Abstract
Although aberrant protein aggregation has been conclusively linked to dozens of devastating amyloid diseases, scientists remain puzzled about the molecular features that render amyloid fibrils or small oligomers toxic. Here, we report a previously unobserved type of amyloid fibril that tests as cytotoxic: one in which the strands of the contributing β-sheets are out of register. In all amyloid fibrils previously characterized at the molecular level, only in-register β-sheets have been observed, in which each strand makes its full complement of hydrogen bonds with the strands above and below it in the fibril. In out-of-register sheets, strands are sheared relative to one another, leaving dangling hydrogen bonds. Based on this finding, we designed out-of-register β-sheet amyloid mimics, which form both cylindrin-like oligomers and fibrils, and these mimics are cytotoxic. Structural and energetic considerations suggest that out-of-register fibrils can readily convert to toxic cylindrins. We propose that out-of-register β-sheets and their related cylindrins are part of a toxic amyloid pathway, which is distinct from the more energetically favored in-register amyloid pathway.
Keywords: X-ray crystallography, BAMs
In contrast to infectious and metabolic disorders, for which researchers can usually uncover the causative entity and the pathway to disease, amyloid diseases have challenged scientists to identify the etiologic agents and the initial pathological events (1–3). Part of the difficulty is that pathways of protein aggregation are diverse, leading to multiple species differing in size, structure, lifetime, and cytotoxicity (4–8). As the most-studied aggregation species, amyloid fibrils have long been associated with devastating human pathologies, including Alzheimer’s disease, type II diabetes, and prion disease (2). However, other proteins form amyloid-like aggregates with normal biological functions (9). Studies by NMR, EPR, X-ray diffraction, and scanning mutagenesis have shown that both deleterious and functional amyloid fibrils are made up of extended β-strands running perpendicular to the fibril axis and hydrogen bonded into β-sheets (10–14). The sheets are normally paired into steric zippers, and most often, the strands run parallel to each other and are strictly in-register (10, 11, 15–17). However, in some cases, the strands are antiparallel (18), and some antiparallel fibrils have been found to be more cytotoxic than parallel counterparts (19). Studies of prion (20), HypF-N (21), and Aβ1–40 (22) indicate that different aggregate morphologies have different levels of cytotoxicity. Therefore, it is important to investigate the various amyloid fibrils from different aggregation pathways, especially those fibrils related to toxic amyloid pathogenesis.
Complicating the picture is the variety of oligomers found apparently on the pathways to fibrils, which are more toxic than the fibrils (2, 23, 24). Amyloid oligomers with distinct structural features exhibit different cytotoxicity. A diversity of structural models has been suggested for amyloid oligomers, including β-barrels (25–28), α-helix bundles (29), and parallel in-register β-sheets (30). However, structural characterization of toxic amyloid oligomers at the atomic level is hindered by their highly transient and polymorphic properties. Recently, we reported the atomic structure of cylindrin, a toxic oligomer formed by six identical 11-residue out-of-register antiparallel β-strands in the shape of a barrel (31). Instead of the in-register β-strands typically observed in amyloid fibril structures, cylindrin is formed by out-of-register β-strands. Molecular dynamics (MD) simulation shows that the transition from the out-of-register β-strands of cylindrin to the in-register strands of fibrils is energetically very costly, suggesting that they might be involved in distinct aggregation pathways (31).
In this study, we found that, in addition to rolling into cylindrin (31), out-of-register β-strands can also pack into a fibril structure that is toxic to mammalian cells. To further investigate how out-of-register β-strands mediate amyloid oligomer and fibril formation, we designed a chemical model system—β-sheet amyloid mimics (BAMs). These BAMs are unable to self-assemble into in-register β-strands, and instead, they form out-of-register, cylindrin-like oligomers and fibrils that are confirmed by crystal structures. On the basis of biochemical and computational studies, we believe that the cylindrin-like oligomer represents an on-pathway intermediate of out-of-register amyloid fibril formation. Together, out-of-register oligomers and fibrils define a distinct toxic amyloid aggregation pathway.
Results
KDWSFY (β2-microglobulin58–63) Protofilaments Feature Out-of-Register β-Strands and Are Cytotoxic.
Here, we report a rare out-of-register amyloid-like protofilament formed from residues 58–63 of the protein β2-microglobulin (β2m) with sequence KDWSFY, termed KDWSFY (β2m58–63), differing fundamentally from the several dozen in-register protofilaments that we have previously characterized from other proteins (10). Earlier, we observed by EM that the segment KDWSFY (β2m58–63) forms amyloid-like fibrils, and in the context of the full protein, it forms the spine of one polymorphic form of fibrillar β2m (32). The crystal structure of KDWSFY (β2m58–63) reveals that it forms an out-of-register protofilament consisting of two out-of-register β-sheets mating at a dry, highly complementary interface (Fig. 1 A–C and Table S1), similar to the structures of the numerous in-register protofilaments (10). Whereas the in-register β-strands observed in previous protofilament structures are perpendicular to the fibril axis and all of the residues in β-strands engage in interstrand hydrogen bonds (Fig. 1D), the β-strands in the KDWSFY structure are not perpendicular to the fibril axis. Instead, each β-strand is at an angle of ∼50° from the perpendicular, and β-strands from adjacent sheets form a crossing angle of ∼80° instead of the 0° angle found for in-register sheets (Fig. 1A). In contrast to the identical hydrogen-bonding interfaces of in-register sheets, each antiparallel out-of-register β-sheet possesses two hydrogen-bonding interfaces: a strong interface with six hydrogen bonds and a weak interface with two hydrogen bonds (Fig. 1B). Therefore, unlike in-register sheets, out-of-register sheets leave unsatisfied hydrogen bond donors and acceptors at the ends of the strands that are not engaged in interstrand hydrogen bonds but to some extent, are satisfied by water molecules observed in the crystal structure. These substantial differences in hydrogen bonding indicate that out-of-register fibrils form through a separate aggregation pathway from in-register fibrils. Pertinent to understanding the cytotoxicity of amyloid, MTT [3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide]-based cell viability assays showed that the KDWSFY (β2m58–63) fibrils are toxic to mammalian cells in a dose-dependent manner (Fig. 1E). As a control, fibrils formed by in-register sheets exhibited little or no toxicity (Figs. S1 and S2 and Table S2). The comparison of toxicity between in-register and out-of-register fibrils suggests that out-of-register fibrils may play a role in the etiology of aggregation diseases.
