Abstract
Despite the fact that the genetic code is known to vary between organisms in rare cases, it is believed that in the lifetime of a single cell the code is stable. We found Acetohalobium arabaticum cells grown on pyruvate genetically encode 20 amino acids, but in the presence of trimethylamine (TMA), A. arabaticum dynamically expands its genetic code to 21 amino acids including pyrrolysine (Pyl). A. arabaticum is the only known organism that modulates the size of its genetic code in response to its environment and energy source. The gene cassette pylTSBCD, required to biosynthesize and genetically encode UAG codons as Pyl, is present in the genomes of 24 anaerobic archaea and bacteria. Unlike archaeal Pyl-decoding organisms that constitutively encode Pyl, we observed that A. arabaticum controls Pyl encoding by down-regulating transcription of the entire Pyl operon under growth conditions lacking TMA, to the point where no detectable Pyl-tRNAPyl is made in vivo. Pyl-decoding archaea adapted to an expanded genetic code by minimizing TAG codon frequency to typically ∼5% of ORFs, whereas Pyl-decoding bacteria (∼20% of ORFs contain in-frame TAGs) regulate Pyl-tRNAPyl formation and translation of UAG by transcriptional deactivation of genes in the Pyl operon. We further demonstrate that Pyl encoding occurs in a bacterium that naturally encodes the Pyl operon, and identified Pyl residues by mass spectrometry in A. arabaticum proteins including two methylamine methyltransferases.
Keywords: codon reassignment, pyrrolysyl-tRNA synthetase, Desulfitobacterium dehalogenans, Desulfitobacterium hafniense
In most organisms the genetic code of 64 nucleotide triplets is decoded into 20 canonical amino acids, whereas three codons signal translational stop. Across the diversity of life the genetic code has been shown to vary in both size and codon assignment in some organisms, and especially in organelles. It is believed, however, that within a single cell the genetic code remains static (i.e., the number of amino acids encoded is constant) and determined by the aminoacyl-tRNA synthetases and tRNAs encoded in the genome. There are 22 genetically encoded amino acids, including selenocysteine (Sec) and pyrrolysine (Pyl); whereas many organisms have an expanded genetic code with 21 amino acids (either Sec or Pyl), only few organisms have 22 amino acids by using Sec and Pyl (1). In the expansion of the genetic code with selenocysteine, an elaborate recoding machinery is required where RNA secondary structure elements specify certain UGA codons as Sec, whereas in their absence UGA codons remain termination signals (2). Pyl, on the other hand, which is well characterized in archaeal methanogens (3), is encoded by UAG (4, 5), suggesting that any TAG codon in the genome of these organisms could (if expressed) lead to Pyl insertion.
Pyl was first identified in the active site of an archaeal methyltransferase (4, 6), which is required for growth on methylamines. The components needed for recoding UAG from stop to Pyl exist in 24 species usually on a single operon. In addition to encoding tRNAPylCUA and pyrrolysyl-tRNA synthetase (PylRS) (7, 8), this operon encodes for PylB, PylC, and PylD, the only enzymes required for the biosynthesis of free Pyl (9). It has been shown that the Pyl operon can be transferred into Escherichia coli, and is sufficient to promote limited read-through of amber codons (10, 11). A Methanosarcina acetivorans strain lacking tRNAPyl is viable when cells are grown on methanol as a carbon source, but detrimental for growth on methylamines (12). This led to the initial assumption that Pyl was solely required in the catalytic active site of methylamine methyltransferases that are highly up-regulated on methylamines and repressed on methanol (13, 14). We demonstrated, however, that Pyl can be incorporated in tRNAHis guanylyltransferase (Thg1), an enzyme that is not involved in methylamine metabolism, and interestingly, Pyl does not participate in the catalytic activity of this enzyme (15). Additionally it has been shown that tRNAPyl is expressed independently of the carbon source in Methanosarcina mazei (16) and that Pyl-containing proteins were recovered from M. acetivorans cells grown on methanol (15). These studies indicate that the expression of particular Pyl-containing proteins is regulated in archaeal organisms, but Pyl-tRNAPyl formation and encoding of UAG as Pyl is constitutive and has not been shown to be regulated during the lifetime of the cell.
Our current understanding of Pyl decoding is based on the knowledge of 24 known Pyl-decoding organisms, with about half of them in the Methanosarcina family of archaeal methanogens and the others consisting of a diverse group of bacteria. Partly due to the lack of genetic tools (17), Pyl decoding in these bacteria has not been investigated in vivo. PylRS and tRNAPyl from Desulfitobacterium hafniense (18) and from Methanosarcina (19) are well characterized in vitro, and the information gained from the structural data (20, 21) led to the development of many orthogonal tRNA/synthetase pairs used for unnatural amino acid incorporation (22, 23). In a heterologous E. coli context, PylRS and tRNAPyl from D. hafniense are active in UAG suppression in vivo (18). Despite this progress, there has been no observation of Pyl decoding in a bacterium that naturally contains the Pyl operon. Here we investigate in vivo expression of Pyl operon gene products, Pyl-tRNAPyl formation, and the production of Pyl-containing proteins in putative Pyl-decoding bacterial species including Acetohalobium arabaticum, Desulfitobacterium dehalogenans, and D. hafniense.
Results
Diversity of Pyl-Decoding Organisms.
