Significance
Newly differentiated neurons migrate over long distances to reach their proper destination in the developing brain. The mechanisms by which neurons accomplish this translocation remain to be clarified. By analyzing the trajectories of antibody-coated single quantum dots bound to specific plasma membrane proteins, we found that membrane proteins of migrating cultured cerebellar granule cells exhibited net forward translocation in a form of biased drift, which is superimposed upon Brownian motion, and that this biased drift appears to be driven by myosin II-dependent active transport processes. Thus, plasma membrane proteins undergo forward translocation in unison with cytoplasmic components in migrating neurons.
Keywords: membrane flow, neuronal migration, protein transport in membrane
Abstract
Directional flow of membrane components has been detected at the leading front of fibroblasts and the growth cone of neuronal processes, but whether there exists global directional flow of plasma membrane components over the entire migrating neuron remains largely unknown. By analyzing the trajectories of antibody-coated single quantum dots (QDs) bound to two membrane proteins, overexpressed myc-tagged synaptic vesicle-associated membrane protein VAMP2 and endogenous neurotrophin receptor TrkB, we found that these two proteins exhibited net forward transport, which is superimposed upon Brownian motion, in both leading and trailing processes of migrating cerebellar granule cells in culture. Furthermore, no net directional transport of membrane proteins was observed in nonmigrating cells with either growing or stalling leading processes. Analysis of the correlation of motion direction between two QDs on the same process in migrating neurons also showed a higher frequency of correlated forward than rearward movements. Such correlated QD movements were markedly reduced in the presence of myosin II inhibitor blebbistatin, suggesting the involvement of myosin II-dependent active transport processes. Thus, a net forward transport of plasma membrane proteins exists in the leading and trailing processes of migrating neurons, in line with the translocation of the soma.
Neuronal migration is an important step in the development of brain architecture (1). The cellular mechanisms associated with cytoskeleton reorganization and nucleus translocation during neuronal migration have been studied extensively (2, 3). However, how plasma membrane components undergo redistribution in migrating neurons remains largely unclear. The topography of membrane components, including cell surface adhesion molecules, growth factor receptors, and other signaling molecules, is important for regulating the polarity and motility of the cell. Knowing the redistribution of membrane components during neuronal migration is critical for understanding how a neuron maintains its proper membrane topography during migration. In migrating cerebellar granule cells in culture, there is a forward flow of cortical F-actin that is required for driving the forward soma translocation (4). Through their linkage to cortical F-actins, plasma membrane proteins and associated components may be driven forward, leading to a bulk forward flow of plasma membrane components in unison with the cytoplasm, as suggested by Bray and White (5). On the other hand, preferential membrane insertion via exocytic fusion of membrane precursor vesicles at the growth cone of the leading process (l.p.) along with membrane removal via endocytic uptake at the trailing process (t.p.) also may result in bulk rearward flow of plasma membrane components, as proposed by the polarized endo-exocytic membrane cycling model of cell migration (6). There also is clear evidence for preferential insertion of newly synthesized membrane proteins at the growth cone of advancing axons (7) and for preferential endocytosis proximal to the soma of the cultured cells migrating from the subventricular zone (8). However, the existence of directional transport of membrane components in the plasmalemma of migrating neurons has not been reported.
Prominent rearward flow of membrane components occurs locally at the advancing front of neuronal growth cones of cultured Aplysia neurons (9). In nonmigrating neurons, where the cell bodies remain stationary, bulk forward flow of incorporated fluorescent lipids along rapidly growing neurites was observed in cultured spinal neurons (10), but rearward flow of beads bound to membrane proteins or lipids was found in neurites of cultured dorsal root ganglion neurons (11). The l.p. of migrating neurons exhibits a dynamic advancing growth cone resembling the growth cone of extending neurites of many nonmigrating neurons. Thus, it is likely that global flow of membrane proteins in either the forward or rearward direction exists in the l.p. of migrating neurons.
In the present study we examined whether directional movement of membrane proteins exists in cultured cerebellar granule cells. These cells assume a bipolar morphology consisting of a long l.p. and a short t.p. We monitored the movement of membrane proteins along the surface of these processes by tracing the movement of antibody-coated single quantum dots (QDs) that were bound specifically to the vesicle-associated membrane protein VAMP2-myc and the endogenous neurotrophin receptor TrkB. Quantitative analysis of the QD trajectories showed that these membrane proteins exhibited frequent drift in both the forward and rearward directions with similar speeds superimposed on their Brownian diffusion. However, in migrating neurons, there was a higher frequency of forward than rearward drift in both l.p. and t.p., leading to net forward transport of these proteins along the direction of cell migration. Finally, using the nonmuscle myosin II inhibitor blebbistatin (Blebb) (12), we examined the potential role of myosin II activity in driving this net forward membrane protein transport.
Results
Labeling Plasma Membrane Proteins with QDs.
We first examined the transport of membrane proteins on the surface of cultured cerebellar granule cells that were isolated from the cerebellum of newborn rats at postpartum day (P0–P1) and cultured for 1 d. The cells were transfected by electroporation with a plasmid expressing VAMP2-myc together with a plasmid expressing fluorescent protein eYFP immediately before cell plating. The motion of surface VAMP2-myc molecules was monitored by surface-bound single QDs (as indicated by their blinking behavior), which were coated with anti-myc antibodies (Materials and Methods). Time-lapse imaging of QD movements along the neuritic process was recorded for 10–20 min (300–600 frames, 2-s intervals).
Whether these single QDs were localized on the cell surface was examined by using the membrane-impermeable quencher QSY21 (Materials and Methods). In the normal culture medium, the fluorescence of QDs attached to the cell surface was eliminated totally after bath-addition of QSY21 (4 mM; Fig. S1A), as was that of QDs bound to the culture substrate, indicating that these QDs were located on the external surface of the plasma membrane. Furthermore, after 5-min incubation with a high-K+ (50 mM) solution before the addition of QSY21, we observed some of the unquenched QDs undergoing rapid transport with steady speeds along the neuritic process (Fig. S1B), consistent with the active transport of QDs that were endocytosed because of membrane depolarization (13). Quantitative analysis of the movement of intracellular QDs (n = 27 QDs from 14 cells) showed an average speed of 0.32 ± 0.04 μm/s during the transport phase, close to that found for intracellular VAMP2-GFP puncta in cultured hippocampal neurons (14) and much faster than the net transport of QDs observed on the cell surface (see below).
Net Forward Transport of VAMP2-myc in Migrating Granule Cells.
During the period of observation, the surface movement of QDs appeared to undergo random Brownian motion. To examine qualitatively whether the apparently random movements of VAMP2-QD on the neurite surface undergo a net directional movement during neuronal migration, we monitored the trajectories of a large number of VAMP2-QDs on migrating granule cells. The neuron was considered to be migrating when the center of the soma underwent translocation for ≥4 μm during the 10-min period of observation. Time-lapsed imaging (300 frames, 2-s intervals) showed most surface-attached QDs exhibited extensive Brownian motions over large distances along the l.p. and t.p. Two examples of QDs on the l.p. are shown in Fig. 1 A and B. Tracings of the trajectories of a large number of QDs showed that many QDs exhibited Brownian motion which was superimposed upon a drift toward either the forward or rearward direction (with respect to the direction of soma translocation) over the 6-min duration. However, the average trajectory of all QDs was found to be forward for both l.p. (Fig. 1C) and t.p. (Fig. 1D).