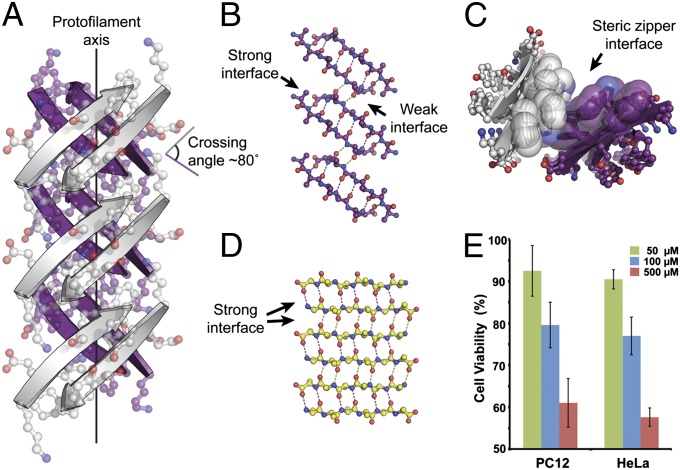
Fig. 1.
Crystal structure of out-of-register, antiparallel protofilament formed by the amyloidogenic peptide KDWSFY (β2m58–63) and its fibril toxicity. (A) View perpendicular to the protofilament axis. The two β-sheets are colored in purple and white. Side chains are shown in ball-and-stick representation. The protofilament axis is denoted by a black line. Note that the β-strands are antiparallel, out-of-register, and not perpendicular to the protofilament axis. The crossing angle between the β-strands of adjacent β-sheets is ∼80°. (B) The geometry of the back (purple) out-of-register β-sheet. For clarity, side chains are omitted. Hydrogen bonds are shown by dotted lines. (C) View down the protofilament axis showing the dry steric zipper interface between the pair of β-sheets. Side chains contributing to this interface are shown in space-filling representation. (D) The geometry of a single in-register antiparallel β-sheet from the KLVFFA crystal structure (Protein Data Bank ID code 2Y2A). (E) Assessment of cytotoxicity of KDWSFY (β2m58–63) fibril samples by MTT-based cell viability assay. KDWSFY (β2m58–63) fibrils show toxicity to both PC12 and HeLa cell lines in a dose-dependent manner. Error bars represent 1 SD (n = 4).
Stability of Out-of-Register Packing.
The reduced number of interstrand hydrogen bonds in out-of-register sheets suggests that they may be energetically less favorable than in-register sheets. This inherent instability may explain why out-of-register sheets are rarely the dominant species in polymorphic fibrils, greatly lowering their chance to be captured by ssNMR or crystallization. In nearly all previously reported structures, β-sheets of fibrils are in-register (10–12). To understand why the KDWSFY fibril is stable with out-of-register packing, we applied Rosetta modeling and MD simulations (33). The results show that the abundance of bulky aromatic side chains in KDWSFY is sterically incompatible with in-register packing, whereas these clashes are avoided by out-of-register packing. Thus, in the case of KDWSFY, the out-of-register fibril represents a lower energy state than the in-register fibril (Fig. S3), explaining how we were able to capture the out-of-register state in the crystal.
Design of Out-of-Register β-Strands.
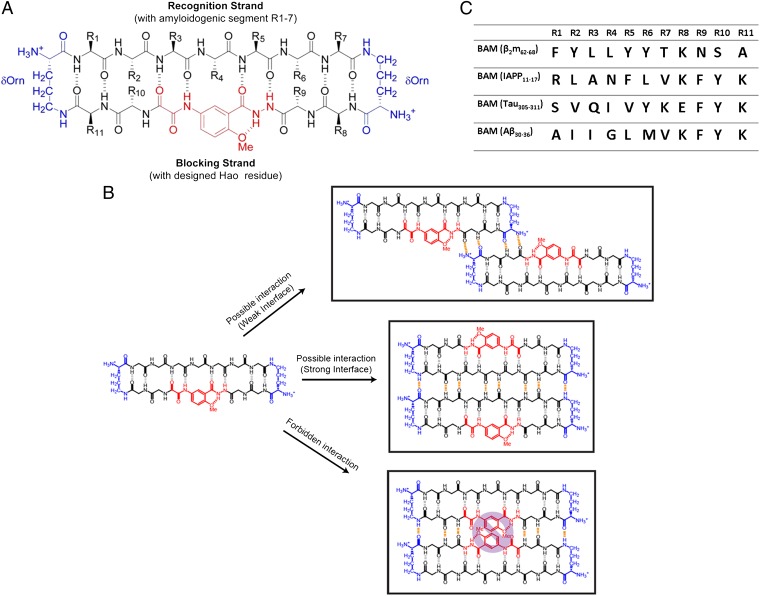
To further study the assembly and toxicity of out-of-register β-sheets from a range of amyloid proteins, we designed peptides that pack exclusively in out-of-register β-sheets. Our design is based on amyloid β-sheet mimics (34); each is a cyclic molecule of two covalently connected antiparallel β-strands (now termed BAMs). In the recognition strand, an amyloidogenic sequence is grafted and restrained to maintain its extended β-strand conformation (Fig. 2A). Each recognition strand is capable of forming an antiparallel β-interaction through hydrogen bonds to the recognition strand of an identical BAM molecule. The second strand in each BAM is termed the blocking strand (35), which contains the nonnatural amino acid Hao flanked by dipeptides. Hao prevents full hydrogen bonding of blocking strands, forcing the formation of an out-of-register β-strand interface with a neighboring BAM. Only the dipeptides on either side of Hao are available to bond to identical molecules (Fig. 2B). Therefore, if a BAM forms an amyloid-like oligomer or fibril, it must form an out-of-register structure with strong hydrogen-bonding interfaces between recognition strands and weak interfaces between blocking strands, similar to the out-of-register assembly observed in the KDWSFY fibril (Fig. 1B).
