Abstract
A significant proportion of colorectal cancer (CRC) patients are resistant to anti-ERBB1 [avian erythroblastic leukemia viral (v-erb-b) oncogene homolog, receptor for EGF] monoclonal antibodies (Mabs). We evaluated both immune and nonimmune effects of cetuximab (anti-ERBB1 Mab), trastuzumab (anti-ERBB2 Mab), pertuzumab (anti-ERBB2 Mab), and lapatinib (dual ERBB1 and ERBB2 tyrosine kinase inhibitor) in a large well-characterized panel of 64 CRC cell lines to find response predictive tumor characteristics. There was a significant correlation between the direct effects of cetuximab and lapatinib. Both agents were associated (P = 0.0004) with “triple’ wild-type status in KRAS, BRAF, and PIK3CA exon 20. Most cell lines were resistant to the direct effects of anti-ERBB2 Mabs, suggesting that the effects of lapatinib might mainly be through ERBB1. Microarray mRNA expression profiles of sensitive and resistant cell lines showed that although ERBB1 receptor or ligand levels did not associate with cetuximab sensitivity, high levels of ERBB2 (P = 0.036) and amphiregulin (P = 0.026) predicted sensitivity to lapatinib. However, higher ERBB1 expression predicted susceptibility to cetuximab-induced antibody-dependent cellular cytotoxicity and occurred independently of KRAS/BRAF/PIK3CA mutations (P = 0.69). Lapatinib may be an effective alternative therapy to cetuximab in triple wild-type tumors. Microarray analysis provides suggestive biomarkers for resistance. ERBB1 levels, independent of mutation status, predict immune killing. Therefore, anti-ERBB1 antibodies may be considered in CRC tumors with higher ERBB1 expression and favorable FcγR polymorphisms.
Keywords: anti-ERBB1 therapy, immune-mediated killing, high throughput screening
Monoclonal antibodies against the epidermal growth factor receptor, ERBB1 [avian erythroblastic leukemia viral (v-erb-b) oncogene homolog, receptor for EGF], including in particular cetuximab, are now commonly used in colorectal cancer (CRC) treatment (1–3). However, only 20% of patients respond to anti-ERBB1 monotherapy (3). The search for predictive markers to improve clinical outcomes (1, 3) has identified that the presence of a KRAS mutation predicts an adverse response leading to routine KRAS testing before anti-ERBB1 therapy (4). However, KRAS mutations are present in only 30–40% of CRC tumors and a significant proportion of KRAS wild-type (WT) patients (50–65%) do not respond to anti-ERBB1 therapy (3). To improve therapeutic outcomes (5), we therefore need to improve understanding of the roles of different antibody-killing mechanisms and the properties of tumors that determine their response to antibody treatment.
The relative contributions of immune [for example antibody-dependant cellular cytotoxicity (ADCC) and antibody-dependant cellular phagocytosis (ADCP)] and nonimmune processes (competitive blocking of receptor-ligand binding) to tumor response following antibody therapy are not yet clear (5, 6). The FcγR polymorphism has been shown to be an independent predictor of response to cetuximab in CRC patients, implicating a role for antibody dependent immune attack in cetuximab therapy (6). Emerging evidence also associates mutations or loss-of-function in genes other than KRAS, for example BRAF, PIK3CA (exon 20), and PTEN with resistance to cetuximab (3, 7–9). Such associations are more likely to be associated with direct rather than immune based effects of antibodies, suggesting that tyrosine kinase inhibitors (TKIs) such as lapatinib, a dual ERBB1 and ERBB2 (v-erb-b2 erythroblastic leukemia viral oncogene homolog 2) receptor inhibitor, might be effective as an alternative therapy (10). There are contradictory reports that ERBB1 receptor overexpression can predict response to cetuximab (11, 12). The expression levels of the ERBB ligands, epiregulin (EREG) and amphiregulin (AREG) have also been associated with differences in response to antibody therapy (13). The objective of our study was to use a large well-characterized panel of more than 60 CRC cell lines to define further the tumor characteristics that predict both the direct and immune mediated responses to cetuximab, and to compare the results with the use of other anti-ERBB antibodies and with the TKI, lapatinib.
Results
Direct Growth Inhibition of CRC Cell Lines by Cetuximab, Trastuzumab, Pertuzumab, and Lapatinib.
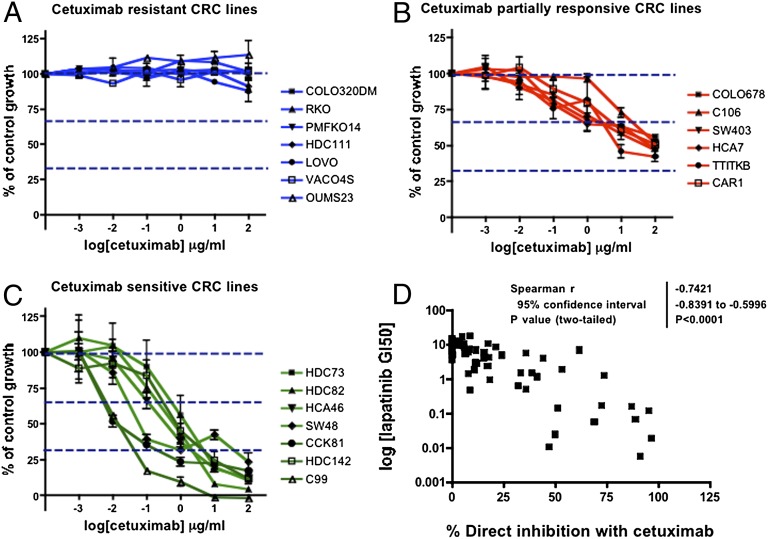
Sixty-four CRC cell lines were screened for direct growth sensitivity to cetuximab (anti-ERBB1 domain 3 Mab that inhibits ligand binding). Figs. 1 A–C show that the cell lines could be grouped into three categories according to their sensitivity to cetuximab, with little or no inhibition of growth (less than 33% of total control growth) (Fig. 1A) in 67% (43 of 64 lines, duplicates excluded) even at antibody concentrations of up to 100 μg/mL. About 20% (13 of 64) of the cell lines showed partial antibody responses (33–66% growth inhibition) (Fig. 1B), but the highest sensitivity to cetuximab (over 66% of growth inhibition) was seen in 12.5% (8 of 64) cell lines (Fig. 1C).