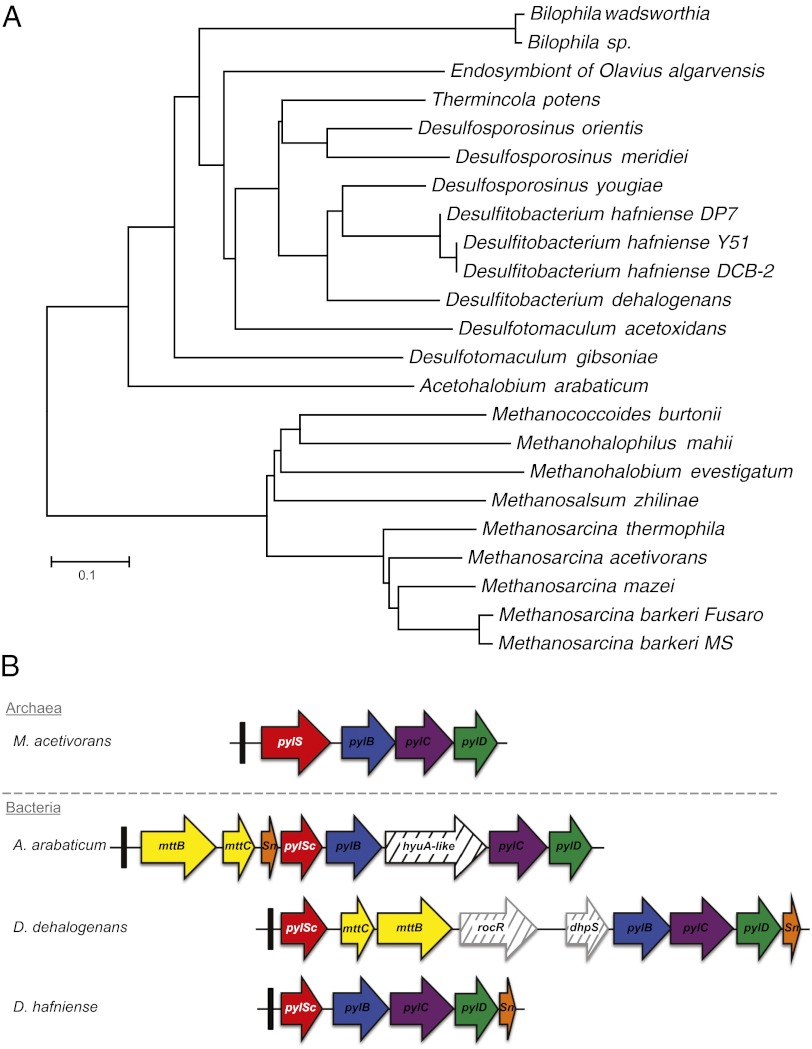
Since the discovery of pyrrolysine in archaea (6), the list of organisms possessing the Pyl operon expanded, representing ∼0.7% of all sequenced genomes. Using BLAST with known PylRS sequences reveals homologs in 24 species including 15 bacteria (Fig. 1A). The gene encoding tRNAPyl (pylT) is present at the 5′ end of the Pyl operon, which is organized differently in archaeal versus bacterial examples (9). Whereas PylRS is encoded by one gene in archaea (pylS), separate pylSn and pylSc genes encode the N-terminal and C-terminal polypeptides of PylRS in bacteria, respectively. Most Pyl-decoding bacteria, except the deepest branching bacterial PylRS from A. arabaticum, encode the N-terminal domain at the 3′ end of the operon. It is still not known how or if these separated domains function in trans. In A. arabaticum, the pylSn is upstream of pylSc (as in archaea) but the two genes are encoded in different reading frames (9). The separated pylSn gene of A. arabaticum may have been inherited or acquired by other bacteria that subsequently rearranged pylSn to the 3′ end of the operon, evolving an operonal organization found in all other known Pyl-decoding bacteria.
Fig. 1.
Phylogenetic distribution of Pyl decoding. (A) To date, the pylRS genes encoding the pyrrolysyl-tRNA synthetase required for pyrrolysine decoding have been found in 24 organisms from archaeal and bacterial domains. Desulfobacterium autotrophicum is not included here because it possesses only a truncated version of pylSc. (B) Genomic arrangement genes required for Pyl decoding; the gene products of pylB, pylC, and pylD biosynthesize pyrrolysine, which is charged onto tRNAPyl (pylT, indicated by the black box) by its cognate pyrrolysyl-tRNA synthetase PylRS.
The bacterial Pyl operon shows additional diversity, in that Pyl operons from A. arabaticum and D. dehalogenans are interrupted by genes for the Pyl-containing trimethylamine methyltransferase (MttB) and its cognate corrinoid protein (MttC) (Fig. 1B). A hydantoinase (HyaA), an arginine pathway regulator (RocR), and a dihydropteroate synthetase (DhpS) are among the gene products encoded within the Pyl operons of bacterial origin. The specific function of these putative proteins and their involvement in the Pyl system remain unknown.
TMA-Dependent Induction of Pyl-tRNAPyl Formation in A. arabaticum.
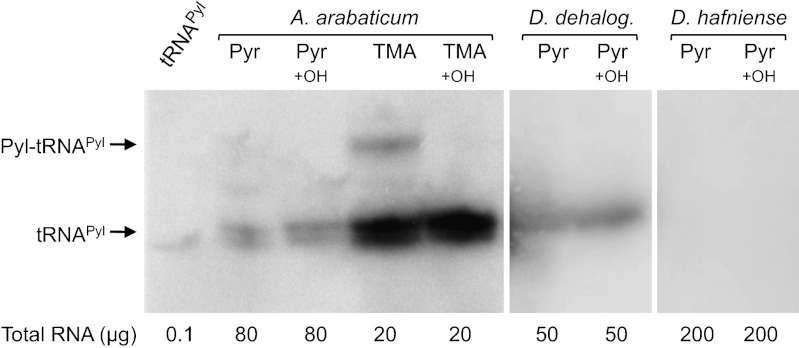
To determine whether tRNAPyl is expressed in putative Pyl-decoding bacteria, total RNA was isolated from cells grown either on pyruvate (Pyr) or trimethylamine (TMA) and analyzed by Northern blot. We focused on three bacterial strains D. hafniense (DCB-2), D. dehalogenans, and A. arabaticum. The Northern blot analysis revealed that Pyl, in the conditions tested, is not encoded in all organisms that contain the Pyl operon in their genome (Fig. 2). We observed that only A. arabaticum and D. dehalogenans expressed tRNAPyl. Interestingly, the level of tRNAPyl expression in A. arabaticum is significantly increased in cells cultivated on TMA. Aminoacylated tRNAPyl was observed in vivo only in A. arabaticum cells cultivated on TMA (Fig. 2). Whereas we found tRNAPyl expression in D. dehalogenans, no expression is seen in D. hafniense. In addition, under the conditions tested, we observed no aminoacyl-tRNAPyl formation in D. dehalogenans or D. hafniense.
Fig. 2.