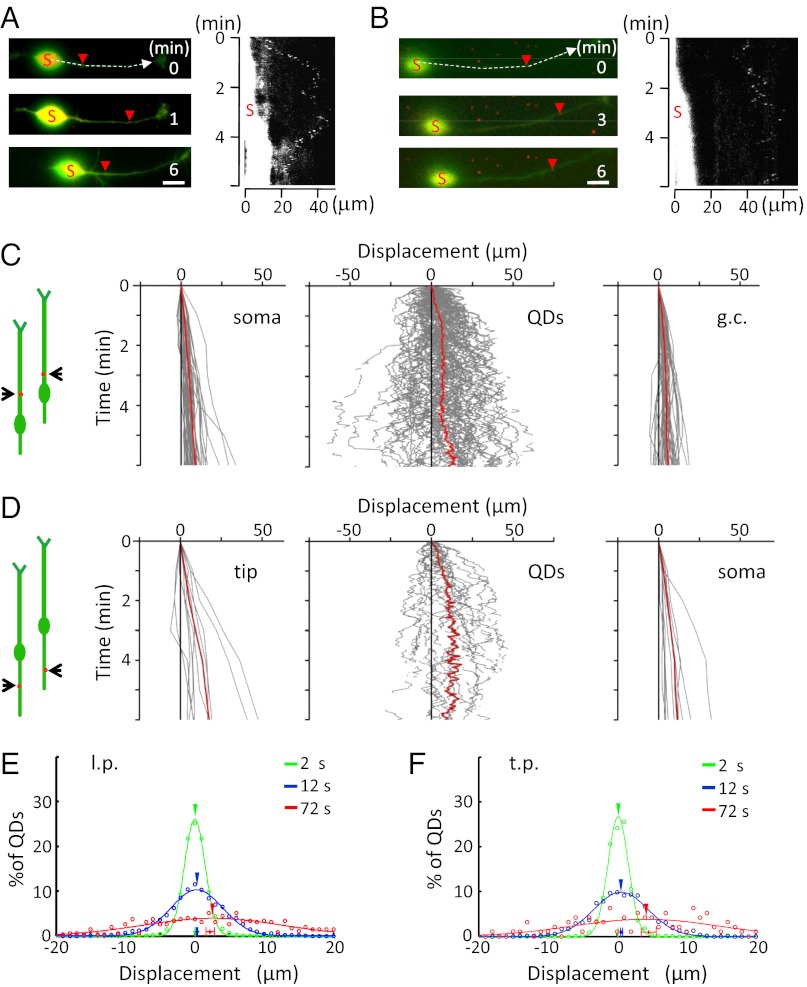
Fig. 1.
Net forward movement of VAMP2-QDs on the surface of migrating granule cells. (A and B) Examples of the surface movement of two VAMP2-QDs in the l.p. of migrating granule cells (Left) and the kymograph of the QD position (Right). Red arrowheads indicate the positions of the same QD at different time points. The soma is marked with an “S,” and the kymograph represents the trajectory of the QD along the neurite at the positions marked by the dashed line. The arrow represents the direction of neuronal migration. The white region marked “S” on the left of the kymograph represents eYFP fluorescence of the soma, revealing forward translocation. (Scale bars, 10 μm.) (C and D) Diagrams on the far left illustrate the migratory behavior of the cell at two different times, and red dots depict QDs. (C) Trajectories of the center of the soma (Left), VAMP2-QDs on the l.p. (Center), and the base of the growth cone of the l.p. (Right) of migrating cells (n = 90 QDs from 45 cells). Red lines represent the average of all trajectories. (D) Trajectories of the tip of the t.p. (Left), VAMP2-QDs on the t.p. (Center), and the center of the soma (Right) of migrating cells (n = 24 QDs from 13 neurons). Red lines indicate the average of all trajectories. (E and F) The displacement of all QDs in the l.p. (E) and t.p. (F), relative to the starting location after 2-s (green), 12-s (blue), and 72-s (red) intervals. Data points with error bars close to the x axis show mean values of the displacement (± SEM). Arrowheads show the center for the fitted Gaussian curve.
We also analyzed the distribution of the QD displacement from the origin (position at the onset time of the observation period) after different defined time intervals for all QDs and found that the distributions for different time intervals were well-fitted by Gaussian curves, consistent with the predominant Brownian motion of QDs. In addition, for the longer time interval of 72 s, the mean value of the distribution was found to shift to the right in both l.p. and t.p. (Fig. 1 E and F), consistent with a forward drift of QDs that was superimposed upon Brownian motion. Furthermore, we noted that the average movement of QDs along the process was close to that of soma translocation, suggesting a coordinated net forward transport of membrane proteins in line with the soma advance.
Analysis of Diffusion Coefficients and Drift Speeds of VAMP2-myc.
Further quantitative analysis of the trajectory of each QD was performed to determine the mean squared displacement (MSD) over different time intervals of QD movements (Materials and Methods). Briefly, we assumed that the QD motion on the neuronal surface reflects Brownian motion that may be superimposed upon a net drift movement in either the forward or rearward direction. The first six data points (12 s) of the MSD-vs.-time plot for each QD trajectory, during which the drift effect was considered negligible (Fig. 1 E and F), were fitted with the equation MSD = 2Dt (where t is the time interval) to determine the diffusion coefficient D. To determine speed (v) and direction of the drift, the QD trajectories were divided into 60-s segments for further analysis. We determined the direction of the drift of each QD by the net translocation in the forward or rearward direction relative to the starting point over the 60-s segment. The MSD-vs.-time plot for each QD trajectory segment then was fitted by the equation MSD = 2Dt + (vt)2 to obtain the best-fit drift speed v, using the D value for the QD obtained from the whole trajectory. For many QDs (∼65%), the MSD vs. t curve obtained by the equation lay below the line defined by MSD = 2Dt (Fig. 2E), consistent with confined diffusion (15). These QDs were not used in the analysis of drift speed.
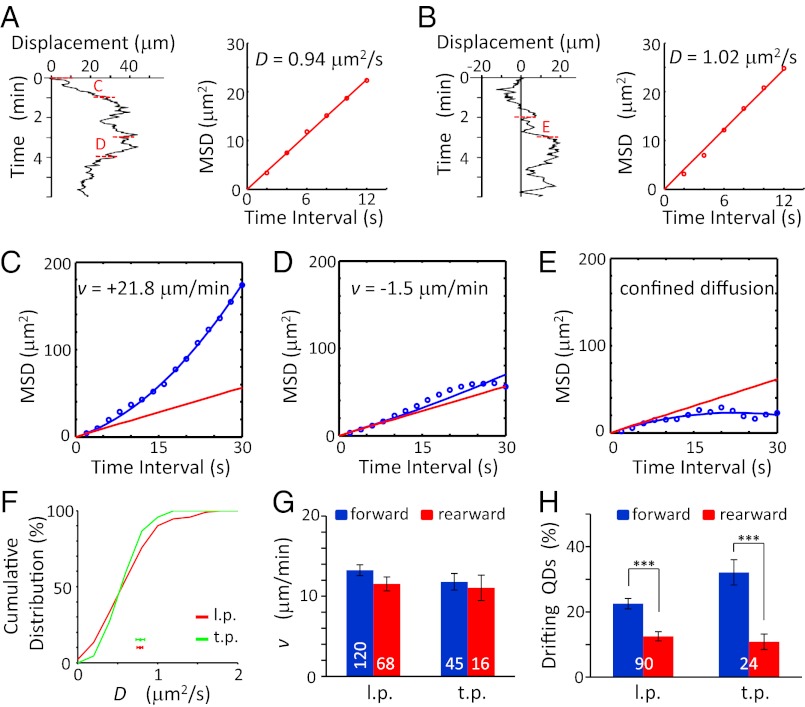
Fig. 2.
Quantitative analysis of surface VAMP2-QDs movements on migrating granule cells. (A and B) (Left) Neurolucida traces of the QD movements shown in Fig. 1 A and B, respectively. (Right) The D value was determined by using the first six data points of the MSD-vs.-time plot, averaged for all starting locations of QDs covered by the trajectory during the 6-min period. The segments marked “C” and “D” in panel A and “E” in panel B represent 60-s segments of the trajectory that were used to calculate the v value. (C–E) The v value was determined by the first 15 points of the MSD vs. the t curve of the 60-s segment. The red line represents the pure diffusion using the D value above, and the blue line represents the best fit of MSD = 2Dt + (vt)2. (F) Cumulative percentage for the distribution of D values. Data points below depict the mean value of D (± SEM) observed for QDs on the l.p. (red trace; n = 90 from 45 cells) t.p. (green trace, n = 24 from 13 cells). The difference between the two curves was not significant [P = 0.75, Kolmogorov–Smirnov (K-S) test]. (G) The mean value of v (± SEM) in migrating cells in the forward (blue) and rearward (red) directions along the l.p. and t.p. The difference was not significant (P > 0.12, t test). (H) The percentage of QDs showing drift in the forward (blue) and rearward (red) directions along the l.p. and t.p. (P < 0.0003, paired t test). Only the percentages of QDs showing drift movement are shown here. The numbers associated with the bars represent the total number of QDs examined. ***P < 0.001. Error bars indicate SEM.
For the examples shown in Fig. 1 A and B, the D values were 0.94 and 1.02 μm2/s, respectively (Fig. 2 A and B). The v values determined for the two sampled 60-s segments of the trajectory were +21.8 and −1.5 μm/min, respectively, with the sign defined by the direction of the drift over the corresponding 60-s segment (Fig. 2 C and D). As suggested by the trajectories of all QDs depicted in Fig. 1C, QDs were found to drift in either forward or rearward direction during the observation period, although the net translocation for all QDs was forward. In total, we measured the trajectories of 90 QDs (from 45 cells) and 24 QDs (from 13 cells) along the l.p. and t.p., respectively. The distributions of D values were identical in l.p. and t.p. (0.78 ± 0.04 and 0.78 ± 0.06 μm2/s, respectively; Fig. 2F). The mean v values for the forward and rearward drift v were similar in the l.p. and t.p. of migrating cells (Fig. 2G), with v values in both directions being much larger than that of the soma translocation (1.5 ± 0.2 μm/min, Fig. 1C). However, for both l.p. and t.p, the percentage of QDs drifting in the forward direction was higher than that of QDs drifting in the rearward direction. (Fig. 2H). Together, these results showed that the net forward transport of VAMP2-myc on the surface of both l.p. and t.p. was caused by more QDs drifting forward than rearward rather than by a higher speed of VAMP2-myc forward drift.
