Abstract
Escherichia coli infections, a leading cause of septic shock, remain a major threat to human health because of the fatal action to endotoxin (LPS). Therapeutic attempts to neutralize endotoxin currently focus on inhibiting the interaction of the toxic component lipid A with myeloid differentiating factor 2, which forms a trimeric complex together with Toll-like receptor 4 to induce immune cell activation. The 1.73-Å resolution structure of the unique endotoxin-neutralizing protective antibody WN1 222-5 in complex with the core region shows that it recognizes LPS of all E. coli serovars in a manner similar to Toll-like receptor 4, revealing that protection can be achieved by targeting the inner core of LPS and that recognition of lipid A is not required. Such interference with Toll-like receptor complex formation opens new paths for antibody sepsis therapy independent of lipid A antagonists.
LPS from Gram-negative bacteria is the major etiological agent of septic shock, which is a serious and often fatal dysregulation of the innate immune response that affects 750,000 people in the United States annually (1). Infection with Escherichia coli, together with Klebsiella, Neisseria, and Pseudomonas, are the most frequent isolates in septic shock (2). A key event initiating the shock cascade is the induction of the innate immune response by the complex formation of a symmetric “m”-shaped multimer composed of two copies of Toll-like receptor 4 (TLR4), myeloid differentiating factor 2 (MD-2), and LPS (3, 4). In a landmark publication, the structure of TLR4-MD-2 bound to LPS (3) was recently described.
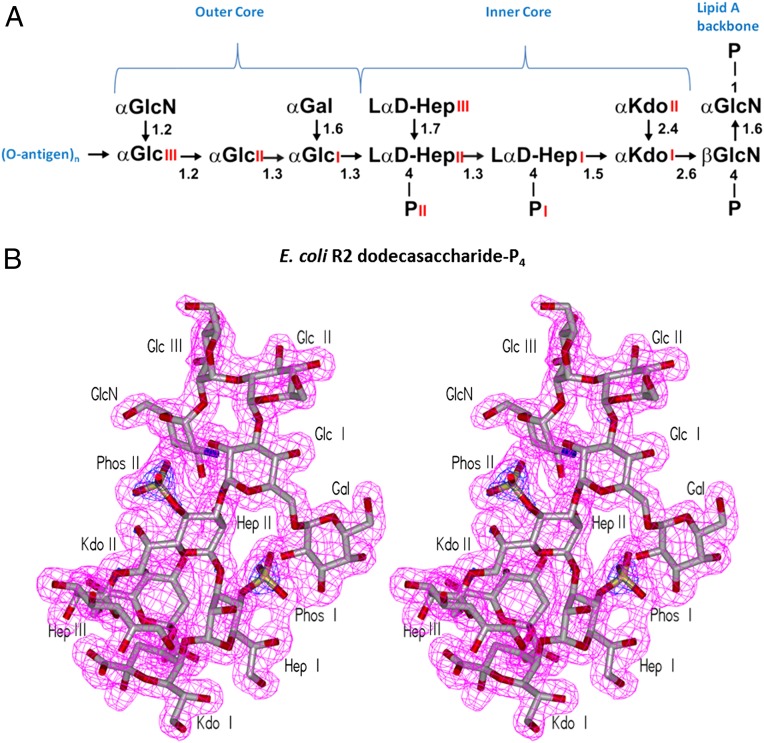
LPS is composed of an acylated glucosamine phosphate disaccharide (i.e., lipid A), which is the endotoxic principle of LPS, a core oligosaccharide (core-OS) and a distal O-polysaccharide (O-PS) often composed of repeating units (Fig. 1A). Whereas the O-PS is structurally heterogeneous, with more than 180 reported E. coli serotypes (5), the core region is composed of a more conserved structure commonly divided into the inner Kdo-heptose and outer hexose regions (6).
Fig. 1.
Structures of LPS and the shape of the combining site. (A) Structure of E. coli R2 dodecasaccharide-P4, representing the core and lipid A of the LPS from Enterobacteria commonly associated with septic shock. (B) Stereo views of electron density corresponding to 10 sugar residues of the core antigen (the lipid A moiety is disordered) contoured at 1.0 σ.
Recognition of LPS leads to a paramount immunological defense reaction caused by the activation of a complex network of immunological mediators. Attempts to control the clinical development of sepsis by neutralizing the most important proinflammatory mediators have failed, including the recent withdrawal of recombinant activated protein C (Xigris). A promising antagonistic lipid candidate called Eritoran (E5564; Eisai) (7) also recently failed in clinical trials, and alternative treatments are urgently needed. The discovery of TLR4 as the principal receptor for endotoxins (8) has stimulated the development of drugs aiming at its down-regulation (9) through interference of LPS–TLR4–MD-2 complex formation (4, 10–12).
Antisera specific for O-PS have been shown to protect against LPS lethality (13); however, the diversity of enterobacterial O-PS together with the rapid onset of septic shock have hindered their introduction into clinical practice (11).
The hypothesis that mAbs specific to the conserved inner core region or lipid A would be protective against a wide range of serovars and even different species was put forward (14) after the discovery of structural similarities within their respective LPSs. WN1 222-5 is the only neutralizing antibody reported to date that displays specificity for an epitope within the structurally conserved region of LPS from a large number of pathogenic E. coli, Salmonella, Shigella, and Citrobacter serovars (15). Further, WN1 222-5 has been shown to inhibit the recognition and uptake of LPS by cells expressing coreceptor mCD14, likely by hindering the transfer of LPS to TLR4–MD-2 (16).
WN1 222-5 has been shown to inhibit the inflammatory cascade in in vivo studies of septic shock, in which it prevents the pyrogenic response in rabbits, inhibits the Limulus amoebocyte lysate assay, and inhibits LPS-induced monokine secretion (15–17).
