Abstract
SUMOylation, an essential posttranslational protein modification, is involved in many eukaryotic cellular signaling pathways. The identification of SUMOylated proteins is difficult, because SUMOylation sites in proteins are hard to predict, SUMOylated protein states are transient in vivo and labile in vitro, only a small substrate fraction is SUMOylated in vivo, and identification tools for natively SUMOylated proteins are rare. To solve these problems, we generated knock-in mice expressing His6-HA-SUMO1. By anti-HA immunostaining, we show that SUMO1 conjugates in neurons are only detectable in nuclei and annulate lamellae. By anti-HA affinity purification, we identified several hundred candidate SUMO1 substrates, of which we validated Smchd1, Ctip2, TIF1γ, and Zbtb20 as novel substrates. The knock-in mouse represents an excellent mammalian model for studies on SUMO1 localization and screens for SUMO1 conjugates in vivo.
Keywords: synapse, affinity purification
SUMOylation is a conserved posttranslational protein modification in eukaryotes, akin to ubiquitylation, and can affect the localization, interactions, function, or stability of substrates (1). SUMOylation processes participate in many cellular signaling pathways, where they intersect with other posttranslational regulatory processes such as phosphorylation, ubiquitylation, or acetylation. Consequently, many cellular processes, from nuclear transport to neuronal synaptic transmission, are controlled by SUMOylation (2), and key SUMOylation substrates or altered SUMOylation are involved in many diseases, from cancer to neurological disorders (3).
Reflecting the prominent nuclear role of SUMOylation, most known SUMOylation substrates are nuclear proteins (1, 2). However, SUMOylation appears to play a much more pervasive regulatory role in cells; recent studies have shown SUMOylation of ion channels, membrane-bound receptors, solute carriers, and mitochondrial or neuronal scaffolding and signaling proteins (4–6), leading to the notion that SUMOylation is a core regulatory process in all cellular subcompartments. This triggered substantial activities to develop tools for the discovery and validation of SUMOylation substrates in cells, which has proven difficult.
Mammalian genomes contain four SUMO genes, encoding SUMO1, SUMO2, SUMO3 (7, 8), and SUMO4, of which SUMO4 is poorly characterized (9). SUMO2 and SUMO3 are almost identical, whereas SUMO1 is 50% homologous to SUMO2 and SUMO3. The 3D structure of SUMOs is similar to that of ubiquitin (10), and like ubiquitylation, SUMOylation involves an E1 activating enzyme, E2 conjugating enzymes, and E3 ligases (1, 2, 7, 8).
SUMOs are conjugated to lysine residues, often within a ϕKxD/E motif (ϕ, hydrophobic residue; x, any amino acid) (1), but in general SUMO acceptor lysines cannot be predicted, which prevents the identification of SUMO substrates by bioinformatics (1). Further, SUMOylated states of proteins are transient in vivo and labile in vitro because of isopeptidases that revert SUMOylation (1), and usually only a small fraction of a given substrate protein is SUMOylated (7), which confounds the discovery and validation of new substrates. Finally, reliable antibody tools for the affinity purification of natively SUMOylated proteins have been lacking. To solve these problems, cell lines, yeast cells, and other models expressing tagged SUMOs in an otherwise wild-type (WT) background were used to affinity-purify SUMOylation substrates and to generate databases of putatively SUMOylated proteins (11–17). However, possible overexpression artifacts and the restriction to cell lines pose limitations to such approaches. Hence, subsequent studies used genetic replacement of endogenous SUMO by affinity-tagged SUMO variants to purify, identify, and characterize novel SUMOylation substrates (18–23). Advantages of such genetic approaches are the close-to-endogenous expression level of the affinity-tagged SUMO, eliminating the risk of overexpression artifacts, and the fact that all cell types of multicellular organisms can be screened. However, corresponding mammalian models are lacking.
We generated a knock-in (KI) mouse line that expresses double affinity-tagged His6-HA-SUMO1 instead of WT SUMO1 from the Sumo1 locus. Focusing on brain tissue, we show that the His6-HA-SUMO1 KI mouse line represents an excellent mammalian model for studies on SUMO1 localization and new SUMO1-conjugated proteins in vivo.
Results
Basic Characterization of His6-HA-SUMO1 KIs.
KI mice expressing N-terminally His6-HA–tagged SUMO1 were generated by homologous recombination in embryonic stem cells (Fig. S1). Heterozygous and homozygous KIs showed no overt phenotypic changes. To generate sufficient numbers of mice for screening experiments, we established homozygous KI and WT mouse lines from heterozygous KI littermates. Mice of the same generations from these lines were used for all experiments.
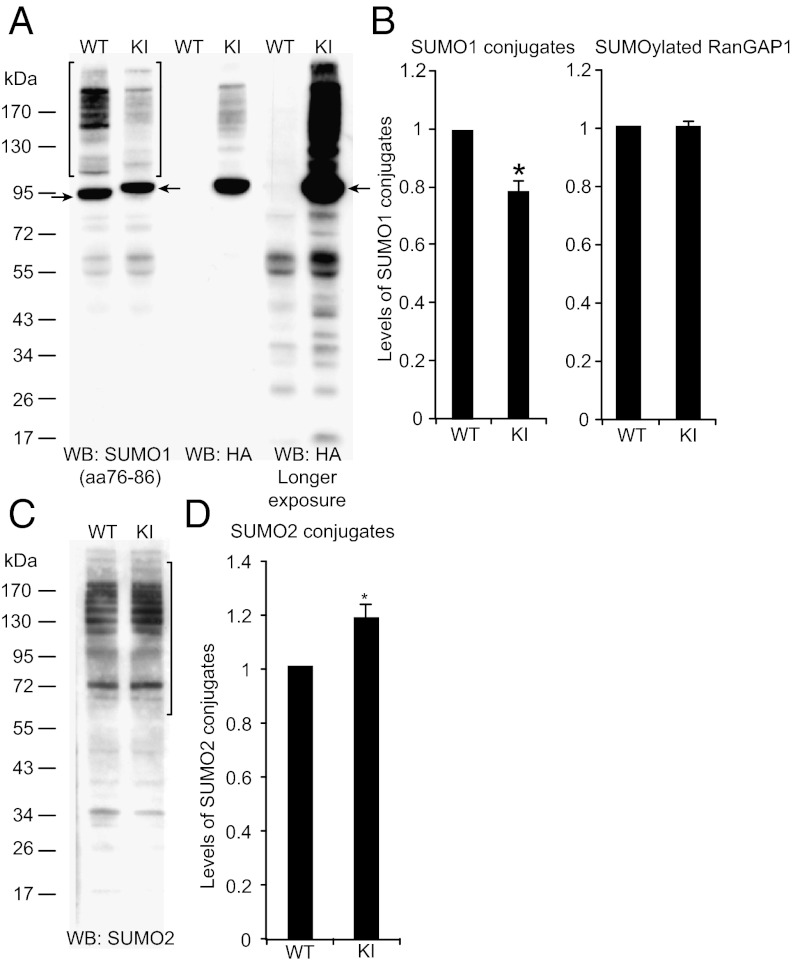
We focused on SUMOylation in the brain because recent evidence indicates that extranuclear brain proteins are SUMOylated and SUMOylation may be involved in various physiological and pathophysiological brain processes (3). Nissl staining of brain sections revealed a normal brain anatomy and cytoarchitecture in adult KIs (Fig. S1D). For the quantification of His6-HA-SUMO1 expression in KIs and the determination of the His6-HA-SUMO1 conjugate complement, we characterized two monoclonal anti-SUMO1 antibodies, anti-SUMO1(21C7) and anti-SUMO1(aa76–86), by Western blotting of mouse brain homogenates. The antibodies yielded similar band patterns, with a prominent 90-kDa band (SUMOylated RanGAP1) and a ladder of larger protein bands typical of SUMO1 conjugates. Because anti-SUMO1(aa76–86) was more specific, with hardly any background detection below 70 kDa (Fig. S1E), it was used further analyses.