No Forward Transport of VAMP2-myc in Nonmigrating Cells.
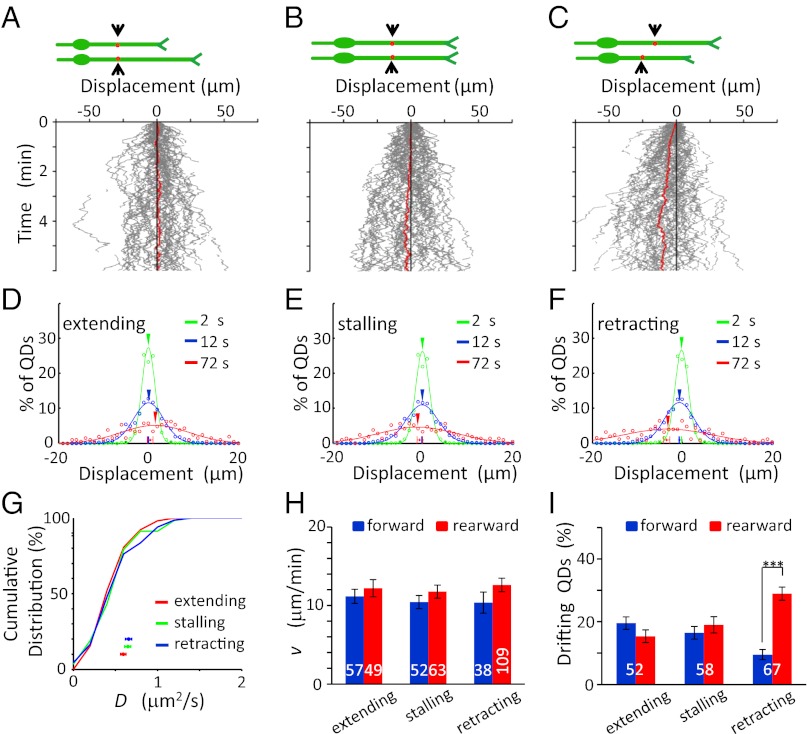
We also analyzed the VAMP2-QD movement on the surface of nonmigrating granule cells with soma translocation less than 1 μm during the 10-min period of observation. These nonmigrating neurons were divided into three groups according to the growth cone motility of their long processes (presumably the l.p.): neurons with extending, stalling, or retracting growth cones. Tracings of VAMP2-QD trajectories on processes with extending growth cones showed that, on the average, there was no bias drift in either the forward (anterograde, away from soma) or rearward (retrograde, toward soma) direction in extending processes (85 QDs in 29 cells; Fig. 3 A and B), although the growth cones of these processes exhibited extension with a rate of 1.7 ± 0.1 μm/min. In long processes with stalling growth cones, similar results were obtained for the QD movement (128 QDs in 57 cells; Fig. 3 C and D). In retracting processes, however, the majority of QDs exhibited rearward drift toward the soma (35 QDs in 17 cells, Fig. 3 E and F). These results supported the notion that the net forward VAMP2-QD transport, i.e., bias drift in the forward direction, occurs only in migrating cells.
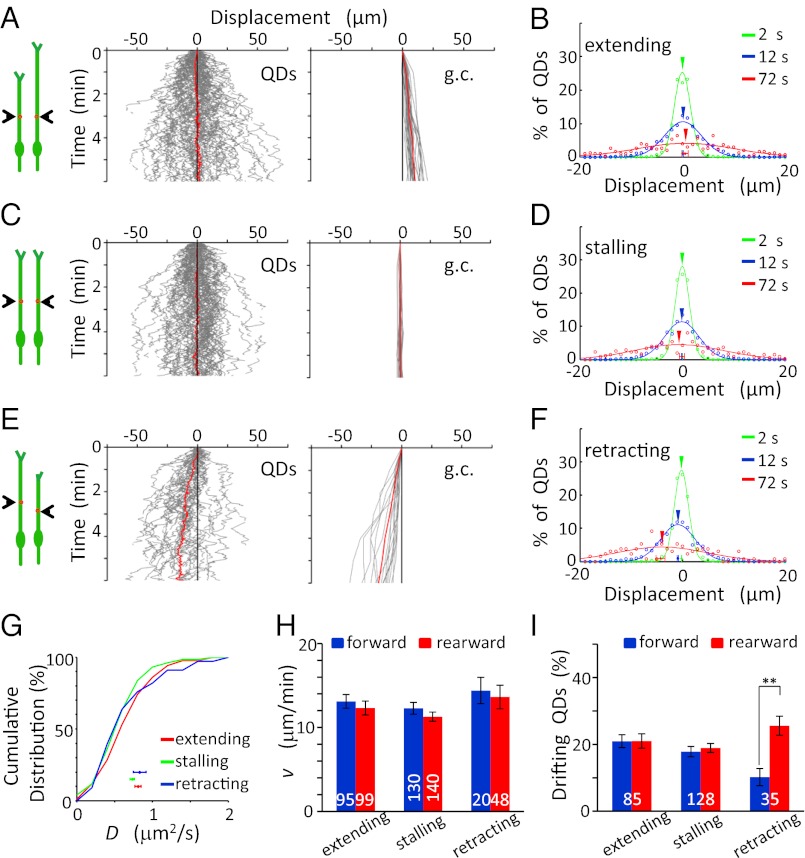
Fig. 3.
No forward transport of surface VAMP2-QDs was seen on nonmigrating cells. (A, C, and E) (Left) Diagrams illustrate the migratory behavior of the cell at two different times, and red dots depict QDs The trajectories of the movement of QDs (Center) and growth cones (g.c.,Right) along the surface of the long process of nonmigrating cells, which were divided into three groups according to the growth cone motility: neurons with extending (A), stalling (C), or retracting (E) growth cones. Red lines represent the average of all trajectories (n = 85, 128, and 35, respectively). (B, D, and F) The distribution of QD movements in the long processes of nonmigrating cells in 2-s (green), 12-s (blue), and 72-s (red) intervals. (G) Cumulative percentage plot for the distributions of D values found along the long process of nonmigrating cells. Data points below depict the mean D values (± SEM). No significant difference was found among all curves (P > 0.44, K-S test). (H) The mean value of the v (± SEM) along the long process of nonmigrating cells. No significant difference was found (P > 0.11, t test). (I) Percentage of QDs in nonmigrating cells showing drift movements in the forward (blue) and rearward (red) directions along the extending, stalling, and retracting processes. The numbers associated with the bars represent the total number of QDs examined. **P < 0.01. Error bars indicate SEM.
Quantitative analysis of QD movements on these nonmigrating cells showed no difference in the D and v values of QDs for cells with three different growth cone conditions. The D values were 0.81 ± 0.04 (extending), 0.73 ± 0.03 (stalling), and 0.80 ± 0.08 μm2/s (retracting) and were not significantly different from that found in migrating cells (P > 0.1, t test; Fig. 3G). The mean value of the v of these QDs (in the range of 11–14 μm/min) was similar to those found in migrating cells (Fig. 3H), and the percentages of QDs drifting in forward and rearward directions were not significantly different in extending and stalling processes of nonmigrating cells (Fig. 3I). Thus, the diffusion and drift speed of QDs along the processes were not affected by growth cone motility. Moreover, the forward movement bias of QDs was associated with the migratory state of these granule cells and was caused not by a higher forward drift speed but by more forwardly drifting QDs.
Net Forward Transport of Endogenous TrkB in Migrating Cells.
To examine further the behavior of endogenous membrane proteins, we monitored the movement of the neurotrophin receptor TrkB on the surface of cultured granule cells, using QDs coated with antibodies against the extracellular domain of TrkB (Fig. 4 A–C). In total, we measured the trajectories of 75 QDs (from 41 cells) and 23 QDs (from nine cells) on the surface of migrating neurons and found that there was a net forward transport of TrkB-QDs in both l.p. and t.p. (Fig. 4 D–G). The distributions of the D values were similar in the l.p. and t.p. of migrating cells (0.64 ± 0.04 and 0.61 ± 0.08 μm2/s, respectively; Fig. 4H), but the D values were smaller than in VAMP2-QD, as is consistent with the larger size of TrkB (92 kDa) as compared with VAMP2 (13 kDa). Quantitative analysis showed that the average speed of forward and rearward drift in l.p. also was similar in the forward and rearward direction (10.6 ± 0.6 and 10.2 ± 0.8 μm/min, respectively; Fig. 4I), again suggesting that the net forward transport of TrkB was caused by more forwardly drifting TrkB (Fig. 4J).
