Abstract
Recent studies have identified a number of transcriptional regulators, including E2A, early B-cell factor 1 (EBF1), FOXO1, and paired box gene 5 (PAX5), that promote early B-cell development. However, how this ensemble of regulators mechanistically promotes B-cell fate remains poorly understood. Here we demonstrate that B-cell development in FOXO1-deficient mice is arrested in the common lymphoid progenitor (CLP) LY6D+ cell stage. We demonstrate that this phenotype closely resembles the arrest in B-cell development observed in EBF1-deficient mice. Consistent with these observations, we find that the transcription signatures of FOXO1- and EBF1-deficient LY6D+ progenitors are strikingly similar, indicating a common set of target genes. Furthermore, we found that depletion of EBF1 expression in LY6D+ CLPs severely affects FOXO1 mRNA abundance, whereas depletion of FOXO1 activity in LY6D+ CLPs ablates EBF1 transcript levels. We generated a global regulatory network from EBF1 and FOXO1 genome-wide transcription factor occupancy and transcription signatures derived from EBF1- and FOXO1-deficient CLPs. This analysis reveals that EBF1 and FOXO1 act in a positive feedback circuitry to promote and stabilize specification to the B-cell lineage.
B-cell development is orchestrated by a complex network of transcription factors whose activities are modulated by a diverse set of signaling modules. The first cells in the bone marrow (BM) primed for a lymphoid cell fate are referred to as lymphoid-primed multipotent progenitors (LMPPs) (1). LMPPs have the capacity to differentiate into common lymphoid progenitors (CLPs), which in turn have the ability to give rise to all lymphoid lineages (2, 3). The CLP compartment is heterogeneous, consisting of cells with different defined lineage potentials, as well as cells already committed to the B-cell fate. The CLP compartment can be segregated into two distinct populations based on the expression of the cell surface marker LY6D (4, 5). During the last two decades, a subset of transcriptional regulators have been identified that specify and promote B-cell fate. Prominent among those are E2A, early B-cell factor 1 (EBF1), and paired box gene 5 (PAX5), as well as effectors acting downstream of the IL7-signaling cascade (6). E2A-deficient mice exhibit a complete block before the expression of LY6D in the CLP compartment (4, 7, 8). In the CLP compartment, the E2A proteins act in concert with HEB to induce the expression of forkhead box O1 (Foxo1) (8). Although it is well established that Foxo1 is critical for normal B-cell development, the dynamic regulation of Foxo1 expression in the CLP compartment suggested roles for Foxo1 at the earliest progenitor cell stage (5, 9–12).
To determine the mechanistic impact of FOXO1 in B-lineage specification, we examined FOXO1-deficient mice for defects in early hematopoiesis. FOXO1-ablated BM exhibited an almost complete lack of CD19+ B cells and an accumulation of cells at the LY6D+ CLP stage, resembling the phenotype observed in EBF1-deficient mice (12–14). Furthermore, we found that FOXO1-deficient LY6D+ CLPs lacked the expression of genes associated with the B-cell lineage and identified striking similarities in the transcription signatures derived from FOXO1- and EBF1-ablated CLPs. Interestingly, we found that FOXO1-deficient progenitors display severely reduced Ebf1 transcript levels, whereas EBF1-deficient progenitors showed reduced Foxo1 transcript abundance. On examination, we found that EBF1 and FOXO1 bind to enhancer regions that interact with promoter regions corresponding to the FOXO1 and EBF1 loci, demonstrating that EBF1 and FOXO1 drive the expression of one another. We have generated a global regulatory network from EBF1 and FOXO1 genome-wide transcription factor occupancy and transcription signatures derived from EBF1- and FOXO1-deficient CLPs. This analysis showed that EBF1 and FOXO1 act in a positive feedback circuitry to promote and stabilize B-cell fate.
Results
B-Lymphoid Development in FOXO1-Depleted Mice Is Arrested at the Common Lymphoid Progenitor Cell Stage.
Previous studies have established that the E2A proteins directly activate Foxo1 expression in LY6D− CLPs (8). To evaluate how activation of Foxo1 expression relates to B-cell specification, we compared Foxo1 mRNA abundance in sorted LY6D− and LY6D+ CLPs. As expected, we observed a substantial and significant increase in Foxo1 expression in the LY6D+ compartment, consistent with the developmental stage at which B-cell specification is initiated (Fig. 1A). To explore the role for FOXO1 in B-cell progenitors in greater detail, we generated mice conditionally deleted for Foxo1 across the entire hematopoietic compartment. Mice carrying a conditional Foxo1 (Foxo1f) allele were bred to mice expressing Cre placed under Vav1-regulatory elements (15). FOXO1f/f Vav1-iCre mice (referred to as FOXO1−/− here) mice did not show gross abnormalities and exhibited normal BM cellularity, whereas the number of cells in the spleen was reduced by a factor of twofold (Fig. 1B; Fig. S1A). Notably, the number of CD19+B220+ cells in the BM was reduced more than 50-fold, and only a few CD19+ cells could be detected in the spleen of FOXO1−/− mice (Fig. 1C; Fig. S1B). The few CD19+ cells identified in the BM of FOXO1−/− lacked expression of IgM, IgD, and CD25, indicating a developmental arrest at the pro-B-cell stage (Fig. S1C). Interestingly, the LY6D+ CLP compartment was substantially increased in the absence of FOXO1 (Fig. 1D).