The difficulties in growing crystals of antibodies in complex with carbohydrate antigens has led to relatively few reported structures (18–21), leading, for example, to increased use of structure prediction tools such as molecular dynamics modeling (22). Thus, in contrast to their great immunological significance during infectious disease, still relatively little is known about carbohydrate recognition by antibodies at the structural level. Whereas cavity- or groove-shaped antibody-combining sites have been observed in most cases, a unique mechanism of binding has been observed for the HIV-1 neutralizing antibody 2G12, binding clusters of carbohydrates from the silent face of gp120 by using “domain swapping” (19, 23, 24).
The structural analysis of antibodies Se155-4 and S20-4 against O-PS of Salmonella enterica and Vibrio cholerae, respectively, have revealed structural insights into the high specificity for a particular serotype (20, 25). However, because of their specificity, antibodies against O-PS are of limited use for the treatment of infectious disease. Nevertheless, structures of antibodies in complex with large carbohydrate antigens have revealed critical insights for vaccine development. The protective antibody F22-4 in complex with an 11-sugar segment from the O-PS of Shigella flexneri serotype 2a (26) allowed the design of new immunogens.
Most attempts in obtaining antibodies that are broadly reactive with a wide variety of LPSs from different Gram-negative bacteria have failed, and epitopes within the deeper core region of LPS have been regarded as not accessible to antibodies in WT LPSs of infectious bacteria. To provide detailed insight on a unique cross-reactive and neutralizing ability, the Fab from WN1 222-5 in complex with a complete core-OS of LPS from E. coli has been crystallized and its structure determined to 1.73-Å resolution.
Results
WN1 222-5 Antigen.
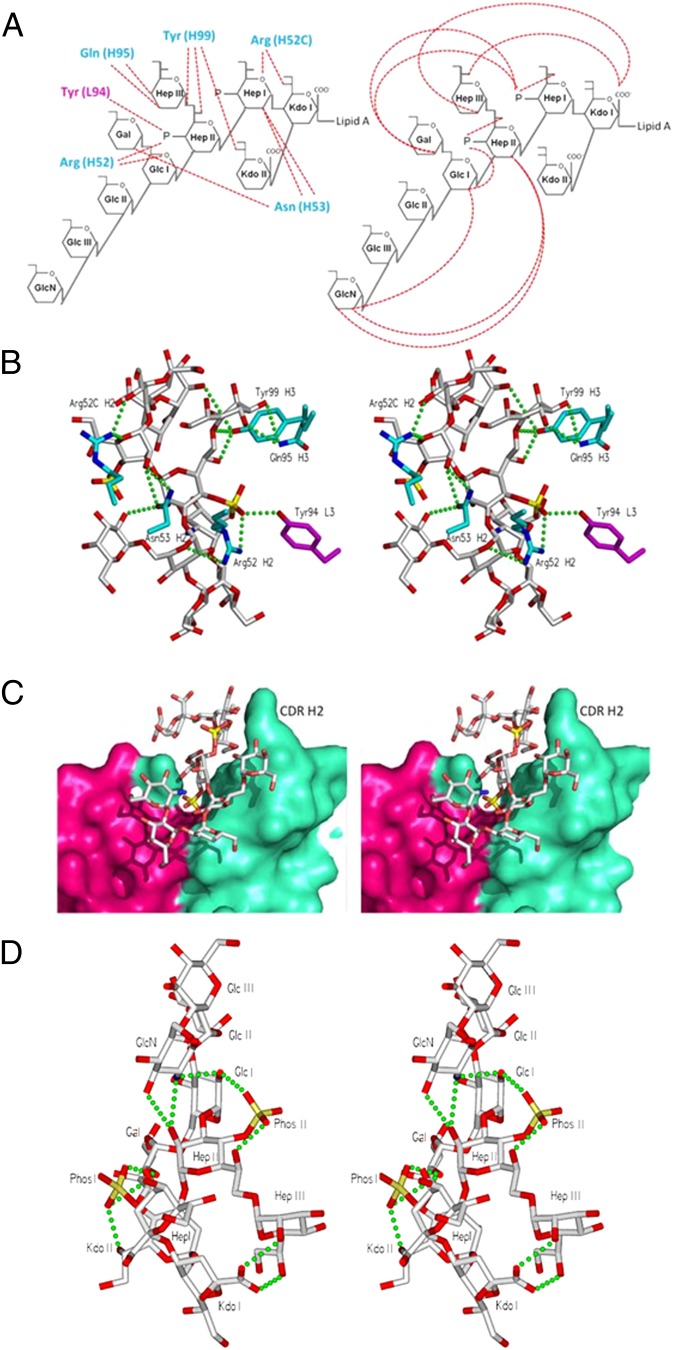
The dodecasaccharide of E. coli R2 LPS has the highest observed affinity of all ligands tested (27) (Fig. S1) and was therefore cocrystallized with WN1 222-5. Seven sugar residues from the ligand form the epitope, including the Hep and Kdo residues of the conserved inner core and the adjacent Glc and branched Gal of the outer core (Fig. 1A). Excellent electron density corresponding to the 10 core sugar residues of the entire LPS core from E. coli was observed in the combining site of the liganded structure (Fig. 1B); however, only diffuse electron density is seen in the area corresponding to the location of the lipid A glucosamine-phosphate backbone. There are 13 hydrogen bonds observed between antibody and antigen, with a strong bias toward the heavy chain (12 hydrogen bonds covering 478 Å2 of buried surface area for the heavy chain vs. a single hydrogen bond covering 47 Å2 for the light chain; Fig. 2). Seven hydrogen bonds stem from complementarity determining region (CDR) H2, whereas five stem from CDR H3.