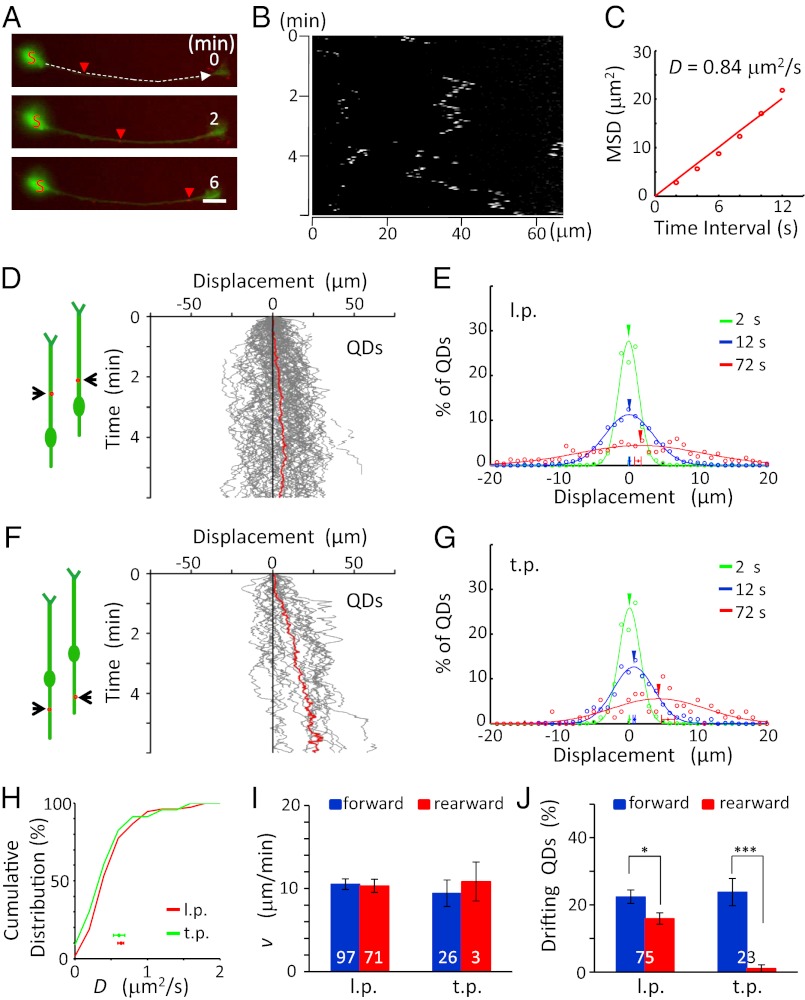
Fig. 4.
Forward transport of surface TrkB-QD in migrating granule cells. (A and B) An example of the surface movement of a TrkB-QD on the l.p. of a migrating granule cell (A) and its kymograph (B). Red arrowheads indicate positions of the same QD at different time points. (Scale bar, 10 μm.) (C) The D value was determined by the first six points of the MSD-vs.-t curve from the start point, averaged for all locations of QDs covered by the trajectory during the 6-min period. (D and F) (Left) Diagrams illustrate the migratory behavior of the cell at two different times, and red dots depict QDs. (Right) The trajectories of the movement of the QDs on the l.p. (D, n = 75 QDs from 40 cells) and t.p. (F, n = 23 QDs from nine cells) of migrating cells during the observation. Red lines represent the average of all trajectories. (E and G) The distribution of QD movement in the l.p. (E) and t.p. (G) in 2-s (green), 12-s (blue), and 72-s (red) intervals. (H) Cumulative percentage for the distributions of D values found alongt he l.p. and t.p. of migrating cells. Data points below depict the mean D values (± SEM). No significant difference was found between the curves (P = 0.21, K-S test). (I) Mean value of the v values (± SEM) along the l.p. and t.p. of migrating cells. There was no significant difference (P > 0.42, t test). (J) Percentage of QDs showing drift movements in migrating cells in the forward (blue) and rearward (red) directions along the l.p. and t.p., respectively (P < 0.04, paired t test). The numbers associated with the bars represent the total number of QDs examined. *P < 0.05; ***P < 0.001. Error bars indicate SEM.
Similar to findings for VAMP2-myc transport, we observed no net transport of TrkB in extending and stalling processes but observed a rearward transport in retracting processes (Fig. 5 A–F). Quantitative analysis of the QD movement on these nonmigrating cells showed no significant difference in both D and v values between nonmigrating and migrating neurons or among nonmigrating neurons with three different behaviors of growth cone motility (Fig. 5 G and H). Furthermore, endogenous TrkB exhibited a higher frequency of forward rearward drift movements on the cell surface of l.p. and t.p. in migrating but not in nonmigrating neurons, similar to findings in VAMP2-myc. Thus, both endogenous and overexpressed exogenous membrane proteins showed net forward transport on the surface of migrating neuron.
Fig. 5.
No forward transport of surface TrkB-QDs on nonmigrating cells. (A–C) (Upper) Diagrams illustrate the migratory behavior of the cell at two different times, and red dots depict QDs. (Lower) The trajectories of QDs along the long process of nonmigrating cells during the observation. Red lines represent the average of all trajectories (n = 52, 58, and 67, respectively). (D–F) The distribution of QD movement in the long process of nonmigrating cells in 2-s (green), 12-s (blue), and 72-s (red) intervals. (G) Cumulative percentage plot for the distribution of D values found along the long process of nonmigrating cells. Data points below depict the mean D values for three groups of neurons with different states of the growth cones. No significant difference was found among all curves (P > 0.09, K-S test). (H) The mean value of v (± SEM) along the long process of nonmigrating cells. No significant difference was found (P > 0.14, t test). (I) Percentages of QDs showing drift movements in the forward (blue) and rearward (red) directions along extending, stalling, and retracting processes, respectively. The numbers associated with the bars represent the total number of QDs examined. ***P < 0.001. Error bars indicate SEM.
Correlation of Surface Movements Between Pairs of QDs.
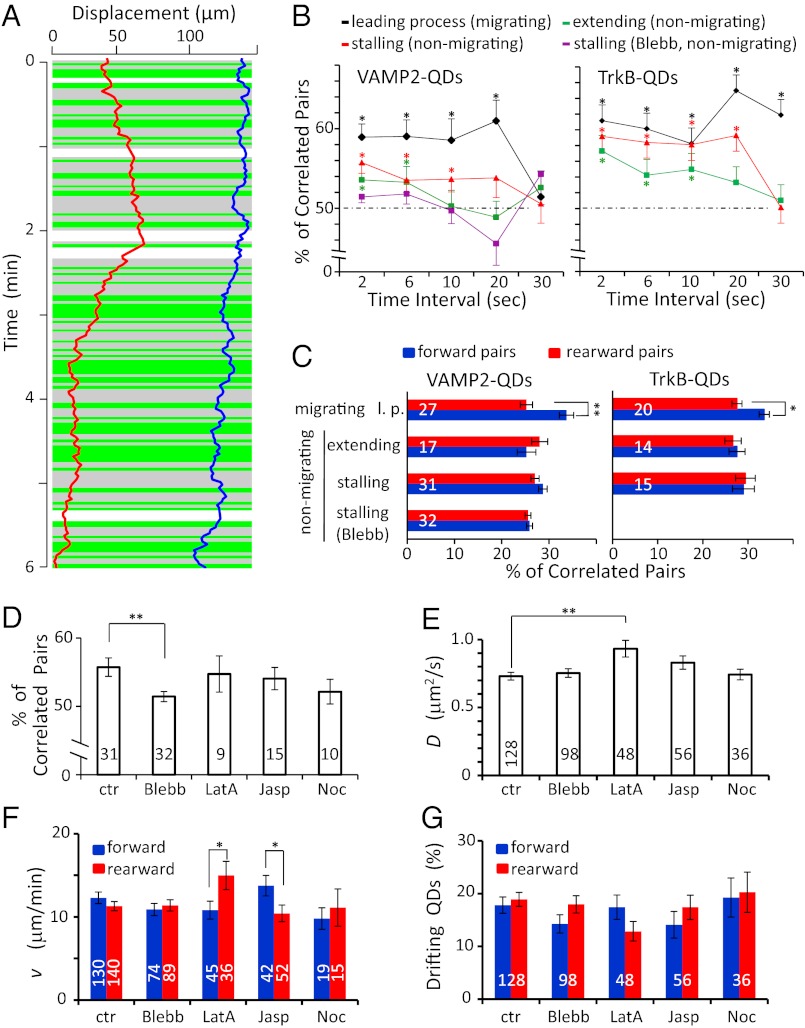
To understand the mechanism underlying the net forward transport of membrane proteins in migrating cells, we further analyzed the correlation in the direction of movements between two QD-labeled membrane proteins in the same processes during the same time intervals (Fig. 6A). We found there was a significant bias toward correlated movement of the pairs (drifting in the same direction) of VAMP2-QD and TrkB-QD pairs in the same neuron when movements were analyzed over successive 2-s and 6-s segments. This bias was seen in both the l.p. of migrating cells and the extending or stalling processes of nonmigrating cells (Fig. 6B), suggesting that there were brief periods (“bursts”) of correlated membrane protein drift in either the forward or rearward direction along the processes. Importantly, in the l.p. of migrating cells but not of nonmigrating cells, the percentage of forward correlated pairs was significantly higher than that of rearward correlated pairs (Fig. 6C), consistent with the idea that the net forward transport of membrane proteins results from a higher frequency of forward burst movement in the migrating cells.