Western blotting of KI mouse brain homogenates with anti-HA antibodies yielded a band pattern that was very similar to the one obtained with anti-SUMO1(aa76–86) (Fig. 1A) but not detected with WT samples (Fig. 1A). This indicates that the substrate specificity of SUMO1 conjugation is largely unperturbed in KI brain. Upon longer blot exposure, smaller proteins (<90 kDa) were detected by anti-HA antibodies (Fig. 1A), which mostly represent nonspecific bands because they appeared at equal intensity in WT and KI samples upon anti-HA detection (Fig. 1A).
Fig. 1.
SUMO1 and SUMO2 conjugation. (A) Homogenates of adult WT and KI brains, analyzed by SDS/PAGE and Western blotting (WB) using anti-SUMO1 and anti-HA antibodies. Brackets indicate the range of SUMO1 conjugates quantified in B. Arrows indicate SUMOylated RanGAP1. (B) Quantification of SUMO1 conjugates (Left; see brackets in A) and SUMO1-conjugated RanGAP1 (Right). Data were normalized to WT levels and are expressed as mean ± SEM (n = 3; *, significant difference between WT and KI, P = 0.015 in Student t test). (C) Homogenates of adult WT and KI brains, analyzed by SDS/PAGE and Western blotting using anti-SUMO2 antibodies. Brackets indicate the range of SUMO2 conjugates quantified in D. (D) Quantification of SUMO2 conjugates (see brackets in C). Data were normalized to WT levels and are expressed as mean ± SEM (n = 3; *, significant difference between WT and KI, P = 0.045 in Student t test).
Whereas the staining intensity of the RanGAP1 band detected with anti-SUMO1(aa76–86) was identical in WTs and KIs, bands of higher molecular weight showed a slightly lower staining intensity in KIs (78 ± 3% of WT levels, P = 0.011, n = 3; Fig. 1 A and B), indicating slightly less efficient SUMOylation with His6-HA-SUMO1. In turn, detection of SUMO2 conjugates yielded similar band patterns in WTs and KIs but slightly higher SUMO2 conjugation levels in KIs (118 ± 6% of WT levels, P = 0.045, n = 3; Fig. 1 C and D), indicating that the reduced SUMO1 conjugation in KIs is compensated by SUMO2 conjugation, as in SUMO1 knock-outs (24). Expression levels of neuronal marker proteins were not different between WTs and KIs (Fig. S1F).
SUMO1 Conjugates in Brain.
Western blotting of brain homogenates from mice of different ages (postnatal days P0–P56) revealed stable, high levels of SUMO1 conjugates between P0 and P15 and decreased levels from P20 onward, when brain cell proliferation and synaptogenesis are largely completed (Fig. S2 A and B). As expected, because most known SUMO1 substrates are transcription factors or nuclear components (1, 8), Western blotting of brain subcellular fractions showed that SUMO1 conjugates are most abundant in nuclei. Substantial amounts of SUMO1 conjugates were also detected in cytosol, synaptic cytosol, or synaptic vesicle fractions (Fig. S2C). However, synaptic membrane fractions [lysed synaptosomal membrane (LP1) and synaptic plasma membrane (SPM)] were devoid of SUMO1 conjugates, except for some RanGAP1, which likely represents a contamination (Fig. S2C).
Because biochemically purified subcellular fractions do not unequivocally report subcellular protein distribution, we used the HA tag of His6-HA-SUMO1 to localize SUMO1 conjugates in brain tissue and cultured neurons by immunostaining. Anti-HA–stained brain sections of KIs revealed a strong nuclear staining in all KI brain regions, whereas WT samples showed no or weak background labeling (Fig. S3). Higher-resolution analyses of His6-HA-SUMO1 localization in the hippocampal CA3 region of KIs showed strong staining of the nuclear envelope and intranuclear structures in pyramidal cells (Fig. S3 D and E). MAP2-positive proximal dendrites of KI CA3 pyramidal cells showed diffuse and punctate staining that was not observed or only weakly present in WTs (Fig. S3E) and not detectable in dendrites of CA1 pyramidal cells or dentate gyrus granule cells (Fig. S3F). Double-labeling experiments showed that the extranuclear His6-HA-SUMO1–positive structures in KIs do not colocalize with the presynapse markers Synapsin (all synapses), VIAAT, or VGluT1 (inhibitory and excitatory synapses, respectively) (Fig. S4), and confocal line scans through cell bodies and dendrites of CA3 pyramidal cells indicated an intracellular localization of these structures within MAP2-positive dendrites (Fig. S5), indicating that extranuclear His6-HA-SUMO1–positive structures are not synaptic.
