Abstract
Basic leucine zipper (bZip) transcription factors regulate cellular gene expression in response to a variety of extracellular signals and nutrient cues. Although the bZip domain is widely known to play significant roles in DNA binding and dimerization, recent studies point to an additional role for this motif in the recruitment of the transcriptional apparatus. For example, the cAMP response element binding protein (CREB)-regulated transcriptional coactivator (CRTC) family of transcriptional coactivators has been proposed to promote the expression of calcium and cAMP responsive genes, by binding to the CREB bZip in response to extracellular signals. Here we show that the CREB-binding domain (CBD) of CRTC2 folds into a single isolated 28-residue helix that seems to be critical for its interaction with the CREB bZip. The interaction is of micromolar affinity on palindromic and variant half-site cAMP response elements (CREs). The CBD and CREB assemble on the CRE with 2:2:1 stoichiometry, consistent with the presence of one CRTC binding site on each CREB monomer. Indeed, the CBD helix and the solvent-exposed residues in the dimeric CREB bZip coiled-coil form an extended protein–protein interface. Because mutation of relevant bZip residues in this interface disrupts the CRTC interaction without affecting DNA binding, our results illustrate that distinct DNA binding and transactivation functions are encoded within the structural constraints of a canonical bZip domain.
Keywords: transcription regulation, cellular signaling, protein–protein interaction
Originally identified in the CCAAT-enhancer binding protein (C/EBP) family of activators, the basic leucine zipper (bZip) domain is harbored by a large and diverse family of nuclear factors in both plants and higher organisms (1). The cAMP response element binding protein (CREB) subfamily of bZip activators [CREB1, activating transcription factor 1 (ATF1), and cAMP response element modulator (CREM)] stimulates cellular gene transcription after its phosphorylation in response to various extracellular hormonal and nutrient cues (2, 3). Elevations in intracellular cAMP trigger the activation of protein kinase A, which phosphorylates CREB at Ser133 and promotes recruitment of the histone acetyltransferase paralogs CREB binding protein (CBP) and p300, leading to increased target gene expression (4).
The CREB-regulated transcriptional coactivators [CRTCs, also referred to as transducer of regulated CREB activity (TORC)s or mucoepidermoid carcinoma translocated protein (MECT)s] have recently emerged as a second family of CREB cofactors (5, 6). Under basal conditions, CRTCs are highly phosphorylated and maintained in the cytoplasm via interactions with 14-3-3 proteins (7). Increases in cAMP and calcium trigger CRTC dephosphorylation and release from 14-3-3 complexes; CRTCs then translocate to the nucleus, where they mediate CREB-dependent transcription.
The CRTC family comprises three members: CRTC1, CRTC2, and CRTC3. CRTC1 is predominantly expressed in brain, whereas CRTC2 and CRTC3 are abundant in most peripheral tissues (5). All three proteins potentiate CREB-dependent transcription; besides a role for CRTC1 in long-term memory (8, 9), both CRTC1 and CRTC3 seem to be important for energy balance (3). CRTC2 is expressed at high levels in liver, where it promotes glucose homeostasis during fasting by inducing the gluconeogenic program (10). Sequestered in the cytoplasm under feeding conditions through phosphorylation by the Ser/Thr kinase SIK2, CRTC2 is dephosphorylated in response to pancreatic glucagon during fasting, when it translocates to the nucleus and stimulates gluconeogenic gene expression.
Recent studies support a conserved role for CRTCs through evolution. Deletion of TORC, the single CRTC homolog in Drosophila, reduces fat stores and increases sensitivity to starvation (11). CRTCs also seem to be important in aging: RNAi-mediated depletion of CRTC1 in Caenorhabditis elegans extends lifespan, in part through down-regulation of the CREB pathway (12).
Unlike CBP/p300, which bind to the kinase-inducible transactivation domain (KID) of CREB harboring phosphoSer133 (13), CRTCs have been proposed to activate transcription by associating with the C-terminal bZip domain, which also mediates CREB dimerization and DNA binding (5). CRTCs have been found to associate with CREB via a conserved N-terminal CREB binding domain (CBD). Indeed, truncated CRTC1 polypeptides containing this N-terminal region seem to function as dominant negative inhibitors of CREB signaling; conversely, fusion of the CBD to unrelated coactivators such as Mastermind converts them into potent CREB cofactors (5, 14). Here we characterize the interaction between CRTC2 and CREB on DNA using relevant interaction domains. These studies provide a conceptual framework to explain how a classic DNA binding motif also functions as a transactivation domain through its association with a coactivator in response to extracellular signals.
Results
CRTC2 Binds CREB in a DNA-Dependent Manner via an N-Terminal CBD.
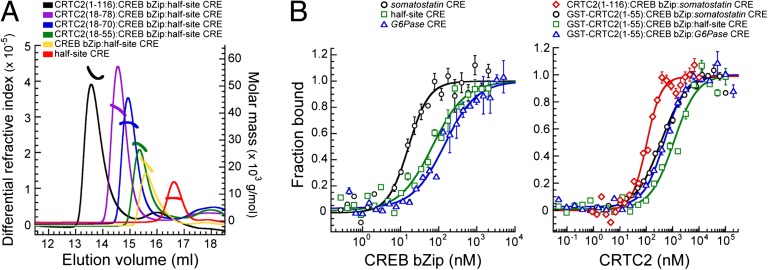
The N-terminal CBD is encoded by a single exon, which is conserved in all CRTC family members. To establish the minimal CBD in CRTC2, we prepared several recombinant CRTC2 polypeptides and evaluated them for CREB bZip binding in the presence of a cAMP response element (CRE). Binding activity was evaluated via high-resolution size exclusion chromatography (SEC) coupled to a multiangle light scattering (MALS) detection system to characterize the molecular weight of each eluting species. SEC-MALS profiles of CRTC2 polypeptides revealed the formation of a long-lived complex, with all three interacting components coeluting together with shorter retention times than that of the constituent apo-proteins/DNA as well as the CREB bZip:CRE binary complex (Fig. 1A). Furthermore, the inferred molecular weight of each coeluting species was consistent with a 2:2:1 stoichiometry for the CRTC2:CREB:CRE complex (henceforth designated the ternary complex; Table S1). A CRTC2 peptide (amino acids 18–55) was sufficient for stable association with CREB in these assays.