Fig. 1.
Direct growth inhibition of CRC cell lines by cetuximab and lapatinib. The SRB assay was used to determine the growth response of CRC cell lines to cetuximab (1 × 10−4 − 1 × 102 μg/mL) over three doubling times. Representative plots for up to seven different cell lines from each cetuximab response category are shown. Three categories were identified: (A) resistant, (B) partially responsive, and (C) sensitive. There was a highly significant correlation (D) between cetuximab and lapatinib response (Spearman correlation, r = 0.7421, P < 0.0001) in the cell-line panel.
The cell lines were also categorized into three groups according to their sensitivity to lapatinib: GI50s less than 0.1 μM (sensitive), between 0.1 and 1 μM (partial responders), and over 1 μM (resistant). Fig. 1D shows that there is a highly significant correlation between cetuximab and lapatinib sensitivities (Spearman correlation, r = 0.7421, P < 0.0001).
The majority of the CRC cell lines were resistant to direct inhibition by trastuzumab (anti-ERBB2 domain 4 Mab) and pertuzumab (anti-ERBB2 domain 2 Mab that inhibits dimerization) (Fig. S1 A–D), with some mild-to-moderate growth inhibition in about 15% of the lines. The same three cell lines (HDC142, C99, and HDC82) were most sensitive to both antibodies (Fig. S1 E and F) but even in these, the combination of cetuximab with trastuzumab or pertuzumab had only a marginal extra effect (Fig. S1 G and H).
Association Between Genetic Mutations and Cetuximab Sensitivity.
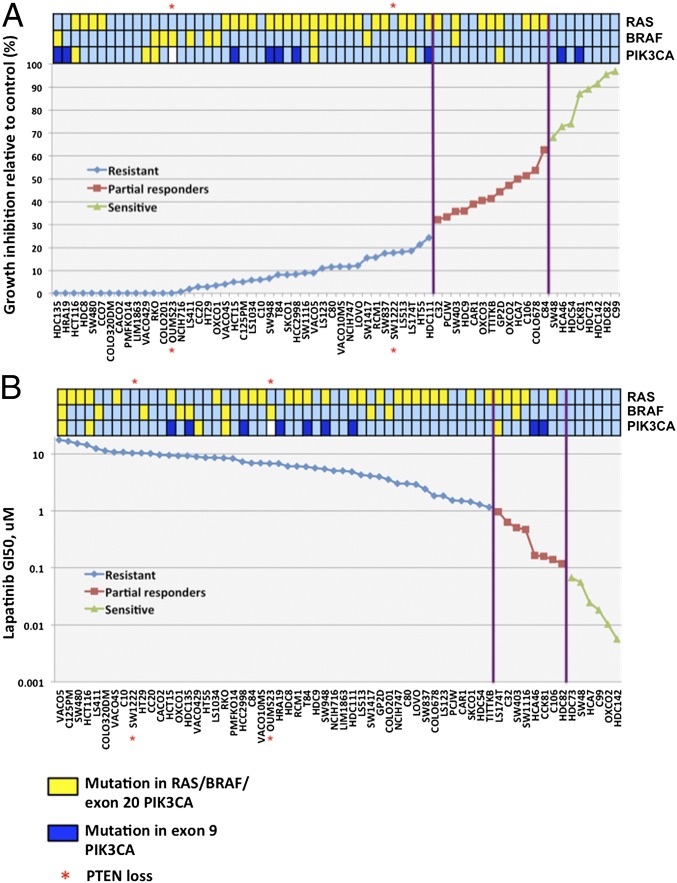
In Fig. 2A, the cell line sensitivities are plotted in rank order from least to most sensitive together with information on mutations in KRAS/NRAS, BRAF, PIK3CA (exons 9 and 20), and PTEN loss. There was a clear highly significant association between the proportions of cell lines with mutations in KRAS/NRAS, BRAF, and PIK3CA exon 20 and sensitivity to the direct effects of cetuximab (Table 1). KRAS/NRAS mutation was, as might be expected, significant on its own. Only one cell line had a NRAS mutation (C32). BRAF on its own was on the margin of significance, although 9 of 10 lines with BRAF V600E mutations were in the resistant group. PIK3CA was in the right direction but with too little data to show significance, and only two cell lines had negligible PTEN mRNA expression and these were both resistant to cetuximab treatment (Table S1).
Fig. 2.
KRAS/BRAF/PIK3CA mutations associate with resistance to cetuximab and lapatinib. Rank plot of the CRC cell-line panel according to sensitivity to (A) cetuximab (calculated as the percentage of growth inhibition relative to control) and (B) lapatinib (calculated as GI50, μM). The presence (yellow shade) or absence (light-blue shade) of mutations in KRAS/NRAS, BRAF (V600E), PIK3CA exon 20 (H1047L/H1047R), and loss of PTEN expression (indicated by asterisk) are also displayed above the plots. PIK3CA exon 9 (E542K/E545K) mutations were not associated with cetuximab resistance (dark-blue shade).
Table 1.
Correlation between genetic data (KRAS, BRAF, PIK3CA exon 20, and PTEN loss) and cetuximab nonimmune response
| KRAS/BRAF/PIK3CA exon 20/PTEN loss | Mutation | WT |
| Resistant | 34 | 9 |
| Partial resistance | 8 | 5 |
| Sensitive | 0 | 8 |
The P value P < 0.0001 is for a χ2 test for trend in a 3 × 2 table.
