Abstract
The closely linked human protocadherin (Pcdh) α, β, and γ gene clusters encode 53 distinct protein isoforms, which are expressed in a combinatorial manner to generate enormous diversity on the surface of individual neurons. This diversity is a consequence of stochastic promoter choice and alternative pre-mRNA processing. Here, we show that Pcdhα promoter choice is achieved by DNA looping between two downstream transcriptional enhancers and individual promoters driving the expression of alternate Pcdhα isoforms. In addition, we show that this DNA looping requires specific binding of the CTCF/cohesin complex to two symmetrically aligned binding sites in both the transcriptionally active promoters and in one of the enhancers. These findings have important implications regarding enhancer/promoter interactions in the generation of complex Pcdh cell surface codes for the establishment of neuronal identity and self-avoidance in individual neurons.
Keywords: chromatin interaction, gene regulation, epigenetic control, DNA methylation, transcriptional hub
The clustered protocadherin (Pcdh) genes are expressed in the nervous system and organized into three closely linked clusters (α, β, and γ) (1–5). The human Pcdhα gene cluster contains 13 highly similar variable first exons (α1 to α13) arrayed in tandem and two more distantly related c-type variable first exons designated αc1 and αc2 (Fig. 1A). The variable first exons encode the extracellular, transmembrane, and juxtamembrane intracellular domains of the Pcdhα proteins. Each of these 15 variable first exons is cis-spliced to a single set of three downstream constant exons that encode a distal intracellular domain (1–3). The human β cluster is located downstream from the α cluster and contains a tandem array of 16 highly similar variable exons but with no constant exons, whereas the γ cluster contains 22 variable first exons arrayed in tandem and divided into three types (γa1 to γa12, γb1 to γb7, and γc3 to γc5) (Fig. 1D). As in the case of the α cluster, each of these 22 γ variable first exons is cis-spliced to a single set of three downstream constant exons, which are distinct from the α constant exons, to generate diverse γ mRNAs (1, 3, 6). Analyses of the α and γ transcripts have revealed that highly similar Pcdh alternate isoforms are expressed in a stochastic fashion, whereas all of the c-type divergent isoforms, αc1 and αc2 in the α cluster and γc3, γc4, and γc5 in the γ cluster, are expressed ubiquitously in all cells (1–3, 5, 7). Hereafter, we refer to the c-type genes as “ubiquitously expressed” in contrast to the “alternately expressed” Pcdh genes (Fig. 1 A and D). A combination of stochastic activation of alternate promoters and constitutive activation of c-type ubiquitous promoters generates enormous single-cell diversity on the surface of individual neurons.
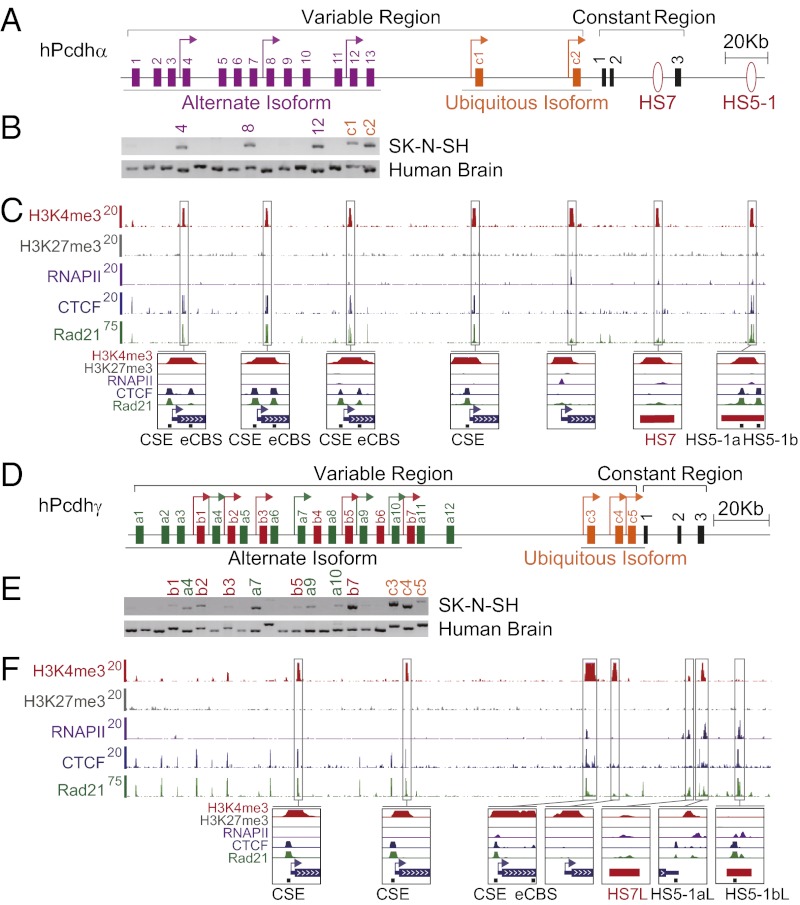
Fig. 1.
SK-N-SH cells as a model system for studying Pcdh promoter choice. (A) Organization of the human Pcdhα cluster. The variable regions encode 13 alternate isoforms (purple) and 2 c-type ubiquitous isoforms (yellow). (B) Expression patterns of three alternate isoforms (α4, α8, and α12) and the two c-type ubiquitous isoforms (αc1 and αc2) in SK-N-SH cells. The location of chromatin marks (H3K4me3 and H3K27me3), and binding of RNAPII, CTCF, and Rad21 in the Pcdh α (C) and γ (F) clusters in SK-N-SH cells. (D) Organization of the human Pcdhγ cluster. The variable region encodes 19 alternate isoforms, which are subdivided into two groups of 12 a-type (γa1 to γa12) (green), 7 b-type (γb1 to γb7) (red), and 3 c-type ubiquitous isoforms (γc3 to γc5) (yellow). (E) Expression patterns of alternate isoforms (γb1, γa4, γb2, γb3, γa7, γb5, γa9, γa10, and γb7) and the three c-type ubiquitous isoforms (γc3, γc4, and γc5) in SK-N-SH cells. Arrow, active promoter.
