Abstract
Autophagy, a cytoplasmic catabolic process, plays a critical role in defense against intracellular infection. In turn, evasion or inhibition of autophagy has emerged as an important virulence factor for intracellular pathogens. However, Anaplasma phagocytophilum, the obligatory intracellular bacterium that causes human granulocytic anaplasmosis, replicates in the membrane-bound compartment resembling early autophagosome. Here, we found that Anaplasma translocated substrate 1 (Ats-1), a type IV secretion effector, binds Beclin 1, a subunit of the class III PI3K and Atg14L, and it nucleates autophagosomes with markers of omegasomes, double FYVE-containing protein 1, Atg14L, and LC3. Ats-1 autophagy induction did not activate the starvation signaling pathway of mammalian target of rapamycin. These autophagy proteins were also localized to the Anaplasma inclusion. Ectopically expressed Ats-1 targeted the Anaplasma inclusions and enhanced infection, whereas host cytoplasmic delivery of anti–Ats-1 or Beclin 1 depletion by siRNA suppressed the infection; beclin 1 heterozygous-deficient mice were resistant to Anaplasma infection. Furthermore, Anaplasma growth arrest by the class III PI3K inhibitor 3-methyladenine was alleviated by essential amino acid supplementation. Thus, Anaplasma actively induces autophagy by secreting Ats-1 that hijacks the Beclin 1-Atg14L autophagy initiation pathway likely to acquire host nutrients for its growth.
Keywords: endoplasmic reticulum, autophagic body, mitochondria
Autophagy is an essential and highly regulated eukaryotic cellular homeostatic process that sequesters and digests intracellular components to recycle (1). Autophagy begins with the nucleation of an isolation membrane that elongates to envelop autophagic targets into double-membrane vacuole autophagosome, which expands and eventually fuses with lysosomes to become degradative autolysosomes (1). Autophagy plays important roles in immunological processes, including direct pathogen elimination, pathogen-associated molecular pattern processing for pattern recognition receptor, inflammasome regulation and autosecretion of alarmins, and cytosolic antigen processing for MHC-II presentation (2). Against intracellular pathogens, such as virus, bacteria, and protozoa, autophagy is one of the primary innate defense mechanisms (1). Intracellular invading bacteria, including Streptococcus pyogenes, Listeria monocytogenes, Salmonella enterica, and Shigella flexneri, have been shown to be ubiquitin-coated for autophagic degradation (1). To fight back, some intracellular pathogens were shown to have evolved strategies to either inhibit or avoid autophagy (1). For example, Shigella and Salmonella cause membrane damage at an early stage of cellular invasion, which inhibits the mammalian target of rapamycin (mTOR) activation of an amino acid starvation-induced autophagy pathway (3). Although Shigella escapes from phagosomes, Salmonella rapidly repairs the membrane damage to block autophagosome development (3). Shigella VirG, an outer membrane protein required for its intracellular actin-based motility, also induces autophagy by binding to Atg5 (4). To escape this autophagy, Shigella secretes a type III secretion effector IcsB, which competitively binds to VirG to evade Atg5 recognition (4). By binding to Beclin 1 (human homolog of yeast Atg6), Herpes simplex virus 1 protein ICP34.5 and γ-herpes virus-encoded viral Bcl-2-like proteins inhibit autophagosome initiation (1), whereas HIV accessory protein Nef or the influenza virus matrix protein 2 blocks autophagosome maturation (1). However, some intracellular bacterial pathogens benefit from autophagy activation, such as Coxiella (5), and Brucella (6), although the detailed mechanisms are not well-understood.
Autophagy has been also observed in obligatory intracellular bacterial infection, including Rickettsia conorii (7) and Orientia (former R.) tsutsugamushi (8). We have been studying the role of autophagy in Anaplasma phagocytophilum (Ap) infection. Ap is a Gram-negative obligatory intracellular bacterium that belongs to the order Rickettsiales (9). Ap infects granulocytes and endothelial cells of various mammalian species (9). In humans, Ap causes an emerging and major tick-borne disease called human granulocytic anaplasmosis, an acute febrile disease that is potentially fatal, especially in elderly or immunocompromised individuals (10). Ap replicates inside the membrane-bound vacuole (inclusion) that is secluded from the endosome–lysosome pathway (11). We previously showed that the Ap inclusion resembles the early autophagosome based on the presence of double-lipid bilayer membranes, endogenous Beclin 1, and GFP-tagged LC3 (human ortholog of yeast Atg8) and the absence of the late endosome–lysosome marker (12). Autophagy becomes evident during the early bacterial growth stage with clumps of Beclin 1 and LC3 and their colocalization with the Ap inclusion (12). However, contrary to the dogma, this autophagy does not seem to be an innate immune response against Ap infection, because inhibition of the autophagy with the class III PI3K (PI3KC3) inhibitor 3-methyladenine (3-MA) does not enhance Ap infection; rather, it arrests its growth (12). Furthermore, although rapamycin induction of autophagy clears Salmonella infection (13), it significantly promotes Ap replication (12). This finding led us to suggest that Ap actively induces inclusion-targeted autophagosome formation to aid its replication.
The type IV secretion (T4S) system is one of the important bacterial secretion systems that transfers bacterial proteins and/or DNA into the eukaryotic host in an ATP-dependent manner to alter critical host cell functions in favor of bacterial growth (14). Ap has genes encoding the T4S machinery, all of which are expressed by Ap in human leukocytes (15). So far, two T4S effectors, an ankyrin repeat-rich protein A (AnkA) and Ap translocated substrate 1 (Ats-1), have been identified experimentally. Secreted AnkA localizes in the host cell cytoplasm and nucleus (16). AnkA is essential for Ap infection, and it binds Abi-1 and the major tyrosine-phosphorylated protein in the infected cells (16). Unlike AnkA, Ats-1 lacks a tyrosine phosphorylation site, but it is imported into the mitochondrial matrix and delays mitochondria-mediated host cell apoptosis to benefit Ap (17). Ats-1 also localizes to Ap inclusions (17). Here, we report that Ats-1 is a unique example of a bacterial Beclin 1 binding protein that hijacks the Beclin 1-Atg14L autophagy initiation pathway likely to provide nutrients for bacterial growth.
Results
Ats-1 Binds Beclin 1 and Induces Extensive Membrane Vesicle Formation.
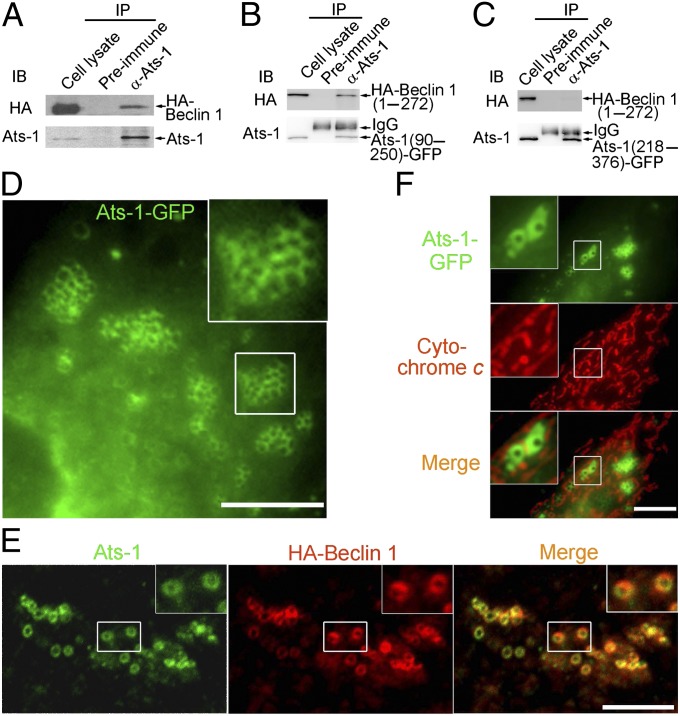
To identify a host cell protein that potentially binds secreted Ats-1, we screened a human bone marrow cDNA prey library by a yeast two-hybrid system using full-length Ats-1 as bait. The result revealed that human Beclin 1 (amino acids 191–327 and 396–450) (Fig. S1A) interacts with Ats-1. Beclin 1 is a core component of the PI3KC3 autophagy initiation complex (18). Ats-1 has a length of 376 aa; no known domain has been identified except the mitochondria-targeting sequence (MTS) in its N terminus. Approximately 60 aa of N terminus are cleaved off by the mitochondrial signal peptidase when Ats-1 is imported into the mitochondrial matrix (17). The T4S substrate signal in the Ats-1 C terminus can be recognized by the VirD4 coupling ATPase for secretion (17). Human Beclin 1 has a length of 450 aa and contains three distinct domains: a Bcl-2 binding domain (amino acids 88–150), a coiled-coil domain (amino acids 144–269), and an evolutionarily conserved domain (amino acids 244–337) (18). The interaction between Ats-1 and Beclin 1 was confirmed in vivo by coimmunoprecipitation (Co-IP) of HA-tagged Beclin 1 (HA-Beclin 1) with Ats-1 (Fig. 1A). The midsection of Ats-1 (amino acids 90–250) but not C-terminal Ats-1 (amino acids 218–376) interacted with the N-terminal one-half of Beclin 1 (amino acids 1–271) (Fig. 1 B and C and Fig. S1D). The reciprocal Co-IP confirmed the interaction between HA-Beclin 1 (amino acids 1–271) and Ats-1 (amino acids 90–250) (Fig. S1B). Strikingly, ectopic expression of Ats-1–GFP or Ats-1 without a tag triggered the formation of numerous cytoplasmic Ats-1–containing puncta and vesicles of ∼0.5–1 μm in diameter in RF/6A cells (Fig. 1 D–F). Individual vesicles were clustered at various degrees (Fig. 1 D–F). These Ats-1 vesicles are distinct from mitochondria, because they did not colocalize with the mitochondria marker cytochrome c (Fig. 1F). Although in control cells, HA-Beclin 1 was diffuse, HA-Beclin 1 localized to Ats-1–containing vesicles in Ats-1–GFP-trasfected cells (Fig. 1E). Ats-1 (amino acids 218–376), which cannot bind Beclin 1, did not induce vesicles (Fig. S1C), suggesting that Ats-1 binding to Beclin 1 is required for vesicle formation.
