Abstract
RNA viruses in insects are targets of an RNA interference (RNAi)-based antiviral immune response, in which viral replication intermediates or viral dsRNA genomes are processed by Dicer-2 (Dcr-2) into viral small interfering RNAs (vsiRNAs). Whether dsDNA virus infections are controlled by the RNAi pathway remains to be determined. Here, we analyzed the role of RNAi in DNA virus infection using Drosophila melanogaster infected with Invertebrate iridescent virus 6 (IIV-6) as a model. We show that Dcr-2 and Argonaute-2 mutant flies are more sensitive to virus infection, suggesting that vsiRNAs contribute to the control of DNA virus infection. Indeed, small RNA sequencing of IIV-6–infected WT and RNAi mutant flies identified abundant vsiRNAs that were produced in a Dcr-2–dependent manner. We observed a highly uneven distribution with strong clustering of vsiRNAs to small defined regions (hotspots) and modest coverage at other regions (coldspots). vsiRNAs mapped in similar proportions to both strands of the viral genome, suggesting that long dsRNA derived from convergent overlapping transcripts serves as a substrate for Dcr-2. In agreement, strand-specific RT-PCR and Northern blot analyses indicated that antisense transcripts are produced during infection. Moreover, we show that vsiRNAs are functional in silencing reporter constructs carrying fragments of the IIV-6 genome. Together, our data indicate that RNAi provides antiviral defense against dsDNA viruses in animals. Thus, RNAi is the predominant antiviral defense mechanism in insects that provides protection against all major classes of viruses.
Keywords: insect immunity, Iridoviridae, siRNA, antiviral immunity, small RNA profiling, next-generation sequencing
Double-stranded RNA (dsRNA) is a danger signal: It cannot be detected in healthy, noninfected cells, but it is produced during infection by many RNA and DNA viruses (1). It is thus not surprising that dsRNA is a central trigger of innate immune responses. In vertebrates, recognition of dsRNA by the cytosolic sensors RIG-I and MDA-5 initiates a cascade of events culminating in the production of type I IFN and the subsequent induction of an antiviral state (2). Likewise, dsRNA triggers a sequence-independent antiviral response in penaeid shrimp that is distinct from the vertebrate IFN pathway (3). In plants, fungi, and arthropods, viral dsRNA triggers an antiviral RNAi response (4, 5).
Studies in insects like Drosophila melanogaster and mosquitoes support a model in which viral dsRNA is processed by Dicer-2 (Dcr-2) into viral small interfering RNAs (vsiRNAs). These vsiRNAs are then incorporated into Argonaute-2 (AGO2) in the RNA-induced silencing complex (RISC), where they guide the recognition and endonucleic cleavage of viral target RNAs (6–8). Indeed, early seminal work in mosquitoes and Drosophila cells directly detected viral small RNAs by Northern blot analysis and demonstrated that knockdown of core RNAi genes resulted in an increase in virus replication (8–10). In accordance, in the genetic model organism D. melanogaster, flies with defects in Dcr-2, R2D2, or AGO2 are unable to control RNA virus replication and, consequently, are hypersensitive to virus infection and succumb more rapidly than their wild-type (WT) controls (11–14).
Small RNA cloning and next-generation sequencing provide detailed insights into vsiRNA biogenesis. In several studies in insects, the polarity of the vsiRNA population deviates strongly from the highly skewed distribution of positive strand (+) over negative (−) viral RNAs that is generally observed in (+) RNA virus infection. Indeed, vsiRNAs mapped in similar proportions to (+) and (−) viral RNA strands in Aedes aegypti, Aedes albopictus, and Culex pipiens mosquitoes infected with a number of arthropod-borne viruses, including Sindbis virus, Semliki Forest virus, West Nile virus, Dengue virus, and Chikungunya virus, as well as in Drosophila infected with (+) RNA viruses from different families (5, 15–24). In addition, in infections of Drosophila with the negative-strand RNA virus vesicular stomatitis virus, similar numbers of (+) over (−) vsiRNAs were recovered (14). These results, together with the distribution of vsiRNAs all along the viral genome, imply that viral replication intermediates of RNA viruses are the main targets for Dcr-2. Also in dsRNA virus infections, similar amounts of vsiRNAs of both polarities were generated, most likely by Dcr-2–dependent processing of viral genomic dsRNA (18). Thus, different classes of viruses seem to be processed by a similar vsiRNA biogenesis pathway. These studies have focused on RNA viruses, because all well-established model viruses of Drosophila and all known mosquito-transmitted viruses are RNA viruses. More recently, a next-generation sequencing approach identified small RNAs derived from a novel densovirus, a single-stranded (ss) DNA virus, in wild-caught C. pipiens molestus (25). These observations suggest that ssDNA viruses are a target for Dicer in mosquitoes, although the biogenesis and function of the viral small RNAs remain unclear.
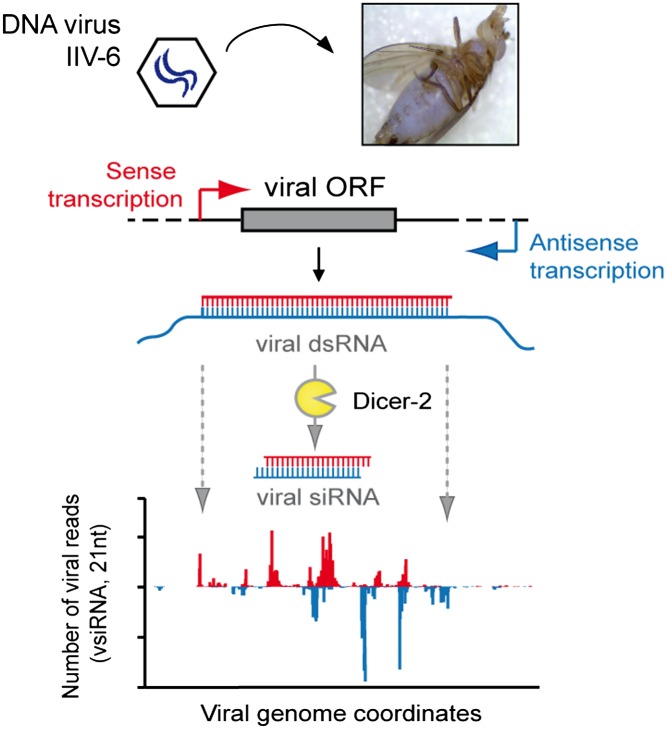
dsDNA viruses produce dsRNA during their replication, presumably due to base pairing of convergent overlapping transcripts from both strands of the DNA genome (26–28). Whether such dsRNA is a bona fide target for Dcr-2 remains to be established. Although a dsDNA virus has recently been identified in wild-caught Drosophila innubila (29), a dsDNA virus that naturally infects D. melanogaster has yet to be discovered. Invertebrate iridescent virus 6 (IIV-6), a member of the Iridovirus genus within the Iridoviridae family, has a broad host range; under experimental conditions it replicates in a number of Dipteran species, including D. melanogaster (30–32). We therefore used IIV-6, also known as Chilo iridescent virus, as a model to analyze the RNAi response against dsDNA viruses in Drosophila. IIV-6 is a large, complex virus with a dsDNA genome of 212,482 bp that encodes 211 putative ORFs distributed along the two strands of the viral genome (33, 34). Here, we report that IIV-6 is a target of the RNAi machinery. We demonstrate that Dcr-2 and AGO2 mutant flies are more sensitive to IIV-6 infection. Moreover, we identified Dcr-2–dependent vsiRNAs that map to both strands of the viral genome, show an uneven distribution across the viral genome, but are remarkably conserved between independent libraries. In accordance, we showed that both sense and antisense transcripts are generated in vivo and in vitro during IIV-6 replication, supporting a model in which viral dsRNA that is produced by bidirectional overlapping transcription is a target for Dcr-2. Together, our results indicate that RNAi provides antiviral defense against DNA viruses in Drosophila.
Results
IIV-6 as a Model to Study Antiviral Immunity Against DNA Viruses in Drosophila.