Significant advances have been made in understanding the mechanisms by which individual neurons express distinct combinations of the clustered Pcdh genes (2, 3, 8–10). Two long-range cis-regulatory elements in the α cluster, HS5-1 and HS7 (hypersensitive sites 5-1 and 7), function as developmental and tissue-specific transcriptional enhancers required for maximal levels of α gene expression (8). HS5-1, which contains two CTCF-binding sites (referred as HS5-1a and HS5-1b), is required for maximal levels of the α6 to α12 and αc1 expression but displays only a moderate role in the regulation of the α1 to α5 genes and no role in αc2 expression (8, 9, 11). In contrast, HS7 regulates gene expression of almost all members of the α cluster, including the αc2 gene (8, 9). Here, we make use of the diploid neuroblastoma cell line SK-N-SH to discover that the active Pcdh promoters and enhancers directly interact with each other through DNA looping, and that this interaction requires CTCF and cohesin. We conclude that CTCF- and cohesin-dependent enhancer/promoter-looping interactions play a central role in the promoter choice required to generate enormous cell surface diversity of Pcdh proteins.
Results
Model Cell Line for Studying Pcdh Promoter Choice.
The complex nature of the single-cell combinatorial expression of the clustered Pcdh genes precludes the possibility of studying the detailed mechanisms of promoter choice in the mammalian brain. Because each cell appears to express a distinct combination of Pcdh isoforms (3, 5–7), it is not possible to correlate the activity of individual alternate isoform promoters with specific protein binding or to examine possible enhancer/promoter interactions in vivo. This problem is made simpler by using cell lines, but most lines are polyploid and, therefore, pose an intermediate level of complexity. However, we find that the human neuroblastoma cell line SK-N-SH is diploid (Fig. S1) (12) and that the pattern of Pcdh expression is what would be expected from a single neuron (6, 7).
As shown in Fig. 1, SK-N-SH cells express a small subset of Pcdhα alternate isoforms (α4, α8, α12) and the ubiquitous isoforms (αc1, αc2) (Fig. 1 A and B). In addition, SK-N-SH cells also express a subset of Pcdhγ alternate isoforms (γb1, γa4, γb2, γb3, γa7, γb5, γa9, γa10, γb7) and the ubiquitous isoforms (γc3, γc4, γc5) (Fig. 1 D and E). This relatively simple pattern of expression is reminiscent of that observed with individual Purkinje cells, which express a small subset of alternate isoforms and the ubiquitous isoforms (6, 7). It appears that the promoter choice made in this clonal line is epigenetically stable, because the same pattern of active isoforms is consistently observed. We have, therefore, used this line to carry out detailed chromatin immunoprecipitation sequencing (ChIP-seq) and in vitro DNA-binding studies, recognizing that the results may not fully reflect the complexity of Pcdh gene regulation in the brain.
RNA Polymerase Binding and Chromatin Marks in SK-N-SH Cells.
To study potential enhancer/promoter interactions in SK-N-SH cells, we characterized specific protein–DNA interactions within the α, β, and γ clusters by ChIP-seq analyses. First, we mapped the active (H3K4me3) and repressive (H3K27me3) histone methylation marks throughout the Pcdh gene clusters. We found that each of the active α promoters and the HS7 and HS5-1 enhancers are marked by H3K4me3 (Fig. 1C). By contrast, neither the active nor inactive promoters nor the enhancers of the Pcdh clusters were marked by the repressive H3K27me3 histone modification (Fig. 1 C and F and Fig. S2A). As a positive control for the antibodies and ChIP-seq protocol, we showed that the promoter of the nonclustered Pcdh1 gene is marked by both H3K27me3 and H3K4me3 (Fig. S2C).
ChIP-seq experiments using a specific antibody against RNA polymerase II (RNAPII) revealed that RNAPII is enriched at the promoter regions of αc2 and γc3 (Fig. 1 C and F), two isoforms that are expressed at high levels in the brain (1, 5). We presume that RNAPII is not detected on the alternate promoters because of the relative low levels of expression compared with the ubiquitously expressed promoters. RNAPII is also enriched at H3K4me3-marked enhancer sites of HS7 and HS5-1 of the α cluster. It is important to note that we find similar enhancer-like sites in the human γ cluster, designated as HS7-like (HS7L), HS5-1a–like (HS5-1aL), and HS5-1b–like (HS5-1bL) [corresponding to the mouse HS17 site (11)], which are enriched in RNAPII and/or H3K4me3 (Fig. 1F). Interestingly, we also identified spliced transcripts that are complementary to the HS7 (AK136271, AK042845), HS7L (DA549378, CF593989), HS5-1aL (DA087514, AI311853), and HS5-1bL (AK094264) sites (all accession numbers from Genbank), suggesting that these enhancers have promoter activities (13). Finally, we note that these α and γ enhancer sites are DNase I–hypersensitive in the Encyclopedia of DNA Elements (ENCODE) database, which is a characteristic of transcriptional enhancers (14).
CTCF and Rad21 Binding Correlates with Promoter Activity.
To investigate the relationship between the level of gene expression of the human α, β, and γ clusters and the binding of CTCF and cohesin subunit Rad21, we examined their binding profiles by ChIP-seq and ChIP–quantitative PCR (ChIP-qPCR) experiments in SK-N-SH cells (Fig. 1 C and F and Fig. S2 A and B). Previous studies of the mouse Pcdhα cluster identified two CTCF-binding sites in the alternate α isoforms, one in the promoter and a second within the downstream exon (10). We observed the same binding pattern in the human α cluster. As shown in Fig. 1C, CTCF binds to both the promoter conserved sequence element (CSE) and the exonic CTCF-binding site (eCBS) in the α4, α8, and α12 genes. This binding profile directly correlates with the high levels of expression of α4, α8, and α12 in SK-N-SH cells (Fig. 1B).
