Abstract
Leaves and flowers begin life as outgrowths from the edges of shoot apical meristems. Stem cell divisions in the meristem center replenish cells that are incorporated into organ primordia at the meristem periphery and leave the meristem. Organ boundaries, regions of limited growth that separate forming organs from the meristem, serve to isolate these two domains and are critical for coordination of organogenesis and meristem maintenance. Boundary formation and maintenance are poorly understood processes, despite the identification of a number of boundary-specific transcription factors. Here we provide genetic and biochemical evidence that the Arabidopsis thaliana transcription factor LATERAL ORGAN BOUNDARIES (LOB) negatively regulates accumulation of the plant steroid hormone brassinosteroid (BR) in organ boundaries. We found that ectopic expression of LOB results in reduced BR responses. We identified BAS1, which encodes a BR-inactivating enzyme, as a direct target of LOB transcriptional activation. Loss-of-function lob mutants exhibit organ fusions, and this phenotype is suppressed by expression of BAS1 under the LOB promoter, indicating that BR hyperaccumulation contributes to the lob mutant phenotype. In addition, LOB expression is BR regulated; therefore, LOB and BR form a feedback loop to modulate local BR accumulation in organ boundaries to limit growth in the boundary domain.
Leaves and flowers are produced from the periphery of the shoot apical meristem, a self-perpetuating structure containing a population of self-renewing stem cells. Stem cell divisions in the meristem center replenish the cells that are incorporated into organ primordia at the meristem periphery and exit the meristem (1). The balance between organogenesis and meristem maintenance is essential for continued organ formation, and the boundary between the meristem and organ primordia plays a key role in maintaining the integrity of the meristem and differentiating organs. Boundary cells are small and divide infrequently relative to cells in the adjacent regions; thus, the boundary is a discrete domain that is distinct from the meristem and organ primordia (2–4). During organ formation, inhibition of growth in the boundary allows formation of a cleft, which results in separation of the forming organ from the meristem. A number of boundary-specific transcription factors in several families act redundantly to specify organ boundary cell fate and meristem maintenance (5–11). Few targets of boundary-specific transcription factors have been identified, and little is known about the physiological and biochemical processes they regulate.
Arabidopsis LATERAL ORGAN BOUNDARIES (LOB) encodes a member of the plant-specific LOB-domain transcription factor family and is expressed specifically in organ boundaries (12). To investigate the developmental function of LOB, we examined the consequence of increased and decreased LOB activity and used expression profiling to identify targets of LOB transcriptional regulation. We show that LOB negatively regulates accumulation of the plant steroid hormone brassinosteroid (BR) in organ boundaries. Loss-of-function lob mutants exhibit organ fusions, whereas ectopic expression of LOB results in reduced BR responses. Microarray analyses demonstrate that LOB regulates expression of BAS1, which encodes a BR-inactivating enzyme, and we demonstrate that BR hyperaccumulation contributes to the lob mutant phenotype. In addition, LOB expression is BR regulated; therefore, LOB and BR form a feedback loop to modulate local BR accumulation to limit growth in the boundary domain.
Results
Loss-of-Function lob Mutants Exhibit Organ Separation Defects.
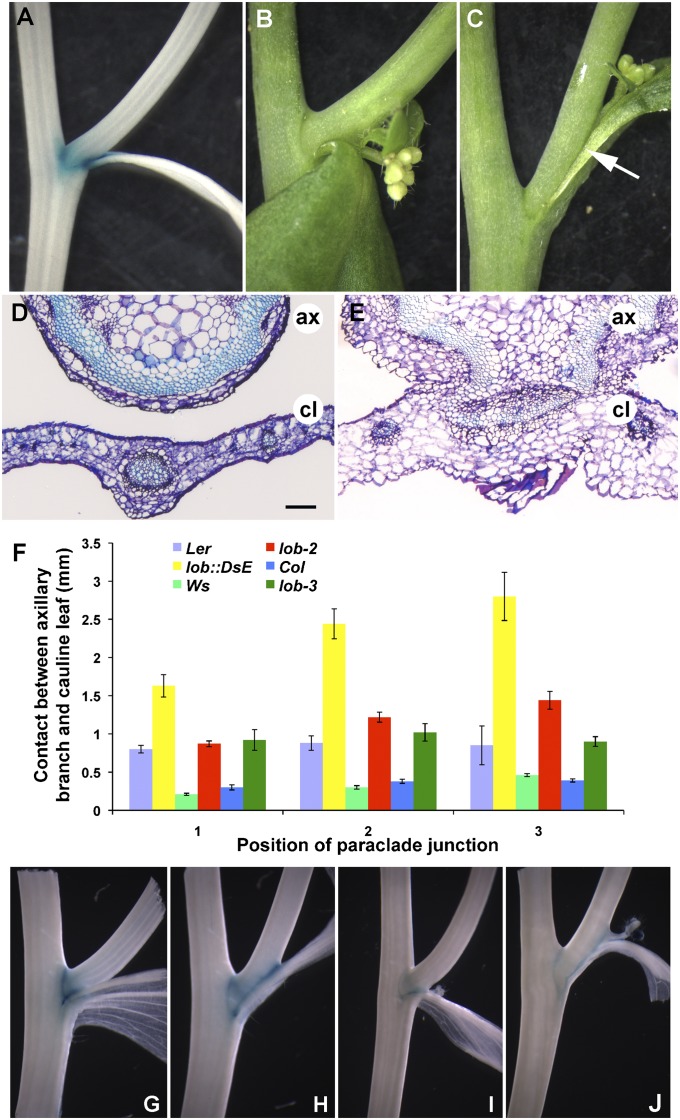
lob mutants exhibited normal seedling and vegetative development as previously reported (12). Inspection of cauline leaf axils, a domain where LOB is expressed (Fig. 1A), revealed that wild-type axillary and accessory branches were well separated from the subtending cauline leaf (Fig. 1B), whereas these structures were fused in loss-of-function lob::DsE mutant plants (Fig. 1C). Three different loss-of-function lob alleles displayed similar fusion phenotypes (Fig. 1F; Fig. S1). The fusion in lob::DsE was significantly more severe than in the other alleles, possibly because of the presence of modifying loci in the Ler ecotype. Introduction of the wild-type LOB gene complemented the lob::DsE mutation (Fig. S1G), demonstrating that the observed fusion was the result of defects in LOB activity.