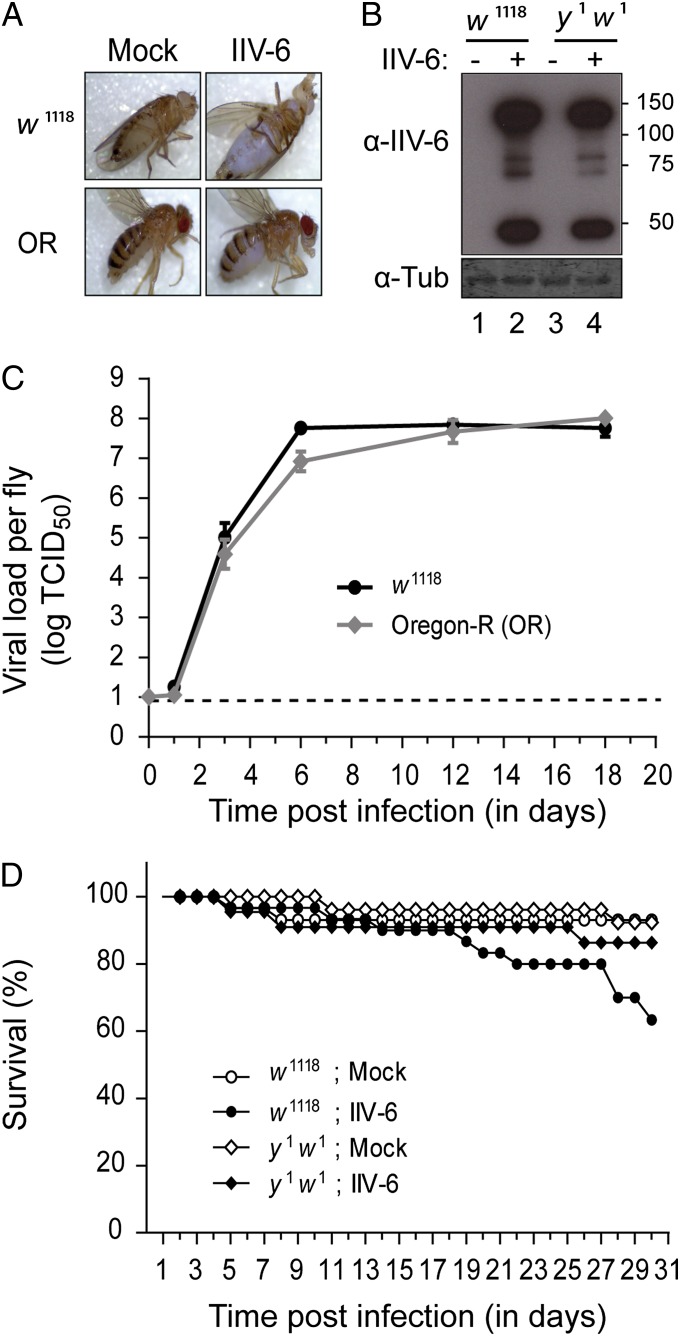
We investigated the replication kinetics of IIV-6 in w1118 and Oregon-R (OR) WT flies. We inoculated adult flies intraabdominally with IIV-6 and monitored survival and viral titers over time. IIV-6 established a productive infection as revealed by iridescence in the eyes, thorax, and abdomen, which is the result of light reflection by assemblies of paracrystalline arrays of IIV-6 particles (35) (Fig. 1A). Accordingly, IIV-6 virion coat proteins were detected by Western blot analysis in total lysates of IIV-6–infected flies (Fig. 1B, lanes 2 and 4). The specificity of the IIV-6 antibody was verified by lack of signal in mock-infected flies (lanes 1 and 3). IIV-6 replicated efficiently, with a rapid 6- to 7-log increase in viral titer over the first 6 days (d) and a relatively stable titer thereafter (Fig. 1C). However, despite these high and stable titers, virus infection did not efficiently kill WT flies over the course of 31 d (Fig. 1D; over 60% survival after follow-up). These results establish IIV-6 as a model to study DNA virus infection in Drosophila.
Fig. 1.
IIV-6 as a model to study antiviral immunity against DNA viruses in D. melanogaster. (A) IIV-6 infection of w1118 and Oregon-R (OR) WT flies results in iridescence in the eyes, thorax, and abdomen. Female flies were inoculated in the abdomen with 14,000 TCID50 units of IIV-6 or with Tris buffer as a control (mock). Representative images of flies at 30 d postinfection are shown. Iridescence becomes apparent as of day 9. (B) Western blot analysis of viral proteins in IIV-6 or mock-infected w1118 and y1w1 WT flies. Female flies were harvested at 12 d postinfection. Polyclonal anti–IIV-6 antibodies were used to visualize virion coat proteins. A polyclonal anti–α-tubulin (α-Tub) antibody was used as a loading control. Molecular mass (kDa) is indicated to the right of the autoradiograph. (C) Viral titers in w1118 (black) and OR (gray) female flies after infection with IIV-6. Three pools of four flies were collected and homogenized at each indicated time point, and viral titer in the homogenate was determined by end-point dilution. Titers represent averages and SEMs of three independent pools of four flies. The dashed line represents the detection limit of the assay. (D) Survival curve of D. melanogaster WT flies after IIV-6 infection. The survival rate of w1118 (circles) and y1w1 (diamonds) female flies was monitored daily after virus infection (filled symbols) or mock infection (open symbols). A representative of three independent experiments is shown.
RNAi Mutant Flies Are More Susceptible to IIV-6 Infection.
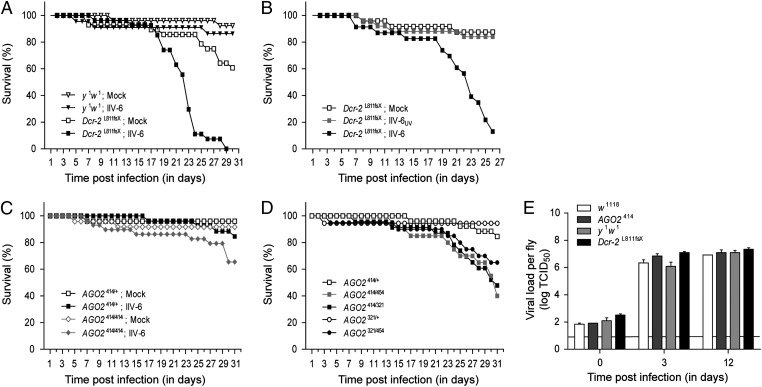
To examine the role of the antiviral RNAi machinery on IIV-6 replication in vivo, we analyzed the outcome of IIV-6 infection in flies that lack core RNAi components. First, we analyzed whether Dcr-2 mutant flies were more sensitive to IIV-6 infection. After an initial stable survival, Dcr-2 mutants died from 18 d postinfection onward, with 100% mortality at 29 d after infection. In contrast, over 85% of y1w1 controls survived over the same time course (Fig. 2A). Survival after inoculation with UV-inactivated IIV-6 was similar to that of mock-infected flies (about 85% survival after follow-up), indicating that mortality is the result of active virus replication (Fig. 2B). To analyze further whether RNAi controls IIV-6 infection in vivo, we monitored survival rates of AGO2 mutant flies. AGO2 homozygous mutants (AGO2414/414) and their heterozygous controls (AGO2414/+) were challenged with IIV-6. Although heterozygous controls were resistant to IIV-6 infection, AGO2 homozygotes were more sensitive to virus infection, with 35% mortality after follow-up for 31 d (Fig. 2C). To exclude the possibility that the susceptibility of AGO2 homozygous mutant flies was due to second-site mutations in the genome of AGO2414 flies, we analyzed survival of flies that carried a combination of two different AGO2 null alleles, and thus do not express AGO2 (AGO2414/454, AGO2414/321, and AGO2321/454 transheterozygotes), and their AGO2 heterozygous controls (AGO2414/+ and AGO2321/+). Because AGO2321 and AGO2454 were reported to be true null alleles (36), we first analyzed survival of these AGO2 transheterozygous mutants in the well-studied Drosophila C virus (DCV) and Cricket paralysis virus (CrPV) models of infection. AGO2 transheterozygous flies are equally sensitive to DCV challenge as AGO2414 homozygous mutant flies, whereas their heterozygous controls displayed a slower mortality rate (Fig. S1A). Similar results were obtained after CrPV challenge (Fig. S1B). These results establish AGO2 transheterozygous mutant flies as a reliable genetic model for the analysis of survival following virus infection. We observed that AGO2 transheterozygous mutants died more rapidly after IIV-6 challenge (40–65% survival after follow-up) compared with their heterozygous controls (over 85% survival) (Fig. 2D) and with their mock-infected controls (over 88% survival). To analyze whether the increased lethality correlates with an increase in viral titers, we analyzed the viral load in RNAi mutant and WT flies. We observed a modest increase in viral titers in RNAi mutant flies compared with WT controls at 3 d postinfection (Fig. 2E). However, viral titers were similar at 12 d after infection. Our results thus imply that RNAi controls IIV-6 infection in Drosophila but that it only modestly affects viral titers.
Fig. 2.
D. melanogaster RNAi mutant flies are susceptible to IIV-6 infection. (A) Survival of Dcr-2 mutant flies after IIV-6 challenge. The survival rate of female Dcr-2L811fsX (squares) and y1w1 control (triangles) flies was monitored daily after IIV-6 (filled symbols) or mock infection (open symbols). A representative of three independent experiments is shown. (B) Survival of Dcr-2L811fsX female flies injected with UV-inactivated IIV-6 (IIV-6UV; gray squares), IIV-6 (black squares), or Tris buffer (white squares). (C) Survival of AGO2414 homozygous (diamonds) and AGO2414/+ heterozygous controls (squares) infected with IIV-6 (filled symbols) or Tris buffer (open symbols). A representative of two independent experiments is shown. (D) Survival of IIV-6–infected AGO2-null mutant flies. AGO2414/454 (gray squares), AGO2414/321 (black squares), and AGO2321/454 (black circles) transheterozygous flies, as well as AGO2414/+ (white squares) and AGO2321/+ (white circles) heterozygous controls, were infected with IIV-6, and survival was monitored daily. A representative of two independent experiments is shown. The experiments in C and D were run in parallel; the survival curve of IIV-6–infected AGO2414/+ flies is depicted in both panels. (E) Viral load in RNAi mutant and WT flies after IIV-6 infection. Dcr-2L811fsX, AGO2414, and their WT controls, y1w1 and w1118, respectively, were infected with IIV-6, and virus production in the flies was monitored over time. At each time point, three pools of four flies were homogenized, and the viral titer in the homogenate was determined by end-point dilution. Bars represent averages and SEMs of three independent pools of four flies. The horizontal line represents the detection limit of the titration.