Fig. 6.
Correlation of the direction of surface movements between pairs of QDs. (A) An example of movement trajectories of 2 VAMP2-QDs during 6 min of observation, with correlation analysis of movement direction for successive 2-s time intervals along the l.p. of a migrating granule cell. Green and gray backgrounds indicate segments showing movements in the same and opposite directions, respectively. The white background marks a period when one or both QDs were not visible. (B) Percentage of VAMP2- and TrkB-QDs pairs showing drift in the same direction when the analysis was done at 2-, 6-, 10-, 20-, and 30-s intervals. The percentages for 2-s and 6-s intervals were found to exhibit significant bias toward correlated movements in the l.p. of migrating cells and in the extending or stalling process of nonmigrating cells (P < 0.05, binomial test). For each cell, we obtained an average percentage of QD pairs showing correlated direction of movement over 6–10 min observation, for all possible pairs of QDs on the same cell. Note that 50% represents correlation by chance, i.e., no correlated drift movement, and that the bias found in the stalling process of nonmigrating cells was abolished by the treatment with blebbistatin (Blebb). (C) The percentage of QD pairs analyzed at 2-s intervals showing correlated movement in the forward and rearward directions, respectively. The remaining percentage comprises pairs showing movements in opposite directions. The percentage of pairs showing forward-correlated movements was significantly higher than the percentage of pairs with rearward-correlated movements in the l.p. of migrating cells (P < 0.02, paired t test) but not in the nonmigrating cells. Numbers associated with the bars represent the total number of cells examined for correlated drift movements. (D) Average percentage of VAMP2-QD pairs showing correlated direction of movement in the stalling processes of nonmigrating neurons in the control condition and after treatment with 50 μM blebbistatin (Blebb, n = 32, P = 0.008, t test, relative to controls), 2 μM latrunculin A (LatA, n = 9), 2 μM jasplakinolide (Jasp, n = 15), and 16 μM nocodazole (Noc, n = 10). (E) Average D value for VAMP2-QDs in stalling processes of nonmigrating neurons in the control condition and after the drug treatments. (LatA, P = 0.004, t test, relative to controls). (F and G) The mean speed of drift movements and the percentage of QDs with drift in the forward (blue) and rearward (red) directions on stalling processes of nonmigrating cells in the control condition and after the drug treatment. The differences in mean v between the forward and rearward directions in panel F were significant after latrunculin A and jasplakinolide treatment (P = 0.05 and 0.04, respectively, t test). The numbers associated with the bars represent the total number of QDs examined. *P < 0.05; **P < 0.01. Error bars indicate SEM.
Because both the membrane protein flow and F-actin flow are driven mainly by actomyosin in the growth cone of cultured neurons (16, 17), we further examined whether myosin II is required for the correlated burst transport of membrane proteins. We found that the percentage of correlated pairs of VAMP2-QD was decreased significantly to 50% after 20 min incubation of the myosin II inhibitor Blebb (50 μM in the bath) in the stalling process of nonmigrating cells (Fig. 6 B–D). Only nonmigrating cells were analyzed for the Blebb effect, because the drug treatment stalled the cell migration. On the other hand, the D values were not affected by the Blebb treatment (Fig. 6E). These results suggest that a myosin II-dependent process is required for the higher frequency of correlated burst movements, presumably because of its action on the cortical F-actin.
We also examined whether F-actin or microtubule contributes to the correlated burst transport of membrane proteins. As shown in Fig. 6D, we found that the percentage of pairs showing correlated movements of VAMP2-QD was not affected after the treatment with latrinculin A (LatA; 2 μM for 30 min) and jasplakinolide (Jasp; 2 μM for 30 min), which are known to cause depolymerization and stabilization of F-actin, respectively. Similarly, disruption of microtubules with nocodazole (Noc; 16 μM for 30 min) had no effect on the percentage of pairs showing correlated movements. The D values were slightly increased after LatA treatment but were not changed after Jasp or Noc treatment (Fig. 6E). On the other hand, we noted that LatA and Jasp treatments resulted in small and opposite effects on the relative drift speeds in the forward and rearward directions, with LatA- and Jasp-treated cells showing higher and lower drift speed in the rearward direction, respectively (Fig. 6F). How the disruption or stabilization of F-actin causes asymmetric modification of the drift movements remains to be elucidated. We note that all these drug treatments led to the stalling of cell migration; thus only QDs on the stalling process of nonmigrating cells could be analyzed. Further analysis of the trajectories of all QDs on the stalling process after drug treatment showed that there was no net forward or rearward transport of QDs (Fig. S2), nor was there any significant difference in the percentage of QDs showing forward vs. rearward drift (Fig. 6G).
Discussion
There is ample evidence for a rearward flow of membrane components on the leading front of migrating fibroblasts and neuronal growth cones (17–21). In this study, we investigated the behavior of membrane proteins on the surface of migrating neurons and its relationship with cortical F-action. Migrating neurons differ from other motile cells by having a long l.p., which exhibits a dynamic growth cone that actively extends filopodia and lamellipodia in a manner similar to that found in migrating fibroblasts and nerve growth cones (4, 22, 23). Neuronal migration involves a sequence of events that include the extension of the leading growth cone, translocation of the soma, and retraction of the rear process (24–26). Because growth cone advance and soma translocation usually are not synchronized, the two events may use distinct mechanisms that are coordinated to achieve the translocation of the entire cell (3). As the structural link between the growth cone to the soma, the l.p. exhibits a forward flow of cortical F-actin during neuronal migration (4), and this F-actin flow could couple the growth cone advance with the soma motility. In the present study, we observed a net forward transport of membrane proteins along the direction of neuronal migration in both l.p. and t.p. but not in those of nonmigrating neurons (Fig. S3A). Such net forward transport of plasma membrane proteins during migration could occur without gross disruption of preexisting membrane topography. This transport may help preserve the existing gradients of guidance-cue receptors in the plasmalemma of the l.p., as found for TrkB in mouse granule cell precursors in vivo (27) and for ROBO2 and ROBO3 in cultured neurons (23, 28). Thus, forward transport of membrane proteins represents a mechanism for plasma membrane reorganization in migrating neurons. Other mechanisms, such as transcytosis of membrane components from the rear end to the leading front of the neuron, also may contribute to membrane reorganization (29–32).
Lateral Diffusion of Membrane Proteins.
We have used VAMP2-myc and TrkB, two membrane proteins with single transmembrane domains, as examples of plasma membrane proteins in migrating neurons. In contrast to VAMP2-myc, which lacks extensive cytoplasmic domain, TrkB has a large cytoplasmic domain that could interact with many cytoplasmic components, including cytoskeleton. The multivalency of the QD antibodies potentially could increase the size of the diffusing QD-bound complex by binding to several VAMP2-myc or TrkB and reduce the rate of lateral diffusion. However, we consistently have observed high lateral diffusion coefficients for both VAMP2-QD and TrkB-QD (around 0.8 and 0.6 μm2/s, respectively, close to the free lateral diffusion of single membrane proteins observed in biological membranes) (15, 33, 34). Thus, it is likely that QDs are bound to single protein in the present study.
