Abstract
Peptide hormones are powerful regulators of various biological processes. To guarantee continuous availability and function, peptide hormone secretion must be tightly coupled to its biosynthesis. A simple but efficient way to provide such regulation is through an autocrine feedback mechanism in which the secreted hormone is “sensed” by its respective receptor and initiates synthesis at the level of transcription and/or translation. Such a secretion–biosynthesis coupling has been demonstrated for insulin; however, because of insulin’s unique role as the sole blood glucose-decreasing peptide hormone, this coupling is considered an exception rather than a more generally used mechanism. Here we provide evidence of a secretion–biosynthesis coupling for glucagon, one of several peptide hormones that increase blood glucose levels. We show that glucagon, secreted by the pancreatic α cell, up-regulates the expression of its own gene by signaling through the glucagon receptor, PKC, and PKA, supporting the more general applicability of an autocrine feedback mechanism in regulation of peptide hormone synthesis.
Keywords: gene expression, signal transduction
Peptide hormones are powerful regulators of various biological processes, including gene expression, metabolism, cell cycle, motility, and apoptosis. To guarantee continuous availability of the hormone and thereby its function, hormone secretion must be tightly coupled to its biosynthesis. A simple but efficient way to provide such regulation is through an autocrine feedback mechanism in which the secreted hormone is “sensed” by its respective receptor and initiates synthesis at the level of transcription and/or translation. We and others have described such an autocrine feedback mechanism for the peptide hormone insulin in both rodent and human pancreatic β cells (1–11). After exocytosis of insulin, a portion of the secreted insulin binds to A-type insulin receptors and, by signaling via PI3 kinase, up-regulates (prepro)insulin gene transcription, which contributes to increased translation.
Given the unique role of insulin as the sole blood glucose-decreasing peptide hormone, it is unclear whether positive autocrine secretion–biosynthesis coupling is a unique phenomenon rather than a more generally used mechanism. We tested the more general role of an autocrine secretion–biosynthesis coupling using glucagon as an example. Glucagon is one of several peptide hormones that increase blood glucose levels. We show that glucagon, secreted by pancreatic α cells, up-regulates the expression of its own gene by signaling through the glucagon receptor, PKC, and PKA, supporting a more generalized applicability of an autocrine feedback mechanism in the regulation of peptide hormone synthesis.
Results and Discussion
Secreted Glucagon Up-Regulates Transcription of Its Own Gene and Renewed Synthesis of Glucagon.
To test whether at all glucagon is able to up-regulate the expression of its own gene, we first stimulated mouse and human islets, as well as clonal glucagon-producing αTC1-9 cells, with exogenous glucagon at a glucose concentration that does not trigger glucagon exocytosis and then analyzed (prepro)glucagon mRNA levels. Stimulation of mouse and human islets for 15 min with 200 nM glucagon led to a greater than threefold increase in (prepro)glucagon mRNA levels by 60 min after the start of stimulation (Fig. 1A). Similarly, a twofold increase in (prepro)glucagon mRNA levels was observed in αTC1-9 cells stimulated with exogenously applied glucagon (Fig. 1C). The increase in (prepro)glucagon mRNA levels was only transient, however, and began to decline at 90 min after the start of stimulation in islets and by 120 min after the start of stimulation in αTC1-9 cells (Fig. 1 B and C).
Fig. 1.
Secreted or exogenously added glucagon stimulates (prepro)glucagon gene transcription and glucagon biosynthesis. (A) (Prepro)glucagon mRNA levels in cultured human and mouse islets at 60 min after the start of stimulation with exogenously added glucagon (200 nM for 15 min). (Prepro)glucagon mRNA levels represent the percentage of mRNA levels of nonstimulated control (given as 100%); n = 5. Data are expressed as mean ± SEM. *P < 0.05; **P < 0.01. (B and C) (Prepro)glucagon mRNA levels in cultured mouse islets (B) and clonal αTC1-9 cells (C) at indicated time points after the start of stimulation with exogenously added glucagon (200 nM for 15 min). (Prepro)glucagon mRNA levels represent the percentage of mRNA levels of nonstimulated control (given as 100%); n = 3. Data are expressed as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001. (D and E) Online monitoring of glucagon promoter-driven DsRed2 expression in transfected αTC1-9 cells. (E) Effects of different amounts of exogenously added glucagon on glucagon promoter-driven and CMV promoter-driven DsRed2 expression. Data represent the ratio of DsRed2 fluorescence values obtained at 240 min and 60 min, expressed as mean ± SEM for at least 15 monitored cells. *P < 0.05; **P < 0.01. (F) Effect of immunoabsorption of secreted glucagon on glucagon promoter-driven DsRed2 expression in transfected αTC1-9 cells. Shown are the effects of glucose at 16.7 mM (1) and 1 mM (2) without immunoabsorption; effects of immunoabsortion with anti-glucagon antibodies at 1 mM glucose (3) and 16.7 mM glucose (4); and effect of control IgG at 1 mM glucose (5). Data represent the ratio of DsRed2 fluorescence values obtained at 240 min and 60 min and are expressed as mean ± SEM for at least 15 monitored cells. (2–5) Significance vs. (1): **P < 0.01. ns: not significant. (3–5) Significance vs. (2): #P < 0.01. ns: not significant. (G) Effects of exogenously added or secreted glucagon on glucagon biosynthesis. αTC1-9 cells were incubated at 16.7 mM glucose without stimulation (1) or stimulated with either 200 nM exogenous glucagon at 16.7 mM glucose (2) or 1 mM glucose (3) for 15 min, and glucagon biosynthesis was analyzed by incorporation of 3H-labeled leucine for 60 min. Data are expressed as mean ± SEM for five independent experiments. **P < 0.01.