In the case of the human β and γ clusters, the binding of CTCF to CSE correlates with the high expression levels of respective isoform promoters (Fig. 1F and Fig. S2A). We do not observe a corresponding eCBS in the members of the β or γ clusters. However, we observed a second site in the ubiquitous γc3 gene that binds to high levels of CTCF (Fig. 1F). The binding profiles of Rad21 in the α, β, and γ clusters are similar to that of CTCF, i.e., Rad21 binds to two sites in active alternate promoters of the α cluster but only one site in the promoters of the β and γ clusters (Fig. 1 C and F and Fig. S2A).
Binding of CTCF and Rad21 to c-Type Promoters.
As was shown in mouse cells, CTCF binds to the CSE of αc1 (Fig. 1C). Thus, in contrast to the alternate isoform promoters, which have two CTCF-binding sites, the ubiquitous αc1 isoform contains only one. In both the mouse and human α clusters, αc2 does not have a CSE and does not bind to CTCF. Thus, both of the constitutively expressed α ubiquitous isoform promoters can be distinguished from the alternate isoform promoters by their interactions with CTCF.
CTCF binds to two sites within the HS5-1 enhancer (Fig. 1C). Reminiscent of the two HS5-1 CTCF sites downstream of the α cluster, we observed high levels of CTCF binding to HS5-1aL and HS5-1bL, downstream of the γ cluster (Fig. 1F). ChIP-seq experiments with SK-N-SH cells revealed that Rad21 is enriched at the ubiquitously expressed αc1 promoter as well as at the enhancer sites HS5-1a and HS5-1b, possibly recruited by the bound CTCF (Fig. 1C). In addition, Rad21 is enriched less so at the ubiquitously expressed αc2 promoter and the enhancer HS7 (Fig. 1C). It is important to note that CTCF does not bind to these two sites, so Rad21 binding at these sites is CTCF-independent.
We also observed Rad21 enrichments in the Pcdhγ cluster at γc3, HS5-1aL, and HS5-1bL in SK-N-SH cells (Fig. 1F), possibly also recruited by the bound CTCF. In addition, Rad21 is enriched to a lesser extent at the promoter regions of γc4 and γc5, as well as HS7L. Interestingly, the location of HS7L in the γ constant regions corresponds to the position of HS7 in the α constant region (Fig. 1F). These CTCF and Rad21-binding patterns strongly suggest that regulatory mechanisms of cell-specific Pcdh gene expression are similar for the α and γ clusters.
In Vitro Binding of Recombinant CTCF to Pcdh Promoters and Enhancers.
To confirm the ChIP-seq data and examine the sequence requirements for CTCF binding, we carried out in vitro DNA-binding experiments with recombinant CTCF. We performed EMSA experiments using recombinant full-length human CTCF proteins and a series of probes each containing the CSE of human α1, α4, α5, α8, α12, αc1, αc2, β1, β3, β9, β15, γa2, γb1, γb5, γa10, γb7, γc3, αc4, and γc5 genes (Fig. S3A). In contrast to a previous report (15), we observed a specific binding of full-length CTCF to every probe except that of αc2, β1, γc4, and γc5 (Fig. S3A), which is consistent with motif predictions (2). The specificity of direct CTCF binding to each probe was confirmed by the detection of a supershifted complex using a CTCF antibody (Fig. S3A). In addition, mutations in either the CGCTG core sequences within the CSE or within the immediate upstream 5 nt abolished the retarded gel-shift complex (Fig. S3B). These data demonstrate that both the CGCTG core sequences and the immediate upstream 5 nt, which correspond to the conserved CCCTC motif within the CTCF consensus (16), are essential for specific binding of CTCF to the CSE.
As shown in Fig. S4A, exonic CTCF-binding sites can be identified in the human α2 to α13 but not α1 genes. EMSA experiments with the recombinant CTCF revealed a shifted complex and specific supershifted band for the eCBS of α4, α5, α8, and α12 but not α1 (Fig. S3A). Moreover, mutation of the eCBS CTCF site abolished the gel-shift complex (Fig. S4B). Thus, CTCF binds directly to two sites in each of human alternate promoters α2 to α13 but only to one site of CSE in the human α1 promoter, consistent with recent findings of two CTCF sites at the 5′ end of each mouse alternate α isoform by ChIP-seq studies (10).
We next carried out EMSA experiments with the HS5-1a and HS5-1b sites, confirming that CTCF binds to both sites within the HS5-1 enhancer (Fig. S3A). Furthermore, mutations of the core consensus for each of these two sites abolish the CTCF binding (Fig. S4 C and D). In addition, we observed a HS5-1a–like CTCF-binding site (HS5-1aL) in the 3′ UTR of the γ constant region (Fig. 1F). EMSA experiments showed the gel-shifted and supershifted complex with the HS5-1aL probe, demonstrating the specific binding of CTCF to HS5-1aL in vitro (Fig. S3A). In addition, mutation of the core CTCF consensus within HS5-1aL also abolishes the CTCF binding in vitro (Fig. S4 E and F). Finally, mutations of the CGCTG or CCCTC core sequences, which disrupt CTCF binding to CSE, result in a significant decrease of the promoter activity for α1, α4, α8, α12, αc1, β3, γb1, γb5, γa10, and γb7 in a luciferase reporter assay (Fig. S5). In summary, we show that CTCF binds directly to a repertoire of regulatory sequences within the Pcdh locus in vitro and in vivo and that this binding correlates with Pcdh gene expression.
Methylation of CSE DNA Blocks CTCF Binding in Vitro.