Fig. 2.
Intermolecular and intramolecular interactions. (A) Schematic diagrams of the antigen show intermolecular (Left) and intramolecular (Right) hydrogen bonds. Stereo diagrams of (B) intermolecular hydrogen bonding; (C) surface of the antibody combining site with the bound 10 sugar core observed, showing predominance of heavy chain (cyan) over light chain (pink) in antigen recognition; and (D) intramolecular hydrogen bonds. There are 10 intramolecular and 13 intermolecular hydrogen bonds, which propagate from the same highly conserved inner core region. Green dots represent intra/intermolecular hydrogen bonds. Ligand shown in gray. Heavy-chain residues are shown in blue, light-chain residues in pink.
The observation that the lipid A backbone was disordered in the crystal structure and formed no part of the epitope to WN1 222-5 was intriguing, as previous studies had indicated that the lipid A backbone and/or Kdo II moieties were critical to binding (27). The results were confirmed by a detailed investigation of binding (Fig. S1 and Table S1).
Ten intramolecular hydrogen bonds exist throughout the ligand. Significantly, all but one of the 10 bonds involve the same phosphorylated heptose trisaccharide region (27) that forms critical parts of the epitope (Fig. 2D).
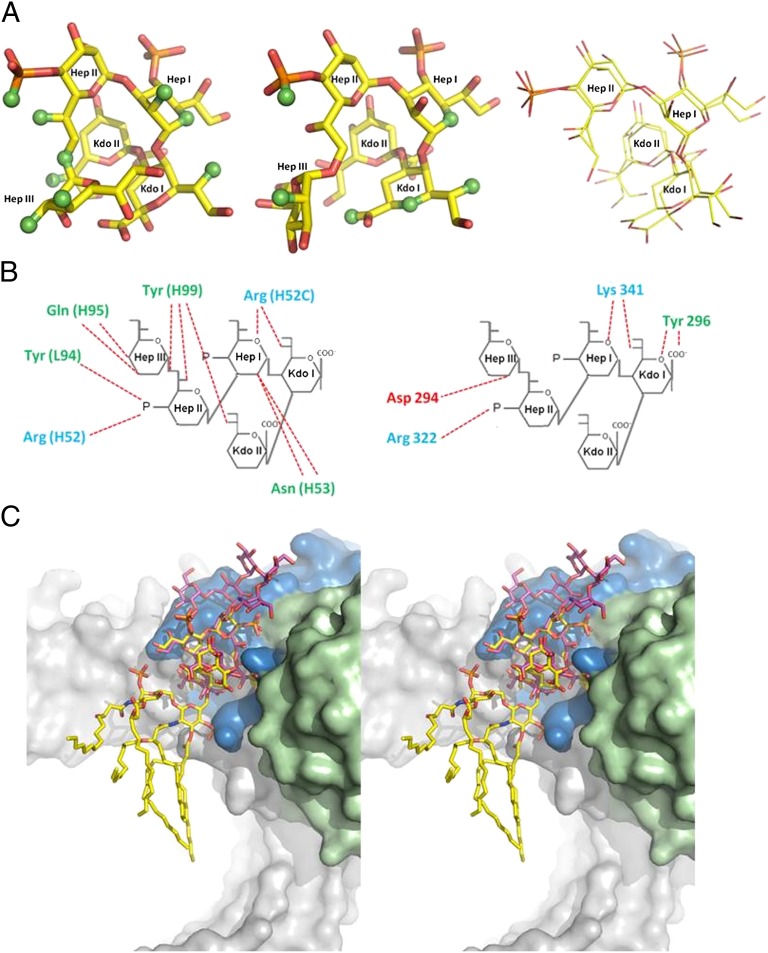
The five inner core sugars form a compact structure centered about Kdo II, whereas the remaining five outer core sugars take on a relatively open structure. Superposition of the inner core with the only other available structure of E. coli LPS inner core [from the LPS–MD-2–TLR4 structure (3); Protein Data Bank (PDB) ID code 3FXI] reveals that the sugars exhibit the same general conformation (Fig. 3A).
Fig. 3.
Complementarity of WN1 222-5 and TLR4 binding of LPS. (A) Inner core sugars from E. coli LPS observed bound to WN1 222-5 combining site (Left) and reported bound to TLR4 (Center) (3), with atoms binding respectively to the antibody and to the receptor in green. Superposition of four of the five inner core sugars (Right) gives an rmsd of 0.6 Å. WN1 222-5 and TLR4 bind the inner core in a similar fashion, where binding occurs along both sides of the ligand. (B) Examination of each ligand’s charge profiles reveals that three critical interactions occur from positively charged residues in each case. Positively charged residues are indicated in blue, neutral in green, negative in red. (C) Stereo diagram shows overlap of TLR4-LPS and WN1 222-5-LPS structures (after superimposition of core LPS). TLR4 is shown in gray, WN1 222-5 in blue (heavy chain) and green (light chain), LPS crystallized with TLR4 (3) in yellow, and LPS crystallized with WN1 222-5 in pink.
Antibody–Antigen Interactions.
The Fab fragment of mAb WN1 222-5 was crystallized unliganded and in complex with the complete core and lipid A backbone structures of LPS from E. coli F576 (Fig. 1A) to 2.13-Å and 1.73-Å resolution, respectively. The rmsd of α-carbon atoms after structural superposition of the variable regions of the liganded and unliganded structures was 0.51 Å, with the largest observed movement of atoms in CDR H2 of 2.31 Å, which is consistent with the antibody’s high affinity (Kd of 32 nM determined by surface plasmon resonance) (27).