Fig. 1.
FOXO1 plays an essential role in B-cell development. (A) LY6D− and LY6D+ CLPs were analyzed by real-time PCR for the abundance of Foxo1 mRNA. Values were normalized to HPRT expression and shown as mean ± SEM, using cells from two independent sorts. (B) Total number of BM cells in sex- and age-matched WT and FOXO1−/− mice. Data shown are pooled from two independent experiments. (C) (Left) Representative FACS plots of CD19+B220+ cells derived from WT and FOXO1−/− BM. (Right) Total numbers of CD19+B220+ B cells isolated from WT and FOXO1−/− BM. Data shown are pooled from two independent experiments. (D) (Left) Representative FACS plots showing gating strategy to identify CLPs. LINneg includes CD11B, GR1, TER119, LY6C, NK1.1, CD3e, CD19, and CD11C. (Right) Numbers of LY6D− and LY6D+ CLPs in WT and FOXO1−/− mice. Data shown are pooled from two independent experiments. (E) Representative FACS plots displaying reconstitution of CD45.2+ cells in CD45.1+ hosts, isolated from WT (Upper) and FOXO1−/− (Lower) BM. (Left) LY6D+/− CLPs. (Center) CD19+B220+ BM B cells. (Right) CD19+B220+ spleen B cells. (F) Diagram displaying the roles of transcription factors in early hematopoiesis and B-cell progenitors.
Because it is well established that FOXO1 plays a critical role in regulating cell cycle progression and cell survival, we examined for viability and the fraction of cycling cells in FOXO1−/− (16). As predicted, FOXO1-ablated CLPs showed a higher percentage of actively cycling cells and lower levels of Annexin V staining (Fig. S1 D and E). Although the use of Vav1-iCre should ensure selective gene deactivation in hematopoietic cells, expression of Foxo1 in multiple other cell types including other hematopoetic cells prompted us to transplant FOXO1-ablated or WT progenitor cells into irradiated recipient mice. We found that BM cells isolated from FOXO1−/− mice were unable to reconstitute the B-lineage compartment on transfer into lethally irradiated hosts, indicating that the requirement for FOXO1 in B-cell development is cell autonomous (Fig. 1E). Taken together, these data indicate that in the absence of FOXO1, B-cell development is arrested in the CLP compartment before commitment to the B-cell lineage (Fig. 1F).
FOXO1 Activity Is Essential to Enforce B-Lineage Restriction.
To investigate the molecular causes underlying the B-cell defect, LY6D+ CLPs derived from FOXO1−/− were sorted and examined for the presence of IgH DH-JH rearrangements. Although BM derived from FOXO1−/− mice exhibited DH-JH rearrangements, the frequency of DH-JH rearrangements in sorted single LY6D+ FOXO1-depeted CLPs was severely reduced compared with WT CLPs (Fig. 2A; Fig. S2A).
Fig. 2.
FOXO1 acts to enforce B-cell fate (A) Graph shows the distribution of DH-JH rearrangements in LY6D+ CLPs. A total of 192 cells/genotype from two independent sorts were assayed. Cells were scored as follows: GL only, germ-line band only; DJ + GL, cells producing one DHJH and one GL DNA fragments; DJ, cells producing one or two DHJH DNA fragments and no GL band; no read out, cells failing to produce detectable PCR products. (B) Graph displays cloning frequency from OP9-coculture experiments using single cell–sorted LY6D+ CLPs derived from WT and FOXO1−/− mice. Cells were cultured in B/NK cell promoting culture conditions. Indicated are percentages of clones containing cells expressing CD19, NK1.1, and/or CD11C. A total of 264 cells per genotype were sorted. (C) Graphs display cloning frequency from OP9-DL1 coculture experiments using single-sorted LY6D− (Left) and LY6D+ (Right) CLPs isolated from WT and FOXO1−/− mice. Shown are the mean ± SEM. A total of 192 cells/genotype were sorted in two independent experiments. (D) Result from microarrays analysis displaying genes that are changed by a factor of twofold between WT and FOXO1−/− LY6D+ CLPs. Displayed data are derived from two microarray replicas using cells from independent sorts. (E) LY6D+ CLPs from WT and FOXO1−/− mice were analyzed by real-time PCR for the abundance of the indicated transcripts. Values were normalized for HPRT expression and are shown as mean ± SEM using mRNA from two independent sorts. (F) Multiplex single cell RT-PCR from LY6D+ CLPs. Graph indicates the percentages of cells expressing the indicated genes. A total of 96 cells were assayed for each genotype.