Fig. 1.
Assembly of a CRTC2:CREB:CRE ternary complex using relevant interaction domains. (A) SEC-MALS profiles of mixtures of various CRTC2 peptides with a CREB bZip:CRE complex. The profiles of the CREB:CRE complex or just the CRE are shown for comparison. The thick lines within or above each peak correspond to the range of molar masses detected for that species. (B) FA curves from titrations conducted with a fluoresceinated double-stranded oligonucleotide harboring various CREs with a CREB bZip peptide (Left). FA curves from titrations of the respective fluoresceinated CRE-bound CREB bZip conducted with increasing amounts of GST-CRTC2 (residues 1–55) or CRTC2 (1–116).
Transcriptional coactivators are generally thought to lack DNA-binding activity; rather they are recruited to the promoter via an association with their transcription factor cognates. Consistent with this notion, we observed no direct interaction between CRTC2 and the CRE, as revealed by SEC experiments in which mixtures of CRTC2 and CRE yielded two separate peaks, with retention times similar to the apo-protein/DNA (Fig. S1). Mixtures of CRTC2 and CREB bZip also yielded two separate peaks, indicating that the association of CRTC2 with CREB requires DNA (Fig. S2). Collectively, these results indicate that CRTC2 lacks DNA-binding activity and binds stoichiometric equivalents of CREB in a DNA-dependent manner.
The physiologically relevant CREs targeted by CREB fall broadly into three categories: (i) palindromic CREs like those found in the somatostatin promoter (5′-TGACGTCA-3′), (ii) palindromic (variant) CREs such as those in the G6Pase promoter (5′-TTACGTAA-3′), and (iii) a nonpalindromic 5-bp sequence termed the half-site CRE (5′-TGACG-3′ or 5′-CGTCA-3′ (15). To quantify the affinity of the CRTC2:CREB interaction and to test whether the stability of the ternary complex differs between these CREB binding sites, we conducted fluorescence anisotropy (FA) assays by titrating fluoresceinated DNA duplexes containing the somatostatin, G6Pase, or half-site CREs with the CREB bZip peptide (Fig. 1B). The peptide bound all three CREs with affinities (EC50) in the nanomolar range, although binding to the half-site CRE and G6Pase CREs were 6- and 12-fold lower, respectively, than to the somatostatin CRE (EC50 ∼12 nM; Table S2).
Titration of these bZip:DNA complexes with CRTC2 peptides produced additional FA enhancements indicative of the formation of a higher molecular weight ternary complex (on all three CRE sites; Fig. 1B). Consistent with its inability to bind the CRE directly, titration of CRTC2 with fluoresceinated DNA had no effect on FA (Fig. S3). A CRTC2 peptide (amino acids 1–55) bound CREB efficiently, although a longer peptide (amino acids 1–116) bound slightly better (Table S1). Interestingly, the stability of the CRTC2–CREB interaction was somewhat enhanced on palindromic CREs relative to half-site CRE (∼360–500 nM vs. ∼1.1 μM). Taken together, these results indicate that the principal affinity determinants for CREB binding reside within the N-terminal 55 residues of CRTC2 and that palindromic, variant, and half-site CREs are capable of supporting moderate-affinity interactions between CREB and CRTC2. Note that although a GST tag was used for FA studies of CRTC2 (1–55), no such tag was used for CRTC2 (1–116) for these studies or for any of the CRTC2 peptides in the SEC-MALS studies that could bias the results.
Cysteine Residues Within the CREB bZip Domain Modulate Interactions with CRTC2.
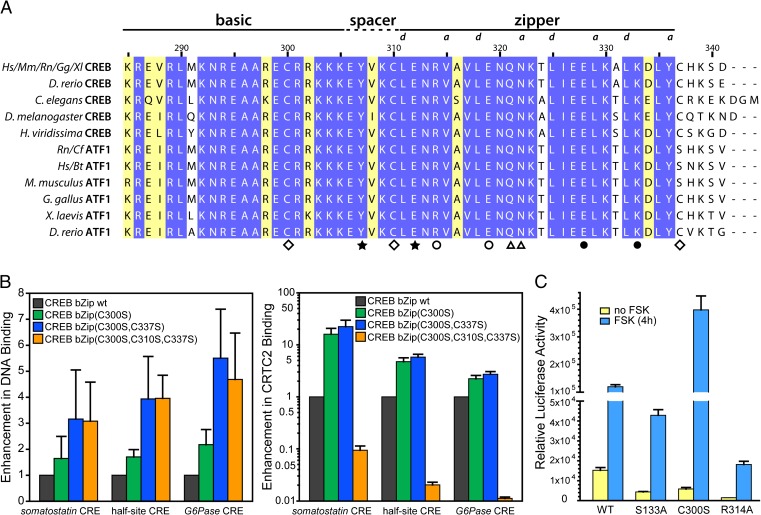
The bZip domain of CREB harbors three cysteine residues, two of which (Cys300 and Cys310) are conserved in CREB orthologs and paralogs (Fig. 2A). To preclude protein oxidation and cross-linking via cysteine disulfide bridge formation, these cysteine residues have been mutated to Ser in structural studies of the bZip bound to DNA (16). Unexpectedly, the Cys to Ser triple mutant bZip bound to CRTC2 with substantially diminished affinity compared with the wild-type protein, although DNA binding was not adversely affected (Fig. 2 A and B and Table S2). Cys310 seemed to be critical in this regard: a bZip mutant with serine substitutions at only Cys300 and Cys337 actually had higher affinity for CRTC2 relative to wild-type (Fig. 2B and Table S2).