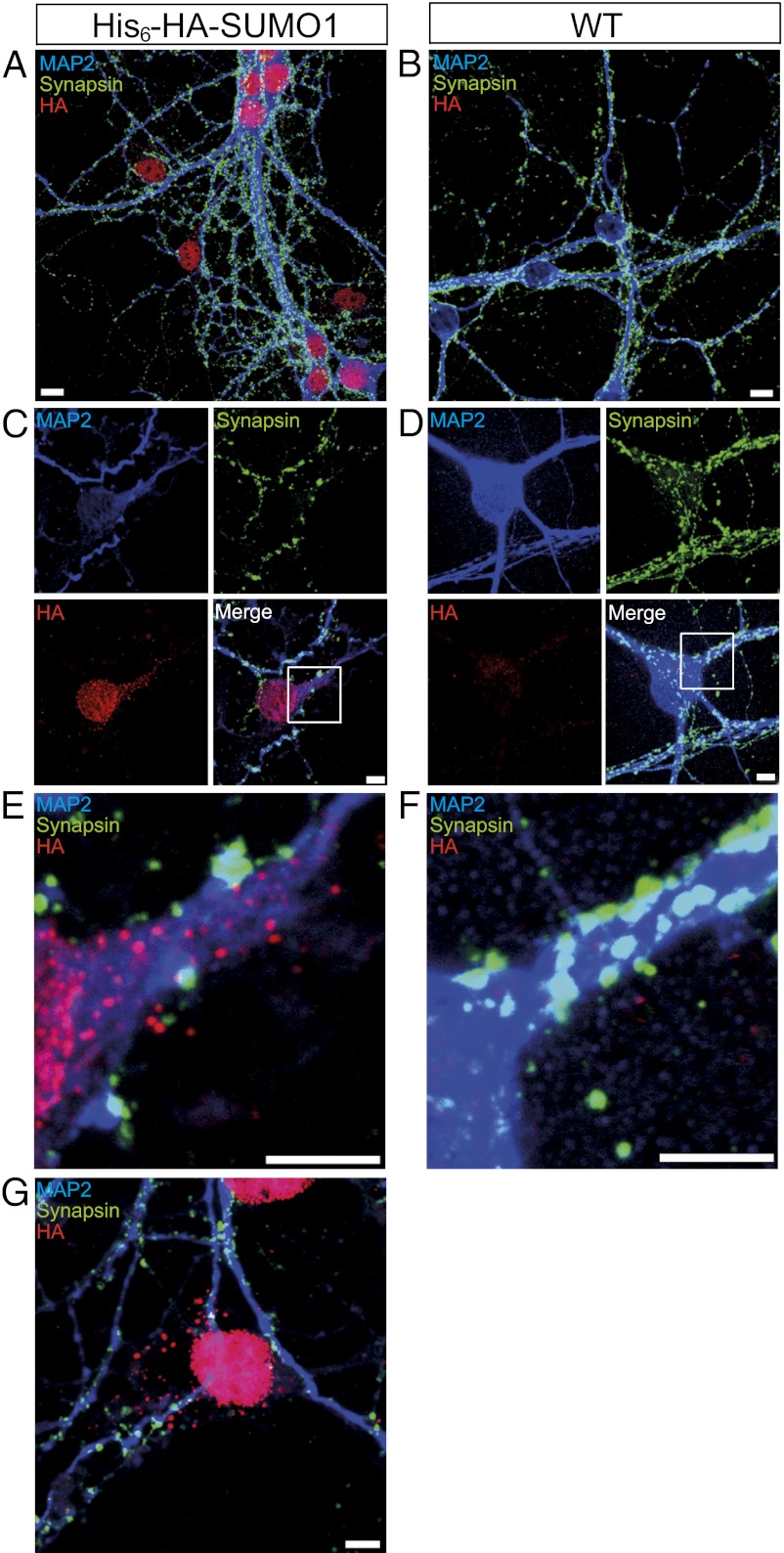
We next turned to hippocampal neuron cultures, which exhibit less background staining and allow the use of a larger marker antibody set. Strong His6-HA-SUMO1 staining was observed in the nuclear envelope and intranuclear structures of KI neurons (Fig. 2 and Fig. S6), which were otherwise indistinguishable from WT cells. In addition, 10% of KI cells showed extranuclear punctate labeling that did not colocalize with Synapsin-positive presynapses, GABAAγ2-positive GABAergic postsynapses, or ProSAP1-positive glutamatergic postsynapses and was absent in WT cells (Fig. 2 and Fig. S6). Also, many MAP2-negative cells, which likely represent astrocytes, showed strong nuclear and distinct extranuclear His6-HA-SUMO1 labeling (Fig. 2G).
Fig. 2.
Subcellular localization of SUMO1 conjugates in cultured hippocampal neurons. Cultured hippocampal neurons were stained for HA (red), MAP2 (blue, somata and dendrites), and Synapsin (green, presynapses). Images are representative of three independent experiments. (A and B) Overview of a KI (A) and a WT culture (B). (Scale bar, 10 μm.) (C and E) Higher-resolution images of a KI cell. Extranuclear, punctate HA-immunopositive staining in the KI cell (red) is prominent in proximal dendrites (blue) but does not colocalize with Synapsin-positive presynapses (green). (Scale bar, 5 μm.) (D and F) Higher-resolution images of a WT cell. Conditions as in C and E. No HA-immunopositive (red) signals were detectable. (Scale bar, 5 μm.) (G) HA-immunopositive signals (red) in a MAP2-negative (presumed astrocytic) cell body. (Scale bar, 10 μm.)