Fig. 1.
Ats-1 binds Beclin 1 and triggers vesicle formation. (A–C) Co-IP assay for protein interaction between Ats-1 (without a tag) and HA-Beclin 1 (A), Ats-1(90–250)-GFP and HA-Beclin 1(1–272) (B), and Ats-1(218–376)-GFP and HA-Beclin 1(1–272) (C). Cotransfected HEK293 cells were immunoprecipitated (IP) with preimmune IgG or anti (α)–Ats-1. Precipitates were immunoblotted (IB) with anti-HA and anti–Ats-1. IgG, rabbit IgG heavy chain. (D) Ats-1–GFP forms clusters of vesicles of ∼0.5–1 μm in diameter in transfected RF/6A cells. (E) Localization of HA-Beclin 1 to vesicles containing tag-less Ats-1 in cotransfected RF/6A cells. (F) Ats-1–GFP vesicles are distinct from mitochondria labeled with anticytochrome c. (Scale bars: D–F, 10 μm.) Insets show magnified boxed areas.
Ats-1 Is Sufficient to Nucleate Autophagosomes by Involving Endoplasmic Reticulum Autophagy Proteins.
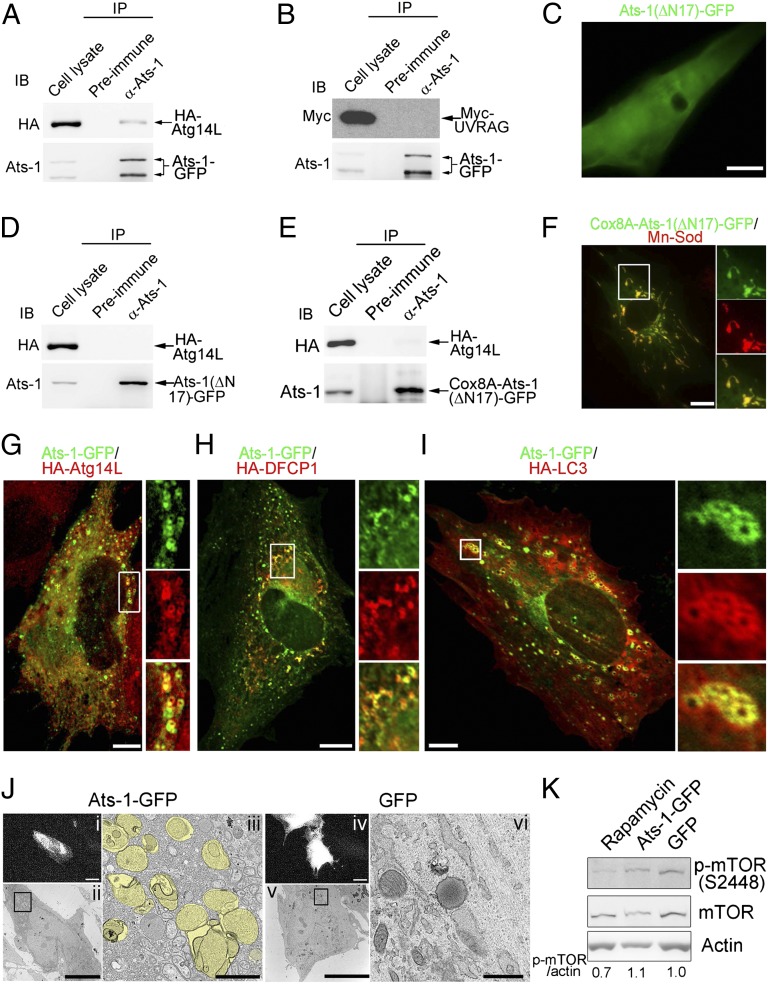
Because Ats-1 binds Beclin 1 and induces Ats-1– and Beclin 1-containing vesicles, we examined the nature of the Ats-1 and Beclin 1 complex and these membrane vesicles. Most cellular Beclin 1 preexists as a complex with Vps34 of PI3KC3 (19), and it is known to bind cytoplasmic proteins critical for initiation and maturation of autophagy (18). Vps34 binds to the evolutionarily conserved domain of Beclin 1 (20), whereas Atg14L/Beclin 1-associated autophagy-related key regulator and UV radiation resistance-associated gene (UVRAG) bind to the coiled-coil domain of Beclin 1 in a mutually exclusive manner (21). Atg14L is an endoplasmic reticulum (ER) protein, and it recruits a subset of the Beclin 1-Vps34 complex to the ER for autophagosome nucleation in response to amino acid starvation (22). Atg14L does not affect the Beclin 1-Vps34 binding but positively regulates PI3KC3 activity (23). However, binding of Beclin 1 to UVRAG promotes autophagosome and endosome maturation to autolysosomes (21, 23). In Ats-1–GFP-expressing cells, anti–Ats-1 antibody pulled down Atg14L but not UVRAG (Fig. 2 A and B), suggesting that Ats-1 binding to Beclin 1 recruits Atg14L to the complex and nucleates autophagosomes. Interestingly, N-terminal 17-aa deletion of Ats-1 and Ats-1(∆N17)-GFP neither recruited Atg14L (Fig. 2D) nor induced vesicles (Fig. 2C). Because Ats-1(∆N17)-GFP also did not target mitochondria (Fig. 2C), we determined whether the mitochondria targeting is required for the Ats-1–Beclin 1 complex to recruit Atg14L or form vesicles. After replacing the N terminus of Ats-1 with the mitochondria-targeting sequence from an unrelated protein, human cytochrome c oxidase subunit VIIIA (Cox8A), Cox8A-Ats-1(∆N17)-GFP localized to mitochondria and was cleaved, but no vesicles were observed in transfected cells (Fig. 2 E and F). Furthermore, Cox8A-Ats-1(∆N17)-GFP poorly recruited Atg14L (Fig. 2E). These results indicate that the mitochondria targeting is not required, but the specific N-terminal sequence of Ats-1 is required for its association with Atg14L and vesicle nucleation. Amino acid sequences, particularly the presence of arginine in MTS, are critical in determining the protein distribution between mitochondria and cytosol (24). Ats-1 N-17 contains four arginines (Fig. S2A). After deletion of the first N-terminal arginine, Ats-1 (∆4R) induced fewer autophagosomes and translocated more to mitochondria than Ats-1 (Fig. S2 B and C), whereas after substitution of the first and second arginines with alanines, Ats-1 (4-5AA) formed more autophagosomes and translocated less to mitochondria than Ats-1 (Fig. S2 B and C). Addition of one more arginine after the second arginine did not have significant difference (Fig. S2 B and C). These results indicate that mitochondria targeting and autophagy are separate activities of Ats-1 influenced by the N-terminal sequence of Ats-1. HA-Atg14L localized to Ats-1–GFP vesicles, which is shown in cotransfected cells (Fig. 2G and Fig. S2D). Interestingly, two patterns of Atg14L localization with Ats-1–GFP vesicles were observed [overlapping with Ats-1–GFP vesicles (Fig. 2G) or laying inside of Ats-1–GFP vesicles as small dots (Fig. S2D)], likely representing different stages of autophagosome nucleation.