Fig. 2.
Conserved cysteines in the CREB bZip domain perform contrasting roles in stabilizing the CRTC2 interaction. (A) A multiple sequence alignment of CREB paralogs and orthologs. Species abbreviations: Bt, Bos taurus; Cf, Canis familiaris; Gg, Gallus gallus; Hs, Homo sapiens; Mm, Mus musculus; Rn, Rattus norvegicus; Xl, Xenopus laevis. The symbols at the bottom identify cysteines (diamonds) and residues that engage in intersubunit hydrogen bonding and/or electrostatic interactions in CREB bZip:CRE complex (16) (Protein Data Bank ID: 1DH3). The “a” and “d”positions of the heptad repeat in the leucine zipper are identified above the alignment. (B) Changes in CRE- and CRTC2-binding affinity (Left and Right, respectively) induced by cysteine mutations in the context of different CREs. Changes in CRTC2-binding affinity shown on log scale. (C) Relative activities of wild-type and mutant CREB polypeptides. CRE-luc reporter activity in HEK293T cells expressing Halo-tagged wild-type, C300S, or R314A CREB. Treatment with FSK (10 μM) or DMSO vehicle (4 h) indicated. Relative luciferase activity normalized to β-galactosidase activity from a cotransfected Rous sarcoma virus (RSV)–β-gal plasmid.
In contrast to the positive role of Cys310, Cys300 in CREB seemed to negatively regulate the CRTC2 interaction as mutation of Cys300 to Ser increased CRTC2 binding 5- to 10-fold (Fig. 2B). On the basis of these observations, we tested whether the Cys300Ser substitution also enhances target gene expression in the context of full-length CREB. Overexpression of wild-type HaloTag-tagged CREB in HEK293T cells increased the activity of a CREB-responsive EVX-luciferase reporter, which harbors two half-site CREs, eightfold after exposure to cAMP agonist (forskolin; FSK); these effects were disrupted in cells expressing nonphosphorylatable (Ser133Ala) CREB (Fig. 2C). Consistent with its effect on CRTC2 interaction, mutation of Cys300 to serine substantially potentiated CREB activity in FSK-stimulated cells relative to the wild-type protein. By contrast, mutation of Arg314 to alanine, which disrupts CREB binding to CRTC2 (see below), correspondingly blocked FSK effects on CREB activity.
Collectively, these results demonstrate that Cys300 and Cys310 modulate CREB activity in part through opposing effects on CRTC2 recruitment.
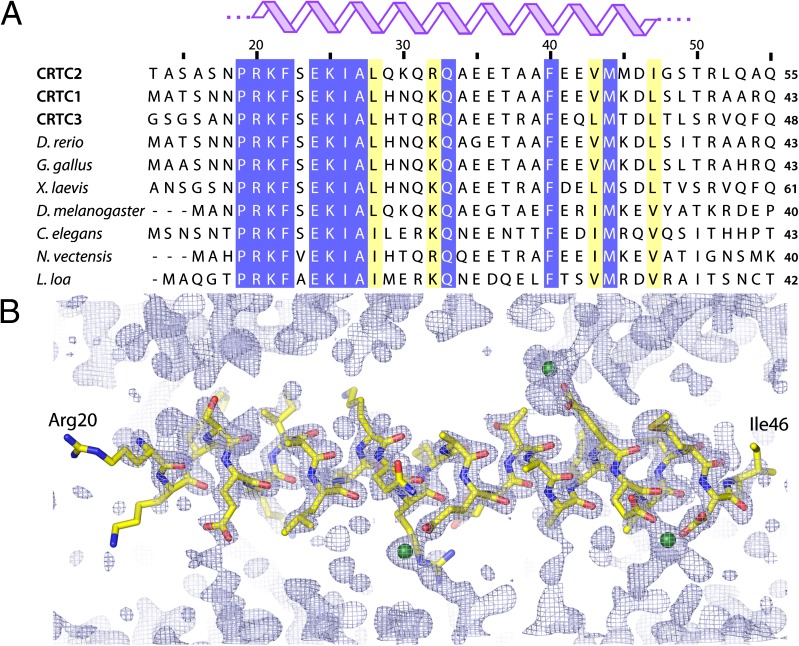
CBD of CRTC2 Adopts a Functionally Important Monomeric Helical Conformation in the Crystal.
The N-terminal 55-residue segment of CRTC2, deemed the minimal CBD, shows a high degree of sequence conservation within the CRTC family across a broad range of species (Fig. 3). The CRTC2 segment spanning residues Pro19 to Ile47 is especially well conserved, hinting at important structural and functional roles for this region. To test this idea, we synthesized a CRTC2 peptide spanning residues Asn18 to Thr50 in the CBD and asked whether this segment could adopt a stable structure. This peptide yielded excellent diffraction-quality crystals, and the structure was solved using single-wavelength anomalous dispersion (Table S3). The 2.0-Å structure of the peptide reveals a single continuous α-helix spanning residues Arg20 and Ile47 (Fig. 3). The helix crystallized as a monomer, and a total of five residues, two at the N terminus and three at the C terminus, are disordered with no interpretable density. We observed no evidence for an oligomeric coiled-coil structure, as proposed previously (Fig. 3) (5).
Fig. 3.
A highly conserved motif in the CBD of CRTC2 forms a helical structure. (A) Multiple sequence alignment spanning the first 55 residues of CRTC2 orthologs and paralogs. The human, mouse, and rat versions of CRTC1, CRTC2, and CRTC3 sequences are identical in this segment. (B) A 2.0-Å crystal structure of a selenomethionine-labeled CRTC2 CBD peptide (residues 18–50) shown along with the 2Fo-Fc electron density map contoured at 1.5 σ. CRCT2 residues Asn18, Pro19, Gly48, Ser49, and Thr50 are disordered in the crystal with no interpretable density; Arg20 through Ile47 form a single continuous α-helix. Green spheres represent Zn2+ ions, presumably from the crystallization buffer.