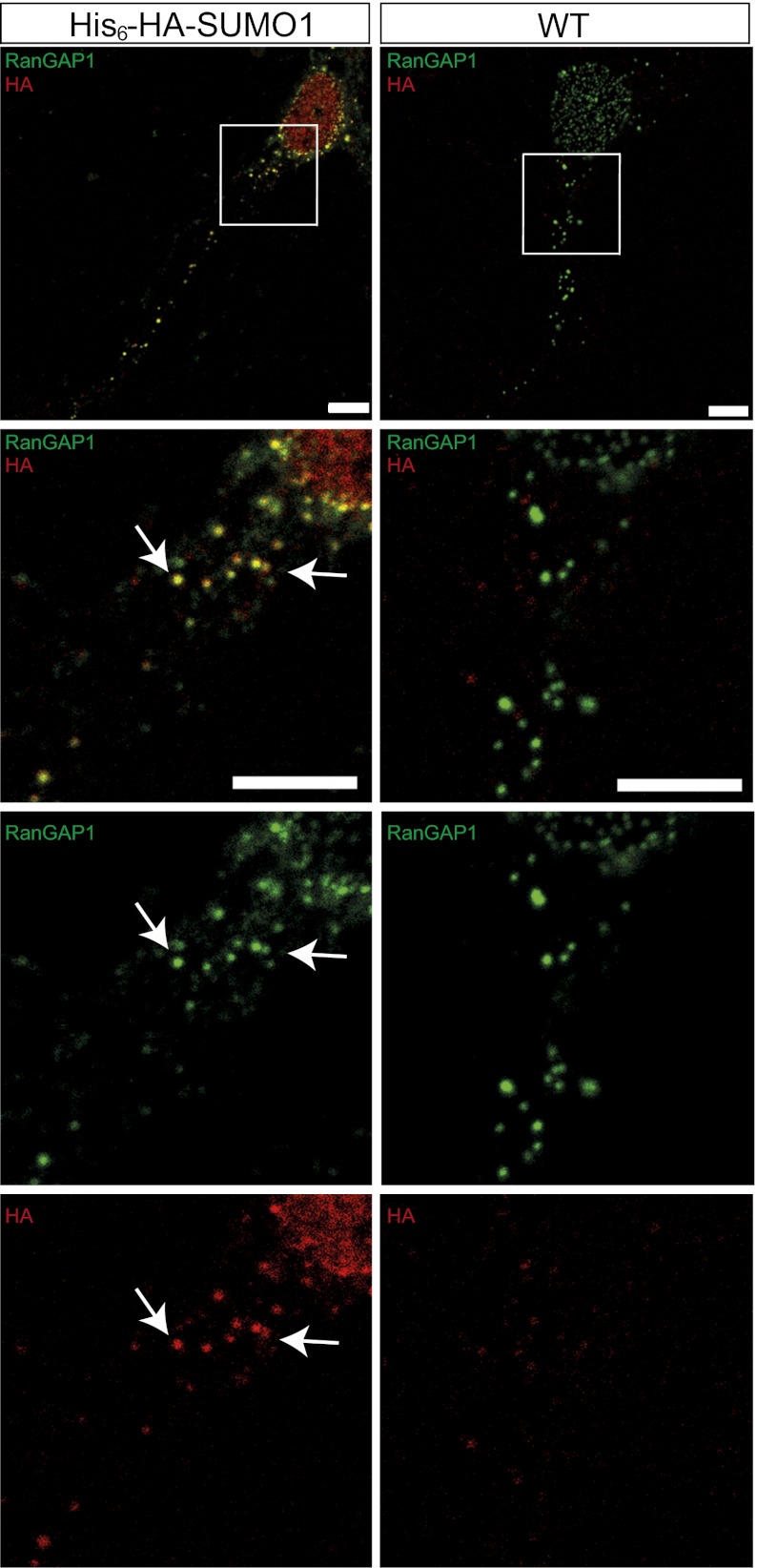
Because our immunolabeling data indicated a nonsynaptic localization of His6-HA-SUMO1–positive extranuclear structures in neurons and their presence in astrocytes, we studied their localization relative to markers of the endoplasmic reticulum (Calnexin), the Golgi complex (GM130), and endosomes (EEA1), which are shared by neurons and glia cells, but did not detect any colocalization (Fig. S6). The nuclear pore components RanGAP1 and RanBP2 are among the most prominent SUMOylated proteins. In many cells, they are not only present in the nuclear envelope but also in extranuclear annulate lamellae (AL), which are thought to serve as reservoirs of excess nuclear membrane components (25). To test whether the extranuclear His6-HA-SUMO1 structures represent AL, we performed double-staining experiments with antibodies to HA, RanGAP1, and RanBP2. We detected abundant RanGAP1- and RanBP2-positive punctate structures in the somatic cytoplasm and proximal dendrites. Most His6-HA-SUMO1–positive extranuclear structures coincided with RanGAP1 (91.34 ± 1.45%, n = 18 cells) (Fig. 3) and RanBP2 (56.2 ± 2.3%, n = 35 cells) staining (Fig. S7), indicating that they represent AL.
Fig. 3.
Colocalization of extranuclear HA-immunopositive signals with markers of annulate lamellae in cultured hippocampal neurons. Neurons from KI (Left) and WT (Right) were stained for HA (red) and RanGAP1 (green). Images are representative of three independent experiments. The bottom panels in each column represent an enlargement of the areas boxed in white above. Arrows indicate colocalization of HA with RanGAP1 in the nuclear envelope and cytoplasm. (Scale bar, 5 μm.)
Purification and Identification of SUMO1 Conjugates in Brain.
In using the KI to affinity-purify SUMO1 conjugates from brain, we opted for a single-step anti-HA affinity purification procedure to maximize the yield and to accelerate the purification procedure and thus minimize deSUMOylation. In addition, Ni-NTA–based affinity purification of His6-SUMO–conjugated proteins from transgenic mice is confounded by the background of brain proteins that bind to Ni-NTA (Ni-nitrilotriacetic acid) (26). Apart from adult brains, we also studied P10 mouse brains because SUMO1 conjugates are abundant in this developmental stage.
Affinity purification experiments were conducted with covalently linked anti-HA antibodies as affinity matrix and HA peptide to elute bound proteins. SDS/PAGE and Western blot analyses of the different fractions obtained during purification showed that His6-HA-SUMO1 conjugates were depleted from the flow-through of the anti-HA column and enriched in the peptide eluate (Fig. S8). However, Coomassie staining of SDS/PAGE gels loaded with eluted material obtained from KIs and WTs also revealed the presence of contaminants, particularly in adult brain samples (Fig. S8), probably because detergent extracts of brain contain many proteins that are known to bind nonspecifically to agarose matrices and large amounts of lipids and DNA that facilitate nonspecific protein interactions with agarose matrices.