Germ-Line Gene Analysis.
The nucleotide sequence of mAb WN1 222-5 (28) was compared with known murine antibody germ-line genes (29). The V and J regions of the heavy chain had 94.9% and 87.0% identity to IGHV7-3*04 and IGHJ4*01, respectively. Two of the mutations from germ line code for residues directly involved in antigen binding (A52cR in H2 and D95Q in H3), and both form two hydrogen bonds to the antigen. There are 18 point mutations in the heavy chain, all but two of which are single point mutations, with one double point mutation. Fifteen of these mutations resulted in amino acid replacements and only three were silent. Four of the replacements were in CDR regions.
The light chain V gene has 95.3% identity to IGKV15-103*01, and the J gene has 94.4% identity to IGKJK1*01. Thirteen single point mutations in the light chain resulted in 7 aa replacements. Three of the somatic mutations occur in the CDR regions—H24R and V31I in L1 and Q89L in L3—but have no apparent influence on antigen binding.
Discussion
WN1 222-5 Does Not Contact Lipid A.
Although lipid A alone is sufficient for the harmful biological activities of LPS, it does not form part of the cognate epitope for this protective antibody. Indeed, well-defined electron density can be seen for every sugar residue on the antigen in the complex except the lipid A backbone disaccharide (Fig. 1B). The inner core is positioned in the combining site such that the lipid A cannot form extensive contact, demonstrating that lipid A binding is not necessarily required for an antibody to be protective against LPS.
Recognition of Conserved Inner Core.
MAb WN1 222-5 is unique in its ability to bind and neutralize LPS of a large number of pathogenic E. coli, Salmonella, Shigella, and Citrobacter serovars involved in septic shock (15, 27). Remarkably, WN1 222-5 binds its LPS core epitope even in the presence of O-PS. By using whole LPS and a number of neoglycoconjugates containing core-OS from all E. coli core types, S. enterica, and the mutant strain E. coli J-5 in ELISA binding studies, we previously identified parts of an inner core epitope accessible to high-affinity binding of mAb WN1 222-5 (27), with an affinity orders of magnitude higher than that determined for mAb Se155-4 against Salmonella group B O-PS and most other carbohydrate binding proteins (30).
The structure is consistent with the ELISA binding studies, which show that features common to all WN1 222-5 antigens include the conserved Glc I, Hep II, Hep I region, and either the 4-phosphate on Hep II or the side chain Hep III. The minimum structure binding with high affinity was octasaccharide-P4 (Fig. S1A), which contained a diphosphorylated heptose trisaccharide, a 2-4 linked Kdo disaccharide, a nonreducing end Glc, and the phosphorylated GlcN lipid A backbone (27). The WN1 222-5 epitope consists of seven sugars composed of the Hep and Kdo residues of the highly conserved inner core and two adjacent sugars Glc and Gal from the outer core.
There is no direct contribution to binding from the terminal three outer core sugars. The first Glc and the branching Gal residues of the outer core bind via Arg H52 and Asn H53, which explains the enhanced affinity observed for oligosaccharides containing an outer core and a branching substitution at position 6 of the first hexose (30). These residues are bound independently of their stereochemistry of ring hydroxyls, which opens the possibility for WN1 222-5 recognition of LPS with other outer core types. Oligosaccharides that contain GlcN at Hep III or do not contain phosphate in position 4 at Hep II are not bound by WN1 222-5. The phosphate is a key residue that is involved in intermolecular binding, and it is also required for stabilizing the bound ligand conformation. A substituent at the side-chain Hep III will prevent interaction for steric reasons.
WN1 222-5 Binds LPS in a Manner Similar to TLR4.
A prerequisite for the induction of septic shock is the specific recognition of LPS and lipid A by the TLR4–MD-2 complex. Many factors are known to affect the initiation and severity of the inflammatory response, including the nature of the inner core and the stereochemical identity of the acylated lipid A moiety, but the precise molecular interactions required to provoke the cascade are not fully understood (31).
To establish an infection, Enterobacteria require the synthesis of a complete LPS containing core-OS and O-PS, which have been shown in clinical studies to prevent the binding of antibodies specific for lipid A or Kdo (32). Significantly, WN1 222-5 recognizes the LPS core types released from most infectious bacteria regardless of the presence of O-PS.
The structure of TLR4–MD-2 reported in complex with E. coli LPS core (PDB ID code 3FXI) (3) contains the same inner core LPS fragment cocrystallized in the present study with antibody WN1 222-5. Superposition of the LPS fragments in the two structures shows that the five inner core sugar residues exhibit the same general conformation (Fig. 3A). Significantly, the paratope of mAb WN1 222-5 mimics the TLR4 inner core receptor site (Fig. 3A), which lies at the foundation of the antibody’s ability to cross-react with and so protect against those variations of LPS from pathogenic species that bind to TLR4. WN1 222-5 shares with the TLR4 receptor site a similar shape and general charge profile with three key hydrogen bond contacts to Kdo I, Hep I, and the phosphate moiety of Hep II of the highly conserved LPS inner core (Fig. 3 B and C).
The lack of contact observed in the crystal structure of WN1 222-5 with lipid A also mimics the binding in the LPS–MD-2–TLR4 structure, in which lipid A recognition takes place almost exclusively through contact of the acyl chains with MD-2 (3).
Kdo II Serves to Stabilize Conformation of Antigen.
The simultaneous removal of the lipid A backbone and Kdo II had been shown to reduce affinity dramatically (by three orders of magnitude) (27). The absence of lipid A from the epitope in the crystal structure underlines the importance of Kdo II, which was subsequently confirmed by detailed binding studies (Fig. S1 and Table S1).