DNA methylation in the Pcdh promoter regions was shown previously to correlate with their activities (3, 17, 18). The core sequences within CSE contain a CpG dinucleotide (2). Recent studies suggested that binding of CTCF to its target site is insensitive to methylation (15); however, CpG methylation has been shown to play an essential role in CTCF-dependent allele-specific expression regulation of imprinted genes (19, 20).
To investigate whether methylation of CpG within the CSE influences CTCF binding, we prepared probes with methylated CpG of both strands for α8, β3, γa2, and γb1, each representing the α, β, γa, and γb groups, respectively. EMSA experiments show that CpG methylation results in a dramatic decrease or complete absence of the CTCF binding (Fig. 2 A and B). In addition, we detected CpG hypomethylation of expressed isoforms and CpG hypermethylation of silenced isoforms in the promoter proximal regions between the two CTCF-binding sites. However, there is constitutive hypermethylation in the promoter distal regions downstream of the eCBS sites in both expressed and silenced isoforms (Fig. 2 C and D). These data, in conjunction with the correlation between Pcdh promoter activity and its hypomethylation in cultured cells, strongly suggest that CpG methylation within the promoter regions plays an important role in the regulation of CTCF-binding and promoter activity.
Fig. 2.
CpG methylation regulates Pcdh gene expression through alteration of CTCF binding. (A) The methylated CSE sequences as representative probes. (B) EMSA experiments with methylated (Me) and unmethylated control (UMe) CSE probes. (C) Schematic of CTCF binding at each alternate promoter region of the Pcdhα cluster. (D) Inverse relationships between Pcdh gene expression and extent of CpG methylation (β score).
CpG methylation may also regulate Pcdh enhancer function. CTCF binds to HS5-1b in every cell line in the ENCODE dataset; however, CTCF binds HS5-1a in only a subset of these cell lines (14). Interestingly, HS5-1a contains a CpG dinucleotide at the position corresponding to the methylation-sensitive CTCF site in CSE, whereas HS5-1b does not contain a CpG site and, thus, cannot be methylated (Fig. S4C). This observation suggests that CTCF binds to HS5-1b constitutively but to HS5-1a in a methylation-sensitive manner. Interestingly, the first site of HS5-1aL contains a CpG dinucleotide at the same position corresponding to that in HS5-1a and CSE (Fig. S4E). Moreover, the CTCF binding to HS5-1aL appears to be regulated, probably in a methylation-sensitive manner, whereas CTCF binding to HS5-1bL appears to be constitutive (14).
DNA Looping Between Enhancers and Active Promoters.
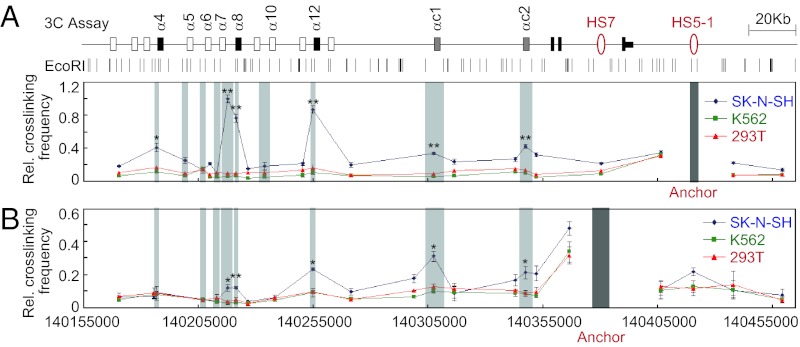
To investigate whether there are long-range DNA-looping interactions between enhancers and promoters of the α cluster, we performed quantitative chromosome conformation capture (3C) assays using SK-N-SH cells. We used two nonneuronal human cell lines K562 and 293T, which do not express Pcdhα isoforms, as controls in the 3C analyses (Fig. S6 A and B). When using an anchor primer matching HS5-1, strong interactions were detected with the active promoters α8 and α12, whereas weak interactions were detected with the active promoters of α4, αc1, and αc2 (Fig. 3A). Importantly, interactions with the inactive promoters of α5, α6, α7, and α10 were not detected (Fig. 3A).
Fig. 3.
The HS5-1 and HS7 enhancers engage in multiple long-range DNA-looping interactions with alternate and ubiquitous promoters. Relative crosslinking frequencies between enhancers HS5-1 (A) or HS7 (B) and promoters of the α cluster were measured by the 3C assay in SK-N-SH (blue), K562 (green), and 293T (red) cells. Data are presented as means ± SEM (n = 3). *P < 0.05; **P < 0.01. Only significance of comparison with K562 is shown. The P value of comparison with 293T is similar to that of K562 for all sites except α4 (P = 0.056).
We note that there is an EcoRI site between the α8 CSE and eCBS, thus making it possible to separate the promoter region into two DNA fragments. Importantly, both fragments form strong DNA-looping interactions with HS5-1 (Fig. 3A). Interestingly, sequencing of the looped-DNA fragments between α8 and HS5-1 revealed that both alleles of the α8 isoform interact with HS5-1 in SK-N-SH cells (Fig. S6 C and D). In addition, sequencing of the α8 mRNA demonstrates that both alleles are expressed in SK-N-SH cells (Fig. S6G). We note that HS5-1 interacts more weakly with the ubiquitously expressed promoters of αc1 and αc2, consistent with the observation that only a minor decrease in the expression of these c-type genes was detected in HS5-1–deletion mice (9, 11).
When we used primers against HS7 as anchors, we observed weak interactions with the active promoters of α8, α12, αc1, and αc2, but not with α4, compared with the control nonneuronal cells of K562 or 293T (Fig. 3B), which do not express the α cluster (Fig. S6 A and B). Similar to the situation with the HS5-1 enhancer, sequencing of the looped-DNA fragments between α8 and HS7 revealed that both alleles of the α8 interact with HS7 in SK-N-SH cells (Fig. S6 C and E). It is important to note that HS7 does not interact with inactive promoters (Fig. 3B). Thus, the pattern of Pcdhα isoform expression observed in SK-N-SH cells strongly correlates with specific enhancer/promoter interactions. We conclude that long-distance (over 250 kb in some cases) DNA looping plays an important role in stochastic promoter choice.