These data clearly show that the mutational status of KRAS, BRAF, PIK3CA exon 20, and PTEN is predictive of the direct effects of cetuximab in CRC cell lines. Thus, mutations in KRAS, BRAF, PIK3CA, or loss of expression in PTEN were present in 79.1% (34 of 43) of the cetuximab-resistant cell lines, but none of the sensitive lines had any of these mutations. The mean level of inhibition was lower in BRAF mutant cell lines (16.0 ± 7.1%) compared with cell lines with either KRAS or PIK3CA mutations (36.7 ± 4.7% and 31.4 ± 13.0%, respectively). These results were all significantly lower than the level of inhibition in triple WT cell lines (80.7 ± 5.1%) (Fig. S2A). Establishing the specific separate effect of PIK3CA exon 20 mutations was difficult because most of the lines with PIK3CA mutations also contained either KRAS or BRAF mutations (five of six).
As might be expected from the strong correlation between response to lapatinib and the direct response to cetuximab (Fig. 1D), the same significant associations with KRAS, BRAF, and PIK3CA were seen for lapatinib response as for the direct effects of cetuximab (Fig. 2B and Table S1). As for cetuximab, the most lapatinib-sensitive cell lines had no mutations in KRAS, BRAF, or PIK3CA and all but one of the BRAF V600E mutants were in the resistant group.
There were no significant associations with any other major genetic or expression changes (loss of heterozygosity) in APC, SMAD4, CDX1, and DCC; RER (replication error defect); mutations in APC, TP53, CTNNB1 (β-catenin), MLH1, MSH2, B2M (β2-microglobulin), and TGFBR2. This finding leaves open the question as to what other features of the cell lines might explain the resistance to the direct effects of cetuximab in the multiple WT cell lines.
Expression of ERBB Receptors and Their Ligands, and Sensitivity to Lapatinib and Cetuximab Direct Effects.
The cell line Affymetrix Gene Microarray (Human Genome U133 Plus 2.0 Array) database was used to query whether the mRNA expression profiles of different ERBB receptors (ERBB1, ERBB2, ERBB3, and ERBB4) and their ligands [EGF, TGFα, AREG, HB-EGF, betacellulin (BTC), EREG, and neuregulin1 (NRG1)] correlated with the direct effects of cetuximab and lapatinib in triple WT CRC cell lines. All of the ERBB receptors and their ligands, except for ERBB4 and EGF, were significantly expressed in at least a subset of the cell lines. The data from a subset of the cell lines (Fig. S2B) show that there is reasonably good agreement between protein and mRNA levels for ERBB1. The mean mRNA expression level differences between the most sensitive and resistant WT cell lines (Table S2) showed only nominally significant associations between lapatinib sensitivity and high ERBB2 (Fig. S2C) and AREG expression (Fig. S2D). There were also suggestive associations for both cetuximab and lapatinib sensitivity with high EREG expression (Fig. S2E) and between lapatinib sensitivity and BTC expression, but none of these associations survive multiple testing significance levels. There were no significant associations between cetuximab or lapatinib sensitivity and ERBB1 or EGF mRNA expression levels (Tables S3 and S4).
Cetuximab-Mediated Effects on Downstream Phosphorylation in Sensitive and Resistant CRC Cell Lines.
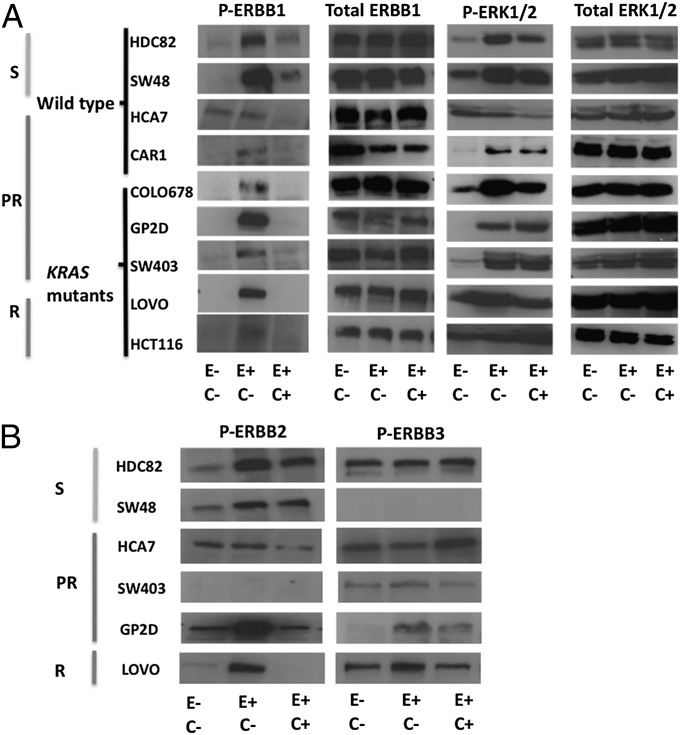
Cetuximab blocking (at a concentration of 20 μg/mL) (Fig. 1C) of EGF-mediated effects on downstream signaling was studied by immunoblot detection of downstream phosphorylation targets.