The antigen is forced to adopt a limited number of conformations by the relatively large number of intramolecular hydrogen bonds and steric crowding within the core region (30, 33) that are seen in the antibody and in the TLR4–MD-2 complexes (3). Kdo II forms only a weak hydrogen bond to Tyr H99 (which also forms two other hydrogen bonds to the antigen), so its importance to antigen recognition probably comes as a steric impediment to conformational rearrangement of the inner core.
Significantly, all but one of the 10 intramolecular hydrogen bonds in the inner core involve the same phosphorylated heptose trisaccharide region (27) that forms the critical part of the epitope (Fig. 2D). This determination represents the structural characterization of the entire core region of nontruncated LPS, and shows that the structure is similar to those predicted through energy minimization (34), where the conserved region is almost globular in shape, and the outer core region bends relative to the inner core domain.
Heavy-Chain Dominance of Antigen Binding Aids Cross-Reactivity.
Previously reported structures [e.g., mAbs Se155-4 (25), S20-4 (20), and S25-2 (35)] show that most antibody–carbohydrate complexes are dominated by the interaction of a single carbohydrate residue buried in a pocket formed at the interface of the light- and heavy-chain CDRs, with each chain contributing ∼150 Å2 of surface contact area.
By contrast, WN1 222-5’s carbohydrate recognition site is dominated by the heavy chain and characterized by an open groove and protruding CDR H2, with a large proportion of the oligosaccharide exposed to bulk solvent. The heavy chain contributes 478 Å2 of buried surface area, vs. 47 Å2 for the light chain (Fig. 2), which is reflected in the observed intermolecular hydrogen bonds, of which 12 of the 13 involve five heavy-chain residues with only one to a single residue on the light chain. CDRs H2 and H3 contribute seven and five hydrogen bonds, respectively, to the antigen.
CDR H3 has long been recognized as having a strong role in the determination of specificity (36, 37), and this is true in WN1 222-5 through specific recognition of the conserved residues Hep III and Kdo II (Fig. 2 A and B) (27, 38).
The dominance of the heavy chain in the paratope is seen in a few other antibodies, such as mAb F22-4 against the Shigella serotype 2a O-PS (26) and the anti-Ley mAb BR96 (21), but not to the extreme shown by WN1 222-5. This reliance on the heavy chain for antigen recognition aids the antibody’s observed cross-reactivity, as the composition and orientation of the light-chain CDRs (Fig. 2C) allows space for variations in sequence and conformation in outer core residues of LPS.
WN1 222-5 Generation.
Despite knowledge of the minimal epitope from binding studies, immunization of mice and rabbits with neoglycoconjugates containing the minimal epitope did not lead to the formation of cross-reactive mAb WN1 222-5–type antibodies, reflecting the difficulties generally encountered in obtaining such antibodies (27, 39). This could be a result of the immunodominance of different sugars in fragments of the complete core, or by the lack of the conformational epitope presented by the complete core. The latter is consistent with the failure to raise such an antibody by using the more labile inner core of Rc LPS from E. coli J-5 (Fig. S1), in which the relatively weak binding of WN1 222-5 to Rc LPS from E. coli J-5 could be explained by a significant entropic penalty.
Further insight can be gained by noting the degree of affinity maturation from the germ-line sequence. WN1 222-5 has 12 aa mutations from germ line in the heavy chain alone. It has been demonstrated that high binding affinity can be correlated to a high frequency of somatic mutations (40). There are 18 point mutations in the germ-line genes themselves, all but three of which led to amino acid replacements. This not only is a high number of point mutations, but a high ratio of amino acid replacement to silent mutations in comparison with other antibodies (41). Significantly, a number of amino acid mutations affect residues that are not directly involved in binding, but lie adjacent to those residues that recognize the antigen. For example, residues in CDR H2 (which is involved in a minor induced fit) have mutations Y55D, T57A, N73Y, Q75R, and Y79H. Changes in residue identity of as far as 15 Å from the binding site have been shown to affect antigen binding strongly (42).
Conclusions.
The present report provides the structure of an antibody in complex with the entire core of LPS from a pathogenic strain of E. coli, which shares a common inner core region with Shigella and Salmonella. The specific recognition of the inner core, and of lipid A in particular, from pathogenic species by the TLR4–MD-2 complex is a prerequisite for the induction of septic shock; however, WN1 222-5 achieves its protective ability by targeting just the inner core and not the lipid A, and therefore can neutralize membrane-anchored or free LPS by blocking any further interactions of lipid A in the inflammatory cascade. Significantly, WN1 222-5 binds LPS in a manner that mimics many aspects of binding by TLR4, including a shared stereochemical charge profile and three key interactions, which allows the antibody to select some of the LPS species that pose a risk of inflammation. Targeting the conserved inner core of specific species in this manner overcomes the tremendous diversity of the O-PS that has hindered antibody therapy, and spotlights a promising avenue for the generation of novel antiendotoxic antibody drugs. The paratope of WN1 222-5 created by the heavy-chain variable region of the antibody may serve as a template structure for the generation of single-domain antibodies with endotoxin-neutralizing activity.
Materials and Methods
Production and Purification of WN1 222-5 IgG and Fab Fragments.
The monoclonal antibody WN1 222-5 (IgG2a) was obtained as described previously in detail (15, 43).