Fig. 1.
Mutations in LOB result in organ fusion. (A) GUS activity in pLOB:GUS between the main stem and axillary stem and between the axillary stem and cauline leaf. (B and C) Paraclade junction between main stem, axillary stem, and cauline leaf in wild-type (B) and lob::DsE (C) plants. The axillary stem and cauline leaf are separated in wild type and fused in lob::DsE (white arrow). (D and E) Cross sections through junction between axillary stem (ax) and cauline leaf (cl) in wild type (D) and lob::DsE (E). (F) Length of fused region in wild-type Landsberg erecta, lob::DsE, wild-type Wassilewskija, lob-2, wild-type Columbia, and lob-3. Position 1 corresponds to lowest cauline leaf axil on stem. SEs are indicated; n ≥ 11 for positions 1 and 2, and n ≥ 5 for position 3 (not all plants have three paraclade junctions). (G–J) GUS expression in boundary marker lines ET4016 (11) (G and H) and GT185 (34) (I and J) in wild type (G and I) and lob (H and J). ET4016 reports expression of LOF1 (11) and GT185 reports expression of ORGAN BOUNDARY1 (13). Expression of both markers is extended throughout the fused region in lob mutants, indicating an expansion or overgrowth of the boundary domain. (Scale bar in D, 100 μm for D and E.)
In cross sections of cauline leaf axils, wild-type cauline leaves and axillary stems were completely separated (Fig. 1D), whereas axillary stem and cauline leaf tissues appeared as a single structure in lob::DsE mutants, with continuity between cortex and vascular tissues in the stem and leaf (Fig. 1E). Cells in the fused region appeared larger than in wild type. We examined expression of boundary markers LATERAL ORGAN FUSION1 (11) and ORGAN BOUNDARY1 (13) in lob mutants and observed expansion of beta-glucuronidase (GUS) reporter gene expression into the fused region (Fig. 1 G–J). Together these results suggest that lob mutants have an expansion or overgrowth of the boundary that results in defects in organ separation.
Ectopic LOB Expression Affects BR Responses.
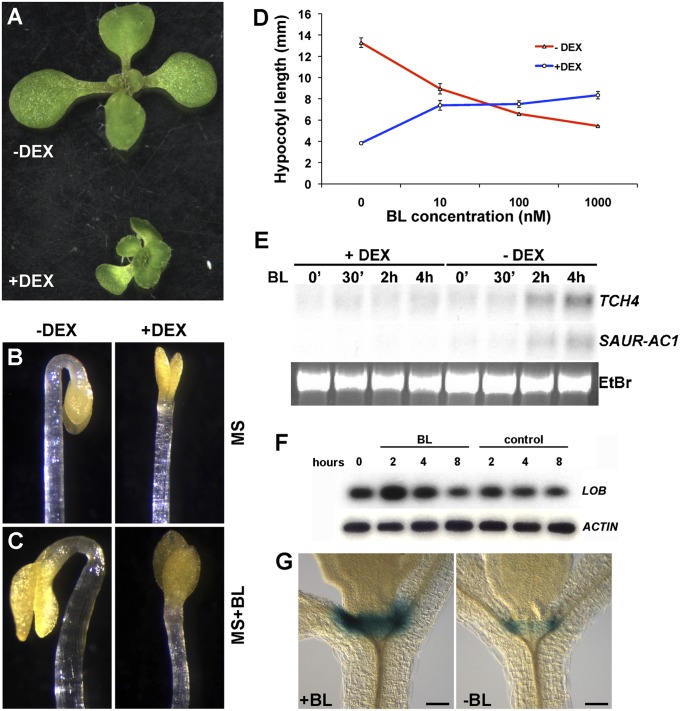
To identify pathways downstream of LOB activity, we constructed an inducible form of LOB, 35S:LOB-GR, generated by fusing LOB to the hormone-binding domain of the glucocorticoid receptor (14), under control of the ubiquitously expressed 35S promoter. Treatment of 35S:LOB-GR plants with the synthetic steroid dexamethasone (DEX) resulted in an overall reduction in growth in a dose-dependent manner, whereas in the absence of DEX, these plants were phenotypically normal (Fig. 2A). 35S:LOB-GR plants grown in the presence of 3 μM DEX resembled 35S:LOB plants, which are dwarf and sterile (12).
Fig. 2.
Ectopic LOB expression disrupts brassinosteroid responses. (A) 35S:LOB-GR plants are dwarfed when grown on 3 μM DEX. (B and C) Dark-grown, 4-d-old 35S:LOB-GR seedlings produce an apical hook when grown on MS medium in the absence of DEX (−DEX) and lack an apical hook when grown in the presence of 3 μM DEX (+DEX). Apical hook formation is not restored by addition of 100 nM epi-brassinolide in the medium (C; MS+BL). (D) Hypocotyl lengths of 35S:LOB-GR seedlings grown in the dark on increasing concentrations of epi-brassinolide (BL) in the presence or absence of 3 μM DEX. SEs (n ≥ 15) are indicated. (E) Northern blot analyses of TCH4 and SAUR-AC1 transcripts. Seven-day-old 35S:LOB-GR seedlings were pretreated overnight in the presence or absence of 3 μM DEX and then incubated in the presence of 1 μM epi-brassinolide (BL) for the indicated times. (F) RT-PCR analysis of LOB transcript levels in 7-d-old wild-type seedlings following treatment with 1 μM epi-brassinolide (BL) for 2, 4, or 8 h. RT-PCR products were detected by blotting and probing with gene-specific probes, following either 15 (LOB) or 12 cycles (ACTIN) of amplification. (G) GUS activity in pLOB:GUS:LOB-3′IGR seedlings after 3-h incubation in liquid MS supplemented with (Left) or without (Right) 1 μM BL. (Scale bar in G and H, 100 μm.)