CTCF and Cohesin Are Required for DNA Looping.
To determine whether CTCF and cohesin are required for enhancer/promoter interactions, we infected SK-N-SH cells with a lentivirus expressing shRNAs directed against GFP (control), CTCF, or Rad21. The shRNA knockdowns led to a decrease in the levels of both CTCF and Rad21 mRNAs and proteins (Fig. S7 A–C). As expected, knockdown of CTCF by shRNA results in a significant decrease of CTCF binding to the CSE and eCBS of α4, α8, and α12; the CSE of αc1, β3, γb1, γb7, and γc3; and the enhancer sites of HS5-1a and HS5-1b, as well as HS5-1aL in SK-N-SH cells (Fig. S8A).
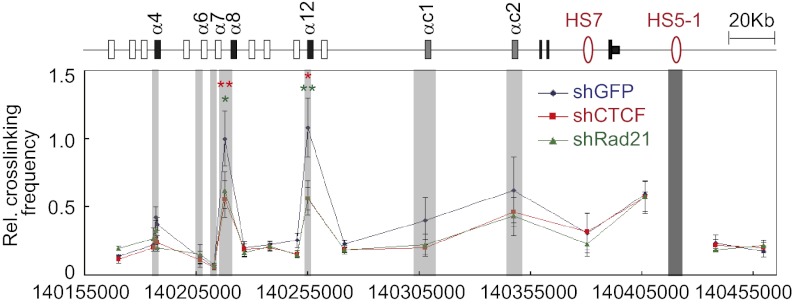
ChIP-qPCR experiments demonstrated that knockdown of CTCF leads to a significant decrease in the levels of the cohesin subunits Rad21 and SMC3, which bind to the CSEs of α4, α8, α12, and αc1, as well as to HS5-1a and HS5-1b (Fig. S7 D and E). Similarly, knockdown of Rad21 results in a significant decrease in the binding of Rad21 and SMC3 to these sites (Fig. S7 F and G). Most importantly, knockdown of CTCF or Rad21 results in a significant decrease in the long-range DNA-looping interactions between HS5-1 and the α8 or α12 promoter (Fig. 4). In addition, CTCF knockdown decreases Rad21 enrichment at the eCBS sites of α8 and α12, as well as CSEs of β3, γb1, γb7, γc3, and HS5-1aL in the β and γ clusters, suggesting that cohesin is recruited by CTCF to these sites (Fig. S8 B and C) (21). We conclude that the binding of CTCF/cohesin to active promoters and enhancers is required for Pcdhα enhancer/promoter interactions through DNA looping.
Fig. 4.
CTCF or cohesin knockdown results in significant decreases of long-range DNA interactions between HS5-1 and promoters. SK-N-SH cells were infected with shGFP (control), shCTCF, or shRad21 lentivirus, and crosslinking frequencies were measured by 3C. Data are presented as means ± SEM (n = 4). *P < 0.05; **P < 0.01.
Interaction of Paired CTCF/Cohesin-Binding Sites.
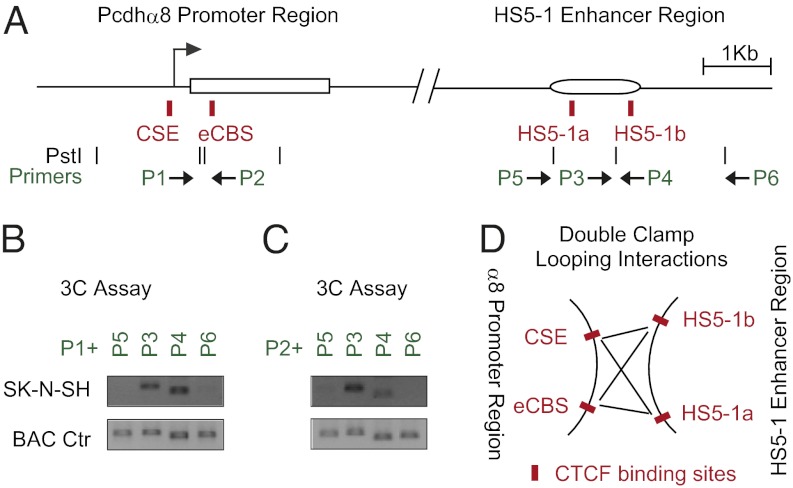
We investigated DNA-looping interactions between the pair of CTCF-binding sites in the Pcdhα8 promoter region (CSE and eCBS) and the pair of binding sites in the HS5-1 enhancer region (HS5-1a, HS5-1b) in SK-N-SH cells. Based on the PstI-digestion patterns, we designed two specific primers, P1 and P2, corresponding to the two fragments containing the CSE and eCBS of α8, respectively. In addition, we designed two primers, P3 and P4, corresponding to the two fragments containing HS5-1a and HS5-1b, respectively. We also designed two primers, P5 and P6, complementary to sequences outside of HS5-1 as negative controls (Fig. 5A).
Fig. 5.
Double clamp of DNA-looping interactions between HS5-1 enhancer and α8 promoter. (A) Schematics of the human α8 promoter and enhancer HS5-1 regions. The positions of the CTCF recognition (red), PstI sites (black), and primers are indicated. (B and C) The 3C assay for DNA-looping interactions between different PstI fragments using an anchor primer P1 (B) or P2 (C) with four enhancer primers P3-P6. (D) Model of DNA-looping interactions between the promoter of α8 and HS5-1 by a double-clamp mechanism. All CTCF-binding sites are highlighted by red boxes.