To determine the lineage potential of FOXO1-deficient CLPs, single cell LY6D+ CLPs were sorted onto OP9 stroma cells and cultured in the presence of IL7, KIT ligand, FLT3 ligand, IL2, and IL15. After 14 d of culture, the majority of WT cells formed colonies containing only CD19+ cells (Fig. 2B). However, in the absence of FOXO1, the majority of colonies lacked CD19 expression but contained cells expressing NK1.1 and/or CD11C (Fig. 2B). Next, we performed T cell–promoting OP9DL-coculture assays. In the uncommitted LY6D− CLP fraction, no difference was observed between WT and FOXO1−/− CLPS (Fig. 2C). However, on comparing WT and FOXO1-deficient progenitors, we found that FOXO1−/− LY6D+ cells exhibited an increased tendency to differentiate into T-lineage cells (Fig. 2C). In sum, these observations indicate that FOXO1 in the CLP compartment acts to enforce commitment to B-cell fate.
FOXO1 Is Critical to Promote B-lineage Specification.
To determine how a deficiency in FOXO1 affects B-cell development, we generated transcription signatures from WT and FOXO−/− CLPs. Specifically, we sorted LY6D− and LY6D+ CLPs from WT and FOXO1−/− BM. RNA was isolated and examined for transcription signatures using microarray analysis. A total of 106 genes were changed more than twofold in LY6D+ CLPs from FOXO1−/− mice compared with WT CLPs (Fig. 2D; Table S1). Conspicuous among the differences was a severe reduction in Ebf1 and Pax5 abundance (Fig. 2 D and E; Table S1). Transcript levels encoding components involved in pre–B-cell receptor signalling (BCR) signaling, including Vpreb1, Vpreb3, Igll1, CD79b, and Blnk, were also severely reduced in FOXO1-deficient CLPs (Fig. 2 D and E; Table S1). Interestingly, transcript abundance encoding for modulators of the PI(3)K pathway including Pten, Blnk, Bank1, and Pik3ip1 exhibited significantly reduced transcript levels in FOXO1−/− CLPs (Fig. 2 D and E; Table S1).
To determine whether the decrease in transcript abundance was caused by a reduction in the number of cells expressing EBF1 or a decrease in Ebf1 mRNA levels in CLPs, we sorted single cells from the LY6D+ compartment from FOXO1−/− and WT mice. RNA was isolated from the sorted cells and examined by multiplex single cell RT-PCR for Hprt, Ebf1, Rag1, Pax5, and Igll1 transcript abundance. As expected, the number of cells expressing Pax5 and Igll1 was severely reduced in LY6D+ CLPs (Fig. 2F; Fig. S2C). Interestingly, however, the number of cells expressing Ebf1 was not significantly altered (Fig. 2F; Fig. S2C). These data indicate that in the CLP compartment, FOXO1 activity is required to induce high levels of Ebf1 abundance to promote the induction of a B lineage–specific program of gene expression.
FOXO1 and EBF1 Share a Spectrum of Target Genes.
Previous observations have indicated that in the absence of EBF1, B-cell development is blocked in the LY6D+ CLP compartment (12–14). Our analysis described above indicates that B-cell development in FOXO1-deficient BM is blocked at the same stage (Fig. 1). To determine how EBF1 and FOXO1 activities are related to promote B-cell fate, we compared our microarray data with expression signatures generated from EBF1-deficient LY6D+ CLPs (17). Both FOXO1 and EBF1 transcript levels increased during the developmental transition from LY6D− to LY6D+ CLPs and further increased at the pro-B-cell stage (Fig. 3A). The up-regulation of Foxo1 and Ebf1 transcript levels correlated well with the expression of early B-cell genes such as Igll1 and Vpreb1, which are well-characterized targets of EBF1 (Fig. 3A). Intriguingly, on assessing Foxo1 and Ebf1 transcript levels in Ebf1−/− and Foxo1−/− LY6D+ CLPs, Foxo1 and Ebf1 transcript levels were significantly decreased, indicating that FOXO1 acts to induce the expression of Ebf1 and vice versa to promote B-cell fate (Fig. 3A).
Fig. 3.
FOXO1 and EBF1 share a common set of target genes. (A) Results from microarray analysis showing the normalized expression of Foxo1, Ebf1, Igll1, and Vpreb1 in Ly6D+/− CLPs and pro-B cells. Genotypes are indicated below graphs. (B) Result from microarrays analysis displaying genes that are changed at least twofold between either WT and FOXO1−/− and/or WT and EBF1−/− LY6D+ CLPs. Data are derived from two microarray replicas using material from independent sorts. (C) Plots show the direction of gene regulation in FOXO1- and EBF1-deficient cells of genes in B. Gene expression was normalized to the average expression of the WT and KO value for each set of WT/KO arrays. (D) Functional classification of genes in B. (E) Distance in the genome from the TSS of genes changed in FOXO1−/− LY6D+ CLPs to closest FOXO1/EBF1 binding sites.