IIV-6 Infection Triggers the Production of Dcr-2–Dependent Viral siRNAs.
dsDNA viruses are known to produce dsRNA during infection (1), presumably as a result of convergent overlapping transcription from both strands of the viral genome. The IIV-6 genome is predicted to encode 211 ORFs, of which 45% and 55% derive from the upper and lower strands, respectively (34). We refer to the viral strand that is transcribed from left to right on the conventional map as the R (upper) strand and to its complement as the L (lower) strand.
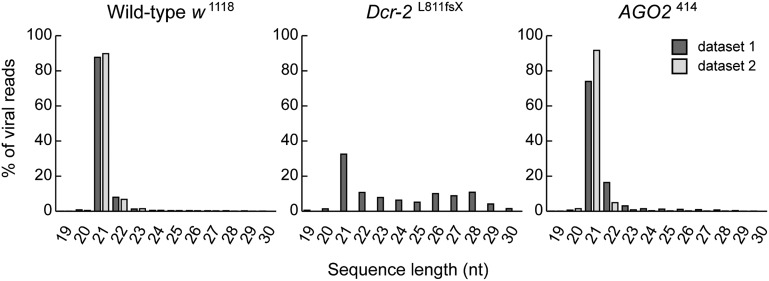
To investigate whether viral dsRNA is processed into vsiRNAs, we sequenced the small RNAs from WT and RNAi mutant flies at 12 d after infection with IIV-6. Small RNAs ranging in size from 19 to 30 nt were cloned and deep-sequenced on an Illumina Genome Analyzer. Two independent libraries of IIV-6–infected WT and AGO2 mutant flies were analyzed (datasets 1 and 2). Small RNAs were first mapped to the Drosophila genome; nonmapping small RNAs were then mapped to the IIV-6 genome (Table 1). The vast majority of IIV-6–derived small RNAs in WT and AGO2 mutant flies were 21 nt long (Fig. 3). These small RNAs were Dcr-2–dependent vsiRNAs, because the normalized levels of vsiRNAs decreased >875-fold in Dcr-2 mutant flies compared with WT flies (Table 1).
Table 1.
Descriptions of small RNA libraries
| w1118 | AGO2414 | Dcr-2L811fsx | |||
| Dataset 1 | Dataset 2 | Dataset 1 | Dataset 2 | Dataset 1 | |
| Drosophila genome, reads | 1,647,783 | 10,588,690 | 7,664,970 | 14,758,882 | 5,371,641 |
| rRNA, %* | 49.0 | 49.9 | 27.7 | 32.1 | 21.5 |
| miRNA, %* | 44.7 | 38.4 | 65.5 | 55.9 | 67.5 |
| IIV-6: 0 mismatch, %* | 8.3 | 8.2 | 2.8 | 10.1 | 0.03 |
| IIV-6: 1 mismatch, %* | 0.4 | 0.3 | 0.2 | 0.5 | 0.001 |
| IIV-6, % to miRNAs† | 17.4 | 20.0 | 3.5 | 17.5 | 0.02 |
*Descriptions of small RNAs in the size range of 19–25 nt; percentage relative to reads mapping to the Drosophila genome.
†Normalized 21-nt viral reads mapping to the IIV-6 genome, allowing 0 or 1 mismatch during alignment; percentage of viral reads relative to total cellular miRNA reads.
Fig. 3.
Production of Dcr-2–dependent viral siRNAs in IIV-6 infection. Size profiles of viral small RNAs mapping to the IIV-6 genome, allowing one mismatch during alignment, in w1118 WT (Left), Dcr-2L811fsX (Center), and AGO2414 (Right) flies are shown. Small RNA libraries were generated from a pool of 15 female flies at 12 d postinfection. Two independent libraries of IIV-6–infected WT flies and AGO2 mutants were analyzed (datasets 1 and 2; dark gray and light gray, respectively).
Dcr-2 generates 21-nt duplex siRNAs in which 19 nucleotides are base-paired, leaving 2-nt 3′ overhangs at each end. We thus analyzed 21-nt viral small RNAs for the presence of this Dcr-2 signature, as previously described (37). In contrast to our observations in infections with Nora virus, a (+) RNA virus (37), we did not observe enrichment for a 19-nt overlap among viral small RNAs mapping to opposite strands of the IIV-6 genome (Fig. S2A). Asymmetrical RISC loading preferentially retains one of the two strands of an siRNA duplex (38). Our results thus suggest that the majority of IIV-6–derived vsiRNAs do not exist as siRNA duplexes but that they are associated with RISC.
Although the size distribution of viral small RNAs was similar between AGO2 mutant and WT flies, the size distribution of the viral small RNAs in Dcr-2 mutants was vastly different. In contrast to the sharp 21-nt peak in WT flies and AGO2 mutants, small RNAs in Dcr-2 mutants showed a broader size distribution, with only a minor enrichment at 21 nt. A similar shift toward small RNAs with sizes other than 21 nt was noted before in Dcr-2 mutant flies infected with vesicular stomatitis virus (14). Recombinant and immunoprecipitated Dicer-1 (Dcr-1) is able to process long dsRNA into siRNA in in vitro assays, albeit with low efficiency (39–41). Moreover, small RNAs derived from an inverted repeat transgene could be detected by Northern blot analysis in Dcr-2 null mutants (42). We thus hypothesize that in the absence of Dcr-2, another nuclease, presumably Dcr-1, processes low levels of viral dsRNA to generate viral small RNAs. The lack of a prominent 22-nt viral RNA peak in Dcr-2 mutants (Fig. 3) suggests that IIV-6 does not produce viral microRNAs (miRNAs). Together, our data indicate that IIV-6 infection results in the production of vsiRNAs in a Dcr-2–dependent manner.
Uneven Distribution of Viral siRNA Profiles Across the IIV-6 Genome.
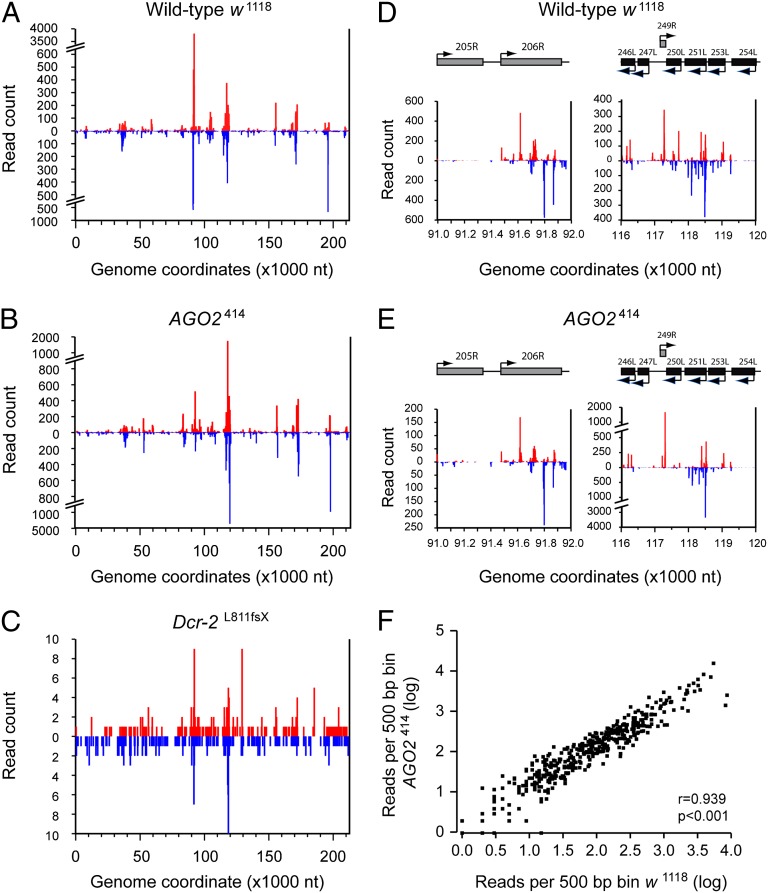
We next analyzed the distribution of vsiRNAs across the viral genome in WT and AGO2 mutant flies (Fig. 4 A and B). A global presentation of vsiRNA coverage over the viral genome indicates that vsiRNAs are derived from both strands of the viral genome. In Dcr-2 mutants (Fig. 4C), the low number of viral small RNAs also mapped to both strands of the viral genome, which is in line with our hypothesis that a low level of viral dsRNA processing may occur in a Dcr-2–independent manner. Although vsiRNAs mapped along the entire viral genome in WT and AGO2 mutant flies, a highly uneven distribution of vsiRNAs with strong clustering in small defined genomic regions (hotspots) and only modest coverage in other regions (coldspots) was observed (Fig. 4 A and B and Fig. S2B). The uneven coverage of vsiRNAs across the genome was highly reproducible between experiments, with a strong correlation between vsiRNA densities in the two independent datasets of WT and AGO2 mutant flies (r = 0.975, P < 0.001 and r = 0.967, P < 0.001, respectively; Fig. S3). Furthermore, the distribution of vsiRNAs across the genome was highly similar between WT and AGO2 mutant flies (Fig. 4 A and B). Indeed, a remarkable congruence of vsiRNA profiles in both genetic backgrounds was apparent in more detailed views of highly covered regions of the genome (representative examples are shown in Fig. 4 D and E). To substantiate the congruence between small RNA profiles in WT and AGO2 mutant flies further, we divided the viral genome into 500-bp bins and compared the number of small RNAs in each bin between these genotypes (Fig. 4F). Indeed, we observed a strong correlation between vsiRNA densities in WT and AGO2 mutant libraries in both independent datasets (r = 0.939, P < 0.001 and r = 0.959, P < 0.001 for dataset 1 and dataset 2, respectively). The concordance of vsiRNA profiles in flies with different genetic backgrounds suggests that the uneven distribution of vsiRNAs and the presence of vsiRNA hotspots in the genome are caused by factors intrinsic to the virus, such as the relative expression of viral transcripts.