TMA-dependent production of Pyl-tRNAPyl. Total RNAs were extracted from A. arabaticum cultivated on Pyr or TMA, and from D. dehalogenans and D. hafniense, both cultivated on Pyr in the presence of 5 mM lysine. Total RNAs were extracted and half of each RNA sample was submitted to alkaline treatment to promote deacylation (indicated by +OH). All RNA samples were separated by acid gel electrophoresis, transferred to a nitrocellulose membrane, and hybridized with a DNA probe specific to each bacterial tRNAPyl. In vitro transcribed tRNAPyl from D. hafniense was loaded to serve as control (lane 1).
Given that Pyl-tRNAPyl formation is dependent on the presence of TMA in the growth medium of A. arabaticum, we tested the ability of D. hafniense or D. dehalogenans to use TMA as growth substrate. Despite the presence of genes encoding putative TMA methyltransferases, these strains were unable to grow on TMA. We also surveyed tRNAPyl expression and Pyl-tRNAPyl formation in these organisms in eight different culture conditions (listed in experimental procedures) using a variety of carbon sources (24). Northern blot analysis revealed that D. dehalogenans expresses tRNAPyl but under no growth condition tested was Pyl-tRNAPyl observed (Fig. S1). In D. hafniense, despite encoding an active PylRS/tRNAPyl pair (18), we found no endogenous expression of tRNAPyl under the conditions tested.
Pyl-Protein Expression in A. arabaticum.
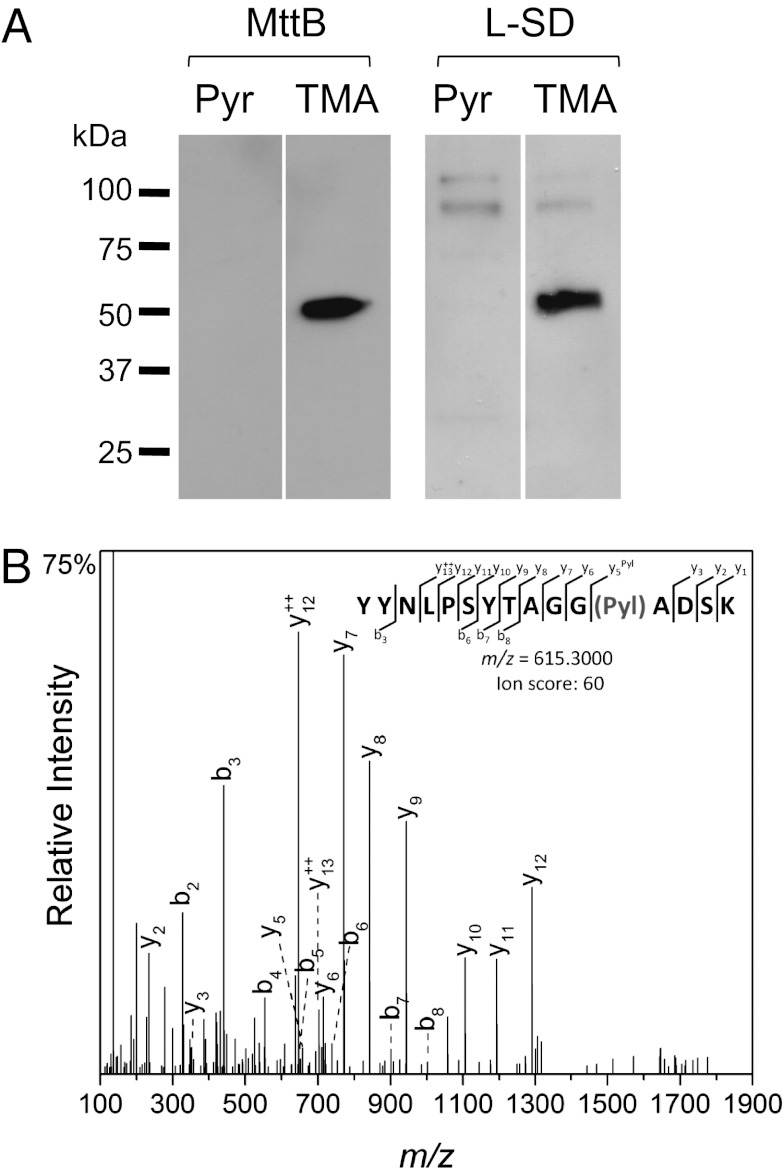
Like its archaeal counterparts, the mttB gene of A. arabaticum contains an in-frame TAG codon. Western blot analysis of crude A. arabaticum cell extract showed that production of full-length MttB was only observed when cells were grown on TMA (Fig. 3A). MS/MS analysis confirmed that a Pyl residue was inserted in response to the UAG codon (Fig. 3B). The expression of an ORF encoding two UAG codons, l-serine dehydratase (l-SD, involved in pyruvate metabolism), was also detected only in the presence of TMA. No truncated version of MttB or l-SD could be found in A. arabaticum cultivated on Pyr. The MS analysis also uncovered expression of a third Pyl-containing protein MtbB, a dimethylamine methyltransferase (Fig. S2).
Fig. 3.
UAG translation in MttB and l-SD in A. arabaticum. (A) Cell extract prepared from A. arabaticum cultivated either on Pyr (lanes 1 and 3) or TMA (lanes 2 and 4) were separated by SDS/PAGE and analyzed by immunoblot using specific antibodies anti–Aar-MttB (lanes 1 and 2) and anti–Aar-l-SD (lanes 3 and 4). (B) Whole cells extracts were separated by SDS/PAGE and the band corresponding to MttB in molecular weight was cut from the gel. Pyrrolysine insertion was detected by mass spectrometry; this graph represents a higher energy collisional dissociation fragment ion spectrum and matched fragment ions showing high scoring peptide identifications: R.YYNLPSYTAGG(Pyl)ADSK.L from MttB.
TMA-Dependent Induction of Genetic Code Expansion.