Ectopic LOB expression resulted in reduced growth, similar to that caused by defects in BR accumulation or response. BRs are plant steroid hormones that regulate cell expansion and cell division (15). Because the boundary cells where LOB is expressed undergo limited division and expansion (2, 3), we considered the possibility that LOB activity might limit growth in organ boundaries by regulating BR responses. We examined a number of BR responses in 35S:LOB-GR plants grown in the presence or absence of DEX. Dark-grown Arabidopsis seedlings produce elongated hypocotyls and form an apical hook, developmental events that require BR signaling (15). When germinated in the dark, DEX-treated 35S:LOB-GR seedlings failed to make an apical hook (Fig. 2B) and had shorter hypocotyls than seedlings grown in the absence of DEX (Fig. 2D). Germination on medium containing epi-brassinolide (BL), a biologically active BR, did not restore hook formation or cause cotyledon expansion as it did in non–DEX-induced plants (Fig. 2C). Furthermore, germination on increasing concentrations of BL resulted in an increase in hypocotyl length in dark-grown 35S:LOB-GR seedlings on DEX, whereas mock-treated seedlings exhibited inhibition of hypocotyl growth (Fig. 2D), similar to that reported for wild type (16). Apical hook formation was not restored by treatment with the phytohormones gibberellic acid (GA), auxin (IAA), or the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC) (Fig. S2). DEX treatment of 35S:LOB-GR plants also resulted in a diminished induction of the BR-response genes TCH4 and SAUR-AC1 (Fig. 2E). Thus, ectopic LOB activity resulted in reduced sensitivity to BR.
To examine the contribution of reduced BR responses to the 35S:LOB-GR phenotype, we crossed 35S:LOB-GR plants to bzr1-1d, a mutant that exhibits constitutive BR responses due to the stabilization of BZR1, a positive regulator of BR signaling (17). Whereas dark-grown 35S:LOB-GR bzr1-1d seedlings lacked an apical hook when grown on DEX, similar to 35S:LOB-GR plants, their hypocotyls were longer than DEX-grown 35S:LOB-GR seedlings (Fig. S3 A–F). Light-grown 35S:LOB-GR bzr1-1d plants were also larger than 35S:LOB-GR plants grown on DEX (Fig. S3 G–I), indicating that the bzr1-1d mutation partially suppressed the growth defects of 35S:LOB-GR plants. That some aspects of the LOB misexpression phenotype were ameliorated by increased BR signaling indicates that reduced BR responses contribute to the phenotype and are consistent with LOB acting upstream of BZR1.
To further examine the relationship between LOB and BR signaling, we investigated the possible regulation of LOB expression by BRs. Treatment of seedlings with exogenous BRs resulted in a transient increase in LOB transcript levels (Fig. 2F). BR treatment also caused an increase in the intensity of GUS staining in pLOB:GUS seedlings; however, no change in the staining pattern was seen (Fig. 2G), indicating that BR influences the level of LOB expression in boundaries.
Identification of LOB-Responsive Transcripts.
We performed microarray experiments to identify genes that were differentially expressed in response to LOB activation. p35S:LOB-GR and wild-type Col-0 seedlings were exposed to DEX or were mock treated for 4 h, and RNA samples from three biological replicates per treatment were hybridized to Affymetrix ATH1 arrays. Following statistical analyses, we identified genes that were differentially expressed in DEX-treated p35S:LOB-GR plants compared with mock treated and not differentially expressed in Col-0 DEX-treated plants compared with mock treated. A total of 288 unique transcripts showed significant changes in response to LOB activation (fold-change ≥2; false discovery rate ≤0.001) (Dataset S1). Differentially expressed genes may be directly or indirectly regulated by LOB activity. LOB and related proteins were shown to bind in vitro to a 6-bp consensus sequence GCGGCG termed the LBD motif (18). Partial LBD motifs (missing the first or last G: GCGGC or CGGCG) are present nearby or within a majority (269/288) of the LOB-regulated genes; therefore, some may be direct LOB targets.
Strikingly, about 60% (175) of the LOB-regulated transcripts were BR modulated in one or more experiments (reviewed in ref. 19), consistent with the hypothesis that LOB contributes to regulation of BR responses (Dataset S1). LOB-regulated genes were also enriched in Gene Ontology (GO) term categories associated with various stimulus responses and cell wall modifications (Dataset S2).