To test the importance of a helical structure in the CRTC2 CBD for its interactions with CREB, we introduced helix-disrupting proline mutations at less conserved positions (Fig. 3A). Relative to wild-type, all three single-site proline mutants including Ser23Pro, Lys30Pro, and Ala38Pro exhibited CREB binding affinities that were almost three or more orders of magnitude lower than the wild-type protein (Fig. 4B and Table S2). Collectively, these results suggest that a helical conformation is accessible to this sequence and is relevant for function.
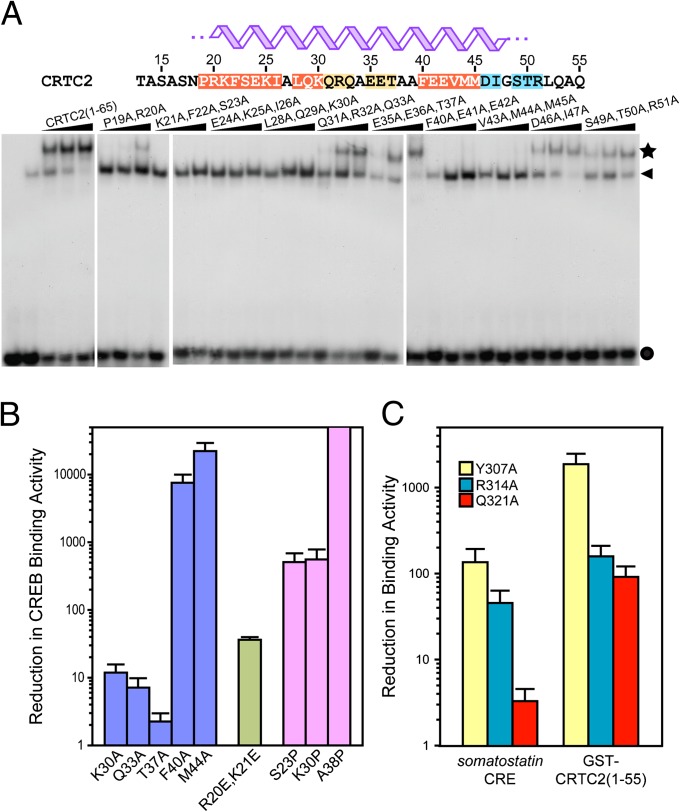
Fig. 4.
CBD in CRTC2 forms an extensive protein–protein interface with the bZip domain in CREB. (A) Multiresidue alanine scanning mutagenesis of residues 19 through 51 of the CRTC2 CBD. The CREB-binding activity of the mutant proteins generated in the GST-CRTC2 (residues 1–65) background was evaluated by gel mobility shift assays. Crescendo bars indicate increasing concentrations of GST-CRTC2 (0.5, 1, and 2 μM). Filled circle, arrowhead, and star identify the locations of the free probe, CREB bZip-bound CRE, and the CRTC2:CREB:CRE complex on the gels, respectively. CRTC2 residues are highlighted according to the effect of the mutations, with light blue, orange, and red denoting small, moderate, and severe reductions, respectively, on CREB binding. (B) Reductions (log scale) in CREB-binding affinity of various CRTC2 alanine, glutamate, and proline mutants in the context of the somatostatin CRE. (C) Reductions (log scale) in CRE- and CRTC2-binding affinity (Left and Right, respectively) of various CREB mutants.
Conserved Residues within CRTC2 CBD Are Important for CREB Interaction.
We performed alanine scanning mutagenesis studies to rapidly identify residues in the CRTC2 CBD helix that stabilize the CREB–CRTC2 interaction. In gel mobility shift assays, addition of CREB or the more readily purified recombinant CREM-α polypeptide, which contains a nearly identical bZip domain to CREB, caused a substantial reduction in the mobility of a probe containing a somatostatin CRE (Fig. 4A). Addition of increasing amounts of GST-CRTC2 (1–55) further reduced the mobility of the CREM:CRE complex, implying formation of a ternary complex. With the exception of the less-conserved segments (Fig. 3A), alanine substitutions in the CBD disrupted ternary complex formation (Fig. 4A), indicating that CRTC2 likely makes multiple contacts with CREB.
Next we evaluated ternary complex formation using single-site alanine mutants of CRTC2 CBD in FA assays. To facilitate reliable measurements of binding affinity, we conducted these studies with the gain-of-function Cys300Ser, Cys337Ser CREB bZip double mutant. Consistent with results from gel shift assays, alanine mutations of Phe40 and Met44—both invariant residues in CRTC orthologs and paralogs—had the most pronounced effects on binding; these Ala substitutions reduced the affinity of CRTC2 for CREB by at least three orders of magnitude (Fig. 4B and Table S2). Alanine mutations of Gln33 and Lys30 moderately reduced bZip binding, whereas an alanine substitution at the poorly conserved Thr37 had negligible effects relative to the wild-type protein, consistent with its noninvolvement in CREB binding. On the other hand, mutation of the well-conserved residues Arg20 and Lys21 to glutamate severely diminished CREB-binding activity by more than 30-fold, implicating them in electrostatic interactions (Fig. 4B and Table S2).
To gain further insight into the CRTC2–CREB interaction, we characterized a CRTC2 CBD peptide (amino acids 18–55) by solution NMR. In contrast to the helical conformation observed in the crystal, the peptide is essentially unstructured in solution, as evidenced by the poor dispersion and the narrow linewidths of the amide proton resonances (Fig. S4). However, only 7 of the 40 expected CRTC2 correlations corresponding to flexible residues could be readily observed in the 32-KDa ternary complex, implicating the remaining 33 residues (approximately the same size as the helix seen in the crystal) in forming a rigid structural unit while engaging CREB (Fig. S4). We obtained similar results for a longer CRTC2 peptide (amino acids 18–78). The presence of alcohol—a well known stabilizer of helical structures—during crystallization likely explains the apparent disparity between solution NMR and crystallographic analyses of the CBD (17). Taken together, these results suggest that the CRTC2 CBD likely undergoes a coil–helix transition upon CREB binding. Indeed, the circular dichroism spectrum of the ternary complex is characterized by a significant increase in helical content (Fig. S5); because CREB bZip is almost exclusively helical when in complex with DNA (16), the increased helicity must be due to the acquisition of stable helical structure in the CBD.