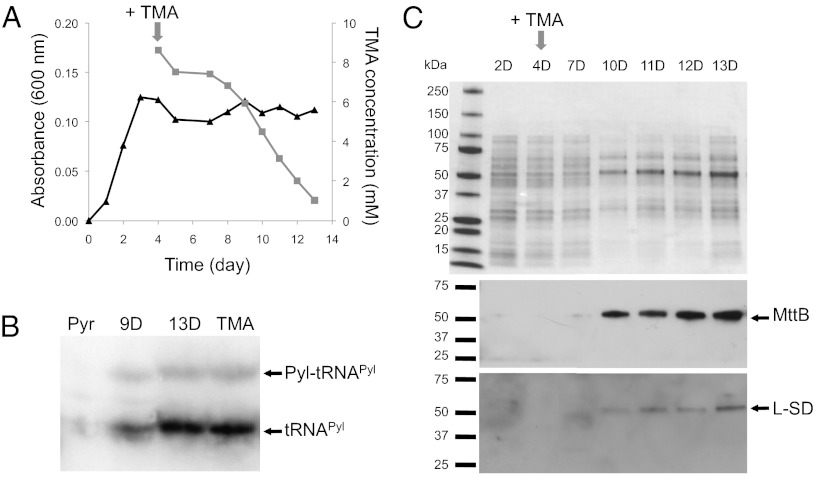
To confirm TMA dependence of Pyl decoding, we followed substrate adaptation in cells actively growing on Pyr then pulsed with TMA. We monitored tRNAPyl and protein expression of MttB and l-SD over a time course of days (Fig. 4). After the addition of TMA, we observed a lag period of ∼4 d before A. arabaticum began to consume TMA (Fig. 4A). The A. arabaticum proteome is qualitatively altered during this adaptation process as we observed on SDS/PAGE from day 2 and day 10 (Fig. 4C). Northern blot analysis showed increasing tRNAPyl expression and Pyl-tRNAPyl formation that correlates with TMA consumption (Fig. 4B). Western blots revealed that MttB protein expression also increased with increasing Pyl-tRNAPyl production (Fig. 4C). l-SD expression followed the same tendency, but with lower expression (Fig. 4C).
Fig. 4.
TMA-dependent induction of Pyl encoding. (A) A. arabaticum cells were grown on Pyr until early stationary phase as measured by A600nm (triangle). TMA then was added to a final concentration of 10 mM and TMA consumption was monitored (square). Data are based on three independent experiments. In the absence of bacterial growth, TMA is stable. Cell samples were taken at several time points and analyzed as described in B and C. (B) Total RNAs were extracted from cells grown on Pyr and TMA and at days 9 and 13 of A. Total RNA was separated by acid gel electrophoresis, transferred to a nitrocellulose membrane, and hybridized with a DNA probe specific to A. arabaticum tRNAPyl. (C) Cell extract prepared from A. arabaticum after a 10-mM TMA pulse were separated by SDS/PAGE and analyzed by immunoblot using specific antibodies anti–Aar-MttB and anti–Aar-l-SD.
Regulation of the Pyl Operon in A. arabaticum and D. dehalogenans.
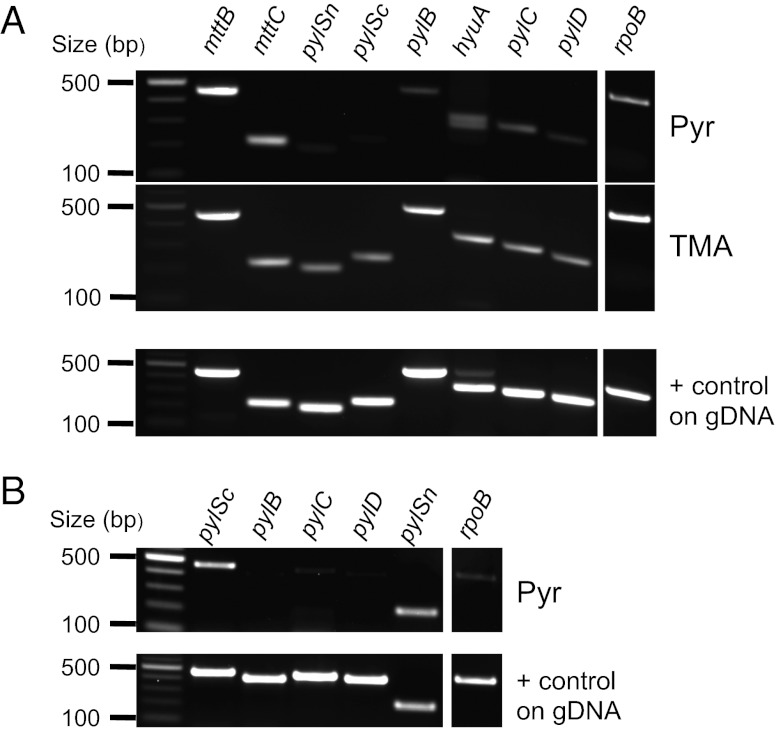
To investigate the transcription of the Pyl operon, we isolated RNA from cells cultivated on TMA or Pyr. RNAs were reverse transcribed and cDNA was amplified using primer pairs specifically targeting each protein encoding gene in the Pyl operon. In A. arabaticum, we observed that pylSn, pylSc, pylB, pylC, and pylD were down-regulated on Pyr compared with TMA (Fig. 5A). Although we see no protein expression of MttB in Pyr grown cells (Fig. 3A), we wondered if transcriptional control is involved. Surprisingly, mttB and mttC transcription was similar under both conditions. Because in the absence of TMA no Pyl-tRNAPyl is made, MttB expression is likely controlled at the translational level. The lack of truncated MttB product despite expression of its mRNA on Pyr may indicate its rapid degradation. In D. dehalogenans cells cultivated on Pyr, transcription of pylBCD is barely detectable, whereas pylSc and pylSn are transcribed (Fig. 5B). The data show that Pyl-tRNAPyl cannot be observed in D. dehalogenans because Pyl is not synthesized under the conditions tested.
Fig. 5.
Expression of the Pyl operon in A. arabaticum and D. dehalogenans. Total RNAs were extracted from cells and reversely transcribed using random hexamer primers. PCR amplifications were then performed from cDNA with primer pairs targeting individual genes as indicated. A positive control of each primer pair was performed on genomic DNA (gDNA). rpoB gene, encoding the beta subunit of the RNA polymerase, was used as internal control for amplification. (A) Gene expression was studied in A. arabaticum cells cultivated in the presence of Pyr or TMA. (B) Gene expression was studied in D. dehalogenans cells cultivated in the presence of Pyr.
Evolution of Pyl-Containing Proteins in Bacteria.