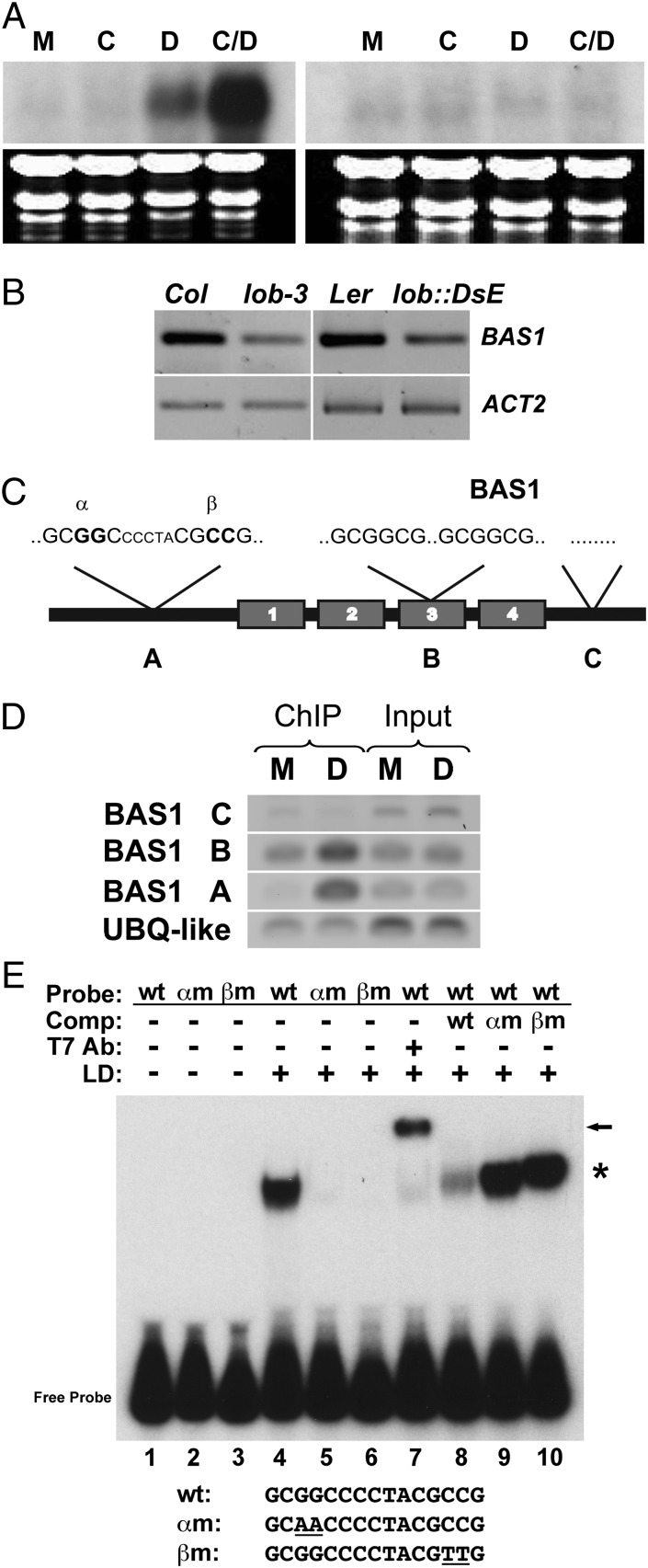
To investigate the significance of LOB regulation of BR-response genes, we characterized one such gene in detail. PHYB ACTIVATION TAGGED SUPPRESSOR1 (BAS1; At2g26710) encodes a cytochrome P450 enzyme that inactivates BRs by C-26 hydroxylation (20) and showed a 9.3-fold increase in transcript abundance in response to DEX induction. In a time-course experiment, an increase in BAS1 transcript levels was observed within 60 min of LOB-GR activation by DEX (Fig. S4). BAS1 transcripts were elevated by DEX treatment in the presence of the protein synthesis inhibitor cycloheximide (Fig. 3A), indicating likely direct regulation by LOB. Additionally, BAS1 transcript levels were reduced in lob mutants (Fig. 3B), consistent with LOB regulation of BAS1 expression.
Fig. 3.
BAS1 is a direct target of LOB. (A) Northern blot analyses of BAS1 transcript levels in 35S:LOB-GR (Left) and Columbia wild-type (Right) 8-d-old seedlings following 4-h mock (M), cycloheximide (C), DEX (D), or cycloheximide plus DEX (C/D) treatment. (B) RT-PCR analysis of BAS1 transcript levels in dissected cauline leaf-axillary stem junctions of Col, lob-3, Ler, and lob::DsE. RT-PCR products were detected by blotting and probing with gene-specific probes, following either 15 (BAS1) or 12 cycles (ACT2) of amplification. (C) Cartoon of the genomic structure of BAS1 showing the locations of LBD motifs and regions tested for enrichment after ChIP. BAS1 A contained two partial LBD sites separated by five nucleotides, 306 bp upstream of the ATG. BAS1 B contained two full LBD sites separated by 46 nucleotides, 1,700 bp downstream of the ATG in the third exon. The BAS1 C region contained no LBD sites and was 2,900 bp downstream of the ATG. (D) PCR products were amplified from DNA obtained before (Input) and after (ChIP) collection of specific LOB-DNA complexes by a LOB 1° antibody. DEX (D) and mock-treated (M) 35S:LOB-GR plants were used in ChIP experiments. BAS1 A and B regions were amplified for 27 cycles, and BAS1 C was amplified for 38 cycles. Thirty cycles of amplification were performed with the control gene UBQ-LIKE (At3g26980). (E) The LOB domain (LD) of LOB was incubated with a 133-bp radiolabeled probe generated from the BAS1 A region and separated on a native polyacrylamide gel. Probes contained unmodified LBD motifs (wt; lanes 1, 4, and 7–10), a mutation in the 5′-most motif in which the central GG residues were mutated to AA (αm; lanes 2 and 5), or a mutation in the 3′-most motif in which the central GG residues were mutated to AA (βm; lanes 3 and 6). Inclusion of T7 Ab against the tag on LD results in a supershift, demonstrating that recombinant LD protein bound the wild-type probe (lane 7). An excess of cold wild-type αm or βm DNA was used in competition experiments to demonstrate specificity of binding (lanes 8–10). The sequence of the central motif in the wt and mutant probes is shown.
LOB Associates with the BAS1 Promoter In Vivo and In Vitro.