Collectively, these structural and mutagenesis studies define the sequence and conformational requirements for efficient interactions of CRTCs with CREB. Within the helical motif of CRTC2 CBD, residues on one face of the helix and at both N and C termini of the helix seem to be critical for engaging CREB (Fig. 5 and Fig. S6), defining an extended protein–protein interface.
Fig. 5.
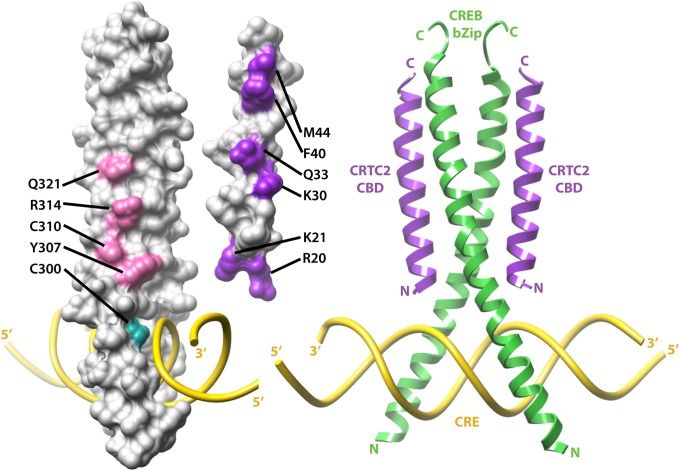
Structural model for the interaction between CRTC2 CBD and CRE-bound CREB bZip. Undocked models of CRE-bound CREB bZip (Left) and CRTC2 (Center), with residues deemed functionally significant by mutagenesis studies colored in magenta and purple, respectively. The cysteine residue (Cys300) in bZip whose mutation to serine enhances the CREB–CRTC2 interaction is shown in cyan. (Right) A manually docked model based on mutational data and the crystal structures of CRTC2 CBD and CREB bZip bound to somatostatin CRE (16) (Protein Data Bank ID: 1DH3).
CRTC2 CBD Targets the Groove Defined by the Monomer Units of the CREB Leucine Zipper Coiled-Coil.
Arg314 in CREB has been shown to promote CRTC–CREB interactions, because mutation of this site disrupted complex formation (7). Because the side chains of Arg314 along with those of Tyr307 and Gln321 are solvent exposed and define, in part, the groove of the bZip coiled-coil (Fig. 2A) (16), we asked whether these residues contributed directly to CRTC2 binding. Individual alanine substitutions at each of these sites reduced the affinity of CREB bZip toward GST-CRTC2 (1–55) by at least two orders of magnitude lower affinity relative to the wild-type protein. The Tyr307Ala mutation appeared particularly deleterious, with CRTC2 binding diminished by 2,000-fold.
Superimposed on their interactions with CRTC2, residues that define the bZip groove are also involved in intersubunit, noncovalent interactions (16). Tyr307 engages in a hydrogen bonding interaction with Glu312, for example, whereas Arg314 forms a salt bridge with Glu319; Gln321 engages in a weaker hydrogen bonding interaction with Asn320. Indeed, single-site alanine mutations of Tyr307 and Arg314 reduced the affinity of the CREB–CRE interaction by 100- and 40-fold, respectively (Fig. 4C), in line with the results of a previous study (18). By contrast, the Gln321Ala mutant bound to the CRE with only slightly diminished affinity as the wild-type protein. Given the much stronger effects of these mutations on CRTC2 binding than DNA binding, these results implicate residues lining the groove between the monomer units of the CREB leucine zipper in direct interactions with CRTC2.
Discussion
Since their discovery in the late 1980s (19), bZip factors have been among the most extensively studied families of transcriptional activators; they have served as the paradigm for understanding the basis for specificity of protein–protein interactions through their differential abilities to form homo- and heterodimers (1). Indeed, the rules for bZip dimer formation are well understood, and mechanisms by which homo-/heterodimerization then modulate DNA-binding preferences are also well documented. Although they often share overlapping DNA-binding specificities, bZip factors nevertheless exert diverse effects on cellular physiology, pointing to the potential involvement of additional coregulators that engage the transcriptional machinery.
Our studies provide insight into the mechanism by which CRTCs associate with CREB on cellular target genes. On the basis of the crystal structures of the apo-CRTC2 CBD and a CREB bZip–CRE complex (16), we propose that the CRTC2 CBD:CREB bZip:CRE ternary complex assembles with a 2:2:1 stoichiometry; each copy of the 28-residue CRTC2 CBD helix targets one of the two grooves formed by the dimeric CREB leucine zipper and the preceding spacer segment (Fig. 5). The N terminus of the CBD helix is positioned proximal to the DNA, rather than distal to it, thus rendering this helix in a parallel orientation relative to the CREB leucine zipper (Fig. 5). We favor this model because the CBD N terminus features several invariant basic residues (Arg20, Lys21, and Lys25 in CRTC2) that could serve to both orient the helix and enhance the stability of this complex by engaging the phosphate groups in DNA in electrostatic interactions. In support of this idea, acetylation of lysine residues in the CBD or their mutation to glutamine has been implicated in diminishing CRTC–CREB interactions (20). The nonspecific interactions with DNA might explain why CRTCs engage CREB bound to diverse CREs with similar affinities. Unlike the basic region of CREB, the CRTC2 CBD harbors substantially fewer basic residues, explaining why it lacks intrinsic DNA-binding affinity.