Furthermore, on aligning the full lists of genes that were changed more than twofold in the absence of either EBF1 or FOXO1, a strikingly similar pattern emerged (Fig. 3B). Specifically, 148 genes were identified whose expression levels were changed by a factor of at least twofold in either or both EBF1- and FOXO1-ablated CLPs, indicating that FOXO1 and EBF1 share a large set of common target genes (Fig. 3 B and C; Table S2). Among the genes whose expression levels declined in the absence of either EBF1 or FOXO1 was a set of genes closely associated with a B lineage–specific program of gene expression (Fig. 3C; Table S2). The majority of these genes showing coordinate regulation by FOXO1 and EBF1 in CLPs were closely associated with cell surface receptors, signaling components, and transcriptional regulators important for specification of B-cell fate (Fig. 3D; Table S2).
Previous studies have established genome-wide E2A, EBF1, and FOXO1 occupancy in pro-B cells (18). As a first approach to explore the possibility that FOXO1 and EBF1 act collaboratively at putative regulatory elements, we used the pro-B-cell ChIPseq data and examined the genomic positions of EBF1 and FOXO1 binding sites relative to the transcription start sites (TSSs). As predicted by a previous study (18), the majority of FOXO1 and EBF1 binding sites appeared to be positioned in distally located regulatory elements (Fig. 3E). However, EBF1 and FOXO1 appeared to occupy different enhancers acting at the same target gene, most prominent among these was Pax5 (Fig. S3 A–C) Taken together, these data raise the possibility that EBF1 and FOXO1 act collaboratively at a subset of distally located regulatory elements to establish B-cell fate.
Intergenic Feedback Circuitry, Involving EBF1 and FOXO1, Establishes B-Cell Identity.
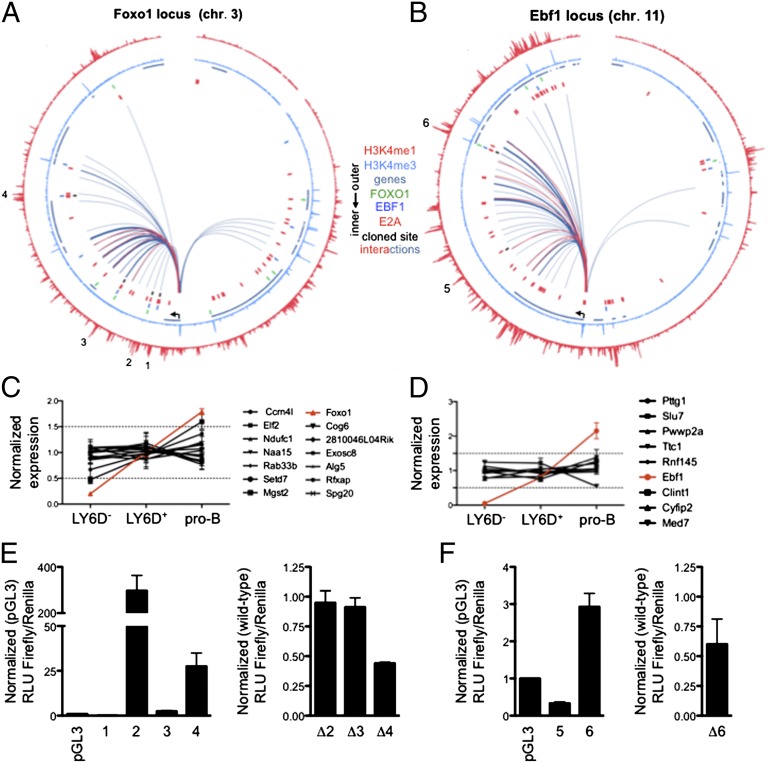
The data described above bring into question as to how EBF1 and FOXO1 act in concert to regulate a B lineage–specific program of gene expression. To link EBF1 and FOXO1 occupancy with the induction of a B lineage–specific program of gene expression, we identified those binding sites that directly interact with promoter regions of either EBF1 or FOXO1. This analysis was performed using pro-B and pre-pro-B-cell interactomes obtained by Hi-C analysis (19). Next, we performed an in silico 4C analysis using the pro-B/pre-pro-B interactome as templates and the EBF1 and FOXO1 promoter regions as baits (Fig. 4 A and B). The data were visualized using Circos diagrams (Fig. 4 A and B). The thicknesses of the lines represent the natural log ratio of observed versus expected interactions (Fig. 4 A and B). Consistent with the requirement for EBF1 and FOXO1 expression, we found significant enrichment for genomic interactions involving regions containing the EBF1 promoter marked by H3K4me3 and regions containing putative enhancer elements marked by H3K4me1 (Fig. 4B). Notably, a region enriched for promoter interactions exhibited FOXO1, EBF, and E2A occupancy (Fig. 4A). Similarly, the FOXO1 promoter showed looping involving multiple enhancer elements that exhibited EBF1, FOXO1, and E2A occupancy (Fig. 4B). These observations directly link EBF1 and FOXO1 occupancy with the promoter regions of the EBF1 and FOXO1 loci.