Examination of the BAS1 sequence revealed four potential LBD motifs. Two partial sites, separated by 5 bp and in inverted orientation relative to one another, were located 306 nucleotides upstream of the BAS1 ATG, and two sites were located in the third exon (Fig. 3C). To determine whether LOB binds to these sites in vivo, we examined the chromatin fragments that immunoprecipitated with a LOB antibody in DEX- and mock-treated 35S:LOB-GR seedlings. In DEX-treated compared with mock-treated samples, we detected enrichment of a fragment spanning the LBD motifs in the BAS1 promoter (Fig. 3D, region A). A lower level of enrichment was observed for a fragment spanning the binding sites in exon 3 (Fig. 3D, region B). No enrichment of a control fragment 3′ to the BAS1 coding sequence (CDS) that lacked LOB binding sites was detected (Fig. 3D, region C). Using EMSAs, we demonstrated that the DNA-binding LOB domain (LD) (18) bound specifically to a fragment from the BAS1 promoter that contained the two partial LBD motifs (Fig. 3E). LD did not bind to fragments in which the two central G residues in either site were mutated to A residues (Fig. 3E, lanes 5 and 6), nor did mutated fragments efficiently compete for LD binding to the wild-type fragments (Fig. 3E, lanes 9 and 10). Thus, both sites are required for LD binding to the BAS1 promoter, consistent with the finding that LOB binds DNA as a homodimer (18). Taken together, these data indicate that LOB associates with LBD motifs in the BAS1 gene to directly regulate BAS1 expression.
BAS1 and LOB Have Overlapping Expression Patterns.
If BAS1 is a direct target of LOB transcriptional regulation, then BAS1 and LOB expression should partially overlap. BAS1 expression was previously shown to be light regulated (21), and publicly available microarray data indicated that BAS1 transcripts were enriched in shoot apices, roots, and immature seeds (22, 23). To characterize BAS1 expression in more detail, we examined expression in transgenic plants carrying a pBAS1:BAS1-GUS reporter construct. pBAS1:BAS1-GUS expression was observed in the basal region of young leaves and in developing seeds (Fig. S5 A and B), consistent with previously reported microarray data (22, 23). In addition, pBAS1:BAS1-GUS expression was detected in the boundary between primary and axillary shoots and weakly between axillary shoots and cauline leaves (Fig. S5C). Thus, expression of BAS1 and LOB overlaps in boundary regions, consistent with LOB functioning in regulation of BAS1 expression.
BAS1 Suppresses Organ Fusion in lob Mutants When Expressed in Organ Boundaries.
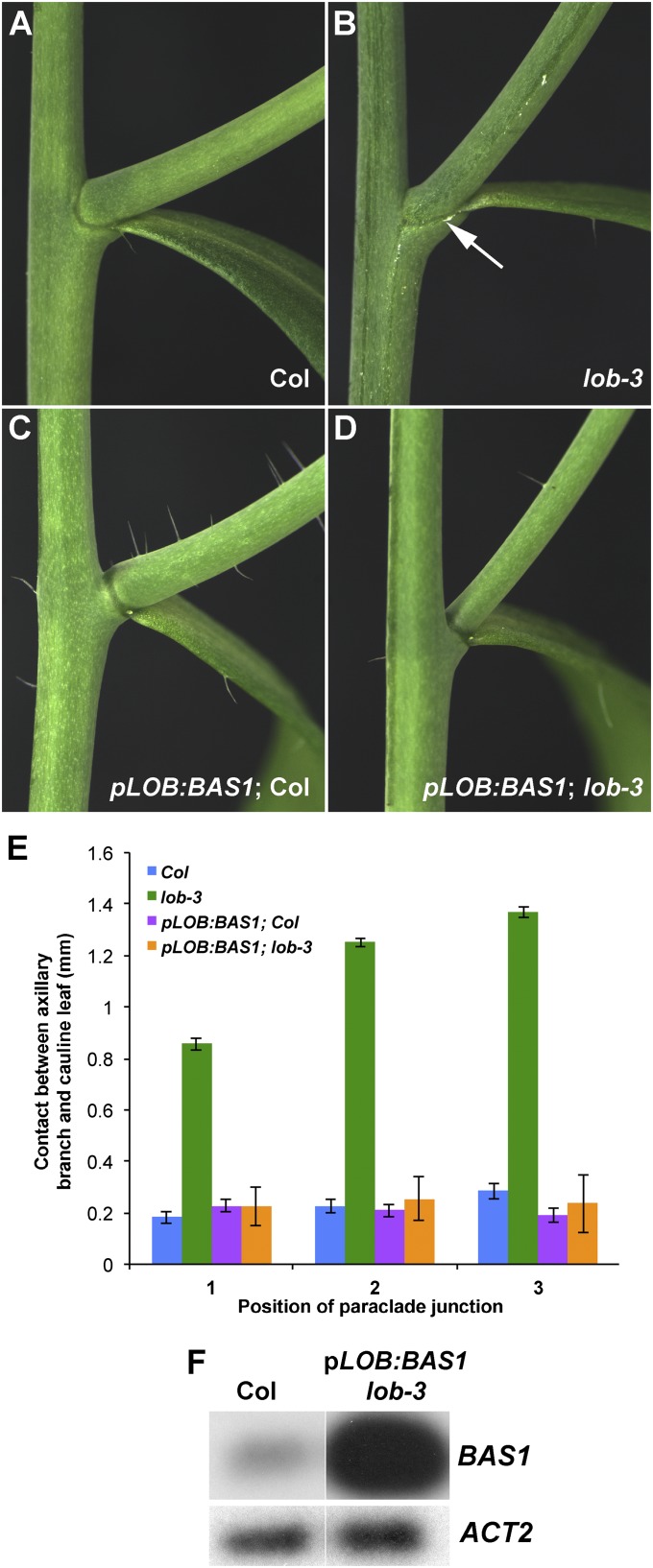
Given that BAS1 is a direct target of LOB transcriptional regulation, 35S:LOB-GR plants are predicted to ectopically express BAS1. The reduced BR sensitivity in LOB-GR plants is consistent with an increase in BAS1 activity. If LOB regulates BAS1 expression in organ boundaries, we hypothesized that reduction in BAS1 expression in the lob mutant may contribute to the fusion phenotype. To test this, we expressed BAS1 under control of the LOB promoter in wild-type and lob-3 mutant plants. In a wild-type background, pLOB:BAS1 plants produced longer pedicels but were otherwise morphologically normal. In the lob mutant background, the fusion between cauline leaves and axillary stems was suppressed by the pLOB:BAS1 construct (Fig. 4). Thus, expression of BAS1 in the LOB domain is sufficient to rescue the lob mutant phenotype, indicating that enhanced BR signaling in lob mutants results in organ fusion. bas1 loss-of-function mutants do not exhibit fusion defects (24); therefore, reduction in BAS1 activity is not the sole cause of fusion in lob mutants. Nearly 300 genes were differentially expressed following DEX induction of 35S:LOB-GR plants, many of them also BR regulated (Dataset S1). Boundary defects in lob mutants likely result from alteration in expression of a suite of genes.