An intriguing feature of the CREB bZip domain is the exceptionally high degree of sequence conservation that extends beyond those residues involved in stabilizing the homodimer and the protein–DNA complex. The involvement of some of these conserved residues in CRTC binding might explain why they are evolutionarily conserved. It also points to the potential involvement of other, as yet uncharacterized, cofactors that further modulate cellular gene expression through an association with the CREB bZip domain.
By binding to the CREB bZip:CRE complex in the manner described above, the CRTC proteins would be predicted to stabilize the complex, decreasing the dissociation kinetics and thereby increasing the residence time of CREB on DNA relative to other bZip factors that are unable to associate with CRTCs. In so doing, CRTC proteins can thus synergize with CBP/p300 coactivators, which target the KID domain of CREB (13), on some promoters. Rather interestingly, wild-type CREB exhibits similar affinities in the micromolar range for CBP/p300 and the CRTCs (21), implying that both sets of coactivators, when operating in isolation, are poised for rapid inactivation.
Our studies reveal that the DNA binding and dimerization properties of the CREB bZip can be uncoupled from its role as a transactivation domain. Cys310 and Gln321 seem to have a direct role in interacting with CRTCs, for example, but they do not seem to be necessary for DNA binding. By contrast, Tyr307 and Arg314 in CREB seem to participate in both DNA binding (by stabilizing the dimeric coiled-coil) as well as CRTC2 binding functions.
Of all of the mutations, the effects produced by the Cys300Ser mutation were the most unexpected, as evidenced by the substantial increase in CRTC2 binding activity relative to the wild-type protein in both transcriptional and in vitro binding assays. The origin of the enhanced CRTC2 binding activity is unclear but could be due to an uncharacterized allosteric effect caused by the Cys to Ser mutation. The contrasting roles of Cys300 and Cys310 in stabilizing the CRTC2 interaction raise the intriguing possibility that these residues might be targeted for redox regulation. In this regard, increases in nitric oxide signaling have been shown to increase CREB target gene expression through the S-nitrosylation of nuclear factors that bind to CREs (22). We also note that Cys300 and Cys310 have both been implicated in functioning as oxygen sensors and modulating both CREB binding to CREs and the resulting transcriptional output in vivo (23–25). Our results suggest that oxidation, S-nitrosylation, or other modifications of CREB on relevant cysteine residues may enhance or diminish target gene expression, in part, through modulation of CRTC recruitment to the promoter.
In summary, we have described how a bZip transcription factor uses a broadly distributed structural motif for its DNA-binding, dimerization, and transactivation functions. Our studies provide structural insights into how other bZip coactivators, such as HTLV-1 Tax, HBV pX, MBF1, and Chameau, could interact with this versatile motif (26–30), thereby promoting the formation of interaction networks that extend to the transcriptional apparatus.
Materials and Methods
Production of CREB and CRTC2 Polypeptides.
The gene sequence encoding CREB bZip (residues 285–341) was inserted into the pMCSG7 vector (31), sequenced to verify integrity, and expressed in Escherichia coli BL21(DE3) cells at 20 °C. The protein was purified via Ni2+-affinity chromatography under denaturing conditions. The His6-tag was cleaved using tobacco etch virus (TEV) protease, and the protein was purified to homogeneity via reversed-phase HPLC. CRTC2 polypeptides spanning residues 1–116, 18–78, and 18–70 were expressed and purified in a similar manner.
The gene sequences for CRTC2 polypeptides spanning residues 1–55 and 1–65 were inserted into the pMCSG10 (31) and pGEX-5x1 vectors, their sequences confirmed by DNA sequencing, and the GST-fusion proteins expressed as discussed above for CREB bZip. Both GST-CRTC2 constructs were purified via glutathione Sepharose-affinity chromatography. Eluted fractions containing the proteins were pooled, dialyzed, snap frozen, and stored at −80 °C.
CREB bZip and CRTC2 mutants for in vitro binding assays were generated using the QuikChange site-directed mutagenesis protocol; the CREB bZip triple Cys to Ser mutant was generated by gene synthesis. All mutations were confirmed by DNA sequencing. The mutant proteins were expressed and purified using the protocols for the wild-type proteins.
FA Assays.
Fluoresceinated oligonucleotides 5′-Fl-CCTGACGTCAGCCCCCTGACGTCAGG-3′ and 5′-Fl-CCTTACGTAAGCCCCCTTACGTAAGG-3′ designed to form hairpins and containing a single somatostatin or G6Pase CRE as well as the complementary oligonucleotides 5′-Fl-ATCTGCGTCAGAGT-3′ and 5′-ACTCTGACGCAGAT-3′ designed to form a duplex containing a half-site CRE found in the inhibin-α promoter (32) were purchased from Integrated DNA Technologies. Protein concentrations were measured spectrophotometrically (33). To minimize cysteine oxidation, all CREB bZip peptides were incubated at 42 °C in 6 M guanidine hydrochloride with 5 mM DTT and rapidly desalted immediately before use. Titrations were performed in 25 mM Tris·HCl buffer (pH 7.6) containing 150 mM NaCl, 5 mM DTT, 5% (vol/vol) glycerol, 0.1 mg/mL BSA, and 5 mM MgCl2. Samples were incubated for 15 min after mixing. Fluorescence data were acquired at room temperature either on a Biotek Synergy HT Microplate reader with excitation and emission wavelengths set to 490 nm and 520 nm, respectively. All titrations were performed at least in triplicate, and the data were fitted via nonlinear regression to standard equations using Kaleidagraph (Synergy Software) (34).
Gel Mobility Shift Experiments.
Gel mobility shift assays were conducted as previously described using a 32P-labeled oligonucleotide probe containing a somatostatin CRE (35), except purified phosphorylated CREM-α (a kind gift from Young-In Chi, University of Minnesota, Austin, MN), GST-CREB bZip, and wild-type/mutant GST-CRTC2 (1–65) peptides were used for these studies.
Tissue Culture and Luciferase Assays.