Fig. 4.
An intergenic feeback circuitry, involving FOXO1 and EBF1, establishes B-cell identity. (A and B) Circos diagram displaying genomic interactions across a 5-Mb genomic region surrounding the (A) Foxo1 and (B) Ebf1 locus (arrows indicate Foxo1/Ebf1 transcriptional start sites). H3K4me1/3 binding patterns and FOXO1/EBF1/E2A binding sites from pro-B cells are indicated. The thicknesses of the connecting lines reflect the natural log ratio of observed versus expected interaction frequency in the Hi-C data sets. Bin size used for analysis of genomic interactions was 50 kb. Blue and red connecting lines represent significant interactions observed in pro-B and pre-pro-B cells, respectively. (C and D) Expression and developmental regulation of genes located within ±3 Mb surrounding the (C) Foxo1 and (D) Ebf1 TSSs, respectively. Expression was normalized to the average of developmental stages displayed. (E) (Left) Transcriptional activity of putative enhancer elements surrounding the Foxo1 locus with associated EBF1 occupancy. (Right) Transcriptional activity after mutation (Δ) of binding sites (Fig. S4). (F) (Left) Transcriptional activity of putative enhancer elements surrounding the Ebf1 locus with associated FOXO1 occupancy. (Right) Transcriptional activity after mutation (Δ) of binding sites (Fig. S5). Luciferase data shown are mean ± SEM derived from two independent experiments.
Because FOXO1 and EBF1 are barely expressed at the LY6D− CLP cell stage, we reasoned that if a gene is not expressed or developmentally regulated during early B-lineage development, it most likely is not associated with enhancer elements containing EBF1 and FOXO1 binding sites. Hence, we screened the microarray expression patterns of genes located ±3Mb away from the TSSs of the Foxo1 and Ebf1 loci in LY6D− CLPs, LY6D+ CLPs, and pro-B cells (Fig. 4 C and D). Although several genes were expressed in early B-cell progenitors, they did not exhibit developmental regulation (Fig. 4 C and D). To determine whether genomic elements that showed deposition of H3K4me1 and EBF1/FOXO1 occupancy indeed function as enhancers, several of putative enhancer regions were inserted into an enhancer luciferase reporter construct and assayed for transcriptional activity (Fig. 4 E and F). As expected, substantial enhancer activity was associated with regions associated with H3K4me1 deposition (Fig. 4 E and F; Figs. S4 and S5; Table S3). These data suggest that enhancers across this region that show EBF1 and FOXO1 occupancy act primarily to induce the expression of either EBF1 or FOXO1, rather than modulating the expression of genes located within close genomic proximity. As expected, mutation of critical binding sites reduced enhancer activity (Fig. 4 E and F; Figs. S4 and S5).
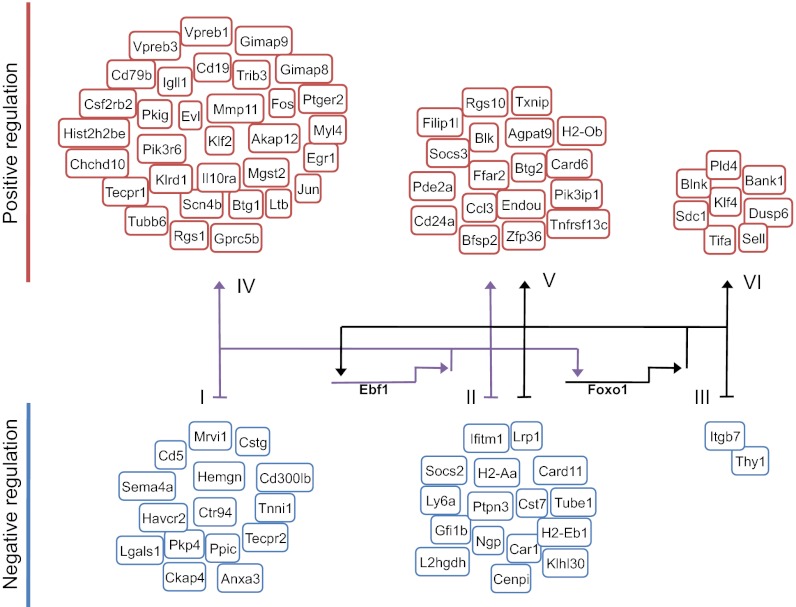
Finally, to link EBF1 and FOXO1 and patterns of gene expression into a common framework, a global network was generated (Fig. 5). To accomplish this, we identified putative regulatory targets of EBF1 and FOXO1. The analysis was based on two criteria. First, genes were selected with a significant change in transcript levels in EBF1 or FOXO1-ablated LY6D+ CLPs (at least a twofold change). Second, we assessed these genes for TSS proximal and distal (<250 kb) EBF1/FOXO1 occupancy (18). We divided genes fulfilling these criteria into six groups: I, negatively regulated by EBF1; II, negatively regulated by both EBF1 and FOXO1; III, negatively regulated by FOXO1; IV, positively regulated by EBF1; V, positively regulated by EBF1 and FOXO1; VI, positively regulated by FOXO1 (Fig. 5). As expected, a large fraction of B cell–associated genes showed associated EBF1 occupancy. Among these are Igll1, Vpreb1, Vpreb3, CD19, and Cd79b, each involved in pre-BCR–mediated signaling. A large fraction of genes displayed both associated EBF1 and FOXO1 binding. This group included both positively regulated (Socs3, Blk, etc.) and transcriptional repressed genes (Gfi1b, Socs2, etc.). Both EBF1 and FOXO1 alone were also associated with genes showing either increased or decreased transcript levels in EBF1- and FOXO1-depleted CLPs. In summary, these observations readily reveal a network in which FOXO1 acts to induce the expression of EBF1 (Fig. 5). Once EBF1 expression is induced, an intergenic feedback circuitry is established, involving the activities of both EBF1 and FOXO1, to promote B-cell fate.