In the absence of EGF ligand, tyrosine-1173 phosphorylation (P-ERBB1) was not seen in the majority of cell lines, apart from HCA7 (Fig. 3A). A possible explanation for this may be autocrine stimulation of ERBB1 by growth factors, such as AREG, which is expressed at high levels in HCA7 (Fig. S2F). With the addition of EGF, P-ERBB1 was induced in all cell lines, whether sensitive or resistant. No additional phosphorylation was seen in HCA7, suggesting that there may be saturation of ERBB1 receptors by endogenous ligands. EGF-induced phosphorylation was reduced by cetuximab in all cases, demonstrating the effectiveness of this antibody to block ERBB1. Downstream ERK1/2 phosphorylation (P-ERK1/2) was also stimulated in the presence of EGF in HDC82, SW48, CAR1, COLO678, GP2D, and SW403 (Fig. 3A). No additional ligand-induced phosphorylation of ERK1/2 was seen in HCT116 and LOVO (both KRAS mutant-resistant cell lines) and HCA7, suggesting that there was maximal constitutive activation of downstream signaling proteins in these cell lines. In no case did EGF stimulation or cetuximab blocking affect overall levels of ERK1/2. Surprisingly, there were significant residual levels of P-ERK1/2 in HDC82, SW48, CAR1, COLO678, GP2D, and SW403 despite cetuximab preincubation. This result may be because of EGF binding to ERBB1-ERBB2 or ERBB1-ERBB3 heterodimers, even in the presence of cetuximab. This mechanism is suggested by the observation that there is induction of ERBB2 or ERBB3 phosphorylation after addition of EGF, and a continued presence of residual phosphorylation after cetuximab preincubation in some of the cell lines (Fig. 3B). Heterodimers can also be stimulated by endogenous ligands, such as EREG and AREG. These findings emphasize the problem of targeting single ERBB receptors, because ligands may activate heterodimers in these receptor systems even in the presence of blocking antibodies for the presumed single ERBB receptor target.
Fig. 3.
Effects of cetuximab on downstream phosphorylation events. Immunoblotting was used to determine the effects of EGF on ERBB1 (tyrosine-1173) and ERK1/2 (threonine 202/tyrosine 204) in nine cell lines with varying sensitivity to cetuximab. HDC82 and SW48 were sensitive (S); HCA7, CAR1, COLO678, GP2D, and SW403 were partial responders (PR); LOVO and HCT116 were resistant (R). HDC82, SW48, HCA7, and CAR1 are WT for KRAS/BRAF/PIK3CA, whereas the remainder have codons 12/13 KRAS mutations. E−, E+, C−, C+ refer to the absence or presence of EGF and cetuximab respectively. (A) EGF increased ERBB1 phosphorylation (P-ERBB1) in all sensitive and resistant cell lines apart from HCA7. Preincubation with cetuximab reduced tyrosine phosphorylation in all cell lines. ERK1/2 phosphorylation (P-ERK1/2) was stimulated in all of the responsive but not resistant cell lines after EGF stimulation. Preincubation with cetuximab reduced P-ERK1/2 only slightly in HDC82, SW48 and COLO678 with no real reduction in CAR1, GP2D, and SW403. No effect was seen in HCT116 and LOVO. (B) The effects of EGF on ERBB2 (tyrosine 1221/1222) and ERBB3 (tyrosine 1289) phosphorylation (P-ERBB2 and P-ERBB3) were also investigated (all cell lines expressed ERBB2 and ERBB3). Addition of EGF resulted in increased P-ERBB2 in HDC82, SW48, GP2D, and LOVO with significant residual activity in the presence of cetuximab in all these cell lines apart from LOVO. A similar effect was seen with P-ERBB3 in GP2D. No P-ERBB3 was seen in SW48, a cell line with low ERBB3 expression.
Genes Differentially Expressed Between KRAS/BRAF/PIK3CA WT CRC Cell Lines Resistant vs. Sensitive to Lapatinib and Cetuximab Direct Effects.
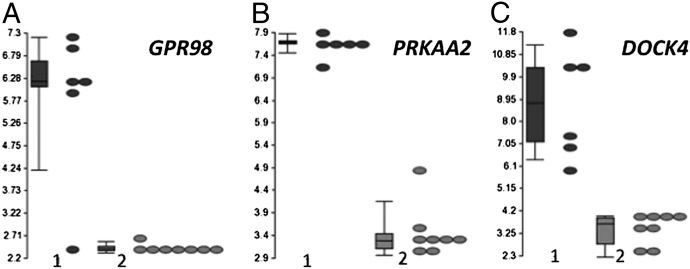
Differentially expressed genes between direct cetuximab- and lapatinib-resistant and sensitive cell lines were ranked by the P values for t tests for differences in expression. Table S5 lists for cetuximab and lapatinib the most differentially expressed genes among the top 25 that were thought to be of most interest functionally. Among these genes, GPR98 (G protein-coupled receptor 98), PRKAA2 (a 5-AMP activated kinase), and DOCK4 (guanine nucleotide exchange factor) are common to both lists and seem likely to be of greatest interest from a functional point of view. These genes were most differentially expressed with minimal mRNA expression in the most sensitive group of cells (Fig. 4). Other genes that are differentially expressed include AMBN, GPR37, ITGA6, and SEL1L3 (Fig. S3). The wide variation in expression in these latter genes seem to suggest two categories (high and low) in the subgroup of triple WT–resistant cell lines. The level of GPR98/PRKAA2/DOCK4 mRNA expression can be used to further classify triple WT tumors according to their response to cetuximab (Table S6). Five of six (83.3%) cell lines with high GPR98/PRKAA2/DOCK4 mRNA expression were cetuximab-resistant. In contrast, 8 of 12 (66.7%) cell lines with low mRNA expression were sensitive. There was a highly significant difference between sensitive, and partial responder or resistant triple WT cell lines with respect to the pattern of expression of these three genes (Table S7) (P = 0.0017).
Fig. 4.
Differential GPR98, PRKAA2, and DOCK4 mRNA expression in CRC cell lines. (A–C) Box-and-whiskers plots of GPR98, PRKAA2, and DOCK4 mRNA expression in triple WT CRC cells that were resistant (group 1) or sensitive (group 2) to the effects of cetuximab. The y axis represents the mRNA level with the values plotted on a log2 scale. The bottom and top of the boxes represent the 25th and 75th percentiles, respectively; the whiskers represent the 10th to the 90th percentiles.