Fig. 4.
Expression of BAS1 under the LOB promoter suppresses fusion in the lob mutant. (A–D) Cauline leaf/axillary stem junctions of Columbia wild type (A), lob-3 (B), transgenic pLOB:BAS1 in Col (C), and pLOB:BAS1 in lob-3 (D). Arrow in B indicates fused region. (E) Length of fused region in Col, lob-3, pLOB:BAS1 Col, and pLOB:BAS1 lob-3. Position 1 corresponds to lowest cauline leaf axil on stem. Expression of pLOB:BAS1 suppresses the fusion in lob-3. SEs (n ≥ 10) are indicated. (F) RT-PCR analysis of BAS1 transcript levels in isolated paraclade junctions of Col and pLOB:BAS1 lob-3 plants. RT-PCR products were detected by blotting and probing with gene-specific probes, following either 15 (BAS1) or 12 cycles (ACT2) of amplification.
Discussion
The establishment and maintenance of adjacent populations of cells with distinctly different cell fates are critical problems in developmental biology. Boundaries between domains play an important role in this process, but the mechanisms controlling boundary formation are relatively poorly understood (25). In plants, cells in the boundary between the meristem and organ primordia are small and divide infrequently. Reduced growth in the boundary allows the forming organ to separate from the meristem. Organ boundaries also play a role in meristem maintenance and are the site of axillary meristem formation; therefore, they contribute to the regulation of overall plant form (4). Despite the identification of a number of boundary-specific transcription factors (5–12), an understanding of the pathways they regulate to specify and maintain boundaries has not been developed.
Here we show that the Arabidopsis transcription factor LOB negatively regulates accumulation of BR in organ boundaries. Our results show that 60% of LOB-regulated genes are also regulated by BR. Furthermore, LOB and BR signaling form a feedback loop involving BR regulation of LOB accumulation and LOB repression of BR accumulation, which functions to limit growth in organ boundaries (Fig. S6). Loss of LOB function results in overgrowth of the boundary region and organ fusions, demonstrating the importance of regulating growth in this domain. Similar fusion defects have been observed in other BR signaling mutants such as bzr1-d (26). In addition, Gendron et al. (26) reported that a BZR1-YFP fusion protein accumulated to low levels in the boundary region compared with the adjacent meristem and primordia, consistent with reduced BR levels in these cells. Moreover, 60% (106) of the 175 genes that are regulated by both LOB and BR are putative BZR1 targets (Dataset S1), raising the possibility that the combined action of LOB and BZR1 influence the expression of a subset of BR-regulated genes.
BRs have long been known to have important functions in plant growth, primarily by promoting cell expansion (15), and have recently been implicated in cell cycle regulation (27–29). Our findings indicate that reduced local BR accumulation and response are critical for patterning shoot architecture. Formation of a region of reduced auxin accumulation has recently been reported to be important for specification of valve margins in the Arabidopsis fruit (30), indicating that the formation of local hormone minima may be a general mechanism for organ patterning in plants.
Materials and Methods
Plant Material, Growth Conditions, and Transformation.
Arabidopsis thaliana plants were grown under standard conditions as previously described (12). Binary T-DNA vectors were introduced into Agrobacterium tumefaciens GV3101, and Arabidopsis plants were transformed by floral dip (31). Transformed plants were selected on Murashige and Skoog medium (32) supplemented with 50 μM kanamycin or 50 μM phosphinothricin. lob::DsE and lob-2 have been previously described (12); lob-3 (SALK_042599) is from the Salk T-DNA collection and is in Columbia-0 (33). Boundary marker lines ET4016 (11) and GT185 (34) are in the Landsberg erecta background.
Phenotypic Analyses.
BR-response assays were conducted on seedlings grown vertically in the dark for 4–7 d on MS medium with or without 3 μM DEX and in the presence of variable concentrations of BL. Hormone concentrations were as follows: BL, 1 nM to 2 μM; IAA, 1 μM; GA3, 10 μM; ACC, 20 μM. Hypocotyls were measured using MCID Elite 7.0 software (Imaging Research). Measurements of contact length between stems and cauline leaf were made using a digimatic caliper (model 700-113; Mitutoyo).
Constructs.
The LOB CDS was amplified with primers that contained introduced restriction sites and subcloned into pBIΔGR (35) to generate the 35S:LOB-GR construct with GR fused in frame to the C terminus of LOB. To construct pLOB:GUS:3′IGR, the 2,557-bp intergenic region 3′ to the LOB stop codon was amplified from genomic DNA using primers LOButrF and LOBigrR, which contained introduced XbaI and PstI restriction sites (Table S1). The amplified fragment replaced the 3′ Octopine synthase terminator in pLOB5.0:GUS (12). To construct pLOB:LOB:3′IGR and pLOB:BAS1:3′IGR, the GUS CDS was replaced with the LOB or BAS1 CDS. The pBAS1:BAS1-GUS construct was modified from a previously described construct that contained a shorter BAS1 promoter (21). pBAS1:BAS1-GUS contains 6,084 bp of genomic DNA upstream of the BAS1 ATG, and the BAS1 gene, including introns, fused, in frame, to GUS.