Fig. 2.
Ats-1 nucleates autophagosomes involving ER autophagy proteins. (A and B) Co-IP assay for protein interaction between Ats-1–GFP and HA-Atg14L (A) or Myc-UVRAG (B). Cotransfected HEK293 cells were immunoprecipitated (IP) with preimmune IgG or anti (α)–Ats-1. Precipitates were immunoblotted (IB) with anti–Ats-1 and anti-HA or anti-Myc. (C) The diffuse pattern of Ats-1(∆N17)-GFP in transfected RF/6A cells. (D and E) Co-IP assay for protein interaction between HA-Atg14L, and Ats-1(∆N17)-GFP (D) or Cox8A-Ats-1(∆N17)-GFP (E). Cotransfected HEK293 cells were IP with preimmune IgG or α–Ats-1. Precipitates were IB with anti–Ats-1 and anti-HA. (F) Localization of Cox8A-Ats-1(∆N17)-GFP (green) with mitochondria labeled with anti–Mn-Sod (red). The boxed area is magnified on the right. (G–I) Localization of HA-Atg14L (G), HA-DFCP1 (H), and HA-LC3 (I) to Ats-1–GFP vesicles. Boxed areas are magnified on the right. (Scale bars: C, E, and G–I, 10 μm.) (J) Correlative light-electron microscopy for RF/6A cells expressing Ats-1–GFP or GFP alone. (i and iv) Fluorescence micrographs of cells expressing Ats-1–GFP and GFP, respectively. (Scale bars: 10 μm.) (ii and v) TEM of the fluorescent cells in i and iv. (Scale bars: 10 μm.) (iii and vi) Magnified boxed area in ii and v. Yellow pseudocolored areas denote autophagosomes. (Scale bars: 1.0 μm.) (K) Phosphorylated mTOR level in Ats-1–GFP-expressing cells. RF/6A cells transfected with plasmid encoding GFP or Ats-1–GFP were subjected to Western blot analysis using antibodies against actin, mTOR, and phosphorylated mTOR (p-mTOR). Rapamycin-treated RF/6A cells were used as positive control. The values under the bands show the relative ratios of band intensities of p-mTOR to actin, with the ratio in the GFP lane set as one.
The earliest stage of autophagosome formation known is omegasome; it is formed on a specialized subdomain of the ER, which serves as a cradle for biogenesis of the isolation membrane (25). The currently known marker for omegasome is double FYVE-containing protein 1 (DFCP1), which has an ER-targeting motif and binds PI3P that is locally produced by the ER-localized Atg14L-Beclin 1-Vps34 PI3KC3 complex (25). ER contains negligible amount of PI3P without autophagy induction (22). Because Atg14L localizes to Ats-1–GFP vesicles, we examined whether Ats-1 nucleates the omegasome. Cotransfection study showed HA-DFCP1 localization to Ats-1–GFP vesicles (Fig. 2H), indicating the involvement of PI3P and omegasomes in Ats-1–containing vesicle formation. LC3 is a canonical autophagy elongation protein (26). Ats-1 vesicles continued to progress to autophagosomes as indicated by acquisition of HA-LC3, especially by relatively larger Ats-1 vesicles (Fig. 2I). The ultrastructural maker of autophagosomes is the presence of double membranes. In agreement with our colocalization experiments, correlative light-electron microscopy (27) showed the appearance of many double membrane-lined autophagosomes with a size of 0.5–1 µm in the cytoplasm of cells expressing Ats-1–GFP but not in the cytoplasm of cells expressing GFP alone (Fig. 2J). Indeed, ectopic expression of Ats-1 induced more conversion of cytosolic endogenous LC3-I to phosphatidylethanolamine-conjugated (membrane-bound) LC3-II, the molecular marker for autophagosome formation (28), compared with ectopic expression of GFP (Fig. S3). Taken together, these results indicate that Ats-1 alone is sufficient to nucleate autophagosomes by recruiting ER-associated autophagy proteins. The phosphorylation of Ser2448 on mTOR is directly related to amino acid and nutrient status (29). mTOR phosphorylation was not reduced in Ats-1–GFP-expressing cells (Fig. 2K), indicating the lack of involvement of the starvation signaling pathway of mTOR in Ats-1–induced autophagy. Rab7 is required for autophagosome maturation (30). Furthermore, we found that Ats-1–GFP vesicles matured to autolysosomes, because some Ats-1–GFP vesicles colocalized with Rab7 and a late endosome/lysosome marker lysosomal-associated membrane protein 1 (LAMP-1) (Fig. S4 A and B). In agreement with this observation, Ats-1–GFP was degraded even faster than GFP in transfected cells (Fig. S4 C and D).
Cytoplasmic Ats-1 Localizes to Anaplasma Inclusions.
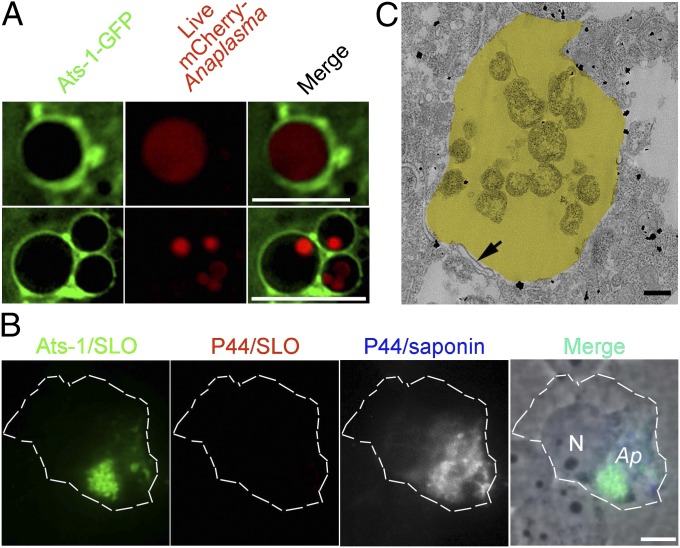
Given autophagosome nucleation by ectopic expression of Ats-1,and the presence of Beclin 1, LC3, and Ats-1 on Ap inclusions (12, 17), we determined whether Ats-1–GFP is recruited to live Ap inclusions. Live image analysis revealed Ats-1–GFP localization to inclusions containing mCherry-expressing Ap (31) (Fig. 3A). To determine whether translocated native Ats-1 localizes on the cytoplasmic face of the inclusion, cells were processed for two rounds of immunostaining: a first round with selective permeabilization of the plasma membrane by streptolysin O (SLO) (32) to detect cytoplasmic Ats-1 but not Ats-1 within inclusions or mitochondria, and after fixation, a second round with saponin permeabilization to detect P44, the major outer membrane protein of Ap within the inclusion. After the first round of staining, clumps of Ats-1 were seen, but P44 was not detectable (Fig. 3B). After saponin permeabilization, Ats-1 and P44 were seen to colocalize, indicating that native Ats-1 localized to the cytoplasmic face of Ap inclusions (Fig. 3B). In agreement with immunofluorescence labeling, transmission EM (TEM) immunogold labeling showed that native Ats-1 localizes to Ap inclusion as well as the cytoplasm of infected HL-60 cells (Fig. 3C).
Fig. 3.
Ats-1 localizes to Ap inclusions. (A) Ats-1–GFP targets Ap inclusions. Live Ats-1–GFP-expressing RF/6A cells were infected with mCherry-Ap. Bacterial inclusions surrounded by Ats-1–GFP in two cells are shown. (Scale bars: 10 μm.) (B) Localization of native Ats-1 on the cytoplasmic side of Ap inclusions. The plasma membrane of infected cells was selectively permeabilized with SLO and labeled with anti–Ats-1 (Ats-1/SLO) and anti-P44 (P44/SLO). After the first round of staining, all cell membranes were permeabilized with saponin, and cells were stained again with anti-P44 (P44/saponin). Secondary antibodies with distinct fluorochromes were used for P44 labeling before (red) and after (blue and gray pseudocolor) saponin treatment. Contours of the HL-60 cell are marked with white dashed lines. Ap, Ap inclusions. N, nucleus. (Scale bar: 5 μm.) (C) Immuno-EM for Ats-1 in infected cells. Yellow pseudocolored area, Ap inclusion. The arrow indicates double membranes. (Scale bar: 0.5 μm.)