Immune-Mediated Killing by Cetuximab and Trastuzumab.
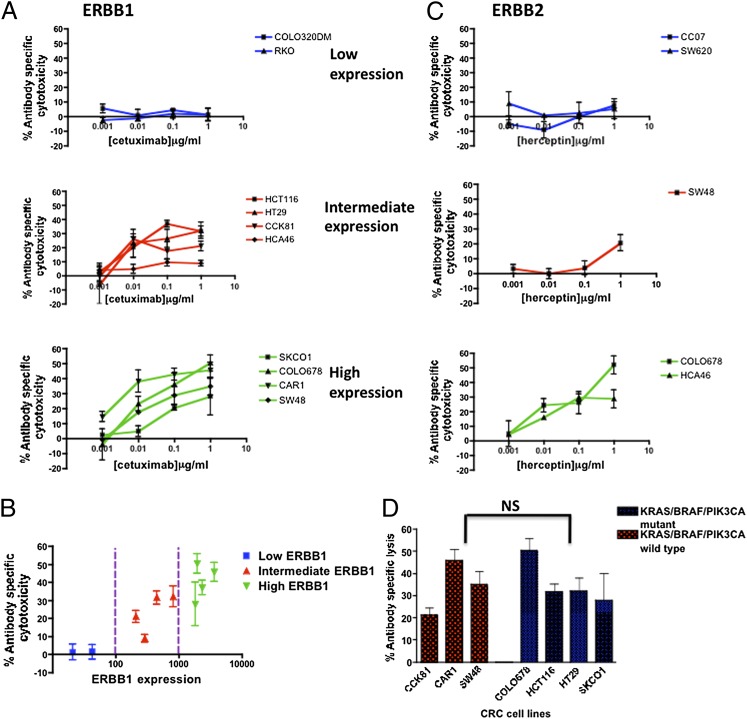
The level of cetuximab-mediated ADCC was evaluated in a selected subset of CRC cell lines, based on their ERBB1 expression levels (Fig. S4A), using a lactate dehydrogenase (LDH) release cytotoxicity assay. Cell lines with negligible ERBB1 levels (COLO320DM and RKO) were resistant to cetuximab-mediated ADCC (Fig. 5 A and B). The high ERBB1-expressing cell lines (SKCO1, SW48, CAR1, and COLO678) were the most susceptible to ADCC, but those with intermediate levels (CCK81, HCA46, HT29, and HCT116) were still sensitive to cetuximab-mediated ADCC, although to progressively lesser extents (Fig. 5 A and B). The efficacy of ADCC by cetuximab is clearly directly related to ERBB1 expression level (Fig. 5B). There was some significant ADCC in nearly all ERBB1+ cells even at concentrations as low as 10 ng/mL, a level that may reflect what is found in the tumor environment in patients.
Fig. 5.
Immune-mediated killing of CRC cell lines in the presence of cetuximab and trastuzumab. (A) Cells with different levels of ERBB1 were used as targets. Unfractionated PBMC cells were used at an effector:target ratio of 40:1. Cetuximab caused ADCC in a concentration dependent manner (0.001–1μg/mL), particularly in ERBB1 high (SW48, CAR1, COLO678, and SKCO1) cell lines. This was also seen in ERBB1 intermediate (HT29, HCT116, CCK81, and HCA46) cell lines but to a lesser degree. The effect was negligible in negative cell lines (COLO320DM and RKO). (B) ADCC strongly correlated with the ERBB1 expression levels. (C) Trastuzumab is able to mediate ADCC of ERBB2+ CRC cell lines (COLO678, HCA46, and SW48) but not in lines that lacked ERBB2-expression (CCO7 and SW620). The y axis represents percentage of specific lysis and the x axis represents log (antibody) concentration (μg/mL). (D) There was no significant (NS) difference in the level of ADCC (1μg/mL cetuximab) measured in KRAS/BRAF/PIK3CA WT and mutant cell lines (Mann–Whitney test, P = 0.69).
Trastuzumab elicited ADCC in ERBB2+ tumor lines with an efficacy related to ERBB2 expression level and absolutely no ADCC in ERBB2− lines (Fig. 5C and Fig. S4 B and C), paralleling the more extensive results for cetuximab and ERBB1 expression.
Copy-number analysis of 39 CRC cell lines on the Welcome Sanger Institute database failed to identify any cell lines that had high levels of gene amplification in ERBB1, ERBB2, or ERBB3. This result rules out gene amplification as a major reason for substantial variation in mRNA levels for these receptors.
There was no significant association between cetuximab-mediated ADCC and the KRAS, BRAF, and PIK3CA mutation status (Fig. 5D). Thus, cetuximab immune-based killing is dependent on ERBB1 expression and not KRAS/BRAF/PIK3CA mutational status. Based on our in vitro results, we predict that at least two-thirds of the CRC cell panel (43 of 64) may be susceptible to immune responses, a significantly higher proportion than are susceptible to the nonimmune direct effects of cetuximab.
Previous evidence shows that the main mediators of ADCC in humans are natural killer (NK) cells (14, 15). We have therefore checked that the cetuximab-mediated ADCC against the cell lines is effective at a NK cell: target cell ratio of 10:1 and is blocked by antibody to the FcγRIII receptor, as would be expected from our own and others previous results with cell line in vitro studies (Fig. S4D). We have also shown that, as in our previous studies with the anticarcinoembryonic antigen (anti-CEA) antibody PR1A3, cetuximab can also mediate ADCP (Fig. S4E) (14).