Expression Analysis.
Total RNA was isolated with TRIzol reagent. RT-PCR was performed as described previously (36). RNA gel blot hybridizations were performed as previously described (37) using gene-specific BAS1, TCH4, and SAUR-AC1 probes. Primers and amplification conditions for ACT2 and LOB were as described previously (12). Primers for BAS1 amplification are shown in Table S1. For DEX treatment in the presence of cycloheximide, DEX was used at 5 µM, and cycloheximide was used at 10 µM.
Histology and Microscopy.
GUS histochemical staining and image capture were performed as previously described (34). Cross sections were performed as previously described (11). Images were captured as previously described (36).
Microarray Experiment.
Nine-day-old 35S:LOB-GR and Col-0 seedlings, grown on MS plates, were flooded with MS medium containing 5 µM DEX or a mock solution and incubated, with shaking, for 4 h. Three independent biological replicates were performed for each treatment. Total RNA was isolated using TRIzol, followed by purification over RNeasy columns (Qiagen). Labeling and hybridization to Arabidopsis ATH1 GeneChips were done at the University of California–Riverside (UCR) Core Instrumentation Facility following the manufacturer’s instructions (Affymetrix). Data were analyzed in R using Bioconductor packages. The Affy package was used for robust multi-array average normalization, and differentially expressed genes were identified using the linear models for microarray data package (38). Differentially expressed genes were identified based on a false-discovery rate (FDR)-adjusted P value ≤ 0.001 (39). GO term enrichment analyses were done with the GOHyperGAll script, which uses hypergeometric distribution (40).
ChIP and EMSA.
For ChIP, 12-d-old 35S:LOB-GR seedlings, grown on MS plates, were induced by flooding with MS medium containing 15 µM DEX or a mock solution and incubated with shaking for 3 h. ChIP was performed as described (41) using an anti-LOB antibody. The ChIP DNA was analyzed by PCR, and LOB binding was calculated as the ratio between the DEX-treated and mock-treated samples. Data were normalized to the control gene At3g26980. Primer sequences are shown in Table S1.
EMSAs were performed with the LD protein as described (18). Probes were generated from a 133-bp fragment corresponding to the upstream region of BAS1 (BAS1 A) and a 110-bp fragment corresponding to the third exon region of BAS1 (BAS1 B). Primer sequences used to generate probes are shown in Table S1. Probe fragments were cloned and sequenced to verify the integrity of the LBD sites. The QuikChange Site Directed Mutagenesis Kit (Stratagene) was used to create mutant α and β versions of the BAS1 A probe according to the manufacturer’s instructions. To create radiolabeled BAS1 A and B probes, fragments were PCR amplified and end-labeled using T4 polynucleotide kinase and 32P-γ-ATP. Cold competition experiments were performed using a 30-fold excess of unlabeled DNA.
Supplementary Material
Acknowledgments
We thank the Arabidopsis Biological Resource Center for providing lob-3 seed, Zhiyong Wang for bzr1-1d seed, Alan Lloyd for pBI-∆GR, and Michelle Brown for assistance with BR response assays. We acknowledge Office of Basic Energy Sciences of the US Department of Energy Grant DE-FG02-05ER15649 (to P.S.S. and T.G.) for funding the microarray studies and BAS1 characterization and National Science Foundation Grant IBN-0420202 (to P.S.S.) for funding LOB phenotypic analyses.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE34209).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1210789109/-/DCSupplemental.
References
- 1.Barton MK. Twenty years on: The inner workings of the shoot apical meristem, a developmental dynamo. Dev Biol. 2010;341(1):95–113. doi: 10.1016/j.ydbio.2009.11.029. [DOI] [PubMed] [Google Scholar]
- 2.Hussey G. Cell division and expansion and resultant tissue tensions in shoot apex during formation of a leaf primordium in tomato. J Exp Bot. 1971;22:702–714. [Google Scholar]
- 3.Breuil-Broyer S, et al. High-resolution boundary analysis during Arabidopsis thaliana flower development. Plant J. 2004;38(1):182–192. doi: 10.1111/j.1365-313X.2004.02026.x. [DOI] [PubMed] [Google Scholar]
- 4.Rast MI, Simon R. The meristem-to-organ boundary: More than an extremity of anything. Curr Opin Genet Dev. 2008;18(4):287–294. doi: 10.1016/j.gde.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 5.Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell. 1997;9(6):841–857. doi: 10.1105/tpc.9.6.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greb T, et al. Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 2003;17(9):1175–1187. doi: 10.1101/gad.260703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vroemen CW, Mordhorst AP, Albrecht C, Kwaaitaal MA, de Vries SC. The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell. 2003;15(7):1563–1577. doi: 10.1105/tpc.012203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hibara K, et al. Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell. 2006;18(11):2946–2957. doi: 10.1105/tpc.106.045716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borghi L, Bureau M, Simon R. Arabidopsis JAGGED LATERAL ORGANS is expressed in boundaries and coordinates KNOX and PIN activity. Plant Cell. 2007;19(6):1795–1808. doi: 10.1105/tpc.106.047159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raman S, et al. Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J. 2008;55(1):65–76. doi: 10.1111/j.1365-313X.2008.03483.x. [DOI] [PubMed] [Google Scholar]
- 11.Lee DK, Geisler M, Springer PS. LATERAL ORGAN FUSION1 and LATERAL ORGAN FUSION2 function in lateral organ separation and axillary meristem formation in Arabidopsis. Development. 2009;136(14):2423–2432. doi: 10.1242/dev.031971. [DOI] [PubMed] [Google Scholar]
