucleotide changes that inactivated D. ezoana 8 are similar to those described recently for D. sechellia E6 (16). It will be interesting to determine whether, despite this sequence divergence, orthologous transcription factors regulate orthologous enhancers and whether, at an even more detailed level of analysis, similar patterns of transcription factor binding site gain or loss have generated developmental parallelism in D. sechellia and D. ezoana.
Materials and Methods
Design of piggyBac Reporter Vectors.
The polylinker-hs43-lacZ region from pCaSpeR-hs43lacZ (33) was PCR-amplified using primers BglII-FW (AGATCTAGATCTTACTAGAATTCGGT) and BglII-RV (AGATCTAGATCTAATTTGCGAGTACG). This 4.6 Kb fragment was cloned into pGEMT (Promega), released by cutting with BglII, and subcloned into the unique BglII site of both piggyBac PB (34) and piggyBac-enhanced yellow fluorescent protein (eYFP) (35), yielding piggyBac-hs43lacZ and piggyBac-eYFP-hs43lacZ, respectively. The difference between these two reporter vectors is the transformation marker; piggyBac-hs43lacZ carries miniwhite, whereas piggyBac-eYFP-hs43lacZ contains Pax6::eYFP.
Reporter Constructs.
The svb gene is located in scaffold 13042 of the D. virilis sequenced genome. The dissected region corresponds to bases 298789–434870 of this scaffold. PCR fragments were amplified from genomic DNA of D. virilis white (Drosophila Species Stock Center strain 15010–1051.53) using the primers listed in Table S1. These fragments were cloned into pGEMT (Promega), excised using NotI, and subcloned into the NotI site of the piggyBac-hs43lacZ polylinker. Fragments that contained internal NotI sites were subcloned using the CloneEZ kit (Genscript). Recombinant plasmids were coinjected with the helper vector pHSPpBac (36) into D. virilis white embryos. At least three independent transgenic lines were established for each construct.
Fragments 3, 8, and 19 from D. ezoana (Drosophila Species Stock Center strain 15010–0971.00) and D. littoralis (Drosophila Species Stock Center strain 15010–1001.00) were amplified using the primers listed in Table S2. Primers were designed based on D. virilis sequence. D. littoralis and D. ezoana fragments 8 and 19 are slightly larger than their D. virilis ortholog; these fragments had to be amplified with primers that flank the enhancers originally defined in D. virilis. Fragments 3, 8, and 19 from D. ezoana and D. littoralis were fully sequenced (GenBank accessions JN792404–JN792409). Primers used to amplify fragments 3, 8, and 19 from D. littoralis and fragments 3 and 8 from D. ezoana had SacII (forward) and SalI (reverse) sites added. These fragments were cloned into pGEMT (Promega), excised with SacII and SalI, and subcloned into piggyBac-eYFP-hs43lacZ (the polylinker was cut with SacII and XhoI). Both primers used to amplify fragment 19 from D. ezoana had XhoI sites added. These fragments were cloned into pGEMT (Promega), excised with XhoI, and subcloned into piggyBac-eYFP-hs43lacZ (the polylinker was cut XhoI).
The D. melanogaster enhancer constructs DG2, DG3, Z, A, E3, E6, and E7 were reported in previous publications (16–18). The image of enhancer 7 in Fig. 2 is from a new 7::luciferase construct that was made by Ella Preger (Janelia Farm Research Campus, Howard Hughes Medical Institute, Ashburn, VA), who kindly provided fixed embryos for our use.
The precise expression domains of the enhancer constructs were determined by double-staining embryos with a mouse anti-βGal antibody (Promega) and a rabbit anti-Dusky-like antibody (37). Fluorescent secondary antibodies were used. Stained embryos were examined with a confocal microscope.
Preparation of Cuticles.
We made overnight embryo collections and transferred embryos to plastic Petri dishes containing distilled water and maintained them in a 25 °C incubator. Two days later, we collected first-instar larvae and incubated them at 60 °C for 4 h. Subsequently, larvae were mounted on a microscope slide in a drop of 1:1 Hoyer’s solution–lactic acid mixture. After overnight drying, the cuticles were imaged with phase-contrast microscopy.