The IgG was purified on a protein A column and the Fab fragment prepared by digestion of the intact IgG with papain. IgG was dialyzed into 20 mM Hepes (Sigma-Aldrich), pH 7.5, diluted to a concentration of 0.5 mg/mL, and 2 mM EDTA (Sigma-Aldrich) and 5 mM DTT (Sigma-Aldrich) were added. The digestion reaction was carried out at room temperature by using a papain (Sigma-Aldrich)-to-IgG ratio of 1:200 for 4 h. The reaction was quenched by the addition of 10 mM iodoacetamide (Sigma-Aldrich) and dialyzed overnight into 20 mM Hepes, pH 7.5. The Fab fragment was purified by cation-exchange chromatography on a CM-825 column (Shodex) by using a linear gradient of 0 to 0.5 M NaCl in 20 mM Hepes, pH 7.5.
ELISA and ELISA Inhibition Experiments.
The binding of mAb WN1 222-5 to the dodecasaccharide-P4 was studied by ELISA and by ELISA inhibition using the immobilized neoglycoconjugate (Fig. S1). For competitive inhibition of binding, LPS of E. coli F576 (R2 core type), F653 (R3 core type), and E. coli J-5 were deacylated by treatment with alkali or mild acid as described earlier (27). Mixtures of purified OS were used as inhibitors and tested in comparison with the purified E. coli R2 core-OS over a concentration range between 240 pM and 500 µM. MaxiSorp microtiter plates (96-well, U-bottom; Nunc) were coated with the neoglycoconjugate (2 pmol ligand per well) in 50 mM carbonate buffer, pH 9.2, overnight at 4 °C, and washed twice in PBS solution (Sigma-Aldrich; all PBS buffers were supplemented with 0.01% thimerosal; Merck) and then blocked with PBS solution supplemented with 2.5% (wt/vol) casein (PBS-C; Sigma-Aldrich) for 1 h at 37 °C on a rocker platform followed by two washings in PBS solution. Serial twofold dilutions of inhibitor in PBS-C supplemented with 5% (wt/vol) BSA were mixed in V-shaped microtiter plates (96-well; Nunc) with an equal volume (30 µL) of mAb WN1 222-5 diluted in the same buffer to give an OD405 of ∼1.5 in ELISA in the absence of inhibitor (mAb concentration of 25 ng/mL, or 170 pM). After incubation for 15 min at 37 °C, 50 µL of the mixtures was transferred to the coated ELISA plates, and incubation was continued for 1 h at 37 °C. After three washings in PBS solution, HRP-conjugated goat anti-mouse IgG(H+L) (Dianova) was added [1:500 diluted in PBS-C supplemented with 5% (wt/vol) BSA], and, after 1 h at 37 °C and three further washings in PBS solution, 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (diammonium salt; Sigma) substrate was added. The substrate solution was freshly prepared by dissolving 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (1 mg/mL final concentration) in 100 mM sodium citrate (Merck) buffer adjusted to pH 4.5 with 2 M citric acid (Merck) and adding hydrogen peroxide (0.0025% final concentration). The reaction was stopped after 30 min incubation at 37 °C by the addition of 2% (wt/vol) aqueous oxalic acid, and absorbance was read at 405 nm. The data were analyzed in Origin 6.0 (OriginLab) by fitting the means of quadruplicate measurements to the built-in logistic function.
Crystallization of WN1 222-5 Fab.
Purified WN1 222-5 was exchanged into 20 mM Hepes, pH 7.5, and concentrated to 12 mg/mL. The Fab was mixed with dodecasaccharide-P4 (5 mM), the major oligosaccharide of the E. coli R2 core type, which had the highest affinity (Kd, 32 nM) observed for all LPS structures tested (27), and equilibrated overnight before the complex was screened by using Crystal Screen I and II (Hampton Research). Crystals (0.5 × 0.5 × 0.1 mm) appeared in Hampton Screen I under condition 47 (0.1 M sodium acetate trihydrate, pH 4.6, 2.0 M ammonium sulfate). Larger crystals (1.0 × 1.0 × 0.2 mm) appeared under similar conditions (0.1 M sodium acetate trihydrate, pH 6.5, 2.0 M ammonium sulfate) using 3-μL drops at 16 °C, with an approximate 250 fold excess of dodecasaccharide to protein. Table S2 provides crystallographic data.
Data Collection and Structure Determination and Refinement.
Crystals were flash-frozen to −160 °C by using a Cryostream 700 crystal cooler (Oxford Cryosystems) using mother liquor supplemented with 25% (vol/vol) (±)-2-methyl-2,4-pentanediol (Sigma-Aldrich) as a cryoprotectant. For the unliganded structure, data were collected on an R-AXIS 4++ area detector (Rigaku) coupled to a MM-002 X-ray generator (CuKα radiation) with Osmic “blue” optics (Rigaku) and processed by using Crystal Clear/d*trek (Rigaku). For the liganded structure, data were collected at the Canadian Macromolecular Crystallography Facility on beamline 08ID-1 (CMCF-ID) of the Canadian Light Source (Saskatoon, SK, Canada) at 0.92-Å wavelength. The structure of WN1 222-5 in complex with its ligand was solved by molecular replacement with Phaser implemented in CCP4 (44) by using the variable domain of the homologous IgG2a antibody D2.3 (PDB ID code 1YEF) as a search model. Manual fitting of σA-weighted Fo-Fc and 2Fo-Fc electron density maps was carried out with Coot (45) and the program SetoRibbon (available on request from S.V.E.). Restrained refinements and translation/libration/screw refinements were carried out with PHENIX (46). Final model and refinement statistics are given in Table S2.
Germ-Line Gene Analysis.
Murine BALB/c germ-line gene segments were compared by using the International ImMunoGeneTics Information System (Marie-Paule Lefranc, Montpellier, France; www.imgt.org) (29, 47).