Fig. 2.
Structure and interaction of BAMs. (A) Structure of BAMs. The macrocyclic ring is connected by two ornithines colored in blue. The Hao residue, which can support intramolecular but not intermolecular hydrogen bonding, is colored in red. All natural amino acids are colored black. In BAM, the recognition strand (upper strand) contains 7 natural aa residues that are replaced with the various amyloidogenic segments in this study. The blocking strand (lower strand) contains 4 natural aa residues to facilitate the folding of the β-conformation and increase the solubility of the molecule. R10 is sometimes replaced with 4-bromo phenylalanine for phase determination of crystal structures. (B) Three types of potential BAM interactions are shown. (Top) Between blocking strands out of register, four intermolecular hydrogen bonds (colored in orange) can be formed. (Middle) Between recognition strands in register, eight intermolecular hydrogen bonds can be formed. (Lower) Between blocking strands in register, six intermolecular hydrogen bonds can be formed. However, the clash between Hao residues precludes this interaction. (C) Sequences of four different BAMs for out-of-register assembly studies.
BAMs Form Toxic Amyloid-Like Oligomers and Fibrils.
We prepared four BAMs with amyloidogenic segments grafted from four different disease-related amyloid proteins (Fig. 2C). These proteins form amyloid-like oligomers and fibrils and exhibit cytotoxicity. Our previous studies showed that macrocyclic peptides form oligomers in solution (36) and mimic amyloid oligomers for structural studies (37). Here, we find that, on incubation at 37 °C for 0.5 h, BAMs form amyloid-like oligomers that are recognized by A11, the polyclonal amyloid–oligomer-specific conformational antibody (Fig. S4A). Diffusion coefficients of BAMs (Aβ30–36) measured by NMR confirmed that they self-associate in solution at millimolar concentrations, consistent with the formation of BAM oligomers. (Fig. S4B). We also observed BAM oligomers to be toxic to mammalian cells (Fig. S4C). On additional incubation at 37 °C with agitation for 4 d, three BAMs form amyloid-like fibrils characterized by transmission EM, cross-β fibril diffraction, and Congo red birefringence (Fig. 3A). The formation of BAM fibrils confirms that fibrils can form even when in-register β-sheet packing is sterically prohibited. MTT assays showed that all of our BAM fibrils are toxic to PC12 cells (Fig. 3B and Table S3). As a control, nonfibril-forming BAMs have little toxic effect, ruling out the possibility that cytotoxicity derives from the macrocyclic platform.
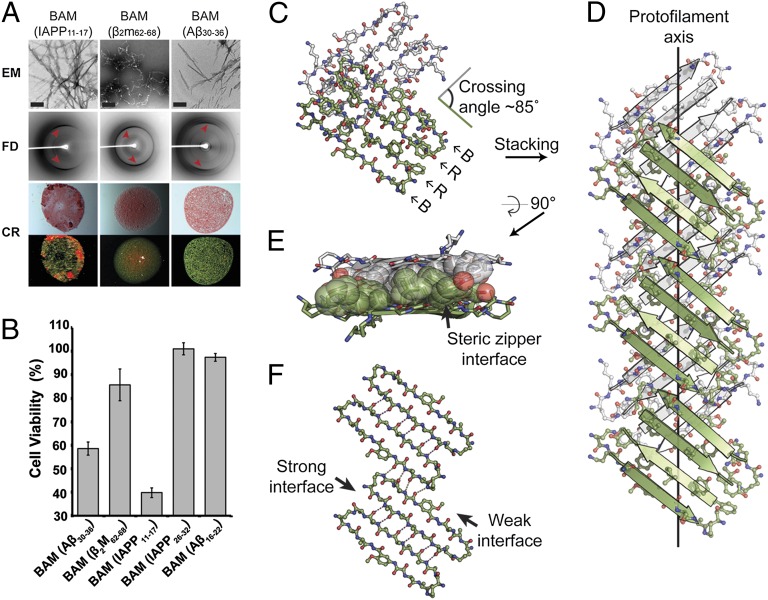
Fig. 3.