Supplementary Material
Acknowledgments
We thank Abe Bassan for assistance with early experiments; Ella Preger for allowing us to use her D. melanogaster7::luciferase reporter construct for Fig. 2; Justin Crocker and Andy Lemire for assistance with the RNAi experiments; and Ella Preger, Justin Crocker, Marcelo Rubinstein, François Payre, Serge Plaza, Jennifer Zanet, Hélène Chanut and Cédric Polesello for helpful comments on the manuscript. This work was supported by The Pew Charitable Trusts Latin American Fellows Program in the Biomedical Sciences Fellowship (to N.F.) and National Institutes of Health Grant GM063622-06A1 and National Science Foundation Grant IOS-0640339 (to D.L.S.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. JN792404–JN792409).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207715109/-/DCSupplemental.
References
- 1.Wood TE, Burke JM, Rieseberg LH. Parallel genotypic adaptation: When evolution repeats itself. Genetica. 2005;123(1-2):157–170. doi: 10.1007/s10709-003-2738-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wlasiuk G, Khan S, Switzer WM, Nachman MW. A history of recurrent positive selection at the toll-like receptor 5 in primates. Mol Biol Evol. 2009;26(4):937–949. doi: 10.1093/molbev/msp018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Streisfeld MA, Rausher MD. Genetic changes contributing to the parallel evolution of red floral pigmentation among Ipomoea species. New Phytol. 2009;183(3):751–763. doi: 10.1111/j.1469-8137.2009.02929.x. [DOI] [PubMed] [Google Scholar]
- 4.Chan YF, et al. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2010;327(5963):302–305. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shapiro MD, Bell MA, Kingsley DM. Parallel genetic origins of pelvic reduction in vertebrates. Proc Natl Acad Sci USA. 2006;103(37):13753–13758. doi: 10.1073/pnas.0604706103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gross JB, Borowsky R, Tabin CJ. A novel role for Mc1r in the parallel evolution of depigmentation in independent populations of the cavefish Astyanax mexicanus. PLoS Genet. 2009;5(1):e1000326. doi: 10.1371/journal.pgen.1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bollback JP, Huelsenbeck JP. Parallel genetic evolution within and between bacteriophage species of varying degrees of divergence. Genetics. 2009;181(1):225–234. doi: 10.1534/genetics.107.085225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J. Parallel adaptive origins of digestive RNases in Asian and African leaf monkeys. Nat Genet. 2006;38(7):819–823. doi: 10.1038/ng1812. [DOI] [PubMed] [Google Scholar]
- 9.Prud’homme B, et al. Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature. 2006;440(7087):1050–1053. doi: 10.1038/nature04597. [DOI] [PubMed] [Google Scholar]
- 10.Sugawara T, et al. Parallelism of amino acid changes at the RH1 affecting spectral sensitivity among deep-water cichlids from Lakes Tanganyika and Malawi. Proc Natl Acad Sci USA. 2005;102(15):5448–5453. doi: 10.1073/pnas.0405302102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sucena E, Delon I, Jones I, Payre F, Stern DL. Regulatory evolution of shavenbaby/ovo underlies multiple cases of morphological parallelism. Nature. 2003;424(6951):935–938. doi: 10.1038/nature01768. [DOI] [PubMed] [Google Scholar]
- 12.Boucher CA, et al. Ordered appearance of zidovudine resistance mutations during treatment of 18 human immunodeficiency virus-positive subjects. J Infect Dis. 1992;165(1):105–110. doi: 10.1093/infdis/165.1.105. [DOI] [PubMed] [Google Scholar]
- 13.Reed RD, et al. optix drives the repeated convergent evolution of butterfly wing pattern mimicry. Science. 2011;333(6046):1137–1141. doi: 10.1126/science.1208227. [DOI] [PubMed] [Google Scholar]
- 14.Dickinson WJ, Tang Y, Schuske K, Akam M. Conservation of molecular prepatterns during the evolution of cuticle morphology in Drosophila larvae. Evolution. 1993;47(5):1396–1406. doi: 10.1111/j.1558-5646.1993.tb02162.x. [DOI] [PubMed] [Google Scholar]
- 15.Sucena E, Stern DL. Divergence of larval morphology between Drosophila sechellia and its sibling species caused by cis-regulatory evolution of ovo/shaven-baby. Proc Natl Acad Sci USA. 2000;97(9):4530–4534. doi: 10.1073/pnas.97.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frankel N, et al. Morphological evolution caused by many subtle-effect substitutions in regulatory DNA. Nature. 2011;474(7353):598–603. doi: 10.1038/nature10200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frankel N, et al. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature. 2010;466(7305):490–493. doi: 10.1038/nature09158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGregor AP, et al. Morphological evolution through multiple cis-regulatory mutations at a single gene. Nature. 2007;448(7153):587–590. doi: 10.1038/nature05988. [DOI] [PubMed] [Google Scholar]