For comparative proteomic analyses, peptide eluates obtained from KI and WT samples were separated by SDS/PAGE, and gel lanes were excised and cut into 23 slices. Each slice was subjected to in-gel tryptic digestion, and the resulting peptides were analyzed by liquid chromatography coupled with mass spectrometry (LC-MS/MS). Reflecting the background binding of WT proteins to the anti-HA affinity matrix as assessed by Coomassie staining of SDS/PAGE gels, we detected a substantial number of proteins in WT samples using mass spectrometry. Therefore, candidate His6-HA-SUMO1 conjugates among the proteins identified in KI samples were initially selected based on their detection in KI samples in at least two out of four independent biological experiments and their absence from WT samples. Several known SUMOylated proteins, such as RanBP2 and RanGAP1, were eliminated by this selection due to their presence in WT samples, which is likely caused by their high abundance and partial nonspecific binding to the affinity matrix. Thus, additional candidate proteins were selected based on their significant enrichment in KI samples using an algorithm for advanced spectral count quantification. In this manner, 147 candidate His6-HA-SUMO1 conjugates from adult KI samples and 304 candidate proteins from P10 KI samples were selected (Dataset S1). Many identified candidate proteins are transcription factors, but in general they are associated with a wide variety of biological processes (Fig. S9A) and show very low connectedness, with only 16 of 147 proteins in the adult brain sample and 33 of 304 proteins in the P10 brain sample exhibiting one or more known interactions with other candidate proteins (Fig. S9B). This indicates that secondary interaction-dependent copurification of non-SUMOylated proteins by our method is negligible, which is supported by the fact that the average clustering coefficient for the networks C(p) is on the order of the average clustering coefficient from 1,000 random subsets from the brain mouse protein interaction network, with C(p) ∼0.07.
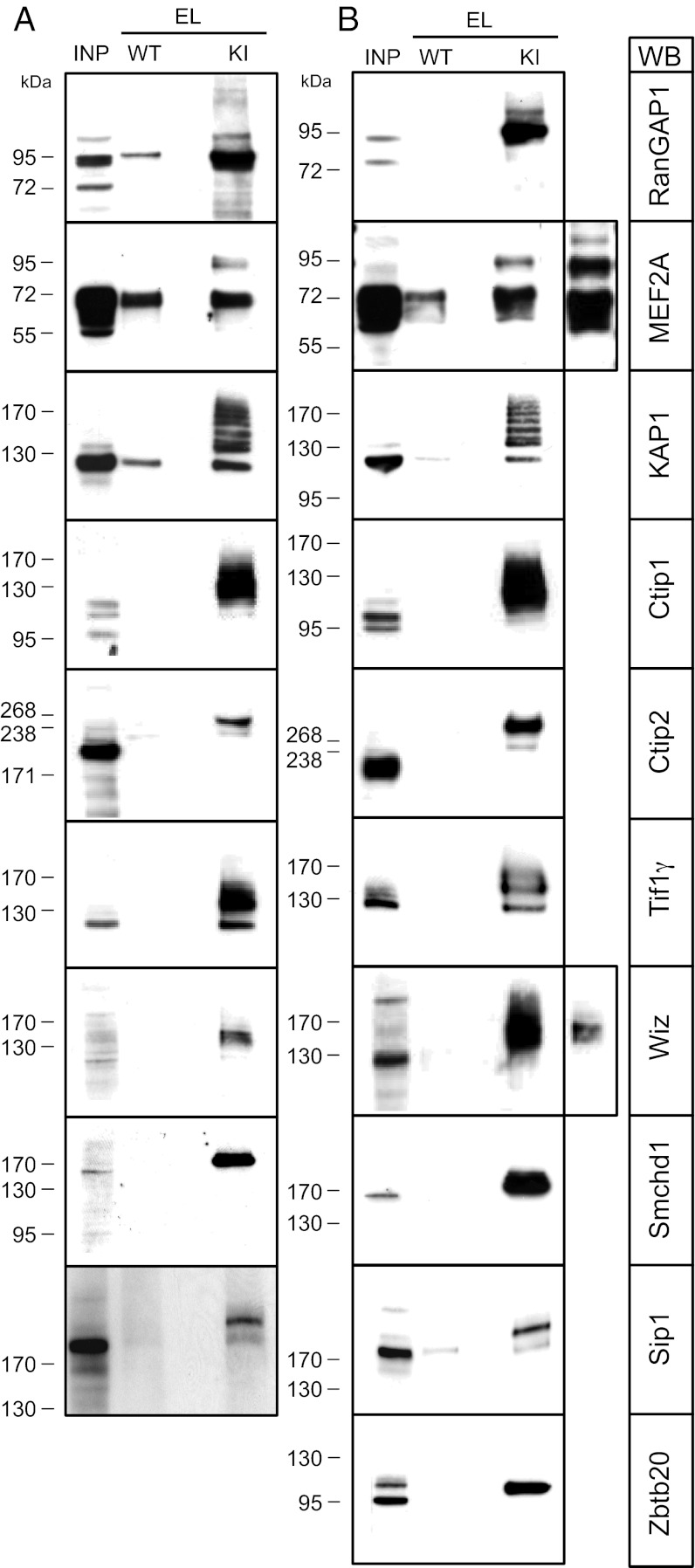
Our selection of candidate proteins contained multiple known or previously proposed SUMO1 substrates (Dataset S1), including RanGAP1 and RanBP2. Using ChopNSpice (27), the SUMO1-conjugated lysine residue of RanGAP1 was identified (Fig. S9C). These findings validate our screening approach. In selected cases, we performed Western blotting analyses of the starting material and HA peptide eluates obtained from KIs and WTs to validate the selective presence of the corresponding known SUMO1 substrates in KI samples. In this manner, we identified SUMOylated RanGAP1 (28), MEF2A (29) (not identified in our proteomic analyses), KAP1 (30), Wiz (31), Sip1 (32), and Ctip1 (33) (only detected in proteomic analyses of P10 KI brain) in brain samples of KI mice, whereas they were either absent or much less abundant in WT (Dataset S1). In all cases, the validated proteins showed a band pattern indicative of conjugation of one or several His6-HA-SUMO1 moieties, which further validates our screening approach (Fig. 4 A and B). Several previously proposed SUMO1 conjugates were not detected in our proteomic screen (e.g., GluK2, CASK, and CtBP1). In analyses of HA peptide eluates from KIs and WTs by Western blotting for GluK2 and CASK, we were unable to demonstrate GluK2 or CASK conjugation with SUMO1. GluK2 bound nonspecifically to the anti-HA affinity matrix, but we found no indication of SUMO1-conjugated GluK2 in HA peptide eluates from KI samples. However, CASK was not immunopurified by the anti-HA affinity matrix at all. Even very long exposures of the corresponding blots showed only trace amounts of CASK. They were present at similar levels in the HA peptide eluates from WT and KI samples, and no indication of SUMO1-conjugated CASK was found (Fig. S10A).
Fig. 4.