Characterization of fibril-forming BAMs showing their amyloid-like character. (A) Negatively stained transmission electron micrographs (EM), fibril diffraction (FD), and Congo red staining (CR) of fibrils formed by BAM (IAPP11–17), BAM (β2m62–68), and BAM (Aβ30–36). Black bars in the EM images represent 200 nm. Meridional reflections corresponding to about 4.7 Å are highlighted by red arrowheads in FD images. CR of BAM fibril pellets shows apple-green birefringence. Pictures of the same view taken under normal (upper row) and polarized light (lower row) are shown. (B) MTT-based cytotoxicity assays of BAMs with mammalian cell lines PC12. Fibril-forming BAMs (left three) show significant cytotoxicity (P < 0.01) compared with nonfibril-forming BAMs [BAM (IAPP26–32) and BAM (Aβ16–22) as determined by one-sided Student t test] (Table S3). Error bars represent 1 SD calculated from four replicates. The concentration of each BAM is 50 μM (monomer equivalence). (C) The asymmetric unit of the BAM (β2m62–68) crystal. Two dimers of macrocycles are shown in green and white. Recognition (R) and blocking strands (B) are labeled. The crossing angle between the green and white strands is ∼85°. (D) View perpendicular to the protofilament axis. Note that the strands are not perpendicular to the protofilament axis and the lack of registration between dimers within a single sheet. (E) A view 90° rotated from D looking down the protofilament axis shows the steric zipper interface. Side chains contributing to the interface are shown in space-filling representation. For clarity, other side chains are omitted. (F) Geometry of a single sheet showing the out-of-register nature of the weak interface. Only backbone atoms are shown. Hydrogen bonds are shown by dotted lines.
Crystal Structure of BAM (Aβ30–36): An Out-of-Register, Cylindrin-Like Oligomer.
In previous work, we determined the crystal structure of BAM (Aβ30–36) in oligomeric form (34), which we now recognize as having cylindrin-like structure and properties. BAM (Aβ30–36) displays β-barrel–like tetramers (Fig. 4 A–D and Fig. S5) in the crystal structure. In agreement with our design, the backbone of the tetramer exhibits an out-of-register β-strand pattern, with both strong and weak hydrogen bonded interfaces (Fig. 4D). However, instead of repeating in 1D, leading to an out-of-register fibril, the macrocyclic rings of BAM (Aβ30–36) roll into a β-barrel. A key driving force of this barrel-like structure is the formation of a dry interface; the barrel core is devoid of water, and instead, it is filled with the side chains of methionine and phenylalanine (Fig. 4C). Structurally, the out-of-register β-barrel of this BAM oligomer is similar to cylindrin—a toxic amyloid oligomer (Fig. 4 E–H) (31). Although similar in basic structure, these two β-barrels differ in shear number and number of β-strands, indicating the possible variety of cylindrin-like assemblies. The fact that both cylindrin and this BAM oligomer are cytotoxic and recognized by the A11 antibody may suggest that the out-of-register β-barrel represents a structural motif common to various toxic amyloid oligomers.
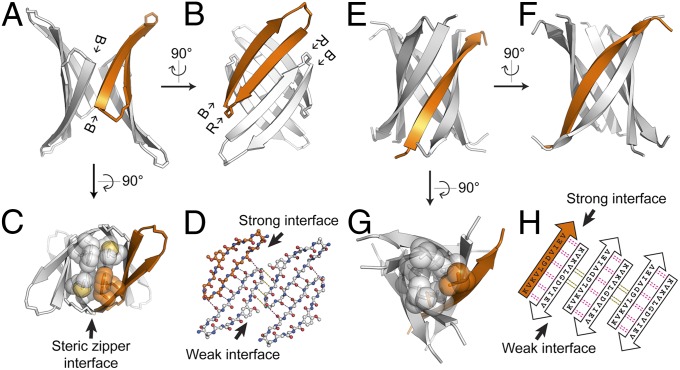
Fig. 4.
Comparison of two cylindrin-like oligomers: crystal structures of BAM (Aβ30–36) and K11V (αB crystallin68–162). (A) The crystal structure of the fibril-forming BAM (Aβ30–36) in cartoon form shows a cylindrin-like oligomer containing four macrocycles, one of which is colored in orange. The view is oriented to show a weak interface between the blocking strands (B). (B) A view 90° rotated from A showing a strong interface between two recognition strands (R). (C) Another view 90° rotated from A showing the steric zipper-like interface in the center of the barrel. Side chains contributing to this interface are shown in space-filling representation. (D) Geometry of the cylindrin-like tetramer. Only backbone atoms are shown. Hydrogen bonds are shown by dotted lines. (E) Cartoon representation of cylindrin structure of K11V (Protein Data Bank ID code 3SGO) from αB crystallin. The view is oriented to show a weak interface analogous to A. (F) A view 90° rotated from E showing a strong interface analogous to B. (G) Another view 90° rotated from E showing the center of the cylindrin. Side chains contributing to the steric zipper-like core are shown in space-filling representation. (H) Geometry of K11V cylindrin. Pairs of hydrogen bonds between corresponding residues are shown by magenta (for strong interface) and black (for weak interface) dotted lines. Notice the out-of-register weak interfaces in both D and H.
Crystal Structures of BAM (β2m62–68) and BAM (Tau305–311): Out-of-Register Protofilaments.
We next determined two crystal structures of BAMs in fibrillar form (Fig. 3 C–F, Fig. S6, and Table S4), finding steric zipper-like structures that were comprised of two β-sheets each. However, in contrast to classic in-register steric zipper protofilaments, both new structures feature out-of-register fibril-like packing reminiscent of the out-of-register structure of KDWSFY. The asymmetric unit of BAM (β2m62–68) crystals contains four macrocyclic rings that form two dimers (Fig. 3C), and each dimer is formed through hydrogen bonding between adjacent recognition strands. The blocking strands hydrogen bond with identical strands on other molecules through the residues flanking Hao resulted in protofilaments that span the entire length of the crystal (Fig. 3 D and F). These bonds create the out-of-register BAM (β2m62–68) protofilament comprised of pairs of β-sheets held together by the dry and highly complementary interfaces typical of steric zippers (Fig. 3E). These features are also observed in the structure of BAM (Tau305–311) (Fig. S6). Energetic analysis reveals that BAMs that form fibrils have tighter intermolecular complementarity and stronger binding energy than those BAMs that do not form fibrils (Table S5).
Out-of-Register β-Sheets Suggest a Distinct Amyloid Aggregation Pathway.