- 12.Shuai B, Reynaga-Peña CG, Springer PS. The LATERAL ORGAN BOUNDARIES gene defines a novel, plant-specific gene family. Plant Physiol. 2002;129(2):747–761. doi: 10.1104/pp.010926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho E, Zambryski PC. ORGAN BOUNDARY1 defines a gene expressed at the junction between the shoot apical meristem and lateral organs. Proc Natl Acad Sci USA. 2011;108(5):2154–2159. doi: 10.1073/pnas.1018542108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Picard D, Salser SJ, Yamamoto KR. A movable and regulable inactivation function within the steroid binding domain of the glucocorticoid receptor. Cell. 1988;54(7):1073–1080. doi: 10.1016/0092-8674(88)90122-5. [DOI] [PubMed] [Google Scholar]
- 15.Clouse SD. Brassinosteroids. Arabidopsis Book. 2011;9:e0151. doi: 10.1199/tab.0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ephritikhine G, Fellner M, Vannini C, Lapous D, Barbier-Brygoo H. The sax1 dwarf mutant of Arabidopsis thaliana shows altered sensitivity of growth responses to abscisic acid, auxin, gibberellins and ethylene and is partially rescued by exogenous brassinosteroid. Plant J. 1999;18(3):303–314. doi: 10.1046/j.1365-313x.1999.00454.x. [DOI] [PubMed] [Google Scholar]
- 17.Wang ZY, et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell. 2002;2(4):505–513. doi: 10.1016/s1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- 18.Husbands A, Bell EM, Shuai B, Smith HM, Springer PS. LATERAL ORGAN BOUNDARIES defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res. 2007;35(19):6663–6671. doi: 10.1093/nar/gkm775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang ZY, Bai MY, Oh E, Zhu JY. Brassinosteroid signaling network and regulation of photomorphogenesis. Annu Rev Genet. 2012;46:701–724. doi: 10.1146/annurev-genet-102209-163450. [DOI] [PubMed] [Google Scholar]
- 20.Neff MM, et al. BAS1: A gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc Natl Acad Sci USA. 1999;96(26):15316–15323. doi: 10.1073/pnas.96.26.15316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turk EM, et al. CYP72B1 inactivates brassinosteroid hormones: An intersection between photomorphogenesis and plant steroid signal transduction. Plant Physiol. 2003;133(4):1643–1653. doi: 10.1104/pp.103.030882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmid M, et al. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37(5):501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- 23.Winter D, et al. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE. 2007;2(8):e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turk EM, et al. BAS1 and SOB7 act redundantly to modulate Arabidopsis photomorphogenesis via unique brassinosteroid inactivation mechanisms. Plant J. 2005;42(1):23–34. doi: 10.1111/j.1365-313X.2005.02358.x. [DOI] [PubMed] [Google Scholar]
- 25.Dahmann C, Oates AC, Brand M. Boundary formation and maintenance in tissue development. Nat Rev Genet. 2011;12(1):43–55. doi: 10.1038/nrg2902. [DOI] [PubMed] [Google Scholar]
- 26.Gendron JM, et al. Brassinosteroids regulate organ boundary formation in the shoot apical meristem of Arabidopsis. Proc Natl Acad Sci USA. 2012;109:21152–21157. doi: 10.1073/pnas.1210799110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheon J, Park SY, Schulz B, Choe S. Arabidopsis brassinosteroid biosynthetic mutant dwarf7-1 exhibits slower rates of cell division and shoot induction. BMC Plant Biol. 2010;10:270. doi: 10.1186/1471-2229-10-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.González-García MP, et al. Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development. 2011;138(5):849–859. doi: 10.1242/dev.057331. [DOI] [PubMed] [Google Scholar]
- 29.Hacham Y, et al. Brassinosteroid perception in the epidermis controls root meristem size. Development. 2011;138(5):839–848. doi: 10.1242/dev.061804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sorefan K, et al. A regulated auxin minimum is required for seed dispersal in Arabidopsis. Nature. 2009;459(7246):583–586. doi: 10.1038/nature07875. [DOI] [PubMed] [Google Scholar]
- 31.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 32.Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant. 1962;15:473–497. [Google Scholar]
- 33.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301(5633):653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 34.Sundaresan V, et al. Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 1995;9(14):1797–1810. doi: 10.1101/gad.9.14.1797. [DOI] [PubMed] [Google Scholar]
- 35.Lloyd AM, Schena M, Walbot V, Davis RW. Epidermal cell fate determination in Arabidopsis: Patterns defined by a steroid-inducible regulator. Science. 1994;266(5184):436–439. doi: 10.1126/science.7939683. [DOI] [PubMed] [Google Scholar]
- 36.Lin WC, Shuai B, Springer PS. The Arabidopsis LATERAL ORGAN BOUNDARIES-domain gene ASYMMETRIC LEAVES2 functions in the repression of KNOX gene expression and in adaxial-abaxial patterning. Plant Cell. 2003;15(10):2241–2252. doi: 10.1105/tpc.014969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martienssen RA, Barkan A, Freeling M, Taylor WC. Molecular cloning of a maize gene involved in photosynthetic membrane organization that is regulated by Robertson’s Mutator. EMBO J. 1989;8(6):1633–1639. doi: 10.1002/j.1460-2075.1989.tb03553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:e3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 39.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc, B. 1995;57:289–300. [Google Scholar]
- 40.Horan K, et al. Annotating genes of known and unknown function by large-scale coexpression analysis. Plant Physiol. 2008;147(1):41–57. doi: 10.1104/pp.108.117366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morohashi K, Xie Z, Grotewold E. Gene-specific and genome-wide ChIP approaches to study plant transcriptional networks. Methods Mol Biol. 2009;553:3–12. doi: 10.1007/978-1-60327-563-7_1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.