Supplementary Material
Acknowledgments
We thank U. Agge, C. Schneider, and V. Susott for technical assistance. This work was supported in part by grants from the Natural Sciences and Engineering Research Council of Canada (to S.V.E.) and the Michael Smith Foundation for Health Research (to S.V.E.), by Deutsche Forschungsgemeinschaft Grant SFB 470/C1 (to S.M.-L. and H.B.), and by Austrian Science Fund (Fonds zur Förderung der wissenschaftlichen Forschung) Grant P22909 (to P.K.). Research described in this work was performed at the Canadian Light Source, which is supported by the Natural Sciences and Engineering Research Council of Canada, National Research Council Canada, the Province of Saskatchewan, Western Economic Diversification Canada, and the University of Saskatchewan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 3V0V (WN1 222-5 unliganded structure) and 3V0W (WN1 222-5 in complex with LPS)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1209253109/-/DCSupplemental.
References
- 1.Li J, Carr B, Goyal M, Gaieski DF. Sepsis: The inflammatory foundation of pathophysiology and therapy. Hosp Pract (Minneap) 2011;39(3):99–112. doi: 10.3810/hp.2011.08.585. [DOI] [PubMed] [Google Scholar]
- 2.Munford RS. Severe sepsis and septic shock: The role of Gram-negative bacteremia. Annu Rev Pathol. 2006;1:467–496. doi: 10.1146/annurev.pathol.1.110304.100200. [DOI] [PubMed] [Google Scholar]
- 3.Park BS, et al. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458(7242):1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 4.Kim HM, et al. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist Eritoran. Cell. 2007;130(5):906–917. doi: 10.1016/j.cell.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Stenutz R, Weintraub A, Widmalm G. The structures of Escherichia coli O-polysaccharide antigens. FEMS Microbiol Rev. 2006;30(3):382–403. doi: 10.1111/j.1574-6976.2006.00016.x. [DOI] [PubMed] [Google Scholar]
- 6.Holst O, Müller-Loennies S. Microbial polysaccharide structures. In: Kamerling JP, editor. Comprehensive Glycoscience. Oxford: Elsevier; 2007. pp. 123–179. [Google Scholar]
- 7.Tidswell M, et al. Eritoran Sepsis Study Group Phase 2 trial of eritoran tetrasodium (E5564), a toll-like receptor 4 antagonist, in patients with severe sepsis. Crit Care Med. 2010;38(1):72–83. doi: 10.1097/CCM.0b013e3181b07b78. [DOI] [PubMed] [Google Scholar]
- 8.Poltorak A, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282(5396):2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 9.Wittebole X, Castanares-Zapatero D, Laterre PF. Toll-like receptor 4 modulation as a strategy to treat sepsis. Mediators Inflamm. 2010;2010:568396. doi: 10.1155/2010/568396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leon CG, Tory R, Jia J, Sivak O, Wasan KM. Discovery and development of toll-like receptor 4 (TLR4) antagonists: A new paradigm for treating sepsis and other diseases. Pharm Res. 2008;25(8):1751–1761. doi: 10.1007/s11095-008-9571-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ianaro A, Tersigni M, D’Acquisto F. New insight in LPS antagonist. Mini Rev Med Chem. 2009;9(3):306–317. doi: 10.2174/1389557510909030306. [DOI] [PubMed] [Google Scholar]
- 12.Tidswell M, LaRosa SP. Toll-like receptor-4 antagonist eritoran tetrasodium for severe sepsis. Expert Rev Anti Infect Ther. 2011;9(5):507–520. doi: 10.1586/eri.11.27. [DOI] [PubMed] [Google Scholar]
- 13.Tate WJ, 3rd, Douglas H, Braude AI, Wells WW. Protection against lethality of E. coli endotoxin with “O” antiserum. Ann N Y Acad Sci. 1966;133(2):746–762. doi: 10.1111/j.1749-6632.1966.tb52403.x. [DOI] [PubMed] [Google Scholar]
- 14.Chedid L, Parant M, Parant F, Boyer F. A proposed mechanism for natural immunity to enterobacterial pathogens. J Immunol. 1968;100(2):292–306. [PubMed] [Google Scholar]
- 15.Di Padova FE, et al. A broadly cross-protective monoclonal antibody binding to Escherichia coli and Salmonella lipopolysaccharides. Infect Immun. 1993;61(9):3863–3872. doi: 10.1128/iai.61.9.3863-3872.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pollack M, et al. Dual effects of LPS antibodies on cellular uptake of LPS and LPS-induced proinflammatory functions. J Immunol. 1997;159(7):3519–3530. [PubMed] [Google Scholar]
- 17.Di Padova F, et al. New anticore LPS monoclonal antibodies with clinical potential. In: Levin J, Alving RS, Munford RS, Stutz PL, editors. Bacterial Endotoxin: Recognition and Effector Mechanisms. Vol 2. Amsterdam: Elsevier; 1993. pp. 325–335. [Google Scholar]
- 18.Cygler M, Wu S, Zdanov A, Bundle DR, Rose DR. Recognition of a carbohydrate antigenic determinant of Salmonella by an antibody. Biochem Soc Trans. 1993;21(2):437–441. doi: 10.1042/bst0210437. [DOI] [PubMed] [Google Scholar]
- 19.Calarese DA, et al. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science. 2003;300(5628):2065–2071. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]
- 20.Villeneuve S, et al. Crystal structure of an anti-carbohydrate antibody directed against Vibrio cholerae O1 in complex with antigen: Molecular basis for serotype specificity. Proc Natl Acad Sci USA. 2000;97(15):8433–8438. doi: 10.1073/pnas.060022997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeffrey PD, et al. The x-ray structure of an anti-tumour antibody in complex with antigen. Nat Struct Biol. 1995;2(6):466–471. doi: 10.1038/nsb0695-466. [DOI] [PubMed] [Google Scholar]