Because BAMs form both cylindrin-like, out-of-register oligomers and out-of-register fibrils, we studied the transition from cylindrin to fibril by both experiments and MD. These studies suggest that, in addition to the well-known in-register pathway to amyloid fibrils, a second amyloid aggregation pathway is possible for out-of-register β-strands. Experiments described above show that BAMs first form oligomers, and then, on additional incubation, they transit to fibrils. MD results are consistent with these experiments; they show that the energy barrier separating BAM (Aβ30–36) oligomers and fibrils is only ∼1.7 kcal/mol-of-tetramer (1 kcal = 4.18 kJ) (Fig. 5). Thus, we expect interchange of out-of-register fibrils and oligomers to be a common occurrence at physiological temperature, needing only a small re-registration of weak interface hydrogen bonding to trigger this transition. For the transition from cylindrin to fibril, the cylindrin sheets flatten and bond face to face to form the out-of-register fibril (Fig. S7). In contrast, interconversion between in-register and out-of-register β-sheets is energetically more expensive, because a complete rearrangement of the hydrogen-bonding network is required (31). Thus, we conclude that there may be separate pathways for formation of in-register and out-of-register fibrils and that the components of the out-of-register pathway are less stable, because they contain unsatisfied hydrogen bonds (Fig. 6).
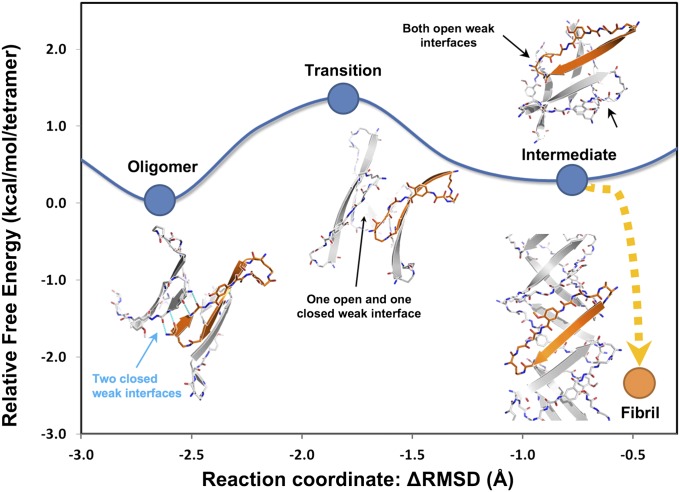
Fig. 5.
Free energy landscape associated with the out-of-register aggregation pathway of BAM (Aβ30–36). The Gibbs free energy landscape of the structural conversion from the cylindrin-like, out-of-register oligomer state to out-of-register transition state to out-of-register intermediate state was explored by MD simulation with explicit water molecules. Representative structural models from the MD trajectories of each state are shown. The reaction coordinate (x axis) is indicated by Δrmsd from the coordinates of the starting and ending states (cylindrin structure and the pair of β-sheets structure). Over the process of the conversion, the hydrogen bonds in the strong interface are maintained. The transition of the conversion is initiated by disrupting the weak interface as shown in the transition state. As the cylindrin unrolls, the weak interface further dissociates (intermediate state), and the interface within the barrel is rearranged to the out-of-register fibril state. When two cylindrin-like oligomers were simulated to form an out-of-register fibril, the free energy dropped by ∼3 kcal/mol-of-tetramer (equivalent to ∼5 RT at 300 K), suggesting a facile conversion between out-of-register oligomers and fibrils. Furthermore, a small free energy difference between the out-of-register oligomer and fibril states indicates that both are relatively stable, consistent with their detection in solution and in crystals. In contrast, the free energy of the KDWSFY out-of-register oligomer is ∼20 kcal/mol-of-monomer higher than the KDWSFY out-of-register fibril (Fig. S3), consistent with our observation that only out-of-register fibrils could be detected in solution and crystals.
Fig. 6.
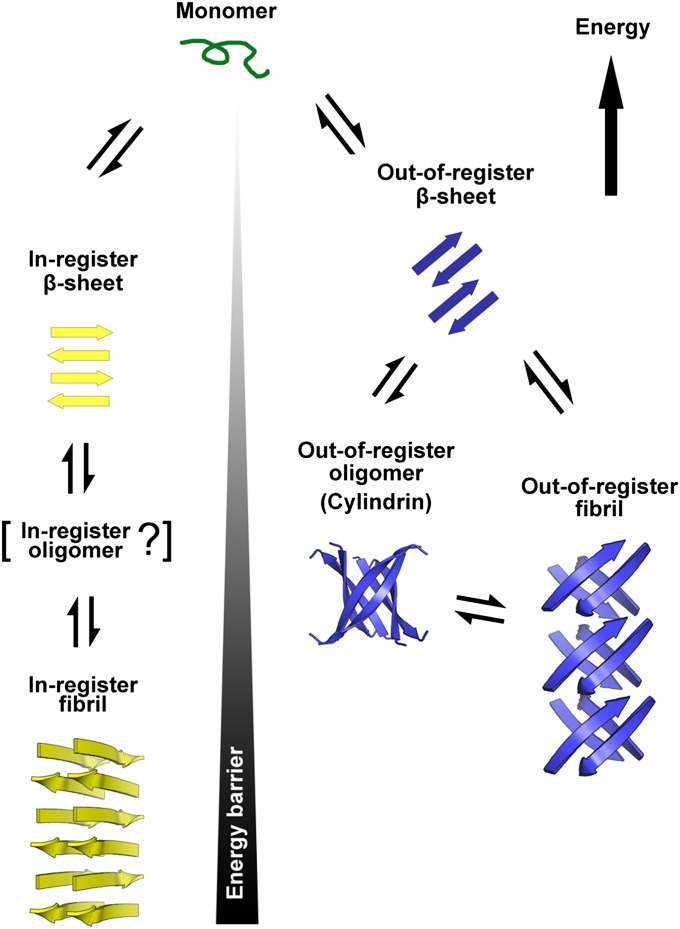
Schematic diagram of two distinct amyloid aggregation pathways. Two distinct pathways branch from monomers to fibrils: one through in-register β-strands (yellow) and the other through out-of register β-strands (blue). The in-register pathway features an in-register β-sheet that serves as the fundamental element, perhaps passing through an in-register intermediate oligomer that has not yet been characterized at the atomic level and then into the in-register fibril, which is widely observed and structurally characterized. The out-of-register pathway features an out-register β-sheet, out-of-register oligomer, and out-of-register fibril. The out-of-register oligomer and out-of-register fibril are structurally related by a common out-of-register β-sheet, suggesting easy interconversion between the two states. A high energy barrier requiring rearrangement of entire hydrogen-bonding networks separates the in-register and out-of-register β-sheet aggregation pathways. Because of their unsatisfied hydrogen bonds, out-of-register fibrils are generally less stable than in-register fibrils, and the energy differences separating out-of-register oligomers and fibrils from monomers are smaller than the corresponding differences for in-register fibrils. Hence, accumulation of in-register fibrils is common, but accumulation of out-of-register fibrils is rare.