ER Autophagy Proteins Localize to Anaplasma Inclusions.
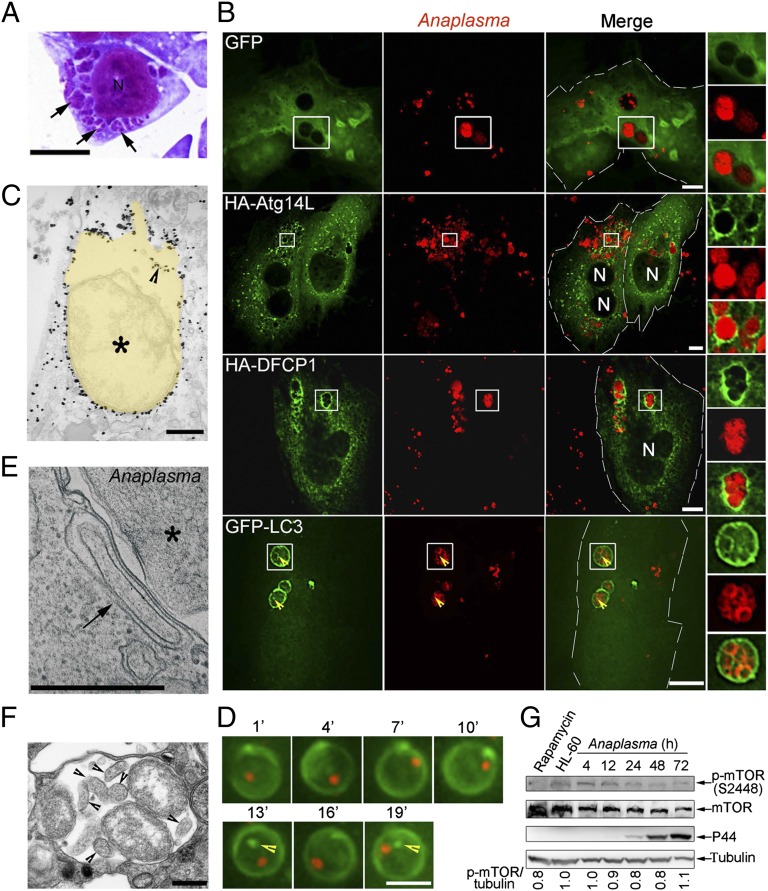
Given the autophagosome characteristics of Ats-1–GFP vesicles and Ats-1–GFP and native Ats-1 targeting to Ap inclusions, we next examined the stages of autophagosome development on Ap inclusions. By light microscopy, Ap inclusions appear as characteristic irregular clumps of various sizes because of dense aggregates of bacteria (a few to >50) inside inclusions called morula (morula means mulberry in Latin) (Fig. 4 A and B) (9). Immunofluorescence labeling showed the localization of Atg14L and DFCP1 to Ap inclusions (Fig. 4B). As a negative control, GFP did not show such localization to Ap inclusions (Fig. 4B). Both outer and inner membranes of autophagosomes are decorated with LC3 (28). Previously, we reported GFP-LC3 on the Ap inclusion membrane by immunofluorescence labeling at a low magnification (12). At a higher magnification using deconvolution microscopy, we found, in addition to the inclusion membrane, some GFP-LC3 clumps inside of Ap inclusions (Fig. 4B). Similarly, TEM immunogold labeling showed the presence of GFP-LC3 on the Ap inclusion and inside of the inclusion (Fig. 4C). This finding is not a fixation permeabilization or sectioning artifact, because GFP-LC3 clumps and puncta were also found inside of dynamic live Ap inclusions without such treatments (Fig. 4D and Movie S1). During the fusion of autophagosomes with vacuoles in yeast, the outer membrane of autophagosomes fuses with the membrane of vacuoles to release single inner membrane vesicles containing the cytoplasm (autophagic body) into vacuoles (33). TEM revealed the outer membrane of autophagosomes closely apposed to the Ap inclusion membrane and the presence of autophagic bodies similar to the host cytoplasmic content within Ap inclusions (Fig. 4 E and F), suggesting that the fusion between autophagosomes and Ap inclusion membranes occurred, releasing autophagic bodies inside inclusions. Taken together, these results indicate that the Ap inclusions contain a similar set of ER autophagy proteins as Ats-1–containing vesicles and acquire host cytoplasmic components such as LC3 and the autophagic body-like structure inside inclusions. mTOR phosphorylation was slightly decreased at the exponential growth stage of Ap (Fig. 4G).
Fig. 4.
Autophagy proteins localize to Ap inclusions. (A) Ap-infected cells at 2 d postinfection (p.i.). Ap morulae/inclusions are indicated with arrows in Diff-Quik staining. N, nucleus. (Scale bar: 5 μm.) (B) Ap-infected RF/6A cells expressing GFP, HA-Atg14L, HA-DFCP1, or GFP-LC3 were immunostained with anti-Ap and anti-HA at 2 d p.i. N, nucleus. Boxed areas are magnified on the right. (Scale bars: 10 μm.) Arrowheads indicate the presence of GFP-LC3 inside of Ap inclusions. (C) Immunogold labeling for GFP-LC3 in Ap-infected GFP-LC3–expressing RF/6A cells. Yellow pseudocolored area, Ap inclusion. An arrowhead indicates the GFP-LC3 inside of Ap inclusion. An asterisk indicates Ap. (D) Selected sequential video images of a single inclusion containing an mCherry-Ap (red) in live RF/6A cells transfected with plasmid encoding GFP-LC3 (green). GFP-LC3 puncta inside of Ap inclusion are indicated with arrowheads. (Scale bar, 4 μm.) (E) An autophagosome (arrow) is closely apposed to the Ap (asterisk) inclusion membrane in HL-60 cells. (F) An Ap inclusion containing autophagic bodies (arrowheads). Note that the content of autophagic vesicles is similar to the host cell cytoplasm. (C, E, and F) TEM. (Scale bars: 0.5 μm.) (G) Phosphorylated mTOR level in Ap-infected HL-60 cells harvested at 4, 12, 24, 48, and 72 h p.i. Western blot analysis using antibodies against α-tubulin, mTOR, and p-mTOR. HL-60, uninfected HL-60 cells. Rapamycin-treated HL-60 cells were used as positive control. The values under the bands show the relative ratios of band intensities of p-mTOR to tubulin, with the ratio in HL-60 lane set as one.
Full-Length Ats-1, LC3-II, and ER Membrane Protein Cofractionate with Anaplasma Inclusions.
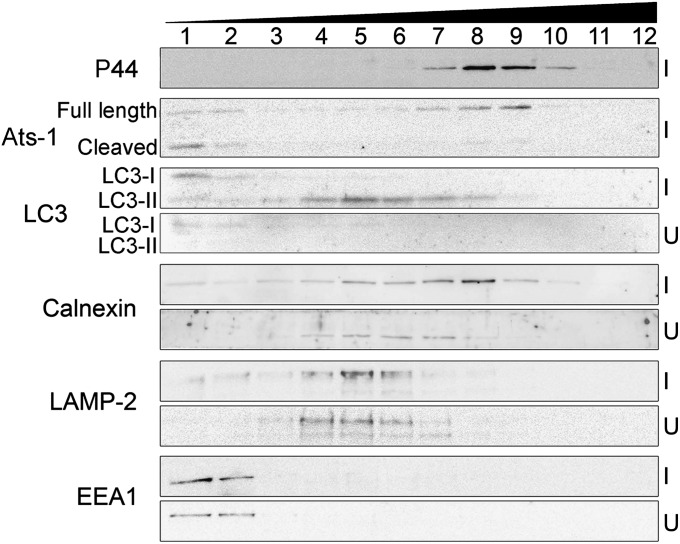
To determine cellular distribution of native Ats-1 and endogenous LC3-II in Ap-infected cells, we fractionated organelles by OptiPrep density gradient centrifugation. By this method, the Ap inclusions remained intact, which was indicated by the lack of digestion of Ap outer membrane protein P44 by pronase (Fig. S5A). Immunoblot analysis of the Optiprep fractions revealed full-length native Ats-1 mostly cofractionated with inclusions (P44 as marker), indicating that inclusion-associated Ats-1 was not cleaved. However, Ats-1 cleaved by the mitochondrial signal peptidase (17) was mostly distributed between fractions 1 and 2 (Fig. 5). In agreement with previous studies (11), the early endosome [early endosome antigen-1 (EEA1)] and lysosome (LAMP-2) did not cofractionate with Ap inclusions, and their distribution was unchanged by infection (Fig. 5). In agreement with our previous work of GFP-LC3 activation in Ap-infected RF/6A cells (12), the endogenous LC3 processing from cytosolic LC3-I to membrane-bound LC3-II and total LC3 up-regulation were evident in Ap-infected HL-60 cells (Fig. 5 and Fig. S5B). Autophagosomes (LC3-II as marker) were, in part, cofractionated with the Ap inclusions (P44 as marker). Furthermore, ER (calnexin as marker) distribution was clearly shifted to the Ap inclusion fraction in infected cells compared with uninfected cells (Fig. 5). In agreement with our morphological observations, these data indicate that Ap-containing autophagosome formation mobilized the autophagy protein and ER but not the endosomes–lysosomes pathway.