In the 24 Pyl-decoding organisms, a total of 67 methylamine methyltransferase genes (mtxB) are identifiable, among them 35 putative Pyl-containing methyltransferases (Fig. S3). The function of non–Pyl-containing MttB homologs is unknown. The methylamine methyltransferases are substrate specific and different variants MtmB, MtbB, and MttB abstract a methyl group from mono-, di-, and trimethylamine, respectively. Among bacteria, only A. arabaticum encodes all three mtxB genes. The A. arabaticum MtxBs are the deepest branching in their phylogenies and most similar to their archaeal relatives. The arrangement of pylSn and pylSc in A. arabaticum is also most similar to the archaeal versions (Fig. 1).
Given MtmB and MtbB are mostly found in archaea, there is support for the notion that Pyl and the MtxBs were horizontally transferred from archaea to bacteria. The fact that the Pyl cassette is easily transmissible to E. coli also supports this idea (10, 11), and in Fig. S3 we find phylogenetic evidence supporting the horizontal gene transfer scenario. In the MtmB clade, the bacterial examples emerge within the archaeal group, which is the phylogenetic signature of horizontal transfer, and suggests A. arabaticum acquired at least mtmB from archaea.
Although l-SD proteins represent a large family, the enzyme is found only in bacteria with no known Pyl-containing homologs. l-SD proteins are composed of two domains (α and β), encoded either by one “full-length” gene or two adjacent genes, dividing the family in two phylogenetic groups (Fig. S4). In A. arabaticum, Pyl incorporation results in read-through of two UAG codons linking the α and β domain to form a single polypeptide. This is the only characterized example of Pyl read-through linking two otherwise separated protein domains and demonstrates that an expanded genetic code can create new protein products and alter the cellular proteome.
Discussion
Origin of Pyl and TMA Metabolism in Bacteria.
A. arabaticum is a member of the phylum Clostridia as are many of the Pyl-decoding bacteria (25). Given the phylogenetic diversity between A. arabaticum and archaeal methanogens, it is interesting to consider how A. arabaticum and other bacteria acquired the Pyl-decoding trait. A. arabaticum has been found in the marine environment alongside these methanogens including the Pyl-decoding Methanohalobium evestigatum, which is also able to survive on methylamine (26). In this hypersaline environment, A. arabaticum participates in the one-carbon metabolism that is important for methanogenesis from methylamines. A. arabaticum can live as a chemolithotroph with H2 + CO2 or CO, a methylotroph on TMA, or as an organotroph using lactate, pyruvate, or betaine, which is an important osmoregulatory compound for many methanogens. Interestingly, betaine is a metabolic precursor of TMA (27). A. arabaticum may live in syntrophy with Pyl-decoding archaea by converting readily available betaine to TMA, a usable carbon source for methanogens. The proximity in the environment of A. arabaticum with methanogens could have facilitated horizontal transfer of the Pyl operon from archaea to bacteria.
Dynamic Modulation of the Genetic Code.
Previous studies in M. acetivorans showed that pylT deletion inhibits growth on methylamines, but cells are viable in growth on methanol, indicating that Pyl is essential only for methylamine metabolism (12). Pyl can nevertheless be incorporated into proteins with the same efficiency in Methanosarcina cells grown either on TMA or methanol, suggesting that growth substrate does not influence Pyl incorporation in archaea (12, 15).
A. arabaticum is the only organism known to modulate the size of its genetic code in response to its energy and carbon sources. If TMA is unavailable in the environment, descendants of Pyl-decoding A. arabaticum could likely eliminate the Pyl cassette from their genomes. The sparse distribution of the Pyl-decoding trait across the diversity of life (in ∼0.7% of known species) indicates that gene loss is a common fate for the Pyl operon. Thus far, experimentally verified natural synthesis of Pyl proteins is restricted to anaerobic organisms that can metabolize TMA, but the diverse arrangement of Pyl operons among bacteria indicates that some organisms may have found unique applications for Pyl.
Adaptation to an Expanded Genetic Code.
As we found in M. acetivorans (15), Pyl is also present in bacterial proteins that have no apparent relation to methylamine metabolism. In A. arabaticum, for example, we observed Pyl in l-serine dehydratase, representing the only known example of a protein containing two Pyl residues. The fact that Pyl-decoding bacteria like A. arabaticum contain in-frame TAGs in ∼20% of their genes, whereas Pyl-decoding archaea contain in-frame TAGs in only ∼5% of their genes (28), indicates that bacteria and archaea adapted differently to the acquisition of the Pyl-decoding trait (Table S1). Perhaps archaea evolved to minimize the frequency of TAGs so that the Pyl operon can be constitutively expressed. In bacteria, possibly like the examples we found here, the Pyl operon is either silenced or controlled so that Pyl-containing proteins are only expressed when needed. It is possible that the separation of the N- and C-terminal domains in bacterial Pyl operons is an additional way to control Pyl incorporation. In the absence of TMA, it might be detrimental for the cell to encode an exceedingly high number of TAGs as Pyl. It has been shown that when the E. coli genetic code is artificially expanded to encode unnatural or noncanonical amino acids, phenotypic defects can be observed and even exacerbated in release factor RF1 deletion strains, which allow high levels of UAG read-through that alter the proteome (29). How release factor variants in Methanosarcina and Acetohalobium differ from those of E. coli to allow efficient UAG suppression is still an open topic (30).
The gradual induction of Pyl-tRNAPyl formation we observed over a period of days following a pulse with TMA, suggests a positive feedback loop involving TMA consumption, Pyl-tRNAPyl formation, subsequent Pyl-protein expression, and the resulting methyltransferase activity. Because mRNA expression levels of the entire Pyl operon in A. arabaticum control Pyl-tRNAPyl formation, a potential feedback loop could be controlled at the transcriptional level. Whether TMA or an MttB reaction product stimulates feedback remains to be seen.