Discussion
We find that out-of-register amyloid aggregates exhibit significant cytotoxicity. The out-of-register hexameric oligomer cylindrin was previously observed to be cytotoxic (31). Here, we find that cylindrin-like, out-of-register BAM oligomers also display toxic effects. In addition, all of the out-of-register fibrils formed by the BAMs and the out-of-register KDWSFY fibrils are toxic to mammalian cells, whereas in-register fibrils formed by various amyloidogenic peptides show little toxicity (Fig. S1). Previous studies have shown that fibrils can serve as reservoirs of toxic oligomers (38, 39). Because of the relative instability of out-of-register fibrils, we hypothesize that they are more prone to release oligomers than in-register fibrils. That is, the toxicity of out-of-register fibrils may arise from their rapid conversion to toxic out-of-register oligomers. In the case of BAMs, MD simulation suggests that the interconversion between out-of-register oligomers and fibrils is rapid, and we find that the toxicity of BAM fibrils is comparable with the toxicity of BAM oligomers (Fig. 3B and Fig. S4C). In contrast, lacking the capacity to convert into a stable cylindrin-like oligomer (Fig. S3) makes the KDWSFY fibril less toxic than BAM fibrils (Fig. 1E).
An important question is whether full-length toxic amyloid proteins, such as Aβ and Tau, are capable of forming out-of-register fibrils and cylindrin-like oligomers. The longer that the amyloidogenic segment involved in self-aggregation is, the more complex that the aggregation pathway may be and the more difficult that it may be to trap intermediate species along the aggregation pathway (4, 8, 40). However, computational simulation offers a way to examine the possibility of out-of-register oligomer and fibril formation of Aβ1–42. Rosetta modeling shows that the C-terminal segment of Aβ1–42 is capable of forming out-of-register β-strands, which can further assemble into either a cylindrin-like oligomer or out-of-register fibril (Fig. S8 A and B). The cylindrin-like oligomer formed by Aβ1–42 C terminus features a highly apolar surface, suggesting a potential role in membrane insertion (Fig. S9). In contract, in-register β-strands formed by the same segment are geometrically prohibited from forming any cylindrin-like structure by steric clashes inside the cylindrin (Fig. S8 C and D).
Thus, although additional experiments are required to understand the mechanism of the toxicity of out-of-register oligomers and fibrils, evidence presented here points to a previously unrecognized out-of-register amyloid aggregation pathway that is separate from the much studied in-register pathway and suggests that the out-of-register pathway may contribute to amyloid pathogenesis.
Materials and Methods
Sample Preparation, Crystallization, Data Collection, and Structure Determination.
Peptide KDWSFY was synthesized by Celtek Bioscience Peptides and dissolved in water at 20 mg/mL. Crystals were grown in 0.1 M bis⋅Tris propane (pH 6.5), 0.2 M sodium fluoride, and 20% (wt/vol) PEG 3,350 at 18 °C using the hanging drop vapor diffusion method. The crystals were allowed to grow for 2 wk before flash-freezing in liquid nitrogen. For iodide derivatives, the crystals were soaked in the crystallization condition with additional 0.5 M potassium iodide for 30 s before flash-freezing in liquid nitrogen. X-ray diffraction data were collected at the Advanced Photon Source (Table S1). Reflections were integrated and scaled using the XDS/XSCALE program packages (41). Experimental phases were obtained by the SIRAS method using HKL2MAP (42). The model was built using COOT (43) and refined using PHENIX (44). Coordinates and structure factor amplitudes have been deposited in the Protein Data Bank with ID code 4E0K.
BAM molecules were synthesized as reported previously (34). The grafted heptapeptides displaying on the recognition strand of nine different BAMs were chosen from various amyloidogenic segments from different amyloid proteins (45–48) (Table S5). For crystal structure determination, lyophilized BAM (β2m62–68) and BAM (Tau305–311) were dissolved in water at 10 mg/mL. Crystals were grown in 35% (vol/vol) 2-methyl-2,4-pentanediol and Na/K phosphate buffer (pH 6.2) at 18 °C using the hanging drop vapor diffusion method. The crystals were allowed to grow for 1 wk before flash-freezing in liquid nitrogen. For iodide derivatives, the crystals were soaked in the crystallization conditions with additional 0.5 M potassium iodide for 30 s before flash-freezing in liquid nitrogen. X-ray diffraction data were collected at the Advanced Photon Source (Table S4). Reflections were integrated and scaled using the XDS/XSCALE program packages (41). Experimental phases were first obtained from BAM (Tau305–311) (form I) using HKL2MAP (42) by the SAD method. BAM (Tau305–311) (forms II and III) and BAM (β2m62–68) were then solved by molecular replacement with the model of BAM (Tau305–311) (form I) using program PHASER (49). All of the models were built using COOT (43) and refined using BUSTER (50). Coordinates and structure factor amplitudes have been deposited in the Protein Data Bank with ID codes 4E0L, 4E0M, 4E0N, and 4E0O.