Fig. 5.
Cellular fractionation of Ap-infected cells. Immunoblotting of fractions (1–12) of Ap-infected HL-60 cells (I) and uninfected HL-60 cells (U) collected after OptiPrep density gradient centrifugation using anti–Ats-1 and various organelle markers: anti-LC3 (autophagosome), anticalnexin (ER), anti–LAMP-2 (lysosomes and late endosomes), anti-EEA1 (early endosomes), and anti-P44 (Ap inclusions).
Ats-1 Promotes Anaplasma Growth.
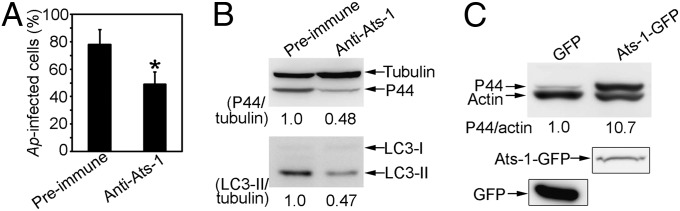
Because Ats-1 nucleates autophagosomes and targets Ap inclusions, we examined whether host cytoplasmic Ats-1 affects Ap replication. Because genetic manipulation of Ap at a specific locus is not possible, we used the Chariot-mediated protein transfection system to deliver affinity-purified anti–Ats-1 antibodies into infected cells to examine the role of native Ats-1 in bacterial replication. Chariot-delivered anti–Ats-1 inhibited Ap replication and infection-induced endogenous LC3-I to LC3-II conversion (Fig. 6 A and B). Conversely, Ap infection was enhanced ∼10-fold in RF/6A cells overexpressing Ats-1–GFP compared with cells expressing GFP alone (Fig. 6C). In contrast, Ats-1–GFP did not enhance the infection of Ehrlichia chaffeensis (Fig. S6), which is closely related to Ap but resides in early endosome-like compartments (11). These results indicate that host cytoplasmic Ats-1 specifically enhances Ap growth.
Fig. 6.
Ats-1 promotes Ap infection. Ap infection (A and B) and autophagy induction (B) in HL-60 cells 2 d after cytoplasmic delivery of anti–Ats-1 or preimmune IgG by the Chariot system. (A) Percent infected cells. Data are shown as the mean ± SD from three independent experiments. *Significantly different (P < 0.01). (B) Immunoblotting using anti-P44, antitubulin, and anti-LC3. The values under the bands show the relative ratios of band intensities, with the ratios of those band intensities from preimmune IgG control set as one. (C) Ap infection in RF/6A cells transfected with plasmid encoding GFP or Ats-1–GFP [0.5 d posttransfection (p.t.), 3 d p.i.]. Immunoblot analysis using anti-P44 and antiactin. The values under the bands show the relative ratios of band intensities, with the ratios of those band intensities from GFP set as one. The expressions of GFP and Ats-1–GFP in transfected cells are shown at the bottom.
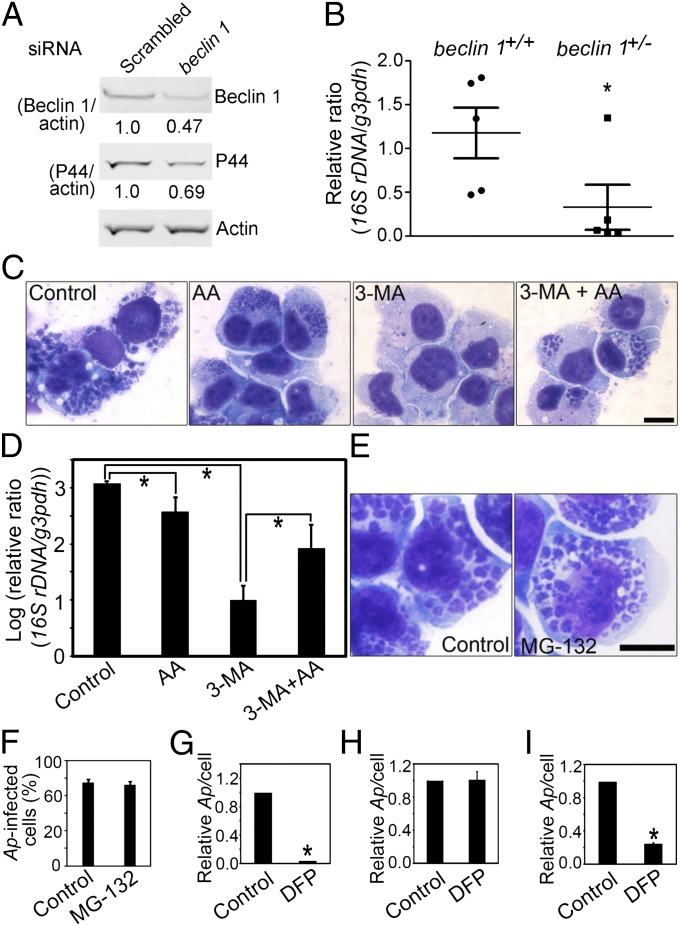
Depletion of Beclin 1 Suppresses Ap Infection in Vitro and in Vivo, and Amino Acid Supplementation Overrides 3-MA Inhibition of Anaplasma Replication.
Because Ats-1 binds Beclin 1 and promotes Ap replication, we next examined, whether Beclin 1 is required for Ap infection. In RF/6A cells in which the Beclin 1 level was reduced by siRNA transfection, Ap replication was significantly reduced compared with cells transfected with a control siRNA (scrambled sequence) (Fig. 7A). Cells from beclin 1 heterozygous-deficient (beclin 1+/−) mice (homozygous deficiency is embryonically lethal) have reduced autophagy (34); beclin 1+/− mice had significantly reduced Ap infection compared with WT mice (Fig. 7B), showing the significant role of Beclin 1-mediated autophagy in Ap infection.
Fig. 7.
Beclin 1 depletion impairs Ap infection, and amino acid supplementation overrides 3-MA inhibition of Ap replication. (A) Immunoblotting of lysates of Ap-infected RF/6A cells transfected with beclin 1 siRNA or scrambled siRNA (2 d p.t., 3 d p.i.) using anti-P44, antiactin, and anti-Beclin 1. The values under the bands show the relative ratios of band intensities, with the ratios of those band intensities from scrambled siRNA set as one. (B) The dot plot of Ap load in the blood from beclin 1+/− and WT mice at 5 d p.i. Quantitative PCR of Ap 16S rDNA normalized to mouse G3PDH DNA. *Significantly different (P < 0.05). (C) Ap-infected cells: untreated (Control), or treated with amino acids supplementation (AA), 3-MA (3-MA), or 3-MA and amino acids (3-MA + AA). Diff-Quik staining. (Scale bar: 10 μm.) (D) Quantitative PCR of Ap 16S rDNA normalized to human G3PDH DNA. *Significantly different (P < 0.05) by ANOVA. (E and F) The effect of MG-132 on Ap growth. Diff-Quik–stained images of Ap in solvent control or MG-132–treated HL-60 cells (E) and the percentage of Ap infected HL-60 cells on 3 d p.i. (F). (Scale bar: 10 μm.) (G–I) The effect of DFP on Ap growth. Host cell-free Ap pretreated with solvent control or DFP was used to infect HL-60 cells (G). HL-60 cells were pretreated with solvent or DFP followed by washing and infection with Ap (H). HL-60 cells were infected with Ap followed by the treatment with solvent or DFP at 1 d p.i. (I). The relative ratios of Ap/cell were shown, with the ratios from control set as one. *Significantly different (P < 0.01).