In the analysis of QD motion, the directional movement deduced by the equation MSD = 2Dt + (vt)2 indicates that different QDs on the same neurite process could drift in opposite directions at different times and locations, suggesting that the local active forces driving the directional motion of membrane proteins vary with time and location and could alternate between forward and rearward directions. Furthermore, our analysis showed that many QDs exhibited confined diffusion on the cell surface, implying the existence of surface domains within which free diffusion of membrane proteins is limited (15, 35–37). A minority (∼35%) of QDs exhibited consistent directional drift movement over long distances, perhaps resulting from the spatial or temporal variation in the intra- and perimembranous structures along neurite processes that led to confined diffusion. In the absence of such confinement, the drift speed may be higher than observed here. Furthermore, under different migratory conditions the D values of VAMP2-myc and TrkB in the l.p. or t.p. were similar, indicating that the lateral diffusion rate of membrane proteins is largely independent of cellular events associated with neuronal migration.
Models of Membrane Flow in Migrating Cells.
Two different models have been proposed to account for the rearward movement of membrane components in the lamellipodia of migrating fibroblasts. In the membrane flow model, the movement of membrane proteins is driven by polarized endo-exocytic membrane recycling (38, 39), in which endocytosis occurs randomly on the cell surface and exocytosis occurs at the cell’s leading front, leading to a rearward membrane flow (6). Our findings, however, do not support the existence of a long-range rearward flow of membrane in migrating neurons, although local membrane flow indeed may occur near the leading growth cone or trailing tip. A cytoskeletal model (5, 40) for rearward membrane flow in fibroblasts or nerve growth cones posits that cortical actin filaments are assembled at the leading edge and disassembled at the rear of cell or growth cone. In the absence of attachment to the substrate via binding to membrane proteins, these actin filaments undergo rearward flow, whereas substrate attachment leads to filopodial extension and forward advance of the leading edge (40, 41). Rearward flow could be observed for membrane components that are bound to those cortical F-actins unrestrained by the substrate attachment (42). In the growth cone of cultured Aplysia neurons, the retrograde flow of both membrane component and F-actin depended on the activity of myosin II (16, 17, 43). The cortical F-actin flow is driven by both myosin II activity and F-actin polymerization at the distal end (44). It is not clear whether such a model could be applied to cortical F-actin dynamics along the neuritic process of these cultured granule cells. In our study, we observed intermittent bursts of drift movements of membrane proteins of similar speed in both forward and rearward directions, but forward drift was more frequent than rearward drift in migrating neurons. If these drift movements were driven by transient attachment of these proteins to underlying cortical F-actin, either through direct binding or via association with other F-actin–interacting proteins, the cortical F-actin in migrating neurons must undergo spatially and temporally heterogeneous movements in both forward and rearward directions but yield a net translocation in the forward direction. Further study of cortical F-actin dynamics in migrating neurons at a high spatiotemporal resolution is required to resolve this issue.
Myosin II Drives the Net Forward Transport.
Myosin II has been identified recently as a key player in neuronal migration (45–47). In many cell types, myosin II-driven flow of cortical F-actin propels cell locomotion, growth cone migration, cytokinesis, and transport of signaling molecules (5). Cortical F-actin is known to flow toward the region of higher actomyosin contractility (5, 48–50). In cultured cerebellar granule cells, a myosin II-dependent forward flow of cortical F-actin has been observed in the l.p., with an average speed similar to that of soma advance, suggesting that such myosin II-dependent cortical F-actin flow provides the traction force for forward soma translocation (4). We note that this cortical F-actin flow also may provide the driving force for the drift of membrane components, perhaps indirectly through creating local bulk membrane flow via proteins that are linked to cortical F-actin. We observed a significantly higher percentage of QD pairs exhibiting correlated drift forward than drift rearward in migrating cells, but not in nonmigrating cells, and this higher percentage of forwardly correlated QDs was abolished by the inhibition of myosin II activity with Blebb in stalling processes, suggesting that the net forward drift of VAMP2 is powered by myosin II. In the cultured cerebellar granule cells, the forward cortical F-actin flow is coupled with a front-high and rear-low distribution of myosin II activity along the l.p. (4). Such polarized distribution of myosin II activity may cause more frequent forward cortical F-actin movements and, hence, a net forward transport drift of membrane proteins (Fig. S3B). In stalling processes of nonmigrating cells we also observed that the Blebb treatment led to the disappearance of correlated movements of QDs on the same process; i.e., the percentage of correlated pairs became 50%. If drift movements are caused by the action of local cortical F-actin, this Blebb effect is consistent with the idea that myosin II activity is responsible for correlated cortical F-actin flow. However, with Blebb treatment, we found both forward and rearward drift movements persisted despite the removal of the forward bias. This unexpected result suggests that local drift movement of the proteins in both directions was driven by motor proteins other than myosin II, although the myosin II activity gradient along the neurite of migrating cells is capable of biasing the drift movements. Alternatively, other mechanisms creating local membrane flow, such as endo- and exocytotic recycling of plasma membrane, may be involved in the stochastic forward and rearward drifts of membrane components.
No Membrane Protein Flow in Growing Neurites of Nonmigrating Cells.
Neurite growth involves an increase in the neurite surface area by incorporating newly synthesized membrane into the plasmalemma via exocytosis of membrane precursor vesicles. Whether there is global flow of membrane components over the entire neurite surface remains controversial. Early experiments by Bray (51) showed stationary membrane surface-bound particles during axon elongation. However, forward global membrane flow along the growing neurites was monitored by fluorescent lipid dye incorporated in the axon membrane in cultured Xenopus spinal neurons (10). In contrast, a rearward flow of plasma membrane components was observed on growing neurites of cultured chicken dorsal root ganglion neurons by using small IgG-coated and lipid-attached beads (11). Our results on extending processes of nonmigrating granule cells showed that QD-labeled membrane proteins diffuse randomly without any detectable net transport, suggesting the lack of directional flow of plasma membrane proteins during neurite elongation in these granule cells. The difference between our results and previous findings may be attributed to the higher adhesion of granule cells to the culture substrate coated with laminin instead of polylysine alone, which may affect membrane dynamics directly by impeding membrane flow or indirectly by suppressing the contractility of cortical cytoskeleton (41, 52–54).
In summary, our work has shown that in migrating neurons plasma membrane proteins undergo net forward movements in both the l.p. and t.p. in accordance with the soma advance. This net protein movement involves a biased forward drift movement driven by the action of myosin II. Such forward plasma membrane protein flow may help preserve membrane protein topography during neuronal migration.
Materials and Methods
Preparation of Cultured Cerebellar Granule Cells.
All the newborn Sprague–Dawley (SD) rats used in the present study were provided by Shanghai Laboratory Animal Care Center. The animal use protocol was approved by the Animal Use Committee of the Institute of Neuroscience, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences. Preparation of cultured cerebellar granule cells was performed as previously described with some modifications (23). Briefly, the cerebellum cortex of P0–P1 SD rats was dissected on ice, digested with 0.125% trypsin for 6–9 min at 37 °C, and dissociated into single cells by gentle trituration. Cells were plated on coverslips coated with 100 μg/mL poly-d-lysine (Invitrogen) and 25 μg/mL laminin (Invitrogen) at a low density in Neurobasal medium (Gibco) supplemented with 10% (vol/vol) FBS (Gibco) and 2% (vol/vol) B27 (NFB medium; Gibco). For neuronal transfection of VAMP2-myc together with eYFP, dissociated cells were electroporated with 1–3 μg of DNA constructs using the Rat Neuron Nucleofector Kit (Amaxa Biosystems) according to the manufacturer’s instructions (23, 55).
DNA Constructs.
For VAMP2-myc plasmid, the cDNA of VAMP2 was cloned from VAMP2-GFP plasmid and constructed into pCS2-myc vector within BamHI and ClaI sites. Plasmids were amplified in Escherichia coli strain DH5α and purified with DNA purification kits (Qiagen).
Living Cell Immunocytochemistry.
Surface protein immunolabeling with QD was performed by methods previously described (56). To label surface VAMP2-myc and TrkB with QD, we incubated cultured cerebellar granule cells with mouse anti-myc antibody (1:1,000; Millipore) and rabbit anti-TrkB antibody (1:500; Abcam), respectively, for 30 min in NFB medium at 37 °C before incubation with Qdot-655 goat anti-mouse IgG conjugate (1:2,000; Invitrogen) and Qdot-655 goat anti-rabbit IgG conjugate (1:2,000; Invitrogen), respectively, in NFB medium for 1–3 min at 37 °C. The cells labeled with QD were rinsed eight times (for 10 s each rinsing) and then were incubated in the NFB medium without phenol red for live-imaging experiments. To quench the fluorescence of external QD, we applied high-K+ (50 mM) artificial CSF solution to the culture for 5 min. Cells were rinsed three times before imaging.