To analyze the mechanisms underlying this positive feedback action in more detail, we performed a reporter gene assay in which expression of the red fluorescent protein DsRed2 was controlled by the rat glucagon promoter (−776/+7 bp). This allowed the analysis of stimulus-induced glucagon promoter activity in living α cells at the single-cell level. Similar to the dynamics described for insulin promoter-driven or c-fos promoter-driven DsRed2 expression/fluorescence in insulin-producing cells (12, 13), DsRed2 fluorescence in glucagon-producing αTC1-9 cells began to increase by 60 min after the start of glucagon stimulation for 15 min and continued to increase up to 240 min (Fig. 1D). The delayed onset of fluorescence increase is related to the time required for protein synthesis of DsRed2 and its maturation into a fluorophore (Fig. S1A). The increase in fluorescence plateaus at 260 min, reflecting a decrease in glucagon promoter activity. A decline in fluorescence does not occur, owing to the long half-life of the fluorescent protein. In contrast to the glucagon promoter-driven expression, no increase in DsRed2 fluorescence was observed in control cells transfected with a cytomegalovirus (CMV) promoter-driven construct. Testing different concentrations of exogenously applied glucagon showed that stimulation with 100 pM glucagon for 15 min allowed maximal activation of the reporter gene (Fig. 1E).
We next tested whether secretion of the hormone leads to up-regulation of glucagon promoter activity. Glucagon-producing αTC1-9 cells secrete glucagon at glucose concentrations below 5 mM (14) (Fig. S1B). Glucagon promoter activity was elevated under conditions that stimulated glucagon secretion (Fig. S1C). Combining glucose-evoked glucagon secretion with immunoabsorption of secreted glucagon by anti-glucagon antibodies prevented the increase in glucagon promoter-driven DsRed2 expression, whereas application of a control IgG had no inhibitory effect (Fig. 1F). Finally, we studied whether the positive feedback action of glucagon would lead to an increase in glucagon biosynthesis. Stimulation of αTC1-9 cells with either 1 mM glucose (which triggers secretion of glucagon) or exogenously applied glucagon (200 nM for 15 min) led to a 30% increase in glucagon biosynthesis within 60 min after the start of stimulation (Fig. 1G).
Glucagon-Stimulated (Prepro)Glucagon Gene Transcription Requires Signal Transduction via Glucagon Receptor, PKC, and PKA.
Recent studies have characterized the expression and functionality of glucagon receptors in pancreatic α cells (15–17). Our data from Western blot (Fig. 2A) and immunohistochemistry analyses (Fig. 2B) confirm the presence of glucagon receptors in α cells in mouse and human pancreatic islets, as well as in glucagon-producing αTC1-9 cells. We next studied whether signal transduction through the glucagon receptor is required to elevate (prepro)glucagon mRNA levels by glucagon. Whereas stimulation with exogenous glucagon (200 nM for 15 min) led to an increase in mRNA levels in αTC1-9 cells and pancreatic islets, the stimulatory effect of glucagon on the transcription of its own gene was abolished when cells or islets were treated with 400 nM glucagon receptor antagonist II (GRA II) (Fig. 2 C and D). Similarly, treatment with GRA II abolished the stimulatory effect of exogenous glucagon (Fig. 2E), as well as that of secreted glucagon after stimulation with 1 mM glucose (Fig. 2F), on glucagon promoter activity. Finally, pretreatment of αTC1-9 cells with 1 µM [des-His1, Glu9]glucagon amide, a glucagon analog that binds to but does not activate the glucagon receptor, abolished glucagon-stimulated activation of the glucagon promoter (Fig. 2G).
Fig. 2.
Glucagon stimulates its own expression by signaling via the glucagon receptor. (A) Identification of the glucagon receptor in lysates of αTC1-9, mouse islets, and control tissues (insulin-producing cell lines HIT-T15, MIN6, and INS1 and mouse muscle) by Western blot analysis. (B) Identification of the glucagon receptor by immunohistochemistry in αTC1-9, mouse islets, and human islets (green). In islets, α cells were identified