Fig. 5.
Global regulatory network that orchestrates B-cell fate. Network is based on the transcription signatures derived from EBF1- and FOXO1-deficient CLPs and genome-wide EBF1 and FOXO1 occupancy in pro-B cells. Intergenic feedback circuitry is indicated. Genes were selected with a significant change in transcript levels in EBF1- or FOXO1-ablated LY6D+ CLPs (at least a twofold change) and with a TSS proximal and distal (<250 kb) EBF1/FOXO1 occupancy. Genes fulfilling these criteria were divided into six groups: I, negatively regulated by EBF1; II, negatively regulated by both EBF1 and FOXO1; III, negatively regulated by FOXO1; IV, positively regulated by EBF1; V, positively regulated by EBF1 and FOXO1; VI, positively regulated by FOXO1.
Discussion
During hematopoiesis, developmental trajectories with multiple branch points have been identified from which progeny cells develop to ultimately become committed to distinct cell lineages. Transcriptional regulators have been characterized that modulate the developmental progression, as well as expansion of the hematopoietic cell lineages. However, it remains largely unknown as to how they are connected to establish a stable lineage- and stage-specific phenotype. Here, we describe a positive feedback circuitry, involving EBF1 and FOXO1, acting to stabilize B-cell fate.
Numerous functional and genome-wide studies have identified E2A, EBF1, and PAX5 as critical factors that establish B-cell identity. Recent studies have also demonstrated that FOXO1 activity is required for proper B-cell development (10). Although the observations indicated a role for FOXO1 at the earliest stages of B-lineage development, it has remained unclear at which developmental checkpoint FOXO1 acts to specify the B-cell fate. Here, we unambiguously demonstrate that the FOXO1 proteins act in the CLP compartment to establish and enforce B-cell identity. FOXO1 acts to specify B-cell fate by activating the expression of EBF1, which in turn, activates the expression of FOXO1 in a positive feedback loop.
Positive feedback circuitries that promote and establish cell fates have been identified in previous studies. These involve developmental progression within the mammalian pancreas, the Drosophila heart, and the skeletogenic micromere in the sea urchin embryo (20, 21). Well-characterized feedback circuitries have also been identified that stabilize gene expression programs in embryonic stem cells, induced pluripotent stem (iPS) cells, and T-cell acute lymphoblastic leukemia (T-ALL) (22–24). We suggest that similarly as described for embryonic and cancer stem cells, the positive intergenic feedback circuitry involving EBF1 and FOXO1 acts to reinforce and maintain a pro-B–specific transcription signature.
It is well established that the nuclear location and abundance of FOXO proteins are regulated by the PI(3)K-AKT pathway (17). Early events in lymphocyte development are modulated by IL7Ra and FLT3-mediated signaling. Both IL7- and FLT3-mediated signaling have been suggested to be involved in the PI(3)K pathways. Thus, we are now faced with the question as to whether IL7- and FLT3-mediated signaling modulates FOXO1 nuclear location and abundance. Recent data have indicated that the main action of IL7 signaling at the CLP stage involves the activation of STAT5 but not AKT-mediated signaling (25, 26). In contrast, FLT3 signaling most likely activates the PI(3)K pathway until the FLT3 receptor is suppressed at later developmental stages by PAX5 (26). These latter observations raise the question as to how FOXO1 protein abundance is maintained in the presence of FLT3-mediated signaling. Our observations indicate that several modulators of the PI(3)K pathway such as Pten, Blnk, Bank1, and Pik3ip1 appear to be regulated by FOXO1 and EBF1. We also note that Blnk and Pik3ip1 are affected in E2A−/− CLPs as well (8). Taken together, we propose that in developing B-cell progenitors, an intricate and tightly controlled regulatory circuitry has been established to ensure the establishment and maintenance of FOXO1 protein abundance in the CLP compartment. We note that it will be important to determine in detail how FOXO1 activity both at the transcriptional and posttranscriptional levels is regulated and how these two circuitries intersect to establish B-cell identity.