- 22.Dyekjaer JD, Woods RJ. Predicting the three-dimensional structures of anti-carbohydrate antibodies: combining comparative modeling and MD simulations. In: Vliegenthar JFG, Woods RJ, editors. NMR Spectroscopy and Computer Modeling of Carbohydrates. Vol 930. Washington, DC: American Chemical Society; 2006. pp. 203–219. [Google Scholar]
- 23.Menendez A, et al. A peptide inhibitor of HIV-1 neutralizing antibody 2G12 is not a structural mimic of the natural carbohydrate epitope on gp120. FASEB J. 2008;22(5):1380–1392. doi: 10.1096/fj.07-8983com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calarese DA, et al. Dissection of the carbohydrate specificity of the broadly neutralizing anti-HIV-1 antibody 2G12. Proc Natl Acad Sci USA. 2005;102(38):13372–13377. doi: 10.1073/pnas.0505763102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cygler M, Rose DR, Bundle DR. Recognition of a cell-surface oligosaccharide of pathogenic Salmonella by an antibody Fab fragment. Science. 1991;253(5018):442–445. doi: 10.1126/science.1713710. [DOI] [PubMed] [Google Scholar]
- 26.Vulliez-Le Normand B, et al. Structures of synthetic O-antigen fragments from serotype 2a Shigella flexneri in complex with a protective monoclonal antibody. Proc Natl Acad Sci USA. 2008;105(29):9976–9981. doi: 10.1073/pnas.0801711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Müller-Loennies S, Brade L, MacKenzie CR, Di Padova FE, Brade H. Identification of a cross-reactive epitope widely present in lipopolysaccharide from enterobacteria and recognized by the cross-protective monoclonal antibody WN1 222-5. J Biol Chem. 2003;278(28):25618–25627. doi: 10.1074/jbc.M302904200. [DOI] [PubMed] [Google Scholar]
- 28.Di Padova F, et al. In: Monoclonal antibodies to endotoxin core as a new approach in endotoxemia therapy Novel Therapeutic Strategies in the Treatment of Sepsis. Morrison DC, Ryan JL, editors. New York: Dekker; 1996. pp. 13–31. [Google Scholar]
- 29.Lefranc MP, et al. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2009;37(database issue):D1006–D1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gourdine JP, et al. High affinity interaction between a bivalve C-type lectin and a biantennary complex-type N-glycan revealed by crystallography and microcalorimetry. J Biol Chem. 2008;283(44):30112–30120. doi: 10.1074/jbc.M804353200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manavalan B, Basith S, Choi S. Similar structures but different roles - An updated perspective on TLR structures. Front Physiol. 2011;2:41. doi: 10.3389/fphys.2011.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aree T, Jacob J, Saenger W, Hoier H. Crystal structure of α-cyclodextrin-acetonitrile-hexahydrate. Carbohydr Res. 1998;307(3-4):191–197. doi: 10.1016/s0008-6215(97)10109-4. [DOI] [PubMed] [Google Scholar]
- 34.Kastowsky M, Gutberlet T, Bradaczek H. Molecular modelling of the three-dimensional structure and conformational flexibility of bacterial lipopolysaccharide. J Bacteriol. 1992;174(14):4798–4806. doi: 10.1128/jb.174.14.4798-4806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brooks CL, et al. Antibodies raised against chlamydial lipopolysaccharide antigens reveal convergence in germline gene usage and differential epitope recognition. Biochemistry. 2010;49(3):570–581. doi: 10.1021/bi9011308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schroeder HW, Jr, Cavacini L. Structure and function of immunoglobulins. J Allergy Clin Immunol. 2010;125(2, suppl 2):S41–S52. doi: 10.1016/j.jaci.2009.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brooks CL, et al. The role of CDR H3 in antibody recognition of a synthetic analog of a lipopolysaccharide antigen. Glycobiology. 2010;20(2):138–147. doi: 10.1093/glycob/cwp150. [DOI] [PubMed] [Google Scholar]
- 38.Müller-Loennies S, Brade L, Brade H. Neutralizing and cross-reactive antibodies against enterobacterial lipopolysaccharide. Int J Med Microbiol. 2007;297(5):321–340. doi: 10.1016/j.ijmm.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 39.Rietschel ET, et al. Bacterial endotoxin: Chemical constitution, biological recognition, host response, and immunological detoxification. Curr Top Microbiol Immunol. 1996;216:39–81. doi: 10.1007/978-3-642-80186-0_3. [DOI] [PubMed] [Google Scholar]
- 40.Weinand RG. Somatic mutation, affinity maturation and the antibody repertoire: A computer model. J Theor Biol. 1990;143(3):343–382. doi: 10.1016/s0022-5193(05)80034-7. [DOI] [PubMed] [Google Scholar]
- 41.Golding GB, Gearhart PJ, Glickman BW. Patterns of somatic mutations in immunoglobulin variable genes. Genetics. 1987;115(1):169–176. doi: 10.1093/genetics/115.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wedemayer GJ, Patten PA, Wang LH, Schultz PG, Stevens RC. Structural insights into the evolution of an antibody combining site. Science. 1997;276(5319):1665–1669. doi: 10.1126/science.276.5319.1665. [DOI] [PubMed] [Google Scholar]
- 43.de StGroth SF, Scheidegger D. Production of monoclonal antibodies: Strategy and tactics. J Immunol Methods. 1980;35(1-2):1–21. doi: 10.1016/0022-1759(80)90146-5. [DOI] [PubMed] [Google Scholar]
- 44.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruiz M, et al. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 2000;28(1):219–221. doi: 10.1093/nar/28.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.