Fig. 4.
Uneven distribution of viral siRNA profiles across the IIV-6 genome. (A–C) Profile of IIV-6–derived vsiRNAs across the IIV-6 genome. Viral siRNAs were aligned to the IIV-6 genome, allowing one mismatch during alignment. The genome coordinates of the 5′ end of each vsiRNA in w1118 WT (A), AGO2414 (B), and Dcr-2L811fsX (C) flies were plotted. Profile of IIV-6–derived vsiRNAs in w1118 WT (D) and AGO2414 (E) flies mapping to genome coordinates 91,000–92,000 (Left) and 116,000–120,000 (Right) that contain regions with a high density of vsiRNAs are shown. ORFs located in the genome region are depicted above the plot, drawn to scale. Viral siRNAs that mapped to the R and L strands of the IIV-6 genome are shown in red and blue, respectively. Small RNA libraries were generated from a pool of 15 female flies at 12 d postinfection. Profiles from dataset 1 are presented; profiles from dataset 2 are provided in Fig. S3. (F) The IIV-6 genome was divided into 500-bp bins, and the number of vsiRNAs per bin in w1118 WT and AGO2414 mutant flies was analyzed in a scatter plot (log-transformed data, dataset 1; r = 0.939, P < 0.001, Pearson’s correlation test).
Convergent Transcripts as a Source of Viral dsRNA in IIV-6 Infection.
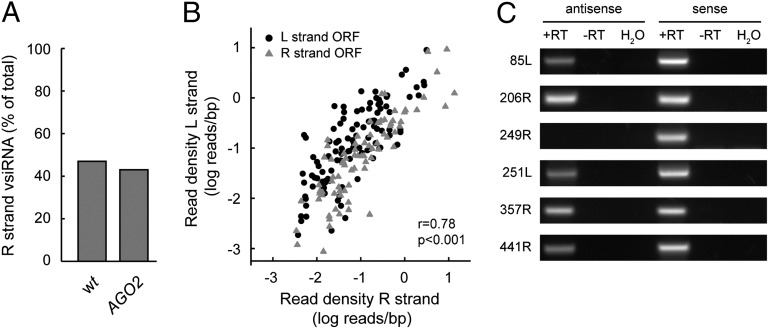
To identify the dsRNA substrate for vsiRNA biogenesis, we analyzed the distribution of the vsiRNAs corresponding to the L and R strands of IIV-6. We found that 47% and 43% of the vsiRNAs mapped to the R strand in WT and AGO2 mutant flies, respectively (Fig. 5A), closely mimicking the distribution of the ORFs over the viral genome strands (45% R strand ORFs). When restricting our analyses to individual ORFs, we also observed that vsiRNAs are derived from both the L and R strands. Moreover, the density of vsiRNAs that map to the L strand strongly correlated with the density of reads mapping to the R strand of individual ORFs (Fig. 5B; r = 0.78, P < 0.001). Thus, individual ORFs produced vsiRNAs that map to both strands of the genome, irrespective of the L or R orientation of the ORF. These results suggest that vsiRNAs predominantly derive from overlapping sense and antisense transcripts rather than from intramolecular stem-loop structures in single-stranded viral transcripts.
Fig. 5.
Convergent transcripts are a source of dsRNA during IIV-6 infection. (A) Percentage of the total vsiRNAs that map to the viral R strand in w1118 WT (wt) and AGO2414 mutant flies. (B) Scatter plot of the vsiRNAs from individual ORFs that map to the viral L and R strands in w1118 WT flies. For each ORF, the total number of vsiRNAs derived from each strand was divided by the length of the ORF. The data were log-transformed and presented in a scatter plot. Circles and triangles indicate ORFs on the viral L and R strands, respectively. Correlation between both read densities was evaluated with a Pearson’s correlation test. (C) Ethidium bromide-stained gels of strand-specific RT-PCR assays for the detection of sense and antisense transcripts from the indicated IIV-6 ORFs in infected w1118 WT flies. RT-PCR assays were performed in either the presence (+RT) or absence (−RT) of reverse transcriptase. H2O was included as a nontemplate control during PCR amplification. 85L, 206R, 249R, 251L, and 441R represent ORFs with a high coverage of vsiRNA reads per base pair (bp), whereas 357R represents an ORF with a low coverage of vsiRNAs per bp.
To analyze the occurrence of antisense transcription during IIV-6 infection, we performed strand-specific RT-PCR assays on infected w1118 WT flies. To avoid possible false-positive results due to mispriming during cDNA synthesis, we used strand-specific primers containing a nonviral tag sequence (T7 promoter) at the 5′ end, thereby generating tagged cDNA that can be PCR-amplified using a combination of a virus-specific primer with a T7 primer (43–46). We selected five ORFs with a high density of vsiRNA reads (85L, 206R, 249R, 251L, and 441R) and one ORF with a low vsiRNA density (357R). For five of six selected ORFs, we readily detected sense and antisense transcripts; for ORF 249R, we could not detect antisense transcripts using this method (Fig. 5C). All gene-specific primers efficiently amplified fragments of the selected ORFs in control PCR reactions using viral DNA as a template (Fig. S4), excluding inefficient primer binding as an explanation for our inability to detect antisense ORF 249R transcripts. These results further support the existence of bidirectional overlapping transcription in the IIV-6 genome. Because antisense transcription also occurs in genomic regions with low vsiRNA coverage (ORF 357R), these results suggest that other factors, such as the relative amounts of viral sense and antisense transcripts, may contribute to differences in vsiRNA coverage across the genome.
To confirm the presence of sense and antisense transcripts, we performed Northern blot analysis on mock and IIV-6–infected Drosophila S2 cells using oligonucleotide probes that specifically recognize either viral RNA strand (Fig. S5). For all tested ORFs (85L, 206R, and 441R), sense transcripts that matched the predicted size of the selected ORFs were readily detected. Northern blot analyses using probes that detect antisense transcripts revealed the presence of high-molecular-weight (>9 kb) antisense transcripts for all three ORFs, as well as a small antisense transcript for ORF 441R. The origin and biogenesis of these antisense transcripts await further investigation.
Together, the small RNA profiles, strand-specific RT-PCR assays, and Northern blot analyses indicate that both sense and antisense transcripts are generated during IIV-6 infection. We propose that these viral transcripts form dsRNA that activates the RNAi pathway, resulting in the production of Dcr-2–dependent vsiRNAs.
IIV-6–Derived vsiRNAs Mediate Gene Silencing.
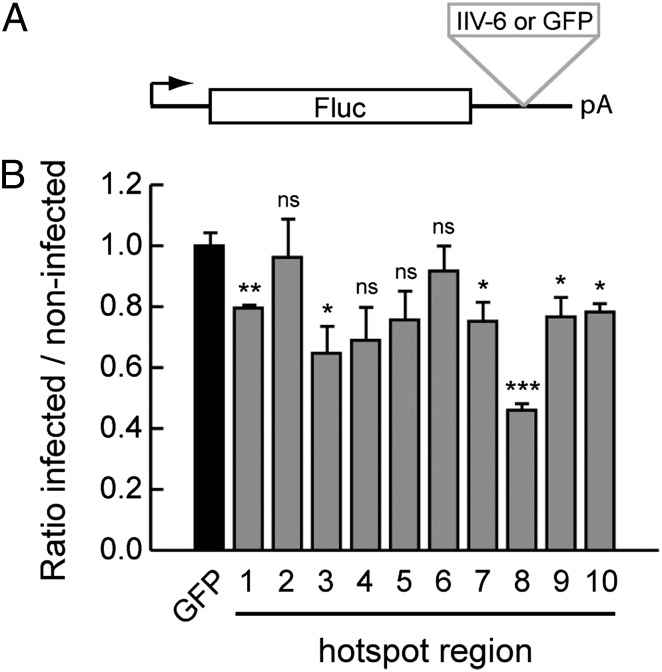
To analyze whether IIV-6–derived vsiRNAs are functional in mediating target gene silencing, we designed luciferase reporter plasmids. To this end, the IIV-6 genome was divided into 500-bp bins, from which we selected 10 hotspot regions that produced the highest number of vsiRNAs (Fig. S2B). These fragments, as well as a nonspecific GFP control sequence, were cloned in the 3′ UTR of a firefly luciferase (FLuc) reporter plasmid (Fig. 6A). Drosophila S2 cells were transfected with these reporter plasmids, together with a plasmid encoding Renilla luciferase (RLuc) as a normalization control. We then infected these cells with IIV-6 and monitored luciferase activity. If vsiRNAs are incorporated in a functional RISC, it is expected that IIV-6 infection will induce specific silencing of the IIV-6 reporters but not the GFP control plasmid. Indeed, we observed significant silencing of six IIV-6 reporters (Fig. 6B). The extent of silencing did not seem to correlate with vsiRNA density. For example, the reporter for hotspot 8 was silenced most efficiently, whereas the reporter for the hotspot with the highest density (hotspot 1) was silenced to a lesser extent. These results are in line with observations in Nicotiana benthamiana, in which no clear correlation was observed between vsiRNA density and efficiency of silencing of a sensor construct (47). Strikingly, in mosquito cells, hotspot vsiRNAs were found to be less efficient at mediating antiviral RNAi than coldspot vsiRNA, which was hypothesized to reflect a decoy strategy to evade the antiviral RNAi response (22). Whether a similar mechanism plays a role in IIV-6 infection remains to be established.