HEK293T cells were grown at 5% CO2 and 37 °C in DMEM with 10% heat-inactivated FBS and 100 mg/mL penicillin/streptomycin. Transient assays were performed with a cAMP responsive pXP2-EVX luciferase reporter (50 ng) cotransfected with wild-type or mutant CREB expression vectors (50 ng) containing an N-terminal HaloTag epitope tag alongside RSV–β-galactosidase vector (20 ng) using Effectene transfection reagent. Twenty-four hours after transfection, cells were treated with 10 µM FSK or DMSO (vehicle) for 4 h. Lysates were assayed for luciferase and β-galactosidase activities as previously described (5). Comparable expression of wild-type and mutant CREB polypeptides was verified by Western blot assay.
Crystallography.
A CRTC2 (18–50) peptide harboring selenomethionine (instead of methionine) residues at positions 44 and 45 was synthesized chemically using automated procedures and purified by reversed-phase HPLC. The identity of the peptide was verified by mass spectrometry. Crystals of the peptide were grown at 22 °C in 100 mM Mes (pH 6), 150 mM zinc acetate, 3 M NaCl, and 12% ethanol using the hanging-drop vapor diffusion method. Crystals were transferred to the reservoir solution supplemented with 20% (vol/vol) glycerol. Diffraction data were acquired on the LS-CAT beamline 21-ID-F at the Advanced Photon Source and were processed using HKL2000 (36). The structure was solved using single-wavelength anomalous dispersion method. Two selenium atoms were located, and phases were calculated by using SHARP/autoSHARP (37), followed by automated model building using ARP/wARP (38). The model was further built, refined, and analyzed in Coot (39), REFMAC (40), and PROCHECK (41). Data collection and refinement statistics are listed in Table S3.
NMR Spectroscopy.
NMR samples were prepared in 10 mM sodium phosphate (pH 6.2) buffer containing 50 mM NaCl, 0.7 mM MgCl2, and 0.2% sodium azide. All data were acquired on a Varian Inova 600-MHz spectrometer equipped with a cold probe at 25 °C. Backbone resonances were assigned by analyzing 3D CBCA(CO)NH and HNCACB spectra acquired for uniformly 15N,13C-labeled samples of CRTC2.
Circular Dichroism Spectroscopy.
CD samples were prepared in 10 mM sodium phosphate (pH 6.4) buffer containing 0.02 mM MgCl2. All data were recorded at 25 °C on a JASCO J-815 instrument using a 10-mm pathlength dual-chamber cuvettete.
SEC with inline MALS.
SEC experiments were performed using a Superdex 200 HR column attached to an inline Wyatt Dawn Heleos II MALS detector and an Optilab T-rEX differential refractometer. Samples of proteins and protein–DNA mixtures at final concentrations of 100 μM were injected, and the scattering and refractive index profiles of the various molecular species were recorded. The experiments were conducted in 50 mM Mes buffer (pH 6.5) containing 150 mM NaCl, 1 mM tris(2-carboxyethyl)phosphine, and 1 mM MgCl2. The refractive index increment (dn/dc) of the solute/solvent system was calibrated with known standards before analysis. The molar mass of each resolved species was quantified using the ASTRA software provided by the manufacturer, Wyatt Technology.
Supplementary Material
Acknowledgments
We thank Lu Sun and Jackie Winters for their contributions, and the Lurie Comprehensive Cancer Center at Northwestern for their support of structural biology research. Funding for this work was provided by American Diabetes Association Grant 1-12-BS-168 (to I.R.) and National Institutes of Health Grants U54 HD41957 (to K.E.M. and I.R.) and R01 DK049777, R01 DK083834, and R01 DK091618 (to M.M.). This work was also funded by the Clayton Foundation for Medical Research, the Kieckhefer Foundation, and the Leona M. and Harry B. Helmsley Charitable Trust (M.M.). K.V. was funded by the Department of Biomedicine, University of Bergen, Norway.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (CRTC2 CBD; PDB ID code 4HTM).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1219028109/-/DCSupplemental.
References
- 1.Vinson C, Acharya A, Taparowsky EJ. Deciphering B-ZIP transcription factor interactions in vitro and in vivo. Biochim Biophys Acta. 2006;1759(1-2):4–12. doi: 10.1016/j.bbaexp.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 2.Montminy M. Transcriptional regulation by cyclic AMP. Annu Rev Biochem. 1997;66:807–822. doi: 10.1146/annurev.biochem.66.1.807. [DOI] [PubMed] [Google Scholar]
- 3.Altarejos JY, Montminy M. CREB and the CRTC co-activators: Sensors for hormonal and metabolic signals. Nat Rev Mol Cell Biol. 2011;12(3):141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chrivia JC, et al. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365(6449):855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 5.Conkright MD, et al. TORCs: Transducers of regulated CREB activity. Mol Cell. 2003;12(2):413–423. doi: 10.1016/j.molcel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 6.Iourgenko V, et al. Identification of a family of cAMP response element-binding protein coactivators by genome-scale functional analysis in mammalian cells. Proc Natl Acad Sci USA. 2003;100(21):12147–12152. doi: 10.1073/pnas.1932773100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Screaton RA, et al. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell. 2004;119(1):61–74. doi: 10.1016/j.cell.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 8.Zhou Y, et al. Requirement of TORC1 for late-phase long-term potentiation in the hippocampus. PLoS ONE. 2006;1:e16. doi: 10.1371/journal.pone.0000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kovács KA, et al. TORC1 is a calcium- and cAMP-sensitive coincidence detector involved in hippocampal long-term synaptic plasticity. Proc Natl Acad Sci USA. 2007;104(11):4700–4705. doi: 10.1073/pnas.0607524104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koo SH, et al. The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature. 2005;437(7062):1109–1111. doi: 10.1038/nature03967. [DOI] [PubMed] [Google Scholar]