Transmission EM.
Five microliters each sample were spotted directly onto freshly glow-discharged carbon-coated EM grids (Ted Pella). After 3 min of incubation, grids were rinsed two times with 5 μL distilled water and stained with 1% (wt/vol) uranyl acetate for 1 min. Specimens were examined in a JEM1200-EX electron microscope at an accelerating voltage of 80 kV. Images were recorded digitally by wide-angle (top-mount) BioScan 600 W 1 × 1-K digital camera (Gatan).
Congo Red Staining.
Fibril samples were spun down at 20,000 × g. The pellet was resuspended in 100 μL 0.1 mg/mL Congo red dissolved in 10 mM Tris (pH 8.0) and 0.15 M NaCl and incubated for 30 min at room temperature. The samples were spun down again at 20,000 × g. The pellet was resuspended in 100 μL water, and the process was repeated until the supernatant was not red. The final pellet was resuspended with 5–7 μL water, pipetted onto a glass coverslip, and dried at room temperature for 15–30 min. The resulting stains were then examined by a ZEISS (SteREO Discovery.V20) microscope under normal and polarized light.
Supplementary Material
Acknowledgments
We thank beam-line staff members at Argonne Photon Source Northeastern Collaborative Access Team Beam Lines 24-ID-E/C for data collection. We also thank Drs. Dan Li and Howard Chang for useful discussions and careful reading of the manuscript. This work was supported by National Institutes of Health Grants AG-029430 and GM-097562, the National Science Foundation, and The Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4E0K, 4E0L, 4E0M, 4E0N, and 4E0O).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218792109/-/DCSupplemental.
References
- 1.Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148(6):1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 3.Härd T, Lendel C. Inhibition of amyloid formation. J Mol Biol. 2012;421(4–5):441–465. doi: 10.1016/j.jmb.2011.12.062. [DOI] [PubMed] [Google Scholar]
- 4.Eichner T, Radford SE. A diversity of assembly mechanisms of a generic amyloid fold. Mol Cell. 2011;43(1):8–18. doi: 10.1016/j.molcel.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 5.Ma B, Nussinov R. Selective molecular recognition in amyloid growth and transmission and cross-species barriers. J Mol Biol. 2012;421(2–3):172–184. doi: 10.1016/j.jmb.2011.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Straub JE, Thirumalai D. Principles governing oligomer formation in amyloidogenic peptides. Curr Opin Struct Biol. 2010;20(2):187–195. doi: 10.1016/j.sbi.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glabe CG. Structural classification of toxic amyloid oligomers. J Biol Chem. 2008;283(44):29639–29643. doi: 10.1074/jbc.R800016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toyama BH, Weissman JS. Amyloid structure: Conformational diversity and consequences. Annu Rev Biochem. 2011;80:557–585. doi: 10.1146/annurev-biochem-090908-120656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maji SK, et al. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science. 2009;325(5938):328–332. doi: 10.1126/science.1173155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sawaya MR, et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447(7143):453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 11.Margittai M, Langen R. Fibrils with parallel in-register structure constitute a major class of amyloid fibrils: Molecular insights from electron paramagnetic resonance spectroscopy. Q Rev Biophys. 2008;41(3-4):265–297. doi: 10.1017/S0033583508004733. [DOI] [PubMed] [Google Scholar]
- 12.Lührs T, et al. 3D structure of Alzheimer’s amyloid-beta(1-42) fibrils. Proc Natl Acad Sci USA. 2005;102(48):17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sunde M, et al. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J Mol Biol. 1997;273(3):729–739. doi: 10.1006/jmbi.1997.1348. [DOI] [PubMed] [Google Scholar]
- 14.Shivaprasad S, Wetzel R. Analysis of amyloid fibril structure by scanning cysteine mutagenesis. Methods Enzymol. 2006;413:182–198. doi: 10.1016/S0076-6879(06)13010-4. [DOI] [PubMed] [Google Scholar]
- 15.Benzinger TL, et al. Propagating structure of Alzheimer’s beta-amyloid(10-35) is parallel beta-sheet with residues in exact register. Proc Natl Acad Sci USA. 1998;95(23):13407–13412. doi: 10.1073/pnas.95.23.13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tycko R. Solid-state NMR studies of amyloid fibril structure. Annu Rev Phys Chem. 2011;62:279–299. doi: 10.1146/annurev-physchem-032210-103539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kajava AV, Baxa U, Steven AC. Beta arcades: Recurring motifs in naturally occurring and disease-related amyloid fibrils. FASEB J. 2010;24(5):1311–1319. doi: 10.1096/fj.09-145979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiang W, Yau WM, Luo Y, Mattson MP, Tycko R. Antiparallel β-sheet architecture in Iowa-mutant β-amyloid fibrils. Proc Natl Acad Sci USA. 2012;109(12):4443–4448. doi: 10.1073/pnas.1111305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berthelot K, Ta HP, Géan J, Lecomte S, Cullin C. In vivo and in vitro analyses of toxic mutants of HET-s: FTIR antiparallel signature correlates with amyloid toxicity. J Mol Biol. 2011;412(1):137–152. doi: 10.1016/j.jmb.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 20.Chien P, Weissman JS, DePace AH. Emerging principles of conformation-based prion inheritance. Annu Rev Biochem. 2004;73:617–656. doi: 10.1146/annurev.biochem.72.121801.161837. [DOI] [PubMed] [Google Scholar]
- 21.Campioni S, et al. A causative link between the structure of aberrant protein oligomers and their toxicity. Nat Chem Biol. 2010;6(2):140–147. doi: 10.1038/nchembio.283. [DOI] [PubMed] [Google Scholar]