Fig. 6.
IIV-6–derived vsiRNAs mediate gene silencing. (A) Schematic representation of reporter plasmids. Sequences (500 bp) of IIV-6 genome or GFP (control) were cloned into the 3′ UTR of a copper-inducible FLuc plasmid to generate IIV-6 and GFP reporter plasmids. (B) RNAi reporter assay in Drosophila S2 cells. IIV-6 or control reporter plasmids were transfected together with RLuc plasmid 1 d before mock or IIV-6 infection. Two days after transfection, expression of FLuc and RLuc was induced, and luciferase activity was measured the day thereafter. FLuc expression was normalized to RLuc expression and presented as a ratio of infected over noninfected cells relative to the values for GFP control plasmid (set at 1). Bars represent the averages and SEMs of three independent experiments. P values were calculated using a Student’s t test. *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant.
Having established that IIV-6–derived vsiRNAs are incorporated into a functional RISC, we analyzed their effect on viral gene expression. To this end, we compared viral transcript levels in IIV-6–infected w1118 WT and AGO2414 mutant flies. We selected five ORFs with a high coverage of vsiRNA reads per base pair and analyzed expression levels using strand-specific quantitative RT-PCR (qRT-PCR) assays. Although we observed a modest, but not statistically significant, 1.6- to 1.8-fold increase in antisense and sense transcripts for one ORF (227L), we were unable to detect a general increase in viral transcript levels in AGO2 mutant flies (Fig. S6). Because we analyzed viral transcript levels at a single time point and viral gene expression is likely to be under tight temporal control, we cannot exclude the possibility that the pathology in RNAi mutant flies is due to an early disequilibrium of transcript levels (i.e., high levels of specific viral transcripts in AGO2 mutants at a specific time in infection). Future genome-wide transcriptome analyses will be required to analyze the dynamics of viral gene expression and the role of the RNAi machinery in the posttranscriptional regulation thereof.
Discussion
RNA viruses in insects are targets of an RNAi-based antiviral immune response, in which viral replication intermediates or viral dsRNA genomes are processed by Dcr-2 into vsiRNAs. In this study, we established IIV-6 as a model to study the role of RNAi in a productive but nonlethal infection by a dsDNA virus. Our results demonstrate that RNAi targets dsDNA viruses in vivo, further extending the role of RNAi as an antiviral defense mechanism.
The production of vsiRNAs during RNA virus infection is a key feature of an antiviral RNAi response. The detection of Dcr-2–dependent vsiRNAs in infected flies provides direct evidence that IIV-6 is a target of the Drosophila RNAi machinery. We observed a highly uneven distribution of vsiRNAs across the IIV-6 genome, with similar levels of vsiRNAs mapping to both strands of the viral genome. Accordingly, analysis of individual ORFs revealed a strong correlation in vsiRNA reads derived from the R and L strands of the viral genome. Furthermore, using strand-specific qRT-PCR and Northern blot analysis, we demonstrate the presence of sense and antisense viral transcripts derived from different regions in the IIV-6 genome. Finally, we show that IIV-6–derived vsiRNAs mediate gene silencing in reporter assays in S2 cells. These results support a model in which dsRNA that is generated by bidirectional overlapping transcription is the major substrate for vsiRNA biogenesis during dsDNA virus infection.
Antisense transcription is prevalent among different families of DNA viruses, and it has been suggested to affect viral gene expression (48–52). Antisense transcripts have the potential to form dsRNA by base-pairing with sense transcripts. Indeed, dsRNA can readily be detected in infections by DNA viruses in mammalian cells (1). However, although mammalian DNA viruses encode miRNAs, they do not seem to produce Dicer-dependent vsiRNAs (53, 54). For example, a recent report detected widespread antisense transcription in lytic Kaposi’s sarcoma-associated herpesvirus infection but failed to identify abundant vsiRNAs (51). Similarly, antisense transcription is very abundant throughout the genome of human cytomegalovirus (50). However, deep sequencing of the small RNAs in infected cells readily identified viral miRNAs but detected only a limited number of other viral small RNAs. These small RNAs mapped to a restricted number of defined genomic loci, and whether they are bona fide Dicer products remains unclear (55). In plants (Arabidopsis), the pararetrovirus Cauliflower mosaic virus (CaMV) also produces long antisense transcripts (56, 57). Furthermore, CaMV vsiRNAs mapped to both strands of the genome, indicating that antisense transcripts base-pair with sense transcripts to produce dsRNA substrates for members of the Dicer-like protein family. Formation of dsRNA by convergent transcription in Cassava mosaic virus, an ssDNA virus of the Geminiviridae family, was also postulated to be the source of vsiRNA production in tobacco (N. benthamiana) and cassava (Manihot esculenta) plants (58, 59).
Thus, DNA viruses in plants, insects, and mammals have an enormous potential to form dsRNA, but only in plants and insects is this dsRNA processed into vsiRNAs. The reason for this discrepancy is not obvious because in vitro and in vivo studies imply that mammalian Dicer is capable of processing long stretches of dsRNA into siRNAs (60–63). Strikingly, humans only encode a single Dicer protein, whereas plants and Drosophila encode multiple Dicer family members (two and four in Drosophila and Arabidopsis, respectively). Perhaps, the diversification of the Dicer gene family in plants and insects allowed the functional specialization and more efficient processing of viral dsRNA for antiviral defense.
Another remarkable difference between vertebrate DNA viruses and IIV-6 is the notable absence of viral miRNAs in IIV-6. Mammalian DNA viruses, including herpesviruses and polyomaviruses, produce viral miRNAs that are capable of modulating viral or host gene expression (64). Individual herpesviruses may encode multiple miRNAs; for example, human cytomegalovirus encodes 22 miRNAs in its 230-kb genome (55). In Drosophila, miRNA biogenesis involves the sequential processing of primary miRNA transcripts into pre-miRNAs by Drosha and its cofactor Pasha (together known as the Microprocessor complex) in the nucleus and the cleavage of pre-miRNA into mature miRNAs by cytoplasmic Dcr-1. Mature miRNAs are then incorporated into an Argonaute-1–containing RISC, where they trigger translational repression or degradation of the target mRNA (65). Iridoviruses are nucleocytoplasmic viruses with an initial stage of replication in the nucleus, where early viral transcripts are generated from the input virion DNA template by host RNA Polymerase II (30). These viral transcripts may follow the canonical miRNA processing pathway of cellular miRNAs. Indeed, 11 viral miRNAs were identified in Singapore grouper iridovirus, a member of the Iridoviridae family (Ranavirus genus) that infects fish (66). Analysis of our deep-sequencing data revealed the lack of a prominent 22-nt RNA peak in Dcr-2 mutants. Furthermore, we were unable to predict computationally pre-miRNA–like stem-loop structures in regions of the viral genome that give rise to the most abundant viral small RNAs. Together, these results suggest that IIV-6 does not produce viral miRNAs. However, the lack of viral miRNAs is not a general attribute of insect DNA viruses, because miRNAs have been identified in other large invertebrate DNA viruses, Heliothis virescens ascovirus and Bombix mori nucleopolyhedrosis virus (67, 68).
Our model postulates that base-pairing of overlapping converging transcripts generates the viral dsRNA substrates for vsiRNA production by Dcr-2. Consequently, vsiRNAs are 100% complementary to viral transcripts and should be able to target them for degradation. Indeed, we observed an enhanced susceptibility of RNAi mutant flies to IIV-6 infection and observed that vsiRNAs mediate silencing of reporter constructs. Nevertheless, we only observed a modest increase in viral load at early time points in RNAi mutant flies. Several hypotheses could explain this apparent paradox. First, vsiRNAs may target viral genes that are involved in viral pathogenesis but not viral replication per se. Consequently, in the absence of a functional RNAi response, expression of these putative pathogenicity factors will be increased, resulting in a more rapid onset of disease. Second, the observed IIV-6–associated mortality may be due to invasion or high-level replication in specific critical tissues. Perhaps RNAi pathway components have tissue-specific functions during IIV-6 replication that could explain the observed mild differences in viral titers in whole flies. Third, the susceptibility of RNAi mutant flies could also be attributed to additional (direct or indirect) functions of Dcr-2 and AGO2, other than controlling viral RNA levels. For example, the putative antiviral effector Vago is induced on RNA virus infection in a Dcr-2–dependent but AGO2-independent manner (69). We note that Dcr-2 mutants seem to be more severely affected by IIV-6 infection than AGO2 mutants, which would fit a Dcr-2–dependent induction of a disease-modifying activity.
It is well established that the RNAi machinery targets endogenous retroviruses and dsRNA viruses, as well as viruses with (+) or (−) RNA genomes. We now show that dsDNA virus-derived siRNAs are produced by Dcr-2 and that these vsiRNAs provide defense against infection. Thus, RNAi in insects is the predominant antiviral defense mechanism that provides protection against all major classes of viruses.