3-MA severely inhibits Ap growth (12). Although fivefold excess of the 12 essential amino acid mixture slightly inhibited Ap growth (by inhibiting the starvation-induced autophagy), it partially alleviated 3-MA–induced growth inhibition (Fig. 7 C and D). Unlike 3-MA, the vacuolar-type ATPase inhibitor Bafilomycin A1 or the proteasome inhibitor MG-132 had no effect on Ap infection (only MG-132 data are shown) (Fig. 7 E and F), suggesting a lack of involvement of lysosomal or proteasomal proteolysis in Ap growth.
Our previous proteomics work revealed several proteases, including serine protease, on the surface of Ap and E. chaffeensis (35). These bacterial surface proteases may be sufficient to degrade host cytoplasmic proteins after they are delivered inside inclusions. To test this hypothesis, we first pretreated Ap with the membrane-permeable serine protease inhibitor diisopropylfluorophosphate (DFP). This treatment completely prevented Ap infection, whereas the pretreatment of host cells with DFP had no effect on Ap infection. Furthermore, DFP added at 1 d postinfection arrested Ap growth, suggesting Ap serine protease involvement in Ap growth likely by digesting autophagic substrates (Fig. 7 G–I).
Discussion
The present findings show that the T4S effector Ats-1 nucleates autophagosomes. So far, only a few non-T4S related bacterial products, including cytolysin from Vibrio cholerae, vacuolating cytotoxin from Helicobacter pylori, and VirG from S. flexneri, have been reported to induce autophagosome formation (36). Although Beclin 1 functions as an interacting hub for several proteins to regulate autophagy in mammalian cells (18), no bacterial protein has been reported to bind Beclin 1. In contrast, several viral proteins are known to bind Beclin 1 (1). Because all known viral Beclin 1 binding proteins inhibit autophagy rather than induce autophagy, Ats-1 is a unique example of a pathogen-derived Beclin 1 binding protein that drives autophagosome formation. Furthermore, Ats-1 is a pathogen-derived molecule that enhances pathogen growth by inducing autophagosomes. Our current study showed that the Ats-1 binding partner Beclin 1 is, indeed, required for in vitro and in vivo Ap infection. Autophagy favors replication of C. burnettii in phagolysosomes (5), whereas autophagy initiation, but not autophagy elongation, proteins are required for B. abortus to complete its intracellular cycle and spread (6). Unlike C. burnetii or Ap, autophagy does not affect replication of B. abortus. The detailed mechanism, particularly the Coxiella and Brucella factor critical for autophagy induction, is currently unknown. Recently, it was reported that infection by Shigella and Salmonella causes host cell membrane damage, leading to down-regulation of the mTOR activity, and autophagy induction through the starvation signaling pathway (3). Our results showed that mTOR Ser2448 phosphorylation was not reduced by Ats-1–GFP transfection. Thus, Ats-1 autophagy induction likely begins on Beclin 1 binding, bypassing the starvation signaling pathway of mTOR. Compared with Shigella and Salmonella, Ap grows much more slowly in the host cells, normally taking 3–4 d to complete intracellular infection cycle. We do not know whether Ap causes host cell membrane damage during entry like Shigella and Salmonella (3), but punctate LC3 was not found in the host cell until 1 d postinfection, when Ap exponential growth begins (12). Moreover, in contrast to Salmonella, which only transiently colocalizes with LC3 at the early stage (1–2 h) of infection (3), Ap inclusions persistently accumulate LC3 during infection. However, we cannot deny the possibility that Ap may cause amino acid starvation in host cells during its growth, leading to concurrent autophagy induction through the starvation pathway, because we found that phosphorylated mTOR was slightly reduced at the exponential growth stage of Ap infection. Ats-1–induced autophagy likely synergizes with the starvation-induced autophagy to promote Ap growth, because rapamycin treatment enhances Ap growth (12). Because rapamycin treatment clears Salmonella (13), this result also supports that Ats-1–induced autophagosomes are distinct from membrane damage-induced autophagosomes. An additional important distinction from other pathogen-induced autophagosomes is that the autophagosomes recruited to Ap inclusions are not matured to autolysosomes, which was evidenced by the lack of LAMP-1 and LAMP-3 and the persistence of GFP-LC3 punctates on Ap inclusions (11, 12).
During the nucleation of autophagosomes, PI3P, generated by the activated Beclin 1-Vps34 complex, marks the omegasome formation site on the ER, which is decorated with DFCP1 (25). Treatment with the PI3KC3 inhibitor 3-MA blocks DFCP1 clumping on the specialized domain of ER in amino acid starvation-induced autophagy (25). Our result of DFCP1 localization to Ap inclusions indicates PI3P on the Ap inclusion membrane. The result is consistent with our previous observation of localization of Beclin 1 puncta to Ap inclusions and 3-MA inhibition of Ap replication (12). This finding seems to be a unique feature of Ap autophagosomes, because DFCP1 has not been reported on any other vacuoles associated with intracellular infection. Similarly, Atg14L was not reported to localize to any intracellular pathogen-associated compartment, although Atg14L is required for completion of the intracellular life cycle of Brucella (6). Atg14L accumulates on highly curved PI3P-enriched autophagic membrane through its C-terminal Beclin 1-associated autophagy-related key regulator/Atg14L autophagosome targeting sequence domain (37). It was suggested that, when autophagosomes expand by membrane fusion, PI3P is dephosphorylated, which leads to the dissociation of the Atg14L-PI3KC3 complex from autophagosomes (37). Furthermore, a study proposed that Atg14L and PI3KC3 dissociation from autophagosome might cause membrane curvature instability and prime the membrane for fusion with endosomes–lysosomes (37). Prevention of lysosomal fusion with Ap autophagosomes is one of the activities essential for intracellular survival of Ap. It is possible that retention or delayed dissociation of Atg14L and DFCP1 (perhaps because of prolonged PI3P accumulation) on Ap inclusions is one of the mechanisms to block lysosomal fusion. UVRAG was reported to be involved in autophagosome maturation (21). Because Atg14L prevents UVRAG from binding to the Ats-1–Beclin 1 complex, it also leads to a lack of lysosomal fusion with Ap inclusions. However, inhibition of lysosomal fusion seems to require additional Ap proteins, because Ats-1–GFP vesicles mature to autolysosomes.
The presence of Atg14L and DFCP1, which have ER localization domains, on Ap inclusions and cofractionation of the ER marker calnexin with the Ap inclusion suggest the involvement of ER in Ap autophagosome and inclusion biogenesis. ER is the major source for autophagosome membrane in amino acid starvation-induced autophagy (22, 25). Alternative or additional membrane sources for autophagosomes reported are mitochondria, plasma membrane, Golgi, endosomes, and the nuclear membrane (26). From mitochondria, mitochondrial outer membrane lipids and proteins are translocated to autophagosomes (38). The N-terminal mitochondria-targeting sequence of Ats-1 is essential for Ats-1–GFP autophagosome formation. However, the lack of Ats-1–GFP vesicle colocalization with mitochondria and the failure of Cox8A-Ats-1(∆N17)-GFP to form autophagosomes suggest that mitochondria may not be involved in Ats-1–GFP autophagosome formation. Results of our MTS mutation experiment showed critical roles of arginines in Ats-1 (N17) in determining distribution of Ats-1 between mitochondria and autophagosomes. Our studies suggest that mitochondria-targeting and autophagy-inducing activities of Ats-1 are competitive. In addition, host cell conditions influence the cellular distribution of Ats-1. When exponentially growing host cells that are used for transfection, Ats-1 primarily targets mitochondria (17). In contrast, less Ats-1 is localized to mitochondria in cells at high confluence, which is shown in the present study. The translocase of the mitochondrial outer membrane complex is the main import pore for nuclear-encoded proteins into mitochondria, and the mitochondrial membrane potential is required to import proteins into mitochondria (39). Cells at high confluence have lower density of the translocase of the mitochondrial outer membrane 20 cluster and lower mitochondrial membrane potential than cells at low confluence (40). This finding suggests that, during infection, host cell physiologic conditions, including the mitochondrial membrane potential, influence the distribution of Ats-1 between mitochondria and autophagosomes and consequently, Ap growth. Although functions of a number of autophagy proteins have been characterized in mammalian cells, the molecular events underlying autophagy nucleation are not fully understood, in part because of complex autophagy inducers and unpredictable cellular sites of autophagosome formation. In this aspect, a single-molecule Ats-1–induced autophagy may serve as a simple model to study the details of autophagosome initiation.