Discussion
Profiling KRAS status is now standard before anti-ERBB1 therapy (1). In keeping with clinical findings, our data demonstrate a strong association between KRAS status and nonimmune response to cetuximab and lapatinib in a CRC cell-line panel (3, 9). We have previously shown a strong association between 5FU and replication error status in this panel, thus mirroring what is found clinically (16). These data further support the use of a well-characterized panel of cell lines to determine clinically relevant molecular markers (16–19). The large number of cell lines in our panel explains why we have found an association with KRAS in contrast to other CRC cell line studies (20, 21). We have also demonstrated that the direct effects of cetuximab are largely replicated with lapatinib, lending support to its potential role as an alternative form of therapy.
Unlike KRAS, mutations in BRAF and PIK3CA are less clearly associated with cetuximab response and are not tested routinely before anti-ERBB1 therapy in patients (3, 22). However, our results emphasize the potential value of using the mutation status in multiple genes (KRAS, BRAF, and PIK3CA exon 20) to further increase the accuracy of predicting resistance to anti-ERBB1 therapy (3, 9). Nearly 80% of the lines in the group directly resistant to cetuximab had at least one of these mutations compared with no mutations in the most sensitive group. We also show that in addition to KRAS, BRAF may strongly confer resistance to anti-ERBB1 therapy. BRAF V600E mutation has been associated with poor clinical outcome but has not yet been shown to be strongly predictive of clinical response to cetuximab and FOLFIRI (2). Previous in vitro studies have shown that BRAF V600E mutant cell lines (HT29 and COLO205) are resistant to cetuximab treatment (23, 24). Our data agree with these findings based on an additional eight BRAF V600E mutant cell lines that were mostly resistant to cetuximab. In addition, five of six cell lines with PIK3CA exon 20 mutations were fully cetuximab resistant. However, all but one of these cell lines also had concomitant mutations in KRAS or BRAF, making it difficult to conclude what impact a PIK3CA exon 20 mutation alone has on resistance (20). Our results show that PIK3CA exon 9 mutations on their own do not confer resistance. Two cell lines with these mutations were clearly responsive to cetuximab, an observation that has also been reported in patients (9). The overall data thus clearly support the use of KRAS, BRAF, and PIK3CA exon 20 profiling for predicting responses to anti-ERBB1 therapy (7, 9, 23).
There is a need to identify additional characteristics that predict response in patients because a significant proportion of KRAS/BRAF/PIK3CA WT tumors fail to respond to cetuximab (3). Reports have suggested that AREG and EREG levels may predict cetuximab response (13, 25). We found a suggestive association between EREG levels and cetuximab direct effects and a strong association between AREG and ERBB2 levels and lapatinib response (Fig. S2 C–E). We have identified a list of differentially expressed genes that distinguish subsets of KRAS/BRAF/PIK3CA (triple) WT tumors according to their cetuximab response. This list includes DOCK4, GPR98, and PRKAA2 that were all overexpressed in the resistant cell lines. In particular, DOCK4 and PRKAA2 seem to be of functional significance. DOCK4 is a guanine nucleotide exchange factor that is involved in exchanging GTP for GDP in Rap1 GTPase. This latter protein regulates RAS/RAF and has also been associated with ERK activation independent of RAS (26). PRKAA2 encodes a 5-AMP activated kinase that has been implicated as a sensor of energy status during cellular metabolic stress. These genes do not come up in the list of genes that have been identified as potential predictive “classifiers” from a clinical study looking at KRAS WT patients treated with cetuximab (25). However, this latter study selected a limited number of genes that did not include any of the top candidate genes that were differentially expressed in our study. Once validated, these candidate genes could be used to select those tumors that are likely to respond to cetuximab treatment.
There are conflicting clinical reports regarding the influence of ERBB1 receptor expression in tumor response to anti-ERBB1 antibodies (12, 27, 28). Our results suggest that cetuximab can act via both immune and nonimmune mechanisms but that ERBB1 expression—and not KRAS, BRAF, or PIK3CA WT—strongly correlates with greater immune-mediated killing. ERBB1 expression shows no association with nonimmune growth inhibition. Differing relative contributions of the two killing mechanisms may explain the conflicting results of clinical studies. A number of studies suggest that increased ERBB1 copy number and expression correlate closely with improved responses to cetuximab in patients, implying that immune-mediated mechanisms may perhaps be important (11, 27). However, there has been reported activity of cetuximab in ERBB1− tumors where immune killing would be a negligible component of response (12, 28). It is likely that these tumors express ERBB1 at a level that is undetectable by immunohistochemistry. In addition, clinical response to panitumumab has not been found to be significantly different between tumors with low/negative (1–9%/<1%) or high (>10%) ERBB1 expression (12). Panitumumab is an IgG2 antibody with reduced affinity for FcγRIIIa, and thus has a reduced ability to evoke ADCC compared with IgG1 antibodies, such as cetuximab. The strongest evidence for nonimmune action in patients is the significant association between cetuximab response and KRAS mutation status (3, 9).
Our data show that cetuximab can block EGF binding to its receptor in both sensitive and resistant cell lines, thus reducing ERBB1-phosphorylation, an observation that has been reported previously with anti-ERBB1 antibodies in CRC, head/neck squamous, and nonsmall cell lung cancer cell lines (21, 29). EGF-induced downstream ERK1/2 phosphorylation was incompletely abolished by 20 μg/mL of cetuximab in vitro, a concentration, however, that is likely to be much higher than that present in a tumor in a clinical setting. Ligand-induced activation of ERBB2 and ERBB3 (through dimerization) was incompletely inactivated by cetuximab and this may be a major cause of at least partial resistance to anti-ERBB1 therapy. Only relatively small amounts of administered antibody reach the tumor, and it is therefore unlikely, in general, that the antibody would saturate enough receptors to completely block ligand binding (30). These data suggest that targeting multiple ERBB receptors may be more effective than monotherapy (31).
In keeping with our work on anti-CEA antibodies and with other studies, we have shown that immune mediated cetuximab killing of ERBB1+ CRC cell lines that express moderate to high levels of ERBB1 receptor occurs both by ADCC and ADCP (14, 15).