Materials and Methods
Virus Titration.
IIV-6 was kindly supplied by C. Joel Funk (US Department of Agriculture-Agricultural Research Service Western Cotton Research Laboratory, Phoenix, AZ) and was propagated in Galleria mellonella (greater wax moth) larvae and purified by 25–65% (wt/vol) sucrose density gradient centrifugation as described previously (70, 71). IIV-6 UV-inactivated virus was produced by exposing the virus stock to a total of 12,000 mJ of UV light in four intervals of 3 min (72).
Viral titers were determined by end-point dilution. Drosophila S2 cells (Invitrogen) were seeded in 96-well plates at a density of 2.5 × 104 cells per well. Subsequently, cells were infected with 10-fold serial dilutions of fly homogenate or virus suspension. Each infection was performed in quadruplicate. IIV-6 infection was scored by a lack of proliferation of cells and by the presence of cell debris and large cells, presumably syncytia. Viral titers were calculated according to the method of Reed and Muench (73).
For titration of infectious virus in adult flies, three pools of four flies were harvested and stored at −70 °C until further processing. Each pool of flies was homogenized in 150 μL of PBS, and fly debris was removed by centrifugation for 10 min at 16,000 × g at 4 °C. The supernatant was transferred to a fresh tube and, before titration, centrifuged again for 5 min at 16,000 × g at room temperature.
Clearance of Wolbachia- and Virus-Infected Flies and Validation by PCR Assay.
To clean fly stocks from persistent viruses, eggs were collected on apple juice agar plates and treated with 50% (vol/vol) household bleach for 5 min. After three extensive washes with water, eggs were transferred to fresh vials containing standard fly food. After culturing for two subsequent generations, we verified the absence of Nora virus and DCV by RT-PCR assay.
For RT-PCR assays, total RNA was extracted from pools of 15 flies using Isol-RNA Lysis Reagent (5 Prime) according to the manufacturer’s recommendations. cDNA synthesis was performed on 1 μg of DNase-I (Invitrogen)–treated total RNA using random hexamers and Taqman reverse transcriptase (Roche). Primers used for PCR amplification of Nora virus were NoV 1370F (5′-ATGGCGCCAGTTAGTGCAGACCT-3′) and NoV 1780R (5′-CCTGTTGTTCCAGTTGGGTTCGA-3′). DCV primers were DCV 1947F (5′-TTGATCTAGATACTGAAACCGCAAATCGTG-3′) and DCV 2528R (5′-TCGCCCATACGATTAAAGAAA-3′). As positive control for RNA extraction, we used Act42A 19F (5′-GCGTCGGTCAATTCAATCTT-3′) and Act42A 386R (5′-CTTCTCCATGTCGTCCCAGT-3′). The PCR program used was 94 °C for 5 min; 35 cycles of 94 °C for 30 s, 57 °C for 30 s, and 72 °C for 50 s; and 72 °C for 10 min.
To clear fly stocks from Wolbachia infection, flies were raised for two generations using standard fly food containing 0.05 mg/mL tetracycline hydrochloride (Sigma). Clearance of Wolbachia infection was validated by PCR amplification of Wolbachia-specific genes from DNA extracts of adult flies, as described previously (32).
Fly Stocks and Virus Infections.
Virus- and Wolbachia-cleared flies were reared on standard medium at 25 °C with a light/dark cycle of 12 h/12 h. We used the following alleles: AGO2414 (74), AGO2321, AGO2454 (36), and Dcr-2L811fsX (42). We used w1118 and y1w1 flies as WT controls for AGO2 and Dcr-2 mutant flies, respectively. For generation of AGO2 transheterozygous mutant flies, AGO2414 homozygous flies were crossed with AGO2454/TM3Sb or AGO2321/TM3Sb flies, and the F1 progeny was selected based on the absence of the balancer. Likewise, for generation of AGO2321/454 transheterozygous flies, AGO2321/TM3Sb flies were crossed with AGO2454/TM3Sb flies. Heterozygous controls were generated by crossing AGO2 flies with w1118 flies.
Female flies (2–4 d of age) were injected with 50 nL of the appropriate virus dilution [containing 14,000 median (50%) tissue culture infectious dose (TCID50) units of IIV-6, 500 TCID50 units of DCV, or 2,500 TCID50 units of CrPV) in 10 mM Tris⋅HCl (pH 7.5) or with 10 mM Tris⋅HCl (pH 7.5) as a mock infection, as described previously (13). Flies were injected in the anterior ventral abdomen near the dorsal-ventral boundary while anesthetized with CO2 (75). Flies were cultured at 25 °C, and mortality was monitored daily. Every 3 to 4 d, the flies were transferred to fresh food. Fly mortality at day 1 was attributed to damage invoked by the injection procedure and was excluded from survival analysis. In all survival experiments, 20–30 female flies per genotype were injected.
Western Blots.
Pools of five IIV-6–infected or mock-infected female flies were homogenized in lysis buffer [50 mM Tris (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% (vol/vol) Nonidet P-40, 0.05% (wt/vol) SDS, 1 mM DTT, 0.5 mM PMSF, and complete protease inhibitor mixture (Roche)] for 30 min at 4 °C. Fly lysates were centrifuged for 10 min at 16,000 × g at 4 °C. The supernatant was transferred to a fresh tube and centrifuged again for 5 min at 16,000 × g at room temperature. For Western blot analysis, SDS-solubilized proteins were separated on a 10% (wt/vol) denaturing polyacrylamide gel and transferred to a nitrocellulose membrane for detection. For the detection of virus-specific proteins, polyclonal rabbit antibodies raised against IIV-6 virion coat proteins were diluted (1:2,000) in blocking buffer [5% (wt/vol) skimmed milk, 0.1% (vol/vol) Tween 20 in PBS]. The blot was incubated for 1 h at room temperature with the primary antibody. Subsequently, the blot was incubated with a secondary antibody, HRP-conjugated polyclonal goat anti-rabbit IgG (Dako), for 1 h in blocking buffer (1:5,000 dilution). Bound antibodies were detected using chemiluminescence (Roche). As a loading control, the same blot was probed with a rat anti–α-tubulin antibody (1:1,000 dilution; AbD Serotec) as a primary antibody. Goat anti–rat-IRdye680 antibody (1:15,000 dilution; Invitrogen) was used as a secondary antibody. Bound antibodies were visualized on an Odyssey infrared imager (LI-COR Biosciences).
Small RNA Library Preparation and Analysis.
Total RNA was extracted from pools of 15 IIV-6–infected female flies harvested at 12 d postinfection, using Isol-RNA Lysis reagent. Small RNA libraries were prepared as previously described (76) and sequenced on a Genome Analyzer IIx (Illumina). Small RNA libraries were analyzed as described previously (77). Briefly, small RNA reads were first mapped to the D. melanogaster genome, allowing no mismatches during alignment. Nonmapping reads were then aligned to the IIV-6 reference genome (National Center for Biotechnology Information database, accession no. AF303741), allowing one mismatch. The IIV-6 genome annotation as proposed by Eaton et al. (34) was used to analyze read density per ORF. Correlations were evaluated using Pearson’s correlation test as implemented in the statistical package SPSS (IBM SPSS). Sequences were submitted to the Sequence Read Archive at the National Center for Biotechnology (accession no. SRA048623).
Strand-Specific RT-PCR Assays.
For strand-specific RT-PCR assays, a pool of 10 IIV-6–infected w1118 flies (14,000 TCID50 units) was harvested at 12 d postinfection. RNA was isolated from the flies using Isol-RNA Lysis Reagent and treated with DNase-I (Ambion). cDNA synthesis was performed on 400 ng of DNase I-treated RNA using TaqMan Reverse Transcription Reagents in a 20-μL reaction according to the manufacturer’s instructions (Applied Biosystems) utilizing strand-specific primers tagged with a 5′ T7 promoter sequence at a final concentration of 0.2 μM (Table S1). Following cDNA synthesis, PCR analysis was performed using a combination of an IIV-6–specific primer and a primer specific for the T7 promoter sequence (Table S1). One microliter of the cDNA product was used in a 20-μL PCR assay containing 2.5 mM MgCl2, 1× reaction buffer, 0.2 mM dNTPs, 0.5 μM of each primer, and 0.625 U/μL Thermoperfect DNA polymerase (Integro). The PCR program was as follows: 95 °C for 5 min; 40 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s; and 72 °C for 10 min. RT-PCR products were analyzed on 2% (wt/vol) agarose gels. Several control reactions were run in parallel for each sample. cDNA synthesis without reverse transcriptase and PCR amplification without cDNA template were performed to verify the absence of contaminating DNA in RNA preparations and PCR reagents, respectively. A control PCR assay was performed on proteinase K (Qiagen)-treated virus stock to verify primer efficiency (Fig. S4).
For strand-specific qRT-PCR assays, RNA was isolated from three pools of 25 IIV-6–infected w1118 and AGO2414 flies at 12 d postinfection. RNA isolation, DNase-I treatment, and cDNA synthesis with T7 promoter-tagged primers were performed as described above. qPCR analysis using a combination of T7 promoter and IIV-6–specific primers was performed on a LightCycler 480 using LightCycler 480 SYBR Green I Master (Roche). The PCR program was as follows: 95 °C for 5 min and 45 cycles of 95 °C for 5 s, 60 °C for 10 s, and 72 °C for 20 s. The data were normalized to Rp49, for which qRT-PCR assays were run in parallel. The entire experiment was performed three times.