The 3C experiments using the primers P1 and P3 or P4 demonstrated significant DNA-looping interactions between the CSE of α8 to both HS5-1a and HS5-1b (Fig. 5B). Similarly, significant DNA-looping interactions were observed between the eCBS of α8 and both HS5-1a and HS5-1b (Fig. 5C), as demonstrated with primers P2 and P3 or P4. Control PCR experiments with BAC DNA preparations showed that these primers amplify predicted products with similar efficiency (Fig. 5 B and C). ChIP-qPCR experiments with four pairs of specific primers matching the CSE, eCBS, HS5-1a, and HS5-1b sites and six pairs of intervening and flanking primers confirmed the binding specificity of CTCF and Rad21 to these four sites in SK-N-SH (Fig. S9 A and B). The conservation of the position and spacing of the CSE/eCBS sites on the one hand (about 650 bp) and the HS5-1a,b on the other (1,009 bp) and the DNA-looping data of Fig. 5 suggest that DNA looping between the HS5-1 enhancer and active alternate promoters involves a “double clamp” in which the four CTCF/cohesin sites interact simultaneously (Fig. 5D).
Discussion
The remarkable genomic organization of the clustered protocadherin genes (1, 2, 4, 5) and the generation of multiple isoforms through promoter choice and alternative pre-mRNA splicing provide enormous single-cell diversity (3, 6). This diversity appears to play a fundamental role in neural circuit assembly (22–25). Understanding the mechanism by which this diversity is generated is, therefore, fundamental to understanding the development and function of the nervous system. Previous studies have identified Pcdhα isoform promoters and enhancer elements (2–5, 8) and have shown that they bind to CTCF and cohesin (9, 10). However, neither the function of CTCF/cohesin nor their possible role in enhancer/promoter interactions was addressed. Here, we made use of the diploid human neuroblastoma cell line SK-N-SH to address these questions. We find that CTCF and cohesin are required for the long range looping of Pcdhα enhancers and active isoform promoters, and we show that methylation of CpG dinucleotides in the CSEs prevents the binding of CTCF to the CSE in vitro. We find that the two sites in the HS5-1 enhancer form long-range DNA-looping interactions with two sites in the active promoters, α8 and α12, and weak DNA interactions with promoters of α4, αc1, and αc2 in human SK-N-SH cells. These interactions correlate with the requirement of HS5-1 for alternate promoter activity in mice (both quantitatively and qualitatively), except for αc2, which does not require HS5-1 for maximal activity (9, 11). Interestingly, the most distal ubiquitous promoter in the cluster, αc2, does not contain a CSE (2), and is not recognized by CTCF (Fig. S3A). The HS7 enhancer, which is located between constant exons 2 and 3 (8), is also not bound by CTCF (10) but is enriched in RNAPII (Fig. 1C). However, both αc2 and HS7 are bound to Rad21. We found that the HS7 enhancer forms long-range DNA-looping interactions with the α8, α12, αc1, and αc2 promoters in human SK-N-SH cells, which, again, is consistent with requirement of HS7 for alternate and ubiquitous promoter activity in mouse genetic studies (9). Thus, there appears to be CTCF- and cohesin-dependent DNA looping (α1 to α13 and αc1) and CTCF-independent but cohesin-dependent DNA looping (αc2). Taken together, these studies provide conclusive evidence that DNA looping occurs between well-characterized enhancers and multiple active Pcdh promoters in the α cluster, and that this looping requires specific binding of CTCF/cohesin to the transcriptionally active promoters and enhancers.
We propose that CTCF/cohesin-dependent long-range DNA-looping interactions among the putative enhancers (HS7L, HS5-1aL, HS5-1bL) of the γ cluster and its variable promoters occur in a similar manner to that observed for the HS7, HS5-1a, and HS5-1b and variable promoters of the α cluster. The variable region of the human γ cluster contains 19 alternate promoters (12 a-type: γa1 to γa12; and 7 b-type: γb1 to γb7) and 3 c-type (γc3 to γc5) ubiquitous promoters. Similar to the α cluster, each of the 19 alternate promoters and the first ubiquitous promoter (γc3) of the γ cluster contains a CSE (2) that binds to CTCF (Fig. 1F and Fig. S3). The last two c-type ubiquitous promoters (γc4 and γc5), similar to the last ubiquitous α promoter (αc2), do not contain a CSE and cannot bind to CTCF (Fig. 1C and Fig. S3A). Similar to HS7 of the α cluster, we found an HS7-like (HS7L) site in the γ cluster, which also has RNAPII enrichment (Fig. 1F) and is located in a similar position between the constant exons 2 and 3. In addition, we observed two downstream CTCF-binding sites in the γ cluster, HS5-1aL and HS5-1bL, similar to HS5-1a and HS5-1b of the α cluster. Interestingly, the HS5-1aL CTCF-binding site in the 3′ UTR of the γ cluster, similar to the HS5-1a of the α cluster, is also in the opposite orientation to that of the CSE in the Pcdh α and γ promoter regions and also contains CpG sequences (Fig. S4E). Evidence that these putative Pcdhγ sequences function as enhancers is provided by the observation that, like the corresponding sequences in the α cluster, they appear as DNase I–hypersensitive sites in the whole-genome DNase I–mapping experiments (14). These observations strongly suggest that the Pcdh α and γ clusters were generated by a duplication event (1, 4) and that the basic organization and regulatory mechanisms necessary to generate single-cell Pcdh diversity were conserved between the Pcdh α and γ clusters.