It has been shown that engineering the hinge region of antibodies can greatly enhance immune-mediated killing (14, 15, 32). The best strategy for overcoming the limitation of direct cetuximab treatment because of the lack of response of tumors that have mutations in any of KRAS, BRAF, or PIK3CA may, therefore, be the development of antibodies to ERBB1, and possibly also to ERBB2 and ERBB3 (that are much more effective in immune-mediated killing), which is not subject to this limitation (33). From our studies of ERBB1 levels in the cell lines, it would be likely that at least 70–80% of colorectal tumors should be susceptible to such treatment.
In conclusion, we propose the use of combination KRAS/BRAF/PIK3CA profiling to define the group of CRC tumors that will respond to direct cetuximab effects (see Fig. S5 for a suggested algorithm for clinical use). We predict that these triple WT tumors will also respond to TKIs such as lapatinib. We have identified several additional biomarkers that further distinguish WT tumors based on their response to anti-ERBB1 therapy. Importantly, immune-mediated antibody responses occur independently of KRAS/BRAF/PIK3CA status. Therefore, high to moderate ERBB1-expressing tumors in patients with favorable FcγR IIa or IIIa genotypes will be particularly susceptible to an immune response (6). Development of antibodies that are engineered to substantially augment ADCC should further increase the magnitude of therapeutic responses.
Materials and Methods
Detailed methods are available in SI Materials and Methods.
Cells were maintained in culture in either DMEM, Iscoves, or RPMI 1640 media with 2 mM l-glutamine, 10% (vol/vol) FCS, 100 U/mL penicillin, and 0.1 mg/mL streptomycin. Cell lines were incubated at 37 °C in a humidified 5–10% CO2 environment. For the ADCC cytotoxicity assays, the cells were suspended in RPMI medium with 1% FCS. Cetuximab (anti-ERBB1; Merck) and trastuzumab (anti-ERBB2; Genentech) were obtained from the Churchill Hospital Pharmacy department, Oxford, UK. Pertuzumab (humanized IgG1 monoclonal antibody that blocks ERBB2 dimerization) was provided by Roche Research Department. Lapatinib (dual ERBB1/ERBB2 receptor TKI) was provided by GlaxoSmithKline Research Department. PCR amplicons were sequenced directly using the appropriate PCR primers and Big Dye Sequencing kit (Applied Biosystems) on an ABI 377 (Applied Biosystems) sequencer. The KRAS (codons 12, 13, 61, 117, and 146), NRAS (codons 12 and 13), BRAF (codon 600), PIK3CA (codons 542, 545, and 1047) genes were analyzed. Gene microarray expression analyses were performed using the Affymetrix Human genome U133+2 chips. Data analysis was carried out using the Partek Genomics Suite software. ERBB1 levels were determined using a β-galactosidase/anti–β-galactosidase ELISA. SDS/PAGE was used to probe for ERBB1, ERBB2, ERBB3, ERK1/2, Β-tubulin and their phosphorylated derivatives. The sulforhodamine B (SRB) assay was used to measure cell growth. NK cells and peripheral blood mononuclear cells (PBMCs) were isolated from fresh whole blood from healthy volunteers who had given their informed consent. The Roche LDH cytotoxicity kit was used to measure the level of cell killing, as per the manufacturer’s instructions.
Cell lines were classified into three categories (sensitive, over 66% growth inhibition; partially sensitive, 33–66% inhibition; or resistant, less than 33% inhibition) depending on their direct response to cetuximab using SRB assays (31). GI50 values were calculated using XLFit (ID Business Solution) in Microsoft Excel. Antibody-dependent (specific) lysis was calculated as (experimental release – antibody independent release)/(maximum release – antibody independent release) × 100. The SEM of triplicate wells was calculated using Prism GraphPad software. Standard normal distribution tests were performed to test the significance of differences found and a P value of less than 0.05 was used as the significance threshold.
Supplementary Material
Acknowledgments
We thank Dr. Philip Conaghan and Dr. Pablo Umana for help with the immune assays and Dr. Djamila Ouaret for help in editing the manuscript. S.Q.A. was funded by an Academy of Medical Sciences Clinical Lecturer Starter grant and support from the Nuffield Department of Surgical Sciences (Oxford University Hospitals National Health Service Trust, Oxford).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218750110/-/DCSupplemental.