Northern Blots.
S2 cells were either mock-infected or infected with 4.23 × 106 TCID50 units of IIV-6. At 3 d postinfection, RNA was isolated from the cells using Isol-RNA Lysis Reagent. The RNA (10 μg per lane) was separated on 1.5% (wt/vol) agarose/formaldehyde gel and transferred onto Nytran SuPerCharge nylon membranes (Whatman). Blots were hybridized with 32P end-labeled oligonucleotide DNA probes for specific detection of sense and antisense transcripts of selected ORFs. Probes for ORF 85L were 85L antisense (5′-tggcttgcgggatatttggatgcagatggctgcgtc-3′) and 85L sense (5′-gacgcagccatctgcatccaaatatcccgcaagcca-3′). ORF 206R probes were 206R antisense (5′-gggaaactcagaagaaaaaggaaagtggcgagtacg-3′) and 206R sense (5′-cgtactcgccactttcctttttcttctgagtttccc-3′). Probes for ORF 441R were 441 antisense (5′-cctcttatagagacttggcaaagtttgccgatcctg-3′) and 441R sense (5′-caggatcggcaaactttgccaagtctctataagagg-3′). Blots were hybridized in ULTRAhyb-Oligo hybridization buffer (Ambion) according to the manufacturer’s instructions and visualized by autoradiography. Blots of gels with in vitro transcribed RNA (25 ng per lane) were run in parallel as positive controls (Table S2).
IIV-6 Sensor Assay in S2 Cells.
To generate IIV-6 sensor plasmids, we amplified ten 500-bp regions of the IIV-6 genome from proteinase K-treated virus stock by PCR assay using the primers from Table S3. PCR fragments were cloned in the 3′ UTR of the FLuc gene in the pMT-GL3 plasmid (13) using the PspOMI and PmeI restriction sites. As a nonspecific control, a 500-bp fragment of GFP (nucleotides 192–691) was amplified from the pEGFP-N1 plasmid (Clontech) and cloned in a similar manner. The pMT-Ren plasmid (13) was used as a transfection control.
For the RNAi reporter assay, 1.5 × 105 S2 cells were seeded in a 24-well plate. Twenty-four hours postseeding, cells were transfected with 12 ng of pMT-Ren and 50 ng of an IIV-6 sensor or GFP control plasmid, as described (78). Twenty-four hours after transfection, cells were mock-infected or infected with 21,000 TCID50 units of IIV-6. Expression of the FLuc and RLuc reporters was induced at 24 h after infection by adding CuSO4 at a final concentration of 500 μM to the culture supernatant. After incubation for an additional 18 h, luciferase activities were measured using the Dual Luciferase reporter system (Promega).
Supplementary Material
Acknowledgments
We thank Lionel Frangeul for bioinformatic support; and P. Zamore, A. Müller, and the Bloomington Drosophila Stock Center for providing fly stocks. This work was financially supported by a VIDI fellowship (Project 864.08.003), the Open Program of the Division for Earth and Life Sciences from the Netherlands Organization for Scientific Research (Project 821.08.003), a Horizon Breakthrough fellowship from the Netherlands Genomics Initiative (Project 93519018), and a fellowship from the Nijmegen Centre for Molecular Life Sciences (to R.P.v.R.), as well as by Grant ANR-09-394 JCJC-0045-01 from the French Agence Nationale de la Recherche and Grant FP7/2007-2013 ERC 242703 from the European Research Council (to M.-C.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the Sequence Read Archive database (accession no. SRA048623).
See Author Summary on page 20792 (volume 109, number 51).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207213109/-/DCSupplemental.
References
- 1.Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol. 2006;80(10):5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kato H, Takahasi K, Fujita T. RIG-I-like receptors: Cytoplasmic sensors for non-self RNA. Immunol Rev. 2011;243(1):91–98. doi: 10.1111/j.1600-065X.2011.01052.x. [DOI] [PubMed] [Google Scholar]
- 3.Robalino J, et al. Induction of antiviral immunity by double-stranded RNA in a marine invertebrate. J Virol. 2004;78(19):10442–10448. doi: 10.1128/JVI.78.19.10442-10448.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130(3):413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Rij RP, Berezikov E. Small RNAs and the control of transposons and viruses in Drosophila. Trends Microbiol. 2009;17(4):139–178. doi: 10.1016/j.tim.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Kawamata T, Tomari Y. Making RISC. Trends Biochem Sci. 2010;35(7):368–376. doi: 10.1016/j.tibs.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 7.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136(4):642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blair CD. Mosquito RNAi is the major innate immune pathway controlling arbovirus infection and transmission. Future Microbiol. 2011;6(3):265–277. doi: 10.2217/fmb.11.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanchez-Vargas I, et al. RNA interference, arthropod-borne viruses, and mosquitoes. Virus Res. 2004;102(1):65–74. doi: 10.1016/j.virusres.2004.01.017. [DOI] [PubMed] [Google Scholar]
- 10.Li HW, Li WX, Ding SW. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296(5571):1319–1321. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- 11.Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL. Essential function in vivo for Dicer-2 in host defense against RNA viruses in drosophila. Nat Immunol. 2006;7(6):590–597. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- 12.Wang XH, et al. RNA interference directs innate immunity against viruses in adult Drosophila. Science. 2006;312(5772):452–454. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Rij RP, et al. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 2006;20(21):2985–2995. doi: 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mueller S, et al. RNAi-mediated immunity provides strong protection against the negative-strand RNA vesicular stomatitis virus in Drosophila. Proc Natl Acad Sci USA. 2010;107(45):19390–19395. doi: 10.1073/pnas.1014378107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brackney DE, Beane JE, Ebel GD. RNAi targeting of West Nile virus in mosquito midguts promotes virus diversification. PLoS Pathog. 2009;5(7):e1000502. doi: 10.1371/journal.ppat.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brackney DE, et al. C6/36 Aedes albopictus cells have a dysfunctional antiviral RNA interference response. PLoS Negl Trop Dis. 2010;4(10):e856. doi: 10.1371/journal.pntd.0000856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myles KM, Wiley MR, Morazzani EM, Adelman ZN. Alphavirus-derived small RNAs modulate pathogenesis in disease vector mosquitoes. Proc Natl Acad Sci USA. 2008;105(50):19938–19943. doi: 10.1073/pnas.0803408105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu Q, et al. Virus discovery by deep sequencing and assembly of virus-derived small silencing RNAs. Proc Natl Acad Sci USA. 2010;107(4):1606–1611. doi: 10.1073/pnas.0911353107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aliyari R, et al. Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila. Cell Host Microbe. 2008;4(4):387–397. doi: 10.1016/j.chom.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flynt A, Liu N, Martin R, Lai EC. Dicing of viral replication intermediates during silencing of latent Drosophila viruses. Proc Natl Acad Sci USA. 2009;106(13):5270–5275. doi: 10.1073/pnas.0813412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scott JC, et al. Comparison of dengue virus type 2-specific small RNAs from RNA interference-competent and -incompetent mosquito cells. PLoS Negl Trop Dis. 2010;4(10):e848. doi: 10.1371/journal.pntd.0000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siu RW, et al. Antiviral RNA interference responses induced by Semliki Forest virus infection of mosquito cells: Characterization, origin, and frequency-dependent functions of virus-derived small interfering RNAs. J Virol. 2011;85(6):2907–2917. doi: 10.1128/JVI.02052-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morazzani EM, Wiley MR, Murreddu MG, Adelman ZN, Myles KM. Production of virus-derived ping-pong-dependent piRNA-like small RNAs in the mosquito soma. PLoS Pathog. 2012;8(1):e1002470. doi: 10.1371/journal.ppat.1002470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vodovar N, et al. Arbovirus-derived piRNAs exhibit a ping-pong signature in mosquito cells. PLoS ONE. 2012;7(1):e30861. doi: 10.1371/journal.pone.0030861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma M, et al. Discovery of DNA viruses in wild-caught mosquitoes using small RNA high throughput sequencing. PLoS ONE. 2011;6(9):e24758. doi: 10.1371/journal.pone.0024758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar M, Carmichael GG. Antisense RNA: Function and fate of duplex RNA in cells of higher eukaryotes. Microbiol Mol Biol Rev. 1998;62(4):1415–1434. doi: 10.1128/mmbr.62.4.1415-1434.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobs BL, Langland JO. When two strands are better than one: The mediators and modulators of the cellular responses to double-stranded RNA. Virology. 1996;219(2):339–349. doi: 10.1006/viro.1996.0259. [DOI] [PubMed] [Google Scholar]
- 28.Friesen PD, Miller LK. The regulation of baculovirus gene expression. Curr Top Microbiol Immunol. 1986;131:31–49. doi: 10.1007/978-3-642-71589-1_3. [DOI] [PubMed] [Google Scholar]
- 29.Unckless RL. A DNA virus of Drosophila. PLoS ONE. 2011;6(10):e26564. doi: 10.1371/journal.pone.0026564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams T, Barbosa-Solomieu V, Chinchar VG. A decade of advances in iridovirus research. Adv Virus Res. 2005;65:173–248. doi: 10.1016/S0065-3527(05)65006-3. [DOI] [PubMed] [Google Scholar]
- 31.Constantino M, Christian P, Marina CF, Williams T. A comparison of techniques for detecting Invertebrate iridescent virus 6. J Virol Methods. 2001;98(2):109–118. doi: 10.1016/s0166-0934(01)00356-1. [DOI] [PubMed] [Google Scholar]