QD Imaging.
Cells were imaged at 37 °C, 5% (vol/vol) CO2 in a live-cell time-lapse imaging chamber mounted onto an inverted microscope (Nikon TI), which was equipped with a 60× oil-immersion objective (NA = 1.69). The excitation wavelength for QD was 510–560 nm (filtered xenon lamp light), and the emission light was filtered at 660 ± 50 nm. Images were sampled for 200 ms at a frequency of 0.5 Hz. Single QD signals were detected as blinking light on the background of eYFP fluorescence, which revealed neuritic processes. The cell movement was monitored with blue light for 100 ms at 1-min intervals. Immobile QDs observed near the neurons were considered to be substrate-bound QDs. Our analyses were focused on single QDs that underwent Brownian motion on the neuronal surface. Membrane-impermeable quencher QSY21 succinimidyl ester (Invitrogen) added to the solution at 4 mM was used to demonstrate that QDs were bound to the external surface of the process.
QD Tracking and Data Analysis.
The trajectories of QDs along the neurite were monitored and analyzed with ImageJ (National Institutes of Health) and Neurolucida (MBF Bioscience) software, together with custom-made MATLAB programs. For most measurements of QD motion, images were taken (for 200-ms exposure) at 2-s intervals for 10 min. Line scans were performed, and kymographs were generated along the neuritic process from the rear to the front from time-lapse images by using ImageJ. The width of kymograph line covered three pixels, about 0.8 μm, wider than the neurite. The trajectory of QD displacement along the axis of the process with time was made with Neurolucida software. The axonal processes were mostly stationary during the migration. To avoid the boundary effects, we measured QD movements only at the middle region of the processes, at least 5 μm away from the soma and the growth cone. The net movement of the protein was determined by the average trajectory of all QD trajectories for the same protein traced from different neurons exhibiting the same migratory behavior. The displacement of these proteins over different time intervals (2, 12, or 72 s) was analyzed by dividing the entire trajectory into the segments of the chosen interval. The distribution of the displacement was fitted with the Gaussian curve. The shift of the mean value of the displacement from the origin represents the directional drift of the membrane protein.
For quantitative determination of the diffusion coefficient of QD-labeled proteins, we plotted the MSD of QD movements along the neuritic axis measured from the kymographs against the time intervals from 2–12 s for each QD. Each MSD value was obtained from the distance traveled with the starting time of the interval covering all time points over the 10-min duration of observation. We assumed that the QD motion reflects the Brownian motion superimposed upon a net drift movement in either the forward or rearward direction and that the drift effect is insignificant if we consider only the initial segment (first six data points, covering the motion over a duration of 12 s) of the relationship between MSD vs. time intervals (57). The data for each QD were least-square fitted with the equation MSD = 2Dt, where D is diffusion coefficient and t is the time interval. To determine the value and direction of the drift speed (v), QD trajectories were divided into 60-s nonoverlapping segments for further analysis. The MSD-vs.-time plot for each QD trajectory segment then was fitted by the equation MSD = 2Dt + (vt)2 to obtain the best-fit drift speed v, using the D value obtained from the whole trajectory. The choice of the starting point for the division of the 60-s segment did not affect the distribution and mean value of drift speeds. In migrating neurons, the direction of the drift is defined by the net transport over each 60-s segment of QD trajectory, forward when the direction of the drift coincides with that of soma translocation and rearward when the drift and soma translocation are in opposite locations. In nonmigrating neurons, the direction is defined as anterograde when the drift is toward the soma and retrograde when the drift is away from the soma. All QD data for the same condition were averaged to yield the average D and v values for a given membrane protein.
Supplementary Material
Acknowledgments
This work was supported by the National Basic Research Program of China (2011CBA00400).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1219203110/-/DCSupplemental.
References
- 1.Hatten ME. Central nervous system neuronal migration. Annu Rev Neurosci. 1999;22:511–539. doi: 10.1146/annurev.neuro.22.1.511. [DOI] [PubMed] [Google Scholar]
- 2.Li S, Guan JL, Chien S. Biochemistry and biomechanics of cell motility. Annu Rev Biomed Eng. 2005;7:105–150. doi: 10.1146/annurev.bioeng.7.060804.100340. [DOI] [PubMed] [Google Scholar]
- 3.Tsai LH, Gleeson JG. Nucleokinesis in neuronal migration. Neuron. 2005;46(3):383–388. doi: 10.1016/j.neuron.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 4.He M, Zhang ZH, Guan CB, Xia D, Yuan XB. Leading tip drives soma translocation via forward F-actin flow during neuronal migration. J Neurosci. 2010;30(32):10885–10898. doi: 10.1523/JNEUROSCI.0240-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bray D, White JG. Cortical flow in animal cells. Science. 1988;239(4842):883–888. doi: 10.1126/science.3277283. [DOI] [PubMed] [Google Scholar]
- 6.Bretscher MS. Getting membrane flow and the cytoskeleton to cooperate in moving cells. Cell. 1996;87(4):601–606. doi: 10.1016/s0092-8674(00)81380-x. [DOI] [PubMed] [Google Scholar]
- 7.Craig AM, Wyborski RJ, Banker G. Preferential addition of newly synthesized membrane protein at axonal growth cones. Nature. 1995;375(6532):592–594. doi: 10.1038/375592a0. [DOI] [PubMed] [Google Scholar]
- 8.Shieh JC, Schaar BT, Srinivasan K, Brodsky FM, McConnell SK. Endocytosis regulates cell soma translocation and the distribution of adhesion proteins in migrating neurons. PLoS ONE. 2011;6(3):e17802. doi: 10.1371/journal.pone.0017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin CH, Forscher P. Cytoskeletal remodeling during growth cone-target interactions. J Cell Biol. 1993;121(6):1369–1383. doi: 10.1083/jcb.121.6.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Popov S, Brown A, Poo MM. Forward plasma membrane flow in growing nerve processes. Science. 1993;259(5092):244–246. doi: 10.1126/science.7678471. [DOI] [PubMed] [Google Scholar]
- 11.Dai J, Sheetz MP. Axon membrane flows from the growth cone to the cell body. Cell. 1995;83(5):693–701. doi: 10.1016/0092-8674(95)90182-5. [DOI] [PubMed] [Google Scholar]
- 12.Straight AF, et al. Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science. 2003;299(5613):1743–1747. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Q, Li Y, Tsien RW. The dynamic control of kiss-and-run and vesicular reuse probed with single nanoparticles. Science. 2009;323(5920):1448–1453. doi: 10.1126/science.1167373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song AH, et al. A selective filter for cytoplasmic transport at the axon initial segment. Cell. 2009;136(6):1148–1160. doi: 10.1016/j.cell.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 15.Saxton MJ, Jacobson K. Single-particle tracking: Applications to membrane dynamics. Annu Rev Biophys Biomol Struct. 1997;26:373–399. doi: 10.1146/annurev.biophys.26.1.373. [DOI] [PubMed] [Google Scholar]
- 16.Forscher P, Smith SJ. Actions of cytochalasins on the organization of actin filaments and microtubules in a neuronal growth cone. J Cell Biol. 1988;107(4):1505–1516. doi: 10.1083/jcb.107.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forscher P, Kaczmarek LK, Buchanan JA, Smith SJ. Cyclic AMP induces changes in distribution and transport of organelles within growth cones of Aplysia bag cell neurons. J Neurosci. 1987;7(11):3600–3611. doi: 10.1523/JNEUROSCI.07-11-03600.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holifield BF, Ishihara A, Jacobson K. Comparative behavior of membrane protein-antibody complexes on motile fibroblasts: Implications for a mechanism of capping. J Cell Biol. 1990;111(6 Pt 1):2499–2512. doi: 10.1083/jcb.111.6.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marcus PI. Dynamics of surface modification in myxovirus-infected cells. Cold Spring Harb Symp Quant Biol. 1962;27:351–365. doi: 10.1101/sqb.1962.027.001.033. [DOI] [PubMed] [Google Scholar]