Ap has a small genome size of 1.47 Mb with a limited number of genes for biosynthesis and metabolism (41), which obliges Ap to acquire host nutrients for growth; how this acquisition is achieved within the confines of inclusions is not understood. A few decades ago, the concept of autophagy that helps feed an obligatory intracellular pathogen O. tsutsugamushi was introduced but lacked investigation using modern methods (8). The present study showed that Ats-1 induces autophagosome formation on Ap inclusions and enhances bacterial replication, and Ap growth arrest caused by the autophagy inhibitor 3-MA treatment is partially abrogated by amino acid supplementation. Taken together, these data imply that Ats-1–induced autophagy provides nutrients for Ap growth. Importantly, Ap nutrient acquisition does not involve lysosomal of proteasome degradation; instead, a bacterial serine protease seems to be involved. Recently, it was shown that host proteasomal degradation generates amino acids essential for Legionella pneumophila growth (42). Thus, intracellular pathogens evolved diverse mechanisms to obtain required nutrients for replication.
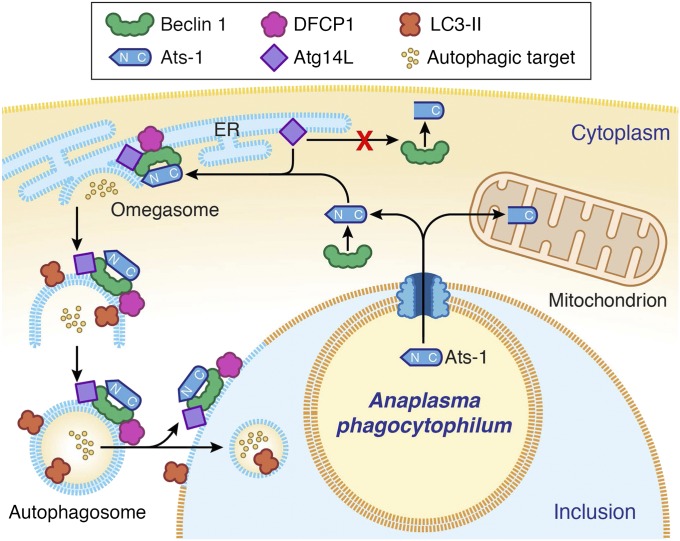
In summary, the present study unraveled a mechanism evolved in an obligatory intracellular pathogen to promote intracellular replication. Based on our previous and current data, our proposed model of autophagosome nucleation by Ats-1 in Ap-infected cells is depicted in Fig. 8. The identified Ats-1 autophagy pathways provide new targets for possible development of intervention strategies against human granulocytic anaplasmosis. In addition, our investigation of autophagy-subverting bacteria and molecule contributes fundamentally to the field of autophagy signaling and regulation.
Fig. 8.
Proposed model of autophagosome nucleation by Ats-1 in Ap-infected cells. Ats-1 is translocated from Ap to the cytoplasm of infected cells by the type IV secretion system. A portion of Ats-1 interacts with the host autophagosome initiation complex (Atg14L-Beclin 1-Vps34), stimulating the formation of omegasomes in ER. Another portion of Ats-1 targets mitochondria, where it exerts antiapoptotic activity. The omegasome is marked with DFCP1. N terminus of Ats-1 is required for Atg14L recruiting and thus, autophagosome formation, but it is cleaved off when Ats-1 is imported into mitochondria. The isolation membrane elongates to envelop the cytoplasmic content into the double-membrane vacuole, the autophagosome, which is decorated with LC3. Ats-1 autophagosomes are recruited to Ap inclusion, and the outer membrane fuses with the Ap inclusion membrane, resulting in the release of the autophagic body-like content into the Ap inclusion.
Materials and Methods
Additional experimental details are provided in SI Materials and Methods, including a summary of primers (Table S1).
Bacteria and Culture.
The Ap HZ strain was cultured in human promyelocytic leukemia HL-60 cells, and E. chaffeensis (Arkansas strain) was cultured in human acute leukemia THP-1 cells. Cells were infected with Ap or E. chaffeensis at a multiplicity of infection of 100 unless otherwise indicated. The detailed method is described in SI Materials and Methods.
Co-IP.
HEK293 cells cotransfected with plasmids expressing Ats-1 and HA-Beclin 1, Ats-1(90–250)-GFP and HA-Beclin 1(1–272), Ats-1(218–376)-GFP and HA-Beclin 1(1–272), Ats-1–GFP and HA-Atg14L, Ats-1(∆N17)-GFP and HA-Atg14L, Cox8A-Ats-1(∆N17)-GFP and HA-Atg14L, or Ats-1–GFP and Myc-UVRAG were resuspended in immunoprecipitation buffer. The cleared lysate was incubated with rabbit anti–Ats-1, rabbit preimmune IgG, mouse anti-HA, or mouse IgG1 isotype control followed by incubation with protein A or protein G agarose beads. The immunoprecipitates were subjected to Western blot analysis using antibodies against Ats-1, HA, and Myc. The detailed protocol is described in SI Materials and Methods.
Colocalization.
RF/6A cells were cotransfected with plasmids encoding Ats-1–GFP, Cox8A-Ats-1(∆N17)-GFP, or Ats-1 with HA-Beclin 1, HA-DFCP1, HA-Atg14L, HA-LC3, or HA-Rab7 and subjected to immunostaining to determine the colocalization of Ats-1 with HA-tagged proteins. RF/6A cells were transfected with plasmid encoding HA-DFCP1, HA-Atg14L, or GFP-LC3 followed by infection with Ap to study the colocalization of Ap inclusions with HA-tagged proteins or LC3. The detailed method is described in SI Materials and Methods.
Localization of Ats-1 in HL-60 Cells Selectively Permeabilized with SLO.
Ap-infected HL-60 cells were treated with DTT-activated SLO on ice for 15 min followed by washing to remove unbound SLO. SLO-treated cells were then incubated at 37 °C for 30 min to allow the formation of pores on the plasma membrane. Cells were fixed and stained with rabbit anti–Ats-1 and mouse anti-P44 followed by incubation with secondary antibodies Alexa Fluor 488–conjugated goat anti-rabbit IgG and Alexa Fluor 555–conjugated goat anti-mouse IgG. After washing, the immunostained cells were incubated with the saponin-containing solution to permeabilize the membranes and labeled with mouse anti-P44 and Alexa Fluor 350-conjugated goat anti-mouse IgG. The detailed method is described in SI Materials and Methods.
Subcellular Fractionation by OptiPrep Density Gradient Centrifugation.
Ap-infected and uninfected HL-60 cells were homogenized in homogenization buffer. After briefly centrifuging to remove unbroken cells and nuclei, the supernatant was loaded onto the top of linear 5–25% (vol/vol) OptiPrep gradient. Fractions (12 total at 1 mL each) were collected from top to bottom sequentially after high-speed centrifugation. Equal aliquots from each fraction were analyzed by immunoblotting using anti-P44, anti-LC3, anticalnexin (Stressgen), anti–LAMP-2, and anti-EEA1. The detailed method is described in SI Materials and Methods.
beclin 1 Heterozygous-Deficient Mice.
Five each beclin 1+/− C57BL/6 mice and congenic WT mice were inoculated intraperitoneally with Ap-infected HL-60 cells. Blood samples were collected at 5 d after inoculation and subjected to quantitative PCR using Ap 16S rDNA and mouse glyceraldehyde 3-phosphate dehydrogenase (G3PDH) gene primers. The detailed method is described in SI Materials and Methods.
Statistical Analysis.
Analyses of significant differences between means were performed using two-tailed Student t and ANOVA tests. Data are shown as mean values ± SDs from three independent experiments. The dot plot for the experiment of beclin 1 heterozygous-deficient mice was drawn using GraphPad Prism 5 program (GraphPad).