References
- 1.Messersmith WA, Ahnen DJ. Targeting EGFR in colorectal cancer. N Engl J Med. 2008;359(17):1834–1836. doi: 10.1056/NEJMe0806778. [DOI] [PubMed] [Google Scholar]
- 2.Van Cutsem E, et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: Updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol. 2011;29(15):2011–2019. doi: 10.1200/JCO.2010.33.5091. [DOI] [PubMed] [Google Scholar]
- 3.De Roock W, et al. KRAS, BRAF, PIK3CA, and PTEN mutations: Implications for targeted therapies in metastatic colorectal cancer. Lancet Oncol. 2011;12(6):594–603. doi: 10.1016/S1470-2045(10)70209-6. [DOI] [PubMed] [Google Scholar]
- 4.Walther A, et al. Genetic prognostic and predictive markers in colorectal cancer. Nat Rev Cancer. 2009;9(7):489–499. doi: 10.1038/nrc2645. [DOI] [PubMed] [Google Scholar]
- 5.Carter PJ. Potent antibody therapeutics by design. Nat Rev Immunol. 2006;6(5):343–357. doi: 10.1038/nri1837. [DOI] [PubMed] [Google Scholar]
- 6.Bibeau F, et al. Impact of FcgammaRIIa-FcgammaRIIIa polymorphisms and KRAS mutations on the clinical outcome of patients with metastatic colorectal cancer treated with cetuximab plus irinotecan. J Clin Oncol. 2009;27(7):1122–1129. doi: 10.1200/JCO.2008.18.0463. [DOI] [PubMed] [Google Scholar]
- 7.Sartore-Bianchi A, et al. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res. 2009;69(5):1851–1857. doi: 10.1158/0008-5472.CAN-08-2466. [DOI] [PubMed] [Google Scholar]
- 8.Negri FV, et al. PTEN status in advanced colorectal cancer treated with cetuximab. Br J Cancer. 2010;102(1):162–164. doi: 10.1038/sj.bjc.6605471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Roock W, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11(8):753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 10.Frank D, et al. A phase II trial of lapatinib and capecitabine for patients with refractory advanced colorectal adenocarcinoma. J Clin Oncol. 2010;28(suppl; abstr):e14092. [Google Scholar]
- 11.Personeni N, et al. Clinical usefulness of EGFR gene copy number as a predictive marker in colorectal cancer patients treated with cetuximab: A fluorescent in situ hybridization study. Clin Cancer Res. 2008;14(18):5869–5876. doi: 10.1158/1078-0432.CCR-08-0449. [DOI] [PubMed] [Google Scholar]
- 12.Hecht JR, et al. Lack of correlation between epidermal growth factor receptor status and response to Panitumumab monotherapy in metastatic colorectal cancer. Clin Cancer Res. 2010;16(7):2205–2213. doi: 10.1158/1078-0432.CCR-09-2017. [DOI] [PubMed] [Google Scholar]
- 13.Jacobs B, et al. Amphiregulin and epiregulin mRNA expression in primary tumors predicts outcome in metastatic colorectal cancer treated with cetuximab. J Clin Oncol. 2009;27(30):5068–5074. doi: 10.1200/JCO.2008.21.3744. [DOI] [PubMed] [Google Scholar]
- 14.Ashraf SQ, et al. Humanised IgG1 antibody variants targeting membrane-bound carcinoembryonic antigen by antibody-dependent cellular cytotoxicity and phagocytosis. Br J Cancer. 2009;101(10):1758–1768. doi: 10.1038/sj.bjc.6605355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lazar GA, et al. Engineered antibody Fc variants with enhanced effector function. Proc Natl Acad Sci USA. 2006;103(11):4005–4010. doi: 10.1073/pnas.0508123103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bracht K, et al. 5-Fluorouracil response in a large panel of colorectal cancer cell lines is associated with mismatch repair deficiency. Br J Cancer. 2010;103(3):340–346. doi: 10.1038/sj.bjc.6605780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma SV, et al. Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nat Rev Cancer. 2010;10(4):241–253. doi: 10.1038/nrc2820. [DOI] [PubMed] [Google Scholar]
- 18.Garnett MJ, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483(7391):570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jhawer M, et al. PIK3CA mutation/PTEN expression status predicts response of colon cancer cells to the epidermal growth factor receptor inhibitor cetuximab. Cancer Res. 2008;68(6):1953–1961. doi: 10.1158/0008-5472.CAN-07-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cunningham MP, et al. Responses of human colorectal tumor cells to treatment with the anti-epidermal growth factor receptor monoclonal antibody ICR62 used alone and in combination with the EGFR tyrosine kinase inhibitor gefitinib. Cancer Res. 2006;66(15):7708–7715. doi: 10.1158/0008-5472.CAN-06-1000. [DOI] [PubMed] [Google Scholar]
- 22.Prenen H, et al. PIK3CA mutations are not a major determinant of resistance to the epidermal growth factor receptor inhibitor cetuximab in metastatic colorectal cancer. Clin Cancer Res. 2009;15(9):3184–3188. doi: 10.1158/1078-0432.CCR-08-2961. [DOI] [PubMed] [Google Scholar]
- 23.Di Nicolantonio F, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26(35):5705–5712. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 24.Prahallad A, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483(7387):100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 25.Baker JB, et al. Tumour gene expression predicts response to cetuximab in patients with KRAS wild-type metastatic colorectal cancer. Br J Cancer. 2011;104(3):488–495. doi: 10.1038/sj.bjc.6606054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Upadhyay G, et al. Molecular association between beta-catenin degradation complex and Rac guanine exchange factor DOCK4 is essential for Wnt/beta-catenin signaling. Oncogene. 2008;27(44):5845–5855. doi: 10.1038/onc.2008.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moroni M, et al. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study. Lancet Oncol. 2005;6(5):279–286. doi: 10.1016/S1470-2045(05)70102-9. [DOI] [PubMed] [Google Scholar]
- 28.Chung KY, et al. Cetuximab shows activity in colorectal cancer patients with tumors that do not express the epidermal growth factor receptor by immunohistochemistry. J Clin Oncol. 2005;23(9):1803–1810. doi: 10.1200/JCO.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 29.Wheeler DL, et al. Mechanisms of acquired resistance to cetuximab: Role of HER (ErbB) family members. Oncogene. 2008;27(28):3944–3956. doi: 10.1038/onc.2008.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Epenetos AA, Snook D, Durbin H, Johnson PM, Taylor-Papadimitriou J. Limitations of radiolabeled monoclonal antibodies for localization of human neoplasms. Cancer Res. 1986;46(6):3183–3191. [PubMed] [Google Scholar]
- 31.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2(2):127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 32.Umaña P, et al. Engineered glycoforms of an antineuroblastoma IgG1 with optimized antibody-dependent cellular cytotoxic activity. Nat Biotechnol. 1999;17(2):176–180. doi: 10.1038/6179. [DOI] [PubMed] [Google Scholar]
- 33.Paz-Ares LG, et al. Phase I pharmacokinetic and pharmacodynamic dose-escalation study of RG7160 (GA201), the first glycoengineered monoclonal antibody against the epidermal growth factor receptor, in patients with advanced solid tumors. J Clin Oncol. 2011;29(28):3783–3790. doi: 10.1200/JCO.2011.34.8888. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.