Recently, transcriptional coactivators such as mediator and TAF3 were shown to mediate DNA-looping interactions via cohesin or CTCF (26, 27). In addition, CTCF was shown to interact directly with RNAPII (28). CTCF was shown to block enhancer- and promoter-looping interactions (19, 20) or to mark the boundary between active and inactive chromatin regions to establish chromatin domains and to insulate neighboring promoters from interfering with each other (29, 30). However, here, we discovered a role of CTCF for promoter activation in the clustered Pcdh genes. Our results reveal that the enhancer-bound CTCF recruits the cohesin complex, which, in turn, brings alternate promoters to the vicinity of cluster-specific enhancers through DNA looping, to facilitate cell-specific gene expression in Pcdh clusters.
Our data demonstrate that both qualitative and quantitative changes of cohesin alter DNA-looping interactions in the Pcdh clusters. Recent studies have revealed a functional role for clustered Pcdh genes in dendritic development and self-avoidance (22–25). Strikingly, a repertoire of heterozygous mutations in chromatid-segregating cohesin (SMC1A, SMC3, RAD21) and cohesin regulators (NIPBL, ESCO2) have been found to cause a class of developmental disorders known as Cornelia de Lange syndrome (CdLS) and Robert syndrome in humans (31). One of the most intriguing unknown aspects of these syndromes is the molecular etiology of CdLS in neurodevelopmental delay and mental retardation. The important role of cohesin in the regulation of clustered Pcdh genes (this study and refs. 10, 32, and 33) suggests a neuropathologic basis for these syndromes caused by human heterozygous cohesin mutations.
The observation that both the HS5-1 and HS7 enhancers interact with and are required for maximal expression of multiple members of the α cluster and that CTCF/cohesin is required for this interaction suggests a complex mechanism of promoter choice. We propose that the DNA-looping interactions recruit the promoters bound to CTCF to enhancers in an active “transcriptional hub” (Fig. 6). This model is based on extensive and highly specific functional and physical interactions between promoters and enhancers, and the fact that the formaldehyde cross-linking used in the ChIP-seq and 3C studies would be expected to crosslink a large complex containing multiple enhancers and promoters. In this model, the DNA-looping interactions between HS5-1 and the promoters of α8 and a12 are formed by a double-clamping mechanism between the HS5-1a/HS5-1b sites of the enhancer and the CSE/eCBS sites of alternate promoters (Fig. 5D). At the same time, HS5-1 must interact with αc1, because HS5-1 is required for its expression (9, 11). In addition, HS7 must directly interact with αc2, as well as with the active alternate promoters and ubiquitous promoters, because this enhancer interacts with and is required for the maximum activation of these promoters (9). Finally, long-range DNA-looping interactions are formed between promoters of αc1 and α8 and both alleles of the α8 promoters form looping interactions with αc1 (Fig. S6 C and F). All of these observations are consistent with the model of Fig. 6, in which the apparent simultaneous interactions between the two enhancers and multiple promoters are the consequence of the formation of a large transcriptional hub. Presumably, promoter/enhancer interactions within this hub must be maintained during DNA replication, because the pattern of Pcdh expression in SK-N-SH cells has been maintained during many rounds of cell division. Moreover, the stability of this complex is required to maintain self-identity during the postmitotic life of individual neurons. Further studies of the mechanisms of Pcdh promoter choice during development and whether and how the choice is maintained should provide important insights into the role of Pcdh diversity in the assembly of neural circuits.
Fig. 6.
Transcriptional hub model for the promoter choice of members of the Pcdhα gene cluster in the SK-N-SH cells. In this model, RNAPII and Rad21 binding to the promoter of the last ubiquitous gene of the α cluster result in the constitutive activation of αc2. Promoter choices are determined through recruiting of CTCF/cohesin to the CSEs of α4, α8, α12, and αc1. CTCF/cohesin-mediated multiple long-range DNA-looping interactions with enhancers (HS7 and HS5-1) bring the chosen promoters into an active transcriptional hub for transcriptional activation.
Materials and Methods
Recombinant Protein Production and EMSA.
Full-length human CTCF cDNA was cloned from total RNA preparations of HEC-1-B by RT-PCR with a pair of specific primers and confirmed by sequencing. The recombinant human CTCF proteins were synthesized from pTNT-CTCF by using TNT T7 System (Promega). All of wild-type (WT) and mutation probes were confirmed by sequencing. See SI Materials and Methods for details.
Methylation Analysis.
To determine CpG methylation state in SK-N-SH cells, genomic DNA was analyzed on Infinium HumanMethylation450 BeadChips (Illumina). CpGs located near each variable exon were identified and grouped by location.
ChIP-qPCR and ChIP-seq.
ChIP was performed by using protein A agarose beads from the Millipore and followed by standard qPCR experiments. The ChIP-seq experiments were performed similarly, except that the precipitated complexes were isolated with protein G agarose beads (Sigma). See SI Materials and Methods for details.
Chromatin Conformation Capture.
The 3C analysis protocols were modified from previous methods. See SI Materials and Methods for details.
Supplementary Material
Acknowledgments
This work was supported by Ministry of Science and Technology of China Grant 2009CB918700, National Natural Science Foundation Grants 31171015 and 30970669, Science and Technology Commission of Shanghai Municipality Grant 09PJ1405300, and State Key Laboratory of Oncogenes and Related Genes Grant 91-10-10 (to Q.W.) and by National Institutes of Health Grant R01NS043915 (to T.M.). Q.W. is a Pujiang Investigator.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1219280110/-/DCSupplemental.