Supplementary Material
Acknowledgments
We thank Drs. Tracy Hagemann at the University of Wisconsin and Beth Levine at the University of Texas Southwestern Medical Center for beclin 1+/− mice. Dr. Ulrike Munderloh at the University of Minnesota provided mCherry-expressing Ap, Dr. Tamotsu Yoshimori at Osaka University provided the pCMV5-Myc-UVRAG plasmid, and Dr. John Brumell at the University of Toronto provided pEGFP-Rab7 plasmid. We thank Mr. Tim Vojt for assistance in preparing Fig. 8. This work was supported by National Institutes of Health Grant R01 AI054476.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218674109/-/DCSupplemental.
References
- 1.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469(7330):323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deretic V. Autophagy: An emerging immunological paradigm. J Immunol. 2012;189(1):15–20. doi: 10.4049/jimmunol.1102108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tattoli I, et al. Amino acid starvation induced by invasive bacterial pathogens triggers an innate host defense program. Cell Host Microbe. 2012;11(6):563–575. doi: 10.1016/j.chom.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 4.Ogawa M, et al. Escape of intracellular Shigella from autophagy. Science. 2005;307(5710):727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- 5.Gutierrez MG, et al. Autophagy induction favours the generation and maturation of the Coxiella-replicative vacuoles. Cell Microbiol. 2005;7(7):981–993. doi: 10.1111/j.1462-5822.2005.00527.x. [DOI] [PubMed] [Google Scholar]
- 6.Starr T, et al. Selective subversion of autophagy complexes facilitates completion of the Brucella intracellular cycle. Cell Host Microbe. 2012;11(1):33–45. doi: 10.1016/j.chom.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walker DH, Popov VL, Crocquet-Valdes PA, Welsh CJ, Feng HM. Cytokine-induced, nitric oxide-dependent, intracellular antirickettsial activity of mouse endothelial cells. Lab Invest. 1997;76(1):129–138. [PubMed] [Google Scholar]
- 8.Rikihisa Y. Glycogen autophagosomes in polymorphonuclear leukocytes induced by rickettsiae. Anat Rec. 1984;208(3):319–327. doi: 10.1002/ar.1092080302. [DOI] [PubMed] [Google Scholar]
- 9.Rikihisa Y. Mechanisms of obligatory intracellular infection with Anaplasma phagocytophilum. Clin Microbiol Rev. 2011;24(3):469–489. doi: 10.1128/CMR.00064-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bakken JS, Dumler S. Human granulocytic anaplasmosis. Infect Dis Clin North Am. 2008;22(3):433–448. doi: 10.1016/j.idc.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 11.Mott J, Barnewall RE, Rikihisa Y. Human granulocytic ehrlichiosis agent and Ehrlichia chaffeensis reside in different cytoplasmic compartments in HL-60 cells. Infect Immun. 1999;67(3):1368–1378. doi: 10.1128/iai.67.3.1368-1378.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niu H, Yamaguchi M, Rikihisa Y. Subversion of cellular autophagy by Anaplasma phagocytophilum. Cell Microbiol. 2008;10(3):593–605. doi: 10.1111/j.1462-5822.2007.01068.x. [DOI] [PubMed] [Google Scholar]
- 13.Homer CR, Richmond AL, Rebert NA, Achkar JP, McDonald C. ATG16L1 and NOD2 interact in an autophagy-dependent antibacterial pathway implicated in Crohn's disease pathogenesis. Gastroenterology. 2010;139(5):1630–1641.e2. doi: 10.1053/j.gastro.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alvarez-Martinez CE, Christie PJ. Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev. 2009;73(4):775–808. doi: 10.1128/MMBR.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin M, Kikuchi T, Brewer HM, Norbeck AD, Rikihisa Y. Global proteomic analysis of two tick-borne emerging zoonotic agents: Anaplasma phagocytophilum and ehrlichia chaffeensis. Front Microbiol. 2011;2:24. doi: 10.3389/fmicb.2011.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rikihisa Y, Lin M, Niu H. Type IV secretion in the obligatory intracellular bacterium Anaplasma phagocytophilum. Cell Microbiol. 2010;12(9):1213–1221. doi: 10.1111/j.1462-5822.2010.01500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niu H, Kozjak-Pavlovic V, Rudel T, Rikihisa Y. Anaplasma phagocytophilum Ats-1 is imported into host cell mitochondria and interferes with apoptosis induction. PLoS Pathog. 2010;6(2):e1000774. doi: 10.1371/journal.ppat.1000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He C, Levine B. The Beclin 1 interactome. Curr Opin Cell Biol. 2010;22(2):140–149. doi: 10.1016/j.ceb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kihara A, Kabeya Y, Ohsumi Y, Yoshimori T. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2001;2(4):330–335. doi: 10.1093/embo-reports/kve061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furuya N, Yu J, Byfield M, Pattingre S, Levine B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy. 2005;1(1):46–52. doi: 10.4161/auto.1.1.1542. [DOI] [PubMed] [Google Scholar]
- 21.Matsunaga K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11(4):385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 22.Matsunaga K, et al. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol. 2010;190(4):511–521. doi: 10.1083/jcb.200911141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong Y, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11(4):468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Regev-Rudzki N, Yogev O, Pines O. The mitochondrial targeting sequence tilts the balance between mitochondrial and cytosolic dual localization. J Cell Sci. 2008;121(Pt 14):2423–2431. doi: 10.1242/jcs.029207. [DOI] [PubMed] [Google Scholar]
- 25.Axe EL, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182(4):685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 27.Polishchuk RS, et al. Correlative light-electron microscopy reveals the tubular-saccular ultrastructure of carriers operating between Golgi apparatus and plasma membrane. J Cell Biol. 2000;148(1):45–58. doi: 10.1083/jcb.148.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kabeya Y, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19(21):5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reynolds TH, 4th, Bodine SC, Lawrence JC., Jr Control of Ser2448 phosphorylation in the mammalian target of rapamycin by insulin and skeletal muscle load. J Biol Chem. 2002;277(20):17657–17662. doi: 10.1074/jbc.M201142200. [DOI] [PubMed] [Google Scholar]
- 30.Gutierrez MG, Munafó DB, Berón W, Colombo MI. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci. 2004;117(Pt 13):2687–2697. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- 31.Felsheim RF, et al. Transformation of Anaplasma phagocytophilum. BMC Biotechnol. 2006;6:42. doi: 10.1186/1472-6750-6-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campbell AM, Kessler PD, Fambrough DM. The alternative carboxyl termini of avian cardiac and brain sarcoplasmic reticulum/endoplasmic reticulum Ca(2+)-ATPases are on opposite sides of the membrane. J Biol Chem. 1992;267(13):9321–9325. [PubMed] [Google Scholar]
- 33.Baba M, Takeshige K, Baba N, Ohsumi Y. Ultrastructural analysis of the autophagic process in yeast: Detection of autophagosomes and their characterization. J Cell Biol. 1994;124(6):903–913. doi: 10.1083/jcb.124.6.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qu X, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112(12):1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ge Y, Rikihisa Y. Identification of novel surface proteins of Anaplasma phagocytophilum by affinity purification and proteomics. J Bacteriol. 2007;189(21):7819–7828. doi: 10.1128/JB.00866-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sumpter R, Jr, Levine B. Autophagy and innate immunity: Triggering, targeting and tuning. Semin Cell Dev Biol. 2010;21(7):699–711. doi: 10.1016/j.semcdb.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan W, Nassiri A, Zhong Q. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L) Proc Natl Acad Sci USA. 2011;108(19):7769–7774. doi: 10.1073/pnas.1016472108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hailey DW, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141(4):656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pfanner N, Truscott KN. Powering mitochondrial protein import. Nat Struct Biol. 2002;9(4):234–236. doi: 10.1038/nsb0402-234. [DOI] [PubMed] [Google Scholar]
- 40.Wurm CA, et al. Nanoscale distribution of mitochondrial import receptor Tom20 is adjusted to cellular conditions and exhibits an inner-cellular gradient. Proc Natl Acad Sci USA. 2011;108(33):13546–13551. doi: 10.1073/pnas.1107553108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dunning Hotopp JC, et al. Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet. 2006;2(2):e21. doi: 10.1371/journal.pgen.0020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Price CT, Al-Quadan T, Santic M, Rosenshine I, Abu Kwaik Y. Host proteasomal degradation generates amino acids essential for intracellular bacterial growth. Science. 2011;334(6062):1553–1557. doi: 10.1126/science.1212868. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.