- 20.Ingram VM. A side view of moving fibroblasts. Nature. 1969;222(5194):641–644. doi: 10.1038/222641a0. [DOI] [PubMed] [Google Scholar]
- 21.Bretscher MS. Fibroblasts on the move. J Cell Biol. 1988;106(2):235–237. doi: 10.1083/jcb.106.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ono K, et al. Filopodia and growth cones in the vertically migrating granule cells of the postnatal mouse cerebellum. Exp Brain Res. 1997;117(1):17–29. doi: 10.1007/pl00005787. [DOI] [PubMed] [Google Scholar]
- 23.Guan CB, Xu HT, Jin M, Yuan XB, Poo MM. Long-range Ca2+ signaling from growth cone to soma mediates reversal of neuronal migration induced by slit-2. Cell. 2007;129(2):385–395. doi: 10.1016/j.cell.2007.01.051. [DOI] [PubMed] [Google Scholar]
- 24.Edmondson JC, Hatten ME. Glial-guided granule neuron migration in vitro: A high-resolution time-lapse video microscopic study. J Neurosci. 1987;7(6):1928–1934. doi: 10.1523/JNEUROSCI.07-06-01928.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Komuro H, Rakic P. Dynamics of granule cell migration: A confocal microscopic study in acute cerebellar slice preparations. J Neurosci. 1995;15(2):1110–1120. doi: 10.1523/JNEUROSCI.15-02-01110.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wichterle H, Garcia-Verdugo JM, Alvarez-Buylla A. Direct evidence for homotypic, glia-independent neuronal migration. Neuron. 1997;18(5):779–791. doi: 10.1016/s0896-6273(00)80317-7. [DOI] [PubMed] [Google Scholar]
- 27.Zhou P, et al. Polarized signaling endosomes coordinate BDNF-induced chemotaxis of cerebellar precursors. Neuron. 2007;55(1):53–68. doi: 10.1016/j.neuron.2007.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katsuki T, Ailani D, Hiramoto M, Hiromi Y. Intra-axonal patterning: Intrinsic compartmentalization of the axonal membrane in Drosophila neurons. Neuron. 2009;64(2):188–199. doi: 10.1016/j.neuron.2009.08.019. [DOI] [PubMed] [Google Scholar]
- 29.Ikonen E, Parton RG, Hunziker W, Simons K, Dotti CG. Transcytosis of the polymeric immunoglobulin receptor in cultured hippocampal neurons. Curr Biol. 1993;3(10):635–644. doi: 10.1016/0960-9822(93)90061-r. [DOI] [PubMed] [Google Scholar]
- 30.Hémar A, Olivo JC, Williamson E, Saffrich R, Dotti CG. Dendroaxonal transcytosis of transferrin in cultured hippocampal and sympathetic neurons. J Neurosci. 1997;17(23):9026–9034. doi: 10.1523/JNEUROSCI.17-23-09026.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yap CC, et al. The somatodendritic endosomal regulator NEEP21 facilitates axonal targeting of L1/NgCAM. J Cell Biol. 2008;180(4):827–842. doi: 10.1083/jcb.200707143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wisco D, et al. Uncovering multiple axonal targeting pathways in hippocampal neurons. J Cell Biol. 2003;162(7):1317–1328. doi: 10.1083/jcb.200307069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poo M, Cone RA. Lateral diffusion of rhodopsin in the photoreceptor membrane. Nature. 1974;247(441):438–441. doi: 10.1038/247438a0. [DOI] [PubMed] [Google Scholar]
- 34.McCloskey M, Poo MM. Protein diffusion in cell membranes: Some biological implications. Int Rev Cytol. 1984;87:19–81. doi: 10.1016/s0074-7696(08)62439-0. [DOI] [PubMed] [Google Scholar]
- 35.Bouchaud JP, Georges A. Anomalous diffusion in disordered media - statistical mechanisms, models and physical applications. Phys Rep. 1990;195:127–293. [Google Scholar]
- 36.Ghosh RN, Webb WW. Automated detection and tracking of individual and clustered cell surface low density lipoprotein receptor molecules. Biophys J. 1994;66(5):1301–1318. doi: 10.1016/S0006-3495(94)80939-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kusumi A, et al. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: High-speed single-molecule tracking of membrane molecules. Annu Rev Biophys Biomol Struct. 2005;34:351–378. doi: 10.1146/annurev.biophys.34.040204.144637. [DOI] [PubMed] [Google Scholar]
- 38.Abercrombie M, Heaysman JE, Pegrum SM. The locomotion of fibroblasts in culture. 3. Movements of particles on the dorsal surface of the leading lamella. Exp Cell Res. 1970;62(2):389–398. doi: 10.1016/0014-4827(70)90570-7. [DOI] [PubMed] [Google Scholar]
- 39.Bretscher MS. Endocytosis: Relation to capping and cell locomotion. Science. 1984;224(4650):681–686. doi: 10.1126/science.6719108. [DOI] [PubMed] [Google Scholar]
- 40.Mitchison T, Kirschner M. Cytoskeletal dynamics and nerve growth. Neuron. 1988;1(9):761–772. doi: 10.1016/0896-6273(88)90124-9. [DOI] [PubMed] [Google Scholar]
- 41.Suter DM, Forscher P. An emerging link between cytoskeletal dynamics and cell adhesion molecules in growth cone guidance. Curr Opin Neurobiol. 1998;8(1):106–116. doi: 10.1016/s0959-4388(98)80014-7. [DOI] [PubMed] [Google Scholar]
- 42.Serge A, Fourgeaud L, Hemar A, Choquet D. Active surface transport of metabotropic glutamate receptors through binding to microtubules and actin flow. J Cell Sci. 2003;116(Pt 24):5015–5022. doi: 10.1242/jcs.00822. [DOI] [PubMed] [Google Scholar]
- 43.Lin CH, Espreafico EM, Mooseker MS, Forscher P. Myosin drives retrograde F-actin flow in neuronal growth cones. Neuron. 1996;16(4):769–782. doi: 10.1016/s0896-6273(00)80097-5. [DOI] [PubMed] [Google Scholar]
- 44.Medeiros NA, Burnette DT, Forscher P. Myosin II functions in actin-bundle turnover in neuronal growth cones. Nat Cell Biol. 2006;8(3):215–226. doi: 10.1038/ncb1367. [DOI] [PubMed] [Google Scholar]
- 45.Schaar BT, McConnell SK. Cytoskeletal coordination during neuronal migration. Proc Natl Acad Sci USA. 2005;102(38):13652–13657. doi: 10.1073/pnas.0506008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsai JW, Bremner KH, Vallee RB. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat Neurosci. 2007;10(8):970–979. doi: 10.1038/nn1934. [DOI] [PubMed] [Google Scholar]
- 47.Vallee RB, Seale GE, Tsai JW. Emerging roles for myosin II and cytoplasmic dynein in migrating neurons and growth cones. Trends Cell Biol. 2009;19(7):347–355. doi: 10.1016/j.tcb.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.White JG, Borisy GG. On the mechanisms of cytokinesis in animal cells. J Theor Biol. 1983;101(2):289–316. doi: 10.1016/0022-5193(83)90342-9. [DOI] [PubMed] [Google Scholar]
- 49.Benink HA, Mandato CA, Bement WM. Analysis of cortical flow models in vivo. Mol Biol Cell. 2000;11(8):2553–2563. doi: 10.1091/mbc.11.8.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Munro E, Nance J, Priess JR. Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans embryo. Dev Cell. 2004;7(3):413–424. doi: 10.1016/j.devcel.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 51.Bray D. Surface movements during the growth of single explanted neurons. Proc Natl Acad Sci USA. 1970;65(4):905–910. doi: 10.1073/pnas.65.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choquet D, Felsenfeld DP, Sheetz MP. Extracellular matrix rigidity causes strengthening of integrin-cytoskeleton linkages. Cell. 1997;88(1):39–48. doi: 10.1016/s0092-8674(00)81856-5. [DOI] [PubMed] [Google Scholar]
- 53.Felsenfeld DP, Choquet D, Sheetz MP. Ligand binding regulates the directed movement of beta1 integrins on fibroblasts. Nature. 1996;383(6599):438–440. doi: 10.1038/383438a0. [DOI] [PubMed] [Google Scholar]
- 54.Thompson C, Lin CH, Forscher P. An Aplysia cell adhesion molecule associated with site-directed actin filament assembly in neuronal growth cones. J Cell Sci. 1996;109(Pt 12):2843–2854. doi: 10.1242/jcs.109.12.2843. [DOI] [PubMed] [Google Scholar]
- 55.Li Y, et al. Essential role of TRPC channels in the guidance of nerve growth cones by brain-derived neurotrophic factor. Nature. 2005;434(7035):894–898. doi: 10.1038/nature03477. [DOI] [PubMed] [Google Scholar]
- 56.Bannai H, Lévi S, Schweizer C, Dahan M, Triller A. Imaging the lateral diffusion of membrane molecules with quantum dots. Nat Protoc. 2006;1(6):2628–2634. doi: 10.1038/nprot.2006.429. [DOI] [PubMed] [Google Scholar]
- 57.Qian H, Sheetz MP, Elson EL. Single particle tracking. Analysis of diffusion and flow in two-dimensional systems. Biophys J. 1991;60(4):910–921. doi: 10.1016/S0006-3495(91)82125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.