- 32.Teixeira L, Ferreira A, Ashburner M. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 2008;6(12):e2. doi: 10.1371/journal.pbio.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jakob NJ, Müller K, Bahr U, Darai G. Analysis of the first complete DNA sequence of an invertebrate iridovirus: Coding strategy of the genome of Chilo iridescent virus. Virology. 2001;286(1):182–196. doi: 10.1006/viro.2001.0963. [DOI] [PubMed] [Google Scholar]
- 34.Eaton HE, et al. Comparative genomic analysis of the family Iridoviridae: Re-annotating and defining the core set of iridovirus genes. Virol J. 2007;4:11. doi: 10.1186/1743-422X-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams T. Iridoviruses of invertebrates. In: Mahy BWJ, Van Regenmortel MHV, editors. Encyclopedia of Virology. 3rd Ed. Oxford: Elsevier; 2008. pp. 161–167. [Google Scholar]
- 36.Hain D, et al. Natural variation of the amino-terminal glutamine-rich domain in Drosophila argonaute2 is not associated with developmental defects. PLoS ONE. 2010;5(12):e15264. doi: 10.1371/journal.pone.0015264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Mierlo JT, et al. Convergent evolution of argonaute-2 slicer antagonism in two distinct insect RNA viruses. PLoS Pathog. 2012;8(8):e1002872. doi: 10.1371/journal.ppat.1002872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwarz DS, et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115(2):199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 39.Jiang F, et al. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev. 2005;19(14):1674–1679. doi: 10.1101/gad.1334005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saito K, Ishizuka A, Siomi H, Siomi MC. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 2005;3(7):e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cenik ES, et al. Phosphate and R2D2 restrict the substrate specificity of Dicer-2, an ATP-driven ribonuclease. Mol Cell. 2011;42(2):172–184. doi: 10.1016/j.molcel.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee YS, et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117(1):69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 43.Purcell MK, Hart SA, Kurath G, Winton JR. Strand-specific, real-time RT-PCR assays for quantification of genomic and positive-sense RNAs of the fish rhabdovirus, Infectious hematopoietic necrosis virus. J Virol Methods. 2006;132(1-2):18–24. doi: 10.1016/j.jviromet.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 44.Plaskon NE, Adelman ZN, Myles KM. Accurate strand-specific quantification of viral RNA. PLoS ONE. 2009;4(10):e7468. doi: 10.1371/journal.pone.0007468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lanford RE, Sureau C, Jacob JR, White R, Fuerst TR. Demonstration of in vitro infection of chimpanzee hepatocytes with hepatitis C virus using strand-specific RT/PCR. Virology. 1994;202(2):606–614. doi: 10.1006/viro.1994.1381. [DOI] [PubMed] [Google Scholar]
- 46.Komurian-Pradel F, et al. Strand specific quantitative real-time PCR to study replication of hepatitis C virus genome. J Virol Methods. 2004;116(1):103–106. doi: 10.1016/j.jviromet.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 47.Szittya G, et al. Structural and functional analysis of viral siRNAs. PLoS Pathog. 2010;6(4):e1000838. doi: 10.1371/journal.ppat.1000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bego M, Maciejewski J, Khaiboullina S, Pari G, St Jeor S. Characterization of an antisense transcript spanning the UL81-82 locus of human cytomegalovirus. J Virol. 2005;79(17):11022–11034. doi: 10.1128/JVI.79.17.11022-11034.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prang N, Wolf H, Schwarzmann F. Latency of Epstein-Barr virus is stabilized by antisense-mediated control of the viral immediate-early gene BZLF-1. J Med Virol. 1999;59(4):512–519. doi: 10.1002/(sici)1096-9071(199912)59:4<512::aid-jmv15>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 50.Zhang G, et al. Antisense transcription in the human cytomegalovirus transcriptome. J Virol. 2007;81(20):11267–11281. doi: 10.1128/JVI.00007-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin YT, et al. Small RNA profiling reveals antisense transcription throughout the KSHV genome and novel small RNAs. RNA. 2010;16(8):1540–1558. doi: 10.1261/rna.1967910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Munroe SH, Zhu J. Overlapping transcripts, double-stranded RNA and antisense regulation: A genomic perspective. Cell Mol Life Sci. 2006;63(18):2102–2118. doi: 10.1007/s00018-006-6070-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Umbach JL, Cullen BR. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 2009;23(10):1151–1164. doi: 10.1101/gad.1793309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pfeffer S, et al. Identification of virus-encoded microRNAs. Science. 2004;304(5671):734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 55.Stark TJ, Arnold JD, Spector DH, Yeo GW. High-resolution profiling and analysis of viral and host small RNAs during human cytomegalovirus infection. J Virol. 2012;86(1):226–235. doi: 10.1128/JVI.05903-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blevins T, et al. Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 2006;34(21):6233–6246. doi: 10.1093/nar/gkl886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blevins T, et al. Massive production of small RNAs from a non-coding region of Cauliflower mosaic virus in plant defense and viral counter-defense. Nucleic Acids Res. 2011;39(12):5003–5014. doi: 10.1093/nar/gkr119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Akbergenov R, et al. Molecular characterization of geminivirus-derived small RNAs in different plant species. Nucleic Acids Res. 2006;34(2):462–471. doi: 10.1093/nar/gkj447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chellappan P, Vanitharani R, Pita J, Fauquet CM. Short interfering RNA accumulation correlates with host recovery in DNA virus-infected hosts, and gene silencing targets specific viral sequences. J Virol. 2004;78(14):7465–7477. doi: 10.1128/JVI.78.14.7465-7477.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002;21(21):5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118(1):57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 62.Billy E, Brondani V, Zhang H, Müller U, Filipowicz W. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc Natl Acad Sci USA. 2001;98(25):14428–14433. doi: 10.1073/pnas.261562698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paddison PJ, Caudy AA, Hannon GJ. Stable suppression of gene expression by RNAi in mammalian cells. Proc Natl Acad Sci USA. 2002;99(3):1443–1448. doi: 10.1073/pnas.032652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cullen BR. Viruses and microRNAs: RISCy interactions with serious consequences. Genes Dev. 2011;25(18):1881–1894. doi: 10.1101/gad.17352611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yan Y, et al. Identification of a novel marine fish virus, Singapore grouper iridovirus-encoded microRNAs expressed in grouper cells by Solexa sequencing. PLoS ONE. 2011;6(4):e19148. doi: 10.1371/journal.pone.0019148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Singh J, Singh CP, Bhavani A, Nagaraju J. Discovering microRNAs from Bombyx mori nucleopolyhedrosis virus. Virology. 2010;407(1):120–128. doi: 10.1016/j.virol.2010.07.033. [DOI] [PubMed] [Google Scholar]
- 68.Hussain M, Taft RJ, Asgari S. An insect virus-encoded microRNA regulates viral replication. J Virol. 2008;82(18):9164–9170. doi: 10.1128/JVI.01109-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Deddouche S, et al. The DExD/H-box helicase Dicer-2 mediates the induction of antiviral activity in drosophila. Nat Immunol. 2008;9(12):1425–1432. doi: 10.1038/ni.1664. [DOI] [PubMed] [Google Scholar]
- 70.Ince IA, Boeren SA, van Oers MM, Vervoort JJ, Vlak JM. Proteomic analysis of Chilo iridescent virus. Virology. 2010;405(1):253–258. doi: 10.1016/j.virol.2010.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Marina CF, Arredondo-Jiménez JI, Castillo A, Williams T. Sublethal effects of iridovirus disease in a mosquito. Oecologia. 1999;119(3):383–388. doi: 10.1007/s004420050799. [DOI] [PubMed] [Google Scholar]
- 72.Hedges LM, Johnson KN. Induction of host defence responses by Drosophila C virus. J Gen Virol. 2008;89(Pt 6):1497–1501. doi: 10.1099/vir.0.83684-0. [DOI] [PubMed] [Google Scholar]
- 73.Reed LJ, Muench H. A simple method of estimating fifty percent endpoints. Am J Hyg. 1938;27(3):493–497. [Google Scholar]
- 74.Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18(14):1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cherry S, Perrimon N. Entry is a rate-limiting step for viral infection in a Drosophila melanogaster model of pathogenesis. Nat Immunol. 2004;5(1):81–87. doi: 10.1038/ni1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gausson V, Saleh MC. Viral small RNA cloning and sequencing. Methods Mol Biol. 2011;721:107–122. doi: 10.1007/978-1-61779-037-9_6. [DOI] [PubMed] [Google Scholar]
- 77.Vodovar N, Goic B, Blanc H, Saleh MC. In silico reconstruction of viral genomes from small RNAs improves viral-derived small interfering RNA profiling. J Virol. 2011;85(21):11016–11021. doi: 10.1128/JVI.05647-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van Cleef KW, van Mierlo JT, van den Beek M, van Rij RP. Identification of viral suppressors of RNAi by a reporter assay in Drosophila S2 cell culture. Methods Mol Biol. 2011;721:201–213. doi: 10.1007/978-1-61779-037-9_12. [DOI] [PubMed] [Google Scholar]