In sum, we propose that, in the CLP compartment, the E2A proteins directly activate the expression of FOXO1, as well as IRF4 and IRF8 (8, 27–29). E2A and FOXO1, in turn, act in concert to directly activate the expression of EBF1 (30–33). The induction of EBF1 expression leads to the establishment of a positive feedback circuitry involving EBF1 and FOXO1. Specifically, we suggest that, on reaching higher levels of FOXO1 and EBF1 than that observed in the CLP compartment, the activities of EBF1 and FOXO1 become interlocked to induce a pro-B cell–specific program of gene expression that must be maintained for a distinct period to establish B-cell identity. It is likely that there will be complementary feedback loops, involving other key regulators, to promote additional steadiness, acting in concert to establish B-cell identity. Describing such circuitries, how they act on each other, and how they are regulated by external cues will eventually permit a description of B-cell fate in mechanistic terms.
Experimental Procedures
Mice and Transfer Experiments.
Lethally irradiated CD45.1 mice were injected with isolated progenitor cells and analyzed 8 weeks after transplantation. For details and mice used see SI Experimental Procedures.
FACS Staining, Purification, and Culture of BM Cells.
For details on antibody/cell cycle/apoptosis staining procedures see SI Experimental Procedures. OP9/OP9DL1 cocultures were performed as previously described (34).
Quantitative RNA and DNA Analysis.
RT-qPCR, single cell RT-PCR, and microarray analysis were performed as previously described (34). Affymetrix data is available through the Gene Expression Omnibus (GEO) database (accession no. GSE41931). Single cell DJ-PCR was performed as previously described (8). For details on RT-qPCR primers/probes used see SI Experimental Procedures.
Luciferase Assay.
Assays were performed with a dual-luciferase reporter assay system (Promega) using 22D6 pro-B cells. For details on procedures and elements investigated see SI Experimental Procedures and Figs. S4 and S5.
In Silico 4C.
Hi-C data describing the pro-B and pre-pro-B-cell interactomes were analyzed to identify genomic interactions involving the Ebf1 and Foxo1 promoters using a 50-kb bin (18, 19). Only interactions showing P < 0.05 were considered. Interaction data was visualized together with ChIPseq binding data using Circos.
Supplementary Material
Acknowledgments
We thank Ron DePhino for providing FOXO1f/f mice and Steve Hedrick (University of California at San Diego) for reagents and scientific advice . We thank Liselotte Lenner for assistance. This work was supported by the National Institutes of Health (R.M., E.W. Y.C.L., J.S.L., C.B., C.K.G., and C.M.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE41931).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1211427109/-/DCSupplemental.
References
- 1.Adolfsson J, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential: A revised road map for adult blood lineage commitment. Cell. 2005;121(2):295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91(5):661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 3.Karsunky H, Inlay MA, Serwold T, Bhattacharya D, Weissman IL. Flk2+ common lymphoid progenitors possess equivalent differentiation potential for the B and T lineages. Blood. 2008;111(12):5562–5570. doi: 10.1182/blood-2007-11-126219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Inlay MA, et al. Ly6d marks the earliest stage of B-cell specification and identifies the branchpoint between B-cell and T-cell development. Genes Dev. 2009;23(20):2376–2381. doi: 10.1101/gad.1836009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mansson R, et al. Single-cell analysis of the common lymphoid progenitor compartment reveals functional and molecular heterogeneity. Blood. 2010;115(13):2601–2609. doi: 10.1182/blood-2009-08-236398. [DOI] [PubMed] [Google Scholar]
- 6.Nutt SL, Kee BL. The transcriptional regulation of B cell lineage commitment. Immunity. 2007;26(6):715–725. doi: 10.1016/j.immuni.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 7.Dias S, Månsson R, Gurbuxani S, Sigvardsson M, Kee BL. E2A proteins promote development of lymphoid-primed multipotent progenitors. Immunity. 2008;29(2):217–227. doi: 10.1016/j.immuni.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Welinder E, et al. The transcription factors E2A and HEB act in concert to induce the expression of FOXO1 in the common lymphoid progenitor. Proc Natl Acad Sci USA. 2011;108(42):17402–17407. doi: 10.1073/pnas.1111766108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amin RH, Schlissel MS. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat Immunol. 2008;9(6):613–622. doi: 10.1038/ni.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dengler HS, et al. Distinct functions for the transcription factor Foxo1 at various stages of B cell differentiation. Nat Immunol. 2008;9(12):1388–1398. doi: 10.1038/ni.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mercer EM, et al. Multilineage priming of enhancer repertoires precedes commitment to the B and myeloid cell lineages in hematopoietic progenitors. Immunity. 2011;35(3):413–425. doi: 10.1016/j.immuni.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zandi S, et al. EBF1 is essential for B-lineage priming and establishment of a transcription factor network in common lymphoid progenitors. J Immunol. 2008;181(5):3364–3372. doi: 10.4049/jimmunol.181.5.3364. [DOI] [PubMed] [Google Scholar]