In Sec-decoding organisms, adaptation to an expanded code is completely distinct because only particular codons are recoded by the Sec insertion sequence (SECIS) element in the mRNA. This regulated recoding allows constitutive formation of Sec-tRNASec without disrupting the proteome by reading through UGA codons intended to stop translation. In bacteria and archaea, Sec encoding is not essential for survival, but formate-dependent growth in Methanococcus maripaludis and E. coli requires Sec insertion into formate dehydrogenase (31, 32). Fascinatingly, whereas Pyl-protein expression in A. arabaticum depends on the presence of TMA, selenoproteins can be overproduced in both bacteria and archaea even when formate is not present in the medium (33). Consequently, in contrast to Pyl decoding in bacteria, Sec encoding does not depend on the carbon or energy source. Nevertheless, selenoprotein production can be regulated in response to Se availability. In human cells when insufficient Se is present, nonessential (stress-related) selenoproteins are down-regulated as a result of deficient methylation of Sec-tRNASec at U34, but the expression of the essential (housekeeping) selenoproteins is maintained by scavenging the available Se (34).
Conclusion
The existence of a carbon-source–dependent genetically encoded amino acid indicates that the genetic code within a single cell can be more dynamic than previously recognized. In archaeal and bacterial Sec-decoding species, Sec-tRNASec is synthesized even in conditions where selenoproteins are nonessential. Despite the fact that no Sec-decoding organisms are known to turn off Sec encoding, it is theoretically possible for these organisms to silence Sec-tRNASec formation. The dynamic nature of the A. arabaticum genetic code also suggests that canonical amino acids might be similarly regulated. Although there are no reports showing that an organism can survive with fewer than 20 amino acids, it has been shown that cells can tolerate mistranslation due to editing-defective tRNA synthetase mutants, a coping mechanism that is advantageous under certain conditions such as amino acid starvation (35–37). Deciphering the transcription factors that regulate Pyl encoding in A. arabaticum is a subject for future work, which will provide further insight into the evolution of the genetic code. By engineering cell survival to depend on essential genes mutated to contain Pyl, the Pyl regulatory system of A. arabaticum can have applications in biological containment of modified or hazardous microbial strains.
Experimental Procedures
Bacterial Strains and Media.
A. arabaticum DSM 5501 was cultivated anaerobically in medium 494 recommended by the Deutsche Sammlung für Mikroorganismen (DSMZ), with an increased NaCl concentration of 3 M. As growth substrate, 25 mM of Pyr or 25 mM TMA supplemented with 2.5 mM acetate as carbon source were used. TMA concentration was measured in the medium by using the Folin-Ciocalteu phenol reagent as described in ref. 38. D. hafniense strain DCB-2 and D. dehalogenans strain JW/IU-DC1 were cultivated anaerobically in medium 717 recommended by the DSMZ with few modifications as described in ref. 39. The strains were grown in the presence of 25 mM pyruvate and also tested with the following combinations of electron donors and acceptors at a final concentration of 25 mM: lactate/fumarate, lactate/thiosulfate, vanillate/fumarate, formate/fumarate, TMA/fumarate, or lactate/TMA.
Cloning, Protein Purification, and Specific Antibody Production.
For antibody production, the N-terminal domain of A. arabaticum l-SD (acear_1197) was cloned between BamHI and NotI into pCDFDuet vector (Novagen) with N-terminal His-tag and transformed into E. coli BL21 (New England Biolabs). A 3-L culture was grown to midlog phase and l-SD expression was induced with 100 mM isopropyl β-d-1-thiogalactopyranoside. Protein was purified by gravity-flow chromatography using Nickel-nitrilotriacetic acid affinity resin (Qiagen). From the purified protein, Cocalico Biologicals generated specific anti–Aar-l-SD rabbit antibodies. For A. arabaticum MttB, an N-terminal peptide (GGDSPDANIDLHDA, position 152–165) predicted to be at the protein surface, was synthetized by Genscript, and injected into a rabbit for specific anti–Aar-MttB antibody production.
Crude Cell Extract Preparation, SDS/PAGE, and Western Blot Analysis.
A. arabaticum cells were harvested by centrifugation at 4 °C and 4,000 g for 10 min. Cells were lysed by osmotic shock in 50 mM Tris buffer, pH 7.5. Protein concentrations were measured by Bradford protein assay (Bio-Rad). Five micrograms of total protein was separated by SDS/PAGE and stained with R-250 Coomassie Blue solution. For Western blot analysis of l-SD and MttB, 500 ng or 50 ng of total protein extract were separated by SDS/PAGE. Proteins were then transferred onto a PVDF membrane. MttB and l-SD were detected by using the respective anti–Aar-MttB and anti–Aar-l-SD antibodies. Western blots shown are representative of at least three independent experiments.
Mass Spectrometric Analysis.
Extracts from A. arabaticum cells cultivated on TMA were treated with sodium cyanoborohydride and fractionated by SDS/PAGE. The band corresponding to the molecular weight of MttB was cut from the gel and subjected to in-gel digestion with trypsin. Capillary LC-MS/MS was carried out on a Orbitrap Velos (ThermoFisher Scientific) using 2 µg digest as described earlier (29). Raw data were processed with MASCOT distiller and searched with MASCOT (version 2.4.0) using a custom database containing 460 proteins from A. arabaticum. The database was modified with the amino acid K at the expected Pyl insertion site. This enables the detection of Pyl-containing peptides by introducing custom modifications for K corresponding to Pyl, hydrolyzed Pyl (PylH2O), and hydrolyzed reduced Pyl (PylH2Ored). Database searches were carried out with full trypsin enzyme specificity and allowing up to two missed cleavages. The precursor ion mass tolerance was set to ±30 ppm for the precursor and ±0.2 Da for fragment ions, respectively. Fixed modifications were propionamide (C), oxidation (M), deamidation (NQ), and the custom modifications for Pyl as described above. Only peptides with an ion score >25, indicating identity or extensive homology, were considered for analysis.
Acid Urea Gel and Northern Blot Analysis.
A. arabaticum, D. hafniense, and D. dehalogenans cells were harvested in exponential phase by centrifugation at 4 °C and 4,000 × g for 10 min, and they were directly subjected to RNA extraction. RNA extractions and acid urea gels were performed according to ref. 40. A total of 20–200 μg of total RNA was gel separated and subsequently transferred onto Hybond-N+ membrane (GE Healthcare). RNAs were cross-linked to the membrane by UV pulse. Membranes were incubated with ULTRAhyb Ultrasensitive buffer (Ambion) for prehybridization at 42 °C for 30 min. DNA oligonucleotides complementary to the acceptor stem and the TψC loop of the respective tRNAPyl were used as probes with a length of 25–26 nucleotides: A. arabaticum tRNAPyl (5′-GGGTTAGAGCCTATGTGATCTTTCCG-3′), D. dehalogenans and D. hafniense tRNAPyl (5′-CGGGGAGTACGGGAGTTTCACCCGC-3′). Probes were 5′ end labeled with [γ-32P]ATP (40) and added to the ULTRAhyb Ultrasensitive buffer (106 cpm/mL buffer) and hybridized overnight at 42 °C. Then the membrane was washed with 2× SSC, 0.1% SDS buffer for 20 min at 42 °C, followed by a wash with 0.1× SSC, 0.1% SDS buffer for 20 min at 42 °C. The Northern blots shown were visualized by PhosphorImaging and are representative of at least two independent experiments.