- 11.Wang B, et al. The insulin-regulated CREB coactivator TORC promotes stress resistance in Drosophila. Cell Metab. 2008;7(5):434–444. doi: 10.1016/j.cmet.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mair W, et al. Lifespan extension induced by AMPK and calcineurin is mediated by CRTC-1 and CREB. Nature. 2011;470(7334):404–408. doi: 10.1038/nature09706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Radhakrishnan I, et al. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: A model for activator:coactivator interactions. Cell. 1997;91(6):741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 14.Rohas LM, et al. A fundamental system of cellular energy homeostasis regulated by PGC-1alpha. Proc Natl Acad Sci USA. 2007;104(19):7933–7938. doi: 10.1073/pnas.0702683104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci USA. 2005;102(12):4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schumacher MA, Goodman RH, Brennan RG. The structure of a CREB bZIP.somatostatin CRE complex reveals the basis for selective dimerization and divalent cation-enhanced DNA binding. J Biol Chem. 2000;275(45):35242–35247. doi: 10.1074/jbc.M007293200. [DOI] [PubMed] [Google Scholar]
- 17.Buck M. Trifluoroethanol and colleagues: Cosolvents come of age. Recent studies with peptides and proteins. Q Rev Biophys. 1998;31(3):297–355. doi: 10.1017/s003358359800345x. [DOI] [PubMed] [Google Scholar]
- 18.Craig JC, et al. Consensus and variant cAMP-regulated enhancers have distinct CREB-binding properties. J Biol Chem. 2001;276(15):11719–11728. doi: 10.1074/jbc.M010263200. [DOI] [PubMed] [Google Scholar]
- 19.Vinson CR, Sigler PB, McKnight SL. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science. 1989;246(4932):911–916. doi: 10.1126/science.2683088. [DOI] [PubMed] [Google Scholar]
- 20.Jeong H, et al. Sirt1 mediates neuroprotection from mutant huntingtin by activation of the TORC1 and CREB transcriptional pathway. Nat Med. 2012;18(1):159–165. doi: 10.1038/nm.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zor T, Mayr BM, Dyson HJ, Montminy MR, Wright PE. Roles of phosphorylation and helix propensity in the binding of the KIX domain of CREB-binding protein by constitutive (c-Myb) and inducible (CREB) activators. J Biol Chem. 2002;277(44):42241–42248. doi: 10.1074/jbc.M207361200. [DOI] [PubMed] [Google Scholar]
- 22.Riccio A, et al. A nitric oxide signaling pathway controls CREB-mediated gene expression in neurons. Mol Cell. 2006;21(2):283–294. doi: 10.1016/j.molcel.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 23.Goren I, Tavor E, Goldblum A, Honigman A. Two cysteine residues in the DNA-binding domain of CREB control binding to CRE and CREB-mediated gene expression. J Mol Biol. 2001;313(4):695–709. doi: 10.1006/jmbi.2001.5064. [DOI] [PubMed] [Google Scholar]
- 24.Abramovitch R, et al. A pivotal role of cyclic AMP-responsive element binding protein in tumor progression. Cancer Res. 2004;64(4):1338–1346. doi: 10.1158/0008-5472.can-03-2089. [DOI] [PubMed] [Google Scholar]
- 25.Meyuhas R, et al. A key role for cyclic AMP-responsive element binding protein in hypoxia-mediated activation of the angiogenesis factor CCN1 (CYR61) in tumor cells. Mol Cancer Res. 2008;6(9):1397–1409. doi: 10.1158/1541-7786.MCR-07-2086. [DOI] [PubMed] [Google Scholar]
- 26.Zhao LJ, Giam CZ. Human T-cell lymphotropic virus type I (HTLV-I) transcriptional activator, Tax, enhances CREB binding to HTLV-I 21-base-pair repeats by protein-protein interaction. Proc Natl Acad Sci USA. 1992;89(15):7070–7074. doi: 10.1073/pnas.89.15.7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baranger AM, et al. Mechanism of DNA-binding enhancement by the human T-cell leukaemia virus transactivator Tax. Nature. 1995;376(6541):606–608. doi: 10.1038/376606a0. [DOI] [PubMed] [Google Scholar]
- 28.Nyborg JK, Egan D, Sharma N. The HTLV-1 Tax protein: Revealing mechanisms of transcriptional activation through histone acetylation and nucleosome disassembly. Biochim Biophys Acta. 2010;1799(3-4):266–274. doi: 10.1016/j.bbagrm.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 29.Palmer CR, Gegnas LD, Schepartz A. Mechanism of DNA binding enhancement by hepatitis B virus protein pX. Biochemistry. 1997;36(49):15349–15355. doi: 10.1021/bi972076m. [DOI] [PubMed] [Google Scholar]
- 30.Miotto B, Struhl K. Differential gene regulation by selective association of transcriptional coactivators and bZIP DNA-binding domains. Mol Cell Biol. 2006;26(16):5969–5982. doi: 10.1128/MCB.00696-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stols L, et al. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr Purif. 2002;25(1):8–15. doi: 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
- 32.Ito M, Park Y, Weck J, Mayo KE, Jameson JL. Synergistic activation of the inhibin alpha-promoter by steroidogenic factor-1 and cyclic adenosine 3′,5′-monophosphate. Mol Endocrinol. 2000;14(1):66–81. doi: 10.1210/mend.14.1.0410. [DOI] [PubMed] [Google Scholar]
- 33.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182(2):319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 34.Chin J, Längst G, Becker PB, Widom J. Fluorescence anisotropy assays for analysis of ISWI-DNA and ISWI-nucleosome interactions. Methods Enzymol. 2004;376:3–16. doi: 10.1016/S0076-6879(03)76001-7. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y, et al. Targeted disruption of the CREB coactivator Crtc2 increases insulin sensitivity. Proc Natl Acad Sci USA. 2010;107(7):3087–3092. doi: 10.1073/pnas.0914897107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 37.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 38.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6(5):458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 39.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 40.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57(Pt 1):122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 41.Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J Mol Biol. 1993;231(4):1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.