- 13.Lin H, Grosschedl R. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 1995;376(6537):263–267. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- 14.Tsapogas P, et al. IL-7 mediates Ebf-1-dependent lineage restriction in early lymphoid progenitors. Blood. 2011;118(5):1283–1290. doi: 10.1182/blood-2011-01-332189. [DOI] [PubMed] [Google Scholar]
- 15.Paik JH, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128(2):309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hedrick SM. The cunning little vixen: Foxo and the cycle of life and death. Nat Immunol. 2009;10(10):1057–1063. doi: 10.1038/ni.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zandi S, et al. Single-cell analysis of early B-lymphocyte development suggests independent regulation of lineage specification and commitment in vivo. Proc Natl Acad Sci USA. 2012;109(39):15871–15876. doi: 10.1073/pnas.1210144109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin YC, et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol. 2010;11(7):635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin YC, et al. Global changes in the nuclear positioning of genes and intra- and interdomain genomic interactions that orchestrate B cell fate. Nat Immunol. 2012 doi: 10.1038/ni.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davidson EH. The Regulatory Genome: Gene Regulatory Networks in Development and Evolution. San Diego: Academic; 2009. [Google Scholar]
- 21.Oliveri P, Tu Q, Davidson EH. Global regulatory logic for specification of an embryonic cell lineage. Proc Natl Acad Sci USA. 2008;105(16):5955–5962. doi: 10.1073/pnas.0711220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8(6):450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 23.Young RA. Control of the embryonic stem cell state. Cell. 2011;144(6):940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanda T, et al. Core transcriptional regulatory circuit controlled by the TAL1 complex in human T cell acute lymphoblastic leukemia. Cancer Cell. 2012;22(2):209–221. doi: 10.1016/j.ccr.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Åhsberg J, et al. Interleukin-7-induced Stat-5 acts in synergy with Flt-3 signaling to stimulate expansion of hematopoietic progenitor cells. J Biol Chem. 2010;285(47):36275–36284. doi: 10.1074/jbc.M110.155531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kikuchi K, Lai AY, Hsu CL, Kondo M. IL-7 receptor signaling is necessary for stage transition in adult B cell development through up-regulation of EBF. J Exp Med. 2005;201(8):1197–1203. doi: 10.1084/jem.20050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holmes ML, Carotta S, Corcoran LM, Nutt SL. Repression of Flt3 by Pax5 is crucial for B-cell lineage commitment. Genes Dev. 2006;20(8):933–938. doi: 10.1101/gad.1396206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Decker T, et al. Stepwise activation of enhancer and promoter regions of the B cell commitment gene Pax5 in early lymphopoiesis. Immunity. 2009;30(4):508–520. doi: 10.1016/j.immuni.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 29.Pongubala JM, et al. Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5. Nat Immunol. 2008;9(2):203–215. doi: 10.1038/ni1555. [DOI] [PubMed] [Google Scholar]
- 30.Treiber T, et al. Early B cell factor 1 regulates B cell gene networks by activation, repression, and transcription- independent poising of chromatin. Immunity. 2010;32(5):714–725. doi: 10.1016/j.immuni.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 31.Roessler S, et al. Distinct promoters mediate the regulation of Ebf1 gene expression by interleukin-7 and Pax5. Mol Cell Biol. 2007;27(2):579–594. doi: 10.1128/MCB.01192-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith EM, Gisler R, Sigvardsson M. Cloning and characterization of a promoter flanking the early B cell factor (EBF) gene indicates roles for E-proteins and autoregulation in the control of EBF expression. J Immunol. 2002;169(1):261–270. doi: 10.4049/jimmunol.169.1.261. [DOI] [PubMed] [Google Scholar]
- 33.Seet CS, Brumbaugh RL, Kee BL. Early B cell factor promotes B lymphopoiesis with reduced interleukin 7 responsiveness in the absence of E2A. J Exp Med. 2004;199(12):1689–1700. doi: 10.1084/jem.20032202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mansson R, et al. B-lineage commitment prior to surface expression of B220 and CD19 on hematopoietic progenitor cells. Blood. 2008;112(4):1048–1055. doi: 10.1182/blood-2007-11-125385. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.