RNA Extraction, Reverse Transcription, and PCR on cDNA.
A. arabaticum and D. dehalogenans cells were harvested in exponential phase by centrifugation at 4 °C and 4,000 g for 10 min and RNA was extracted by using TRIzol as described in ref. 41. Reverse transcription was performed on 1 μg of RNA mixed with 200 ng of random hexamers, in the presence of the SuperScript II Reverse Transcriptase (Invitrogen) according to the manufacturer’s instructions. Gene specific amplifications using forward and reverse primers (listed in Table S2) were carried out with Taq DNA polymerase (New England BioLabs). PCR products were separated and visualized by 1.5% agarose gel electrophoresis.
Phylogenetic Analysis.
PylRS, methylamine methyltransferase, and l-serine dehydratase sequences were downloaded from the National Center for Biotechnology Information and from the Integrated Microbial Genomes at the Doe Joint Genome Institute (42). ClustalX was used for sequence alignment (43). The phylogenetic trees were generated with MEGA5 (44).
Supplementary Material
Acknowledgments
We thank Chris Cho, Michael Hohn, Jiqiang Ling, Yuchen Liu, Julien Maillard, and Noah Reynolds for inspired discussions. I.U.H. was a Postdoctoral Fellow of the Deutsche Forschungsgemeinschaft (HE5802/1-1). This work was supported by grants from the National Institute of General Medical Sciences and the National Science Foundation (to D.S.), and National Institute of Diabetes and Digestive and Kidney Diseases Grant K01DK089006 (to J.R.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218613110/-/DCSupplemental.
References
- 1.Ambrogelly A, Palioura S, Söll D. Natural expansion of the genetic code. Nat Chem Biol. 2007;3(1):29–35. doi: 10.1038/nchembio847. [DOI] [PubMed] [Google Scholar]
- 2.Lobanov AV, Turanov AA, Hatfield DL, Gladyshev VN. Dual functions of codons in the genetic code. Crit Rev Biochem Mol Biol. 2010;45(4):257–265. doi: 10.3109/10409231003786094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaston MA, Jiang R, Krzycki JA. Functional context, biosynthesis, and genetic encoding of pyrrolysine. Curr Opin Microbiol. 2011;14(3):342–349. doi: 10.1016/j.mib.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hao B, et al. A new UAG-encoded residue in the structure of a methanogen methyltransferase. Science. 2002;296(5572):1462–1466. doi: 10.1126/science.1069556. [DOI] [PubMed] [Google Scholar]
- 5.James CM, Ferguson TK, Leykam JF, Krzycki JA. The amber codon in the gene encoding the monomethylamine methyltransferase isolated from Methanosarcina barkeri is translated as a sense codon. J Biol Chem. 2001;276(36):34252–34258. doi: 10.1074/jbc.M102929200. [DOI] [PubMed] [Google Scholar]
- 6.Srinivasan G, James CM, Krzycki JA. Pyrrolysine encoded by UAG in Archaea: Charging of a UAG-decoding specialized tRNA. Science. 2002;296(5572):1459–1462. doi: 10.1126/science.1069588. [DOI] [PubMed] [Google Scholar]
- 7.Blight SK, et al. Direct charging of tRNA(CUA) with pyrrolysine in vitro and in vivo. Nature. 2004;431(7006):333–335. doi: 10.1038/nature02895. [DOI] [PubMed] [Google Scholar]
- 8.Polycarpo C, et al. An aminoacyl-tRNA synthetase that specifically activates pyrrolysine. Proc Natl Acad Sci USA. 2004;101(34):12450–12454. doi: 10.1073/pnas.0405362101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaston MA, Zhang L, Green-Church KB, Krzycki JA. The complete biosynthesis of the genetically encoded amino acid pyrrolysine from lysine. Nature. 2011;471(7340):647–650. doi: 10.1038/nature09918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Longstaff DG, et al. A natural genetic code expansion cassette enables transmissible biosynthesis and genetic encoding of pyrrolysine. Proc Natl Acad Sci USA. 2007;104(3):1021–1026. doi: 10.1073/pnas.0610294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Namy O, et al. Adding pyrrolysine to the Escherichia coli genetic code. FEBS Lett. 2007;581(27):5282–5288. doi: 10.1016/j.febslet.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 12.Mahapatra A, et al. Characterization of a Methanosarcina acetivorans mutant unable to translate UAG as pyrrolysine. Mol Microbiol. 2006;59(1):56–66. doi: 10.1111/j.1365-2958.2005.04927.x. [DOI] [PubMed] [Google Scholar]
- 13.Bose A, Pritchett MA, Rother M, Metcalf WW. Differential regulation of the three methanol methyltransferase isozymes in Methanosarcina acetivorans C2A. J Bacteriol. 2006;188(20):7274–7283. doi: 10.1128/JB.00535-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Longstaff DG, Blight SK, Zhang L, Green-Church KB, Krzycki JA. In vivo contextual requirements for UAG translation as pyrrolysine. Mol Microbiol. 2007;63(1):229–241. doi: 10.1111/j.1365-2958.2006.05500.x. [DOI] [PubMed] [Google Scholar]
- 15.Heinemann IU, et al. The appearance of pyrrolysine in tRNAHis guanylyltransferase by neutral evolution. Proc Natl Acad Sci USA. 2009;106(50):21103–21108. doi: 10.1073/pnas.0912072106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Veit K, Ehlers C, Schmitz RA. Effects of nitrogen and carbon sources on transcription of soluble methyltransferases in Methanosarcina mazei strain Go1. J Bacteriol. 2005;187(17):6147–6154. doi: 10.1128/JB.187.17.6147-6154.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smidt H, van der Oost J, de Vos WM. Development of a gene cloning and inactivation system for halorespiring Desulfitobacterium dehalogenans. Appl Environ Microbiol. 2001;67(2):591–597. doi: 10.1128/AEM.67.2.591-597.