References
- 1.Wu Q, Maniatis T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell. 1999;97(6):779–790. doi: 10.1016/s0092-8674(00)80789-8. [DOI] [PubMed] [Google Scholar]
- 2.Wu Q, et al. Comparative DNA sequence analysis of mouse and human protocadherin gene clusters. Genome Res. 2001;11(3):389–404. doi: 10.1101/gr.167301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tasic B, et al. Promoter choice determines splice site selection in protocadherin alpha and gamma pre-mRNA splicing. Mol Cell. 2002;10(1):21–33. doi: 10.1016/s1097-2765(02)00578-6. [DOI] [PubMed] [Google Scholar]
- 4.Wu Q. Comparative genomics and diversifying selection of the clustered vertebrate protocadherin genes. Genetics. 2005;169(4):2179–2188. doi: 10.1534/genetics.104.037606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zou C, Huang W, Ying G, Wu Q. Sequence analysis and expression mapping of the rat clustered protocadherin gene repertoires. Neuroscience. 2007;144(2):579–603. doi: 10.1016/j.neuroscience.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 6.Wang X, Su H, Bradley A. Molecular mechanisms governing Pcdh-gamma gene expression: Evidence for a multiple promoter and cis-alternative splicing model. Genes Dev. 2002;16(15):1890–1905. doi: 10.1101/gad.1004802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaneko R, et al. Allelic gene regulation of Pcdh-alpha and Pcdh-gamma clusters involving both monoallelic and biallelic expression in single Purkinje cells. J Biol Chem. 2006;281(41):30551–30560. doi: 10.1074/jbc.M605677200. [DOI] [PubMed] [Google Scholar]
- 8.Ribich S, Tasic B, Maniatis T. Identification of long-range regulatory elements in the protocadherin-alpha gene cluster. Proc Natl Acad Sci USA. 2006;103(52):19719–19724. doi: 10.1073/pnas.0609445104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kehayova P, Monahan K, Chen W, Maniatis T. Regulatory elements required for the activation and repression of the protocadherin-alpha gene cluster. Proc Natl Acad Sci USA. 2011;108(41):17195–17200. doi: 10.1073/pnas.1114357108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monahan K, et al. Role of CCCTC binding factor (CTCF) and cohesin in the generation of single-cell diversity of protocadherin-α gene expression. Proc Natl Acad Sci USA. 2012;109(23):9125–9130. doi: 10.1073/pnas.1205074109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yokota S, et al. Identification of the cluster control region for the protocadherin-beta genes located beyond the protocadherin-gamma cluster. J Biol Chem. 2011;286(36):31885–31895. doi: 10.1074/jbc.M111.245605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biedler JL, Roffler-Tarlov S, Schachner M, Freedman LS. Multiple neurotransmitter synthesis by human neuroblastoma cell lines and clones. Cancer Res. 1978;38(11 Pt 1):3751–3757. [PubMed] [Google Scholar]
- 13.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465(7295):182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunham I, et al. ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Golan-Mashiach M, et al. Identification of CTCF as a master regulator of the clustered protocadherin genes. Nucleic Acids Res. 2012;40(8):3378–3391. doi: 10.1093/nar/gkr1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lobanenkov VV, et al. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5′-flanking sequence of the chicken c-myc gene. Oncogene. 1990;5(12):1743–1753. [PubMed] [Google Scholar]
- 17.Dallosso AR, et al. Frequent long-range epigenetic silencing of protocadherin gene clusters on chromosome 5q31 in Wilms’ tumor. PLoS Genet. 2009;5(11):e1000745. doi: 10.1371/journal.pgen.1000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawaguchi M, et al. Relationship between DNA methylation states and transcription of individual isoforms encoded by the protocadherin-alpha gene cluster. J Biol Chem. 2008;283(18):12064–12075. doi: 10.1074/jbc.M709648200. [DOI] [PubMed] [Google Scholar]
- 19.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405(6785):482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 20.Hark AT, et al. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405(6785):486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 21.Xiao T, Wallace J, Felsenfeld G. Specific sites in the C terminus of CTCF interact with the SA2 subunit of the cohesin complex and are required for cohesin-dependent insulation activity. Mol Cell Biol. 2011;31(11):2174–2183. doi: 10.1128/MCB.05093-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suo L, Lu H, Ying G, Capecchi MR, Wu Q. Protocadherin clusters and cell adhesion kinase regulate dendrite complexity through Rho GTPase. J Mol Cell Biol. 2012 doi: 10.1093/jmcb/mjs034. [DOI] [PubMed] [Google Scholar]
- 23.Garrett AM, Schreiner D, Lobas MA, Weiner JA. γ-Protocadherins control cortical dendrite arborization by regulating the activity of a FAK/PKC/MARCKS signaling pathway. Neuron. 2012;74(2):269–276. doi: 10.1016/j.neuron.2012.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen WV, et al. Functional significance of isoform diversification in the protocadherin gamma gene cluster. Neuron. 2012;75(3):402–409. doi: 10.1016/j.neuron.2012.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lefebvre JL, Kostadinov D, Chen WV, Maniatis T, Sanes JR. Protocadherins mediate dendritic self-avoidance in the mammalian nervous system. Nature. 2012;488(7412):517–521. doi: 10.1038/nature11305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kagey MH, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467(7314):430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Z, Scannell DR, Eisen MB, Tjian R. Control of embryonic stem cell lineage commitment by core promoter factor, TAF3. Cell. 2011;146(5):720–731. doi: 10.1016/j.cell.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chernukhin I, et al. CTCF interacts with and recruits the largest subunit of RNA polymerase II to CTCF target sites genome-wide. Mol Cell Biol. 2007;27(5):1631–1648. doi: 10.1128/MCB.01993-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wendt KS, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451(7180):796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 30.Phillips JE, Corces VG. CTCF: Master weaver of the genome. Cell. 2009;137(7):1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nasmyth K, Haering CH. Cohesin: Its roles and mechanisms. Annu Rev Genet. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 32.Kawauchi S, et al. Multiple organ system defects and transcriptional dysregulation in the Nipbl(+/-) mouse, a model of Cornelia de Lange syndrome. PLoS Genet. 2009;5(9):e1000650. doi: 10.1371/journal.pgen.1000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Remeseiro S, Cuadrado A, Gómez-López G, Pisano DG, Losada A. A unique role of cohesin-SA1 in gene regulation and development. EMBO J. 2012;31(9):2090–2102. doi: 10.1038/emboj.2012.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.