Western blot analysis of candidate SUMO1 conjugates. (A and B) Representative detergent extract input sample (INP, from WT) and specific HA-peptide eluates (EL) of HA-immunoaffinity purified samples from WTs and KIs were analyzed by SDS/PAGE and Western blotting (WB) with antibodies to the indicated proteins. Images are representative of three independent experiments. (A) Adult brain. (B) P10 brains.
To identify new SUMOylated proteins, we focused on samples from P10 KI brains because these contained fewer contaminants. We first analyzed Smchd1, Ctip2, TIF1γ, and Zbtb20 (Fig. 4B). All four proteins were selectively enriched in the eluates of KI samples and absent from WT eluates and showed a band pattern indicative of conjugation of one or several His6-HA-SUMO1 moieties, demonstrating that Smchd1, Ctip2, TIF1γ, and Zbtb20 are new SUMO1 conjugates in P10 mouse brain (Dataset S1). SUMOylated Smchd1, Ctip2, and TIF1γ were also selectively detected in HA peptide eluates from adult KI brain samples (Fig. 4A and Dataset S1). The available tools allowed us to immunoprecipitate Ctip2 from adult KI brain and test for the presence of His6-HA-SUMO1–conjugated Ctip2 in the purified fraction. Western blotting showed that the specific immunoprecipitate contains His6-HA-SUMO1–conjugated Ctip2 as judged by immunodetection with anti-HA and anti-SUMO1 antibodies (Fig. S10B), providing independent support for the validity of our screening approach and further establishing Ctip2 as a new SUMO1 substrate in the brain in vivo. However, several candidates that we identified could not be verified by subsequent Western blotting analyses (Dataset S1).
Discussion
To generate a KI mouse expressing tagged SUMO1, we opted for a His6-HA tag because it is known to leave SUMO1 function in vitro unaffected (34, 35) and has been used without negative side effects in multiple cell lines and organisms to purify SUMOylated proteins (11–23). The KIs show no obvious phenotypic changes; KI brains show normal morphology and cell layering; and KI neurons exhibit normal cytoarchitecture, subcellular organization, and expression and localization of marker proteins. Thus, His6-HA tagging of SUMO1 has no deleterious effect on brain or neuronal development and function, which is expected because even SUMO1 loss can be compensated by other SUMOs (24).
KI brains revealed a slight deviation from WT with regard to SUMO1 conjugate levels, which were reduced by 22% along with an up-regulation of SUMO2 conjugation by 18%, indicating that SUMO2 can replace SUMO1 (24). The reduced SUMO1 conjugate levels in KIs may be due to slightly faster deSUMOylation of His6-HA-SUMO1 conjugates, for instance, because interactions that mask covalently bound SUMO1 and protect from deSUMOylation are slightly perturbed. The SUMOylation machinery appears to operate with normal efficiency and specificity in the KI because the pattern of SUMO1-conjugated proteins is unchanged and RanGAP1 is SUMOylated normally. The reduced SUMO1 conjugation levels in KIs may somewhat reduce the threshold for detection of conjugates, but this does not confound the use of the KIs as a tool to localize SUMO1 in situ or to screen for SUMO1-conjugated proteins.
Our KI has several advantages over previously published models. First, our KI strategy excludes overexpression artifacts. In many published cases where expression of tagged SUMOs was used to purify SUMOylated proteins, the tagged SUMOs were expressed in a WT background (11–17). This bears the risk of overexpression artifacts such as “off-target” SUMOylation. Second, extremely well established, highly specific, and commercially readily available anti-HA antibodies can be used for the localization and immunoaffinity purification of SUMOylated proteins from KIs, along with WT mice as an ideal negative control. This is important because specific anti-SUMO1 antibodies are rare and most are unsuitable for affinity purification protocols. Third, our KI is a unique tool for in-depth analysis of specific SUMO1 targets in vivo. By performing immunoprecipitation experiments using specific antibodies, followed by anti-HA Western blotting, it is possible to study the SUMOylation of low-abundance substrates (Fig. S10). Importantly in this context, commercially available anti-HA antibodies have much higher epitope affinity than anti-SUMO1 antibodies. Fourth, our KI is a unique genetic mammalian model for SUMO1 localization and the identification of SUMO1 substrates in vivo. Along with the possibilities of using WT mice as optimal negative control and of crossing the KI mutation into any desired genetic background, the KI allows for systematic analyses of SUMO1 localization and conjugation in any tissue and any physiological or pathophysiological state that can be induced in mice. Thus, our KI is a valid tool to study SUMO1 localization and conjugation in mice in vivo. Data on the subcellular distribution of His6-HA-SUMO1 reflect the native state, and information on candidate SUMO1 substrates purified from KIs is a valuable basis for further studies.
Most SUMO1 substrates, particularly the best-established and most abundant ones, function in the nucleus (1, 2, 7, 8), which is supported by our immunostaining data. Punctate, extranuclear His6-HA-SUMO1 immunopositive structures were apparent in proximal dendrites of neurons and in glia cells. These structures contain RanBP2 and RanGAP1 and likely represent AL (25). Apart from extranuclear SUMO1 in AL, we found no evidence of other neuronal structures containing SUMO1. This is in conflict with published data indicating the presence of the SUMOylation machinery in nonnuclear neuronal compartments such as synapses or the existence of multiple extranuclear SUMO substrates (4), but we cannot exclude that the corresponding SUMOylated proteins escaped our detection. In extranuclear compartments, our immunolabeling mainly detects RanGAP1, which is the most abundant SUMO1 target. Its SUMOylation is stable and resistant to isopeptidases, and SUMOylated RanGAP1 is strongly enriched at nuclear pore complexes, which are part of AL and contain multiple SUMOylated RanGAP1 molecules. In view of this, the fact that we readily detected His6-HA-SUMO1 conjugates only in AL does not unequivocally exclude the possibility that we failed to detect structures containing fewer His6-HA-SUMO1 conjugates, such as synapses. It is also possible that the corresponding extranuclear substrates are only SUMOylated under particular physiological conditions that we did not mimic, or that they are de-SUMOylated swiftly and escape detection by our methods. Finally, we might have been unable to detect certain candidate SUMO1 substrates because they have a different paralog specificity in vivo than the one found in vitro after SUMO1 overexpression. Conversely, most commercially available antibody tools for the detection of SUMOylated proteins have limitations, particularly regarding specificity and the design of stringent controls. In light of this and in view of the present study, it is likely that subsets of previously published data on extranuclear SUMOylation should be reassessed. Here, our KI represents an ideal test system because it can be crossed with SUMO isopeptidase knock-outs, or cells derived from KIs could be treated with isopeptidase inhibitors to localize and identify rare SUMO1 targets.