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herring S, Ambrogelly A, Polycarpo CR, Söll D. Recognition of pyrrolysine tRNA by the Desulfitobacterium hafniense pyrrolysyl-tRNA synthetase. Nucleic Acids Res. 2007;35(4):1270–1278. doi: 10.1093/nar/gkl1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ambrogelly A, et al. Pyrrolysine is not hardwired for cotranslational insertion at UAG codons. Proc Natl Acad Sci USA. 2007;104(9):3141–3146. doi: 10.1073/pnas.0611634104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kavran JM, et al. Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation. Proc Natl Acad Sci USA. 2007;104(27):11268–11273. doi: 10.1073/pnas.0704769104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nozawa K, et al. Pyrrolysyl-tRNA synthetase-tRNA(Pyl) structure reveals the molecular basis of orthogonality. Nature. 2009;457(7233):1163–1167. doi: 10.1038/nature07611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu Rev Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 23.Neumann H. Rewiring translation: Genetic code expansion and its applications. FEBS Lett. 2012;586(15):2057–2064. doi: 10.1016/j.febslet.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 24.Villemur R, Lanthier M, Beaudet R, Lépine F. The Desulfitobacterium genus. FEMS Microbiol Rev. 2006;30(5):706–733. doi: 10.1111/j.1574-6976.2006.00029.x. [DOI] [PubMed] [Google Scholar]
- 25.Sikorski J, et al. Complete genome sequence of Acetohalobium arabaticum type strain (Z-7288) Stand Genomic Sci. 2010;3(1):57–65. doi: 10.4056/sigs.1062906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhilina NT, Zavarzin GA. Extremely halophilic, methylotrophic, anaerobic bacteria. FEMS Microbiol Lett. 1990;87:315–321. [Google Scholar]
- 27.King GM. Methanogenesis from methylated amines in a hypersaline agal mat. Appl Environ Microbiol. 1988;54(1):130–136. doi: 10.1128/aem.54.1.130-136.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Baranov PV, Atkins JF, Gladyshev VN. Pyrrolysine and selenocysteine use dissimilar decoding strategies. J Biol Chem. 2005;280(21):20740–20751. doi: 10.1074/jbc.M501458200. [DOI] [PubMed] [Google Scholar]
- 29.Heinemann IU, et al. Enhanced phosphoserine insertion during Escherichia coli protein synthesis via partial UAG codon reassignment and release factor 1 deletion. FEBS Lett. 2012;586(20):3716–3722. doi: 10.1016/j.febslet.2012.1008.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alkalaeva E, et al. Translation termination in pyrrolysine-utilizing archaea. FEBS Lett. 2009;583(21):3455–3460. doi: 10.1016/j.febslet.2009.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hohn MJ, Palioura S, Su D, Yuan J, Söll D. Genetic analysis of selenocysteine biosynthesis in the archaeon Methanococcus maripaludis. Mol Microbiol. 2011;81(1):249–258. doi: 10.1111/j.1365-2958.2011.07690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sawers G, Heider J, Zehelein E, Böck A. Expression and operon structure of the sel genes of Escherichia coli and identification of a third selenium-containing formate dehydrogenase isoenzyme. J Bacteriol. 1991;173(16):4983–4993. doi: 10.1128/jb.173.16.4983-4993.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mansell JB, Guévremont D, Poole ES, Tate WP. A dynamic competition between release factor 2 and the tRNA(Sec) decoding UGA at the recoding site of Escherichia coli formate dehydrogenase H. EMBO J. 2001;20(24):7284–7293. doi: 10.1093/emboj/20.24.7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carlson BA, Yoo MH, Tsuji PA, Gladyshev VN, Hatfield DL. Mouse models targeting selenocysteine tRNA expression for elucidating the role of selenoproteins in health and development. Molecules. 2009;14(9):3509–3527. doi: 10.3390/molecules14093509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li L, et al. Naturally occurring aminoacyl-tRNA synthetases editing-domain mutations that cause mistranslation in Mycoplasma parasites. Proc Natl Acad Sci USA. 2011;108(23):9378–9383. doi: 10.1073/pnas.1016460108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pezo V, et al. Artificially ambiguous genetic code confers growth yield advantage. Proc Natl Acad Sci USA. 2004;101(23):8593–8597. doi: 10.1073/pnas.0402893101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruan B, et al. Quality control despite mistranslation caused by an ambiguous genetic code. Proc Natl Acad Sci USA. 2008;105(43):16502–16507. doi: 10.1073/pnas.0809179105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ikawa M, Schaper TD, Dollard CA, Sasner JJ. Utilization of Folin-Ciocalteu phenol reagent for the detection of certain nitrogen compounds. J Agric Food Chem. 2003;51(7):1811–1815. doi: 10.1021/jf021099r. [DOI] [PubMed] [Google Scholar]
- 39.Prat L, Maillard J, Grimaud R, Holliger C. Physiological adaptation of Desulfitobacterium hafniense strain TCE1 to tetrachloroethene respiration. Appl Environ Microbiol. 2011;77(11):3853–3859. doi: 10.1128/AEM.02471-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Köhrer C, Rajbhandary UL. The many applications of acid urea polyacrylamide gel electrophoresis to studies of tRNAs and aminoacyl-tRNA synthetases. Methods. 2008;44(2):129–138. doi: 10.1016/j.ymeth.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prat L, Maillard J, Rohrbach-Brandt E, Holliger C. An unusual tandem-domain rhodanese harbouring two active sites identified in Desulfitobacterium hafniense. FEBS J. 2012;279(15):2754–2767. doi: 10.1111/j.1742-4658.2012.08660.x. [DOI] [PubMed] [Google Scholar]
- 42.Markowitz VM, Kyrpides NC. Comparative genome analysis in the integrated microbial genomes (IMG) system. Methods Mol Biol. 2007;395:35–56. doi: 10.1007/978-1-59745-514-5_3. [DOI] [PubMed] [Google Scholar]
- 43.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 44.Tamura K, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.