- 22.Petkova AT, et al. Self-propagating, molecular-level polymorphism in Alzheimer's beta amyloid fibrils. Science. 2005;307(5707):262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 23.Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: Lessons from the Alzheimer’s amyloid beta-peptide. Nat Rev Mol Cell Biol. 2007;8(2):101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 24.Kodali R, Wetzel R. Polymorphism in the intermediates and products of amyloid assembly. Curr Opin Struct Biol. 2007;17(1):48–57. doi: 10.1016/j.sbi.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 25.De Simone A, Derreumaux P. Low molecular weight oligomers of amyloid peptides display beta-barrel conformations: A replica exchange molecular dynamics study in explicit solvent. J Chem Phys. 2010;132(16):165103. doi: 10.1063/1.3385470. [DOI] [PubMed] [Google Scholar]
- 26.Jang H, et al. β-Barrel topology of Alzheimer’s β-amyloid ion channels. J Mol Biol. 2010;404(5):917–934. doi: 10.1016/j.jmb.2010.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cerf E, et al. Antiparallel beta-sheet: A signature structure of the oligomeric amyloid beta-peptide. Biochem J. 2009;421(3):415–423. doi: 10.1042/BJ20090379. [DOI] [PubMed] [Google Scholar]
- 28.Stroud JC, Liu C, Teng PK, Eisenberg D. Toxic fibrillar oligomers of amyloid-β have cross-β structure. Proc Natl Acad Sci USA. 2012;109(20):7717–7722. doi: 10.1073/pnas.1203193109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hebda JA, Miranker AD. The interplay of catalysis and toxicity by amyloid intermediates on lipid bilayers: Insights from type II diabetes. Annu Rev Biophys. 2009;38:125–152. doi: 10.1146/annurev.biophys.050708.133622. [DOI] [PubMed] [Google Scholar]
- 30.Ahmed M, et al. Structural conversion of neurotoxic amyloid-beta(1-42) oligomers to fibrils. Nat Struct Mol Biol. 2010;17(5):561–567. doi: 10.1038/nsmb.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laganowsky A, et al. Atomic view of a toxic amyloid small oligomer. Science. 2012;335(6073):1228–1231. doi: 10.1126/science.1213151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu C, Sawaya MR, Eisenberg D. β₂-microglobulin forms three-dimensional domain-swapped amyloid fibrils with disulfide linkages. Nat Struct Mol Biol. 2011;18(1):49–55. doi: 10.1038/nsmb.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brooks BR, et al. CHARMM: The biomolecular simulation program. J Comput Chem. 2009;30(10):1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng PN, Liu C, Zhao ML, Eisenberg D, Nowick JS. Amyloid β-sheet mimics that antagonize protein aggregation and reduce amyloid toxicity. Nat Chem. 2012;4(11):927–933. doi: 10.1038/nchem.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richardson JS, Richardson DC. Natural beta-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc Natl Acad Sci USA. 2002;99(5):2754–2759. doi: 10.1073/pnas.052706099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khakshoor O, Demeler B, Nowick JS. Macrocyclic beta-sheet peptides that mimic protein quaternary structure through intermolecular beta-sheet interactions. J Am Chem Soc. 2007;129(17):5558–5569. doi: 10.1021/ja068511u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu C, et al. Characteristics of amyloid-related oligomers revealed by crystal structures of macrocyclic β-sheet mimics. J Am Chem Soc. 2011;133(17):6736–6744. doi: 10.1021/ja200222n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cremades N, et al. Direct observation of the interconversion of normal and toxic forms of α-synuclein. Cell. 2012;149(5):1048–1059. doi: 10.1016/j.cell.2012.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shahnawaz M, Soto C. Microcin amyloid fibrils A are reservoir of toxic oligomeric species. J Biol Chem. 2012;287(15):11665–11676. doi: 10.1074/jbc.M111.282533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eichner T, Radford SE. Understanding the complex mechanisms of β2-microglobulin amyloid assembly. FEBS J. 2011;278(20):3868–3883. doi: 10.1111/j.1742-4658.2011.08186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystsllogr. 1993;26:795–800. [Google Scholar]
- 42.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64(Pt 1):112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 43.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 44.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.von Bergen M, et al. Assembly of tau protein into Alzheimer paired helical filaments depends on a local sequence motif ((306)VQIVYK(311)) forming beta structure. Proc Natl Acad Sci USA. 2000;97(10):5129–5134. doi: 10.1073/pnas.97.10.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Platt GW, Routledge KE, Homans SW, Radford SE. Fibril growth kinetics reveal a region of beta2-microglobulin important for nucleation and elongation of aggregation. J Mol Biol. 2008;378(1):251–263. doi: 10.1016/j.jmb.2008.01.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luca S, Yau WM, Leapman R, Tycko R. Peptide conformation and supramolecular organization in amylin fibrils: Constraints from solid-state NMR. Biochemistry. 2007;46(47):13505–13522. doi: 10.1021/bi701427q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vilar M, et al. The fold of alpha-synuclein fibrils. Proc Natl Acad Sci USA. 2008;105(25):8637–8642. doi: 10.1073/pnas.0712179105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bricogne C, et al. Womack BUSTER, Version 2.8.0. Cambridge, United Kingdom: Global Phasing Ltd; 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.