Our screens for His6-HA-SUMO1–conjugated proteins in adult and P10 KI brains yielded a substantial collection of candidate proteins. Analysis of their connectedness indicates that interaction-dependent copurification of non-SUMOylated proteins is negligible. Further, the candidate His6-HA-SUMO1 conjugates did not contain noncovalent interactors of SUMOs, indicating that we mainly detected true conjugates. Our screen identified a large number of known SUMO1 substrates, of which we validated several. Other well-known SUMO substrates (Ubc9, TIF1α, DNA topoisomerase 2β, LARP4B, and BRCA1) were also found but not subjected to validation. Beyond established SUMO substrates, many candidates we identified had been detected in previous proteomics studies (13, 17, 23, 31, 36). These findings provide convincing proof-of-principle for the use of our KI as a discovery tool to identify SUMO1 conjugates.
Our validation studies on new SUMO1 substrates were guided by the spectral count data, the plausibility of the candidates based on SUMOylation of homologs, and the availability of suitable antibodies. We validated Smchd1, Ctip2, TIF1γ, and Zbtb20 as new SUMO1 substrates, which documents the power of our discovery approach. However, we were unable to validate several candidate proteins that we identified, indicating that candidates identified by proteomic screens cannot be taken at face value even if they were purified with rather stringent methods. Conversely, some very well established SUMO1 substrates, such as MEF2A, were not identified in our screen, as was the case for most of the SUMOylated extranuclear proteins described so far (4). Exceptions to the latter include focal adhesion kinase 1 and mGluR7, which were not analyzed further. Thus, the fact that a candidate SUMO1 substrate was not detected in our proteomic screen does not necessarily imply that it is not SUMOylated in vivo. Instead, the corresponding His6-HA-SUMO1 conjugates might be poorly accessible to mass spectrometric analysis or of too low abundance. In the future, it may be possible to circumvent such problems by enriching relevant subcellular fractions for affinity purification of His6-HA-SUMO1 conjugates or by direct testing of candidate proteins. Of note, we directly tested the SUMOylation of selected published candidate substrates (e.g., CASK) in our KI and failed to detect SUMO1-conjugated variants. This is a matter of concern and emphasizes the need for further studies, for which our KI model is ideally suited.
Altogether, our KI is a valuable, universally applicable, and reliable tool to localize SUMO1 conjugates in cells and tissues, to screen for SUMO1 conjugates in vivo and to validate candidate SUMO1 substrates in any mouse tissue. In combination with genetic, pharmacological, or environmental perturbations, the KI can be used to study SUMOylation in a multitude of physiological and pathophysiological contexts.
Materials and Methods
A complete description of the materials and methods used in this study is provided in SI Materials and Methods.
His6-HA-SUMO1 KI Mice.
His6-HA-SUMO1 KI mice were generated by homologous recombination in ES cells (Fig. S1). We generated WT and KI littermates that were used to establish separate WT and KI lines for experiments. All experiments were approved by the responsible local government organization (Bezirksregierung Braunschweig).
Immunohistochemistry and Immunocytochemistry.
Immunohistochemical, immunocytochemical, and Nissl staining experiments were performed on fixed sagittal brain sections of perfusion-fixed mice or on fixed hippocampal primary cells. The antibodies used are listed in Tables S1 and S2.
Immunoaffinity Purification.
For anti-HA immunoaffinity purification, proteins from powdered frozen brains were solubilized with a buffer containing 1% (vol/vol) Triton X-100, 0.5% (wt/vol) sodium deoxycholate, and 0.1% (vol/vol) SDS. HA-tagged proteins were purified using an anti-HA column (Roche), from which bound material was eluted with HA peptide.
Quantitative Western Blotting.
For quantitative Western blotting, proteins were visualized by Fast Green FCF (Sigma) staining of blots and analyzed with an Odyssey reader (LI-COR) for normalization of protein loading. Western blots with secondary antibodies conjugated to IRDye800 (Biomol) were quantified using an Odyssey reader (LI-COR).
Mass Spectrometry and Data Analysis.
Mass spectrometry and data analyses were performed essentially as described previously (27), with the exception that an Orbitrap Velos instrument was used.
Supplementary Material
Acknowledgments
We thank K. Hellmann, F. Benseler, I. Thanhäuser, D. Schwerdtfeger, C. Harenberg, and L. van Werven and the staff of the Max Planck Institute of Experimental Medicine Transgenic Animal Facility for technical support. This work was supported by the Max Planck Society (N.B. and H.U.), the European Commission (EUROSPIN FP7-HEALTH-F2-2009-241498, SynSys FP7-HEALTH-F2-2009-242167 N.B.), the German Research Foundation (Ti601/1-1, SFB 860 INST 186/859-1 to M.T. and H.U.), and an International Max Planck Research School fellowship (to M.N.) from the Molecular Biology doctoral Program at Göttingen University.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1215366110/-/DCSupplemental.
References
- 1.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: A decade on. Nat Rev Mol Cell Biol. 2007;8(12):947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 2.Gareau JR, Lima CD. The SUMO pathway: Emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol. 2010;11(12):861–871. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sarge KD, Park-Sarge OK. SUMO and its role in human diseases. Int Rev Cell Mol Biol. 2011;288:167–183. doi: 10.1016/B978-0-12-386041-5.00004-2. [DOI] [PubMed] [Google Scholar]
- 4.Wilkinson KA, Nakamura Y, Henley JM. Targets and consequences of protein SUMOylation in neurons. Brain Res Brain Res Rev. 2010;64(1):195–212. doi: 10.1016/j.brainresrev.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kantamneni S, et al. Activity-dependent SUMOylation of the brain-specific scaffolding protein GISP. Biochem Biophys Res Commun. 2011;409(4):657–662. doi: 10.1016/j.bbrc.2011.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craig TJ, et al. Homeostatic synaptic scaling is regulated by protein SUMOylation. J Biol Chem. 2012;287(27):22781–22788. doi: 10.1074/jbc.M112.356337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hay RT. SUMO: A history of modification. Mol Cell. 2005;18(1):1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 8.Seeler JS, Dejean A. Nuclear and unclear functions of SUMO. Nat Rev Mol Cell Biol. 2003;4(9):690–699. doi: 10.1038/nrm1200. [DOI] [PubMed] [Google Scholar]
- 9.Owerbach D, McKay EM, Yeh ET, Gabbay KH, Bohren KM. A proline-90 residue unique to SUMO-4 prevents maturation and sumoylation. Biochem Biophys Res Commun. 2005;337(2):517–520. doi: 10.1016/j.bbrc.2005.09.090. [DOI] [PubMed] [Google Scholar]
- 10.Bayer P, et al. Structure determination of the small ubiquitin-related modifier SUMO-1. J Mol Biol. 1998;280(2):275–286. doi: 10.1006/jmbi.1998.1839. [DOI] [PubMed] [Google Scholar]
- 11.Bayona JC, et al. 2011. SUMOylation pathway in Trypanosoma cruzi: Functional characterization and proteomic analysis of target proteins. Mol Cell Proteomics, 10.1074/mcp.M110.007369.