- 19.Mével-Ninio M, Terracol R, Salles C, Vincent A, Payre F. ovo, a Drosophila gene required for ovarian development, is specifically expressed in the germline and shares most of its coding sequences with shavenbaby, a gene involved in embryo patterning. Mech Dev. 1995;49(1–2):83–95. doi: 10.1016/0925-4773(94)00305-7. [DOI] [PubMed] [Google Scholar]
- 20.Payre F, Vincent A, Carreno S. ovo/svb integrates Wingless and DER pathways to control epidermis differentiation. Nature. 1999;400(6741):271–275. doi: 10.1038/22330. [DOI] [PubMed] [Google Scholar]
- 21.McGaughey DM, et al. Metrics of sequence constraint overlook regulatory sequences in an exhaustive analysis at phox2b. Genome Res. 2008;18(2):252–260. doi: 10.1101/gr.6929408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rebeiz M, Posakony JW. GenePalette: A universal software tool for genome sequence visualization and analysis. Dev Biol. 2004;271(2):431–438. doi: 10.1016/j.ydbio.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 23.Mount DW. Using the Basic Local Alignment Search Tool (BLAST) CSH Protoc. 2007;2007:pdb top17. doi: 10.1101/pdb.top17. [DOI] [PubMed] [Google Scholar]
- 24.Hare EE, Peterson BK, Iyer VN, Meier R, Eisen MB. Sepsid even-skipped enhancers are functionally conserved in Drosophila despite lack of sequence conservation. PLoS Genet. 2008;4(6):e1000106. doi: 10.1371/journal.pgen.1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cande J, Goltsev Y, Levine MS. Conservation of enhancer location in divergent insects. Proc Natl Acad Sci USA. 2009;106(34):14414–14419. doi: 10.1073/pnas.0905754106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meireles-Filho AC, Stark A. Comparative genomics of gene regulation-conservation and divergence of cis-regulatory information. Curr Opin Genet Dev. 2009;19(6):565–570. doi: 10.1016/j.gde.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 27.de Rosa R, et al. Hox genes in brachiopods and priapulids and protostome evolution. Nature. 1999;399(6738):772–776. doi: 10.1038/21631. [DOI] [PubMed] [Google Scholar]
- 28.Noordermeer D, et al. Variegated gene expression caused by cell-specific long-range DNA interactions. Nat Cell Biol. 2011;13(8):944–951. doi: 10.1038/ncb2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chanut-Delalande H, Fernandes I, Roch F, Payre F, Plaza S. Shavenbaby couples patterning to epidermal cell shape control. PLoS Biol. 2006;4(9):e290. doi: 10.1371/journal.pbio.0040290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287(5785):795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 31.Stern DL, Orgogozo V. The loci of evolution: How predictable is genetic evolution? Evolution. 2008;62(9):2155–2177. doi: 10.1111/j.1558-5646.2008.00450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stern DL, Orgogozo V. Is genetic evolution predictable? Science. 2009;323(5915):746–751. doi: 10.1126/science.1158997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thummel CS, Pirrotta V. New pCaSpeR P element vectors. Drosoph Inf Serv. 1992;71:150. [Google Scholar]
- 34.Thibault ST, et al. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet. 2004;36(3):283–287. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- 35.Horn C, Wimmer EA. A versatile vector set for animal transgenesis. Dev Genes Evol. 2000;210(12):630–637. doi: 10.1007/s004270000110. [DOI] [PubMed] [Google Scholar]
- 36.Handler AM, Harrell RA., 2nd Germline transformation of Drosophila melanogaster with the piggyBac transposon vector. Insect Mol Biol. 1999;8(4):449–457. doi: 10.1046/j.1365-2583.1999.00139.x. [DOI] [PubMed] [Google Scholar]
- 37.Fernandes I, et al. Zona pellucida domain proteins remodel the apical compartment for localized cell shape changes. Dev Cell. 2010;18(1):64–76. doi: 10.1016/j.devcel.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 38.Bokor P, DiNardo S. The roles of hedgehog, wingless and lines in patterning the dorsal epidermis in Drosophila. Development. 1996;122(4):1083–1092. doi: 10.1242/dev.122.4.1083. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.