- 12.Braun L, et al. The small ubiquitin-like modifier (SUMO)-conjugating system of Toxoplasma gondii. Int J Parasitol. 2009;39(1):81–90. doi: 10.1016/j.ijpara.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 13.Park HC, et al. Identification and molecular properties of SUMO-binding proteins in Arabidopsis. Mol Cells. 2011;32(2):143–151. doi: 10.1007/s10059-011-2297-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosas-Acosta G, Russell WK, Deyrieux A, Russell DH, Wilson VG. A universal strategy for proteomic studies of SUMO and other ubiquitin-like modifiers. Mol Cell Proteomics. 2005;4(1):56–72. doi: 10.1074/mcp.M400149-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vertegaal AC, et al. Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol Cell Proteomics. 2006;5(12):2298–2310. doi: 10.1074/mcp.M600212-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Nie M, Xie Y, Loo JA, Courey AJ. Genetic and proteomic evidence for roles of Drosophila SUMO in cell cycle control, Ras signaling, and early pattern formation. PLoS ONE. 2009;4(6):e5905. doi: 10.1371/journal.pone.0005905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tatham MH, Matic I, Mann M, Hay RT. Comparative proteomic analysis identifies a role for SUMO in protein quality control. Sci Signal. 2011;4(178):rs4. doi: 10.1126/scisignal.2001484. [DOI] [PubMed] [Google Scholar]
- 18.Panse VG, Hardeland U, Werner T, Kuster B, Hurt E. A proteome-wide approach identifies sumoylated substrate proteins in yeast. J Biol Chem. 2004;279(40):41346–41351. doi: 10.1074/jbc.M407950200. [DOI] [PubMed] [Google Scholar]
- 19.Miller MJ, Barrett-Wilt GA, Hua Z, Vierstra RD. Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc Natl Acad Sci USA. 2010;107(38):16512–16517. doi: 10.1073/pnas.1004181107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leach MD, Stead DA, Argo E, Brown AJ. Identification of sumoylation targets, combined with inactivation of SMT3, reveals the impact of sumoylation upon growth, morphology, and stress resistance in the pathogen Candida albicans. Mol Biol Cell. 2011;22(5):687–702. doi: 10.1091/mbc.E10-07-0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaminsky R, et al. SUMO regulates the assembly and function of a cytoplasmic intermediate filament protein in C. elegans. Dev Cell. 2009;17(5):724–735. doi: 10.1016/j.devcel.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denison C, et al. A proteomic strategy for gaining insights into protein sumoylation in yeast. Mol Cell Proteomics. 2005;4(3):246–254. doi: 10.1074/mcp.M400154-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Hannich JT, et al. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J Biol Chem. 2005;280(6):4102–4110. doi: 10.1074/jbc.M413209200. [DOI] [PubMed] [Google Scholar]
- 24.Zhang FP, et al. Sumo-1 function is dispensable in normal mouse development. Mol Cell Biol. 2008;28(17):5381–5390. doi: 10.1128/MCB.00651-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hetzer M, Gruss OJ, Mattaj IW. The Ran GTPase as a marker of chromosome position in spindle formation and nuclear envelope assembly. Nat Cell Biol. 2002;4(7):E177–E184. doi: 10.1038/ncb0702-e177. [DOI] [PubMed] [Google Scholar]
- 26.Krumova P, et al. Sumoylation inhibits alpha-synuclein aggregation and toxicity. J Cell Biol. 2011;194(1):49–60. doi: 10.1083/jcb.201010117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsiao HH, Meulmeester E, Frank BT, Melchior F, Urlaub H. “ChopNSpice,” a mass spectrometric approach that allows identification of endogenous small ubiquitin-like modifier-conjugated peptides. Mol Cell Proteomics. 2009;8(12):2664–2675. doi: 10.1074/mcp.M900087-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88(1):97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 29.Riquelme C, Barthel KK, Liu X. SUMO-1 modification of MEF2A regulates its transcriptional activity. J Cell Mol Med. 2006;10(1):132–144. doi: 10.1111/j.1582-4934.2006.tb00295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X, et al. Role for KAP1 serine 824 phosphorylation and sumoylation/desumoylation switch in regulating KAP1-mediated transcriptional repression. J Biol Chem. 2007;282(50):36177–36189. doi: 10.1074/jbc.M706912200. [DOI] [PubMed] [Google Scholar]
- 31.Galisson F, et al. 2011. A novel proteomics approach to identify SUMOylated proteins and their modification sites in human cells. Mol Cell Proteomics, 10.1074/mcp.M110.004796.
- 32.Long J, Zuo D, Park M. Pc2-mediated sumoylation of Smad-interacting protein 1 attenuates transcriptional repression of E-cadherin. J Biol Chem. 2005;280(42):35477–35489. doi: 10.1074/jbc.M504477200. [DOI] [PubMed] [Google Scholar]
- 33.Kuwata T, Nakamura T. BCL11A is a SUMOylated protein and recruits SUMO-conjugation enzymes in its nuclear body. Genes Cells. 2008;13(9):931–940. doi: 10.1111/j.1365-2443.2008.01216.x. [DOI] [PubMed] [Google Scholar]
- 34.Johnson ES, Blobel G. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J Cell Biol. 1999;147(5):981–994. doi: 10.1083/jcb.147.5.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin SF, Tatham MH, Hay RT, Samuel ID. Quantitative analysis of multi-protein interactions using FRET: Application to the SUMO pathway. Protein Sci. 2008;17(4):777–784. doi: 10.1110/ps.073369608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matic I, et al. Site-specific identification of SUMO-2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Mol Cell. 2010;39(4):641–652. doi: 10.1016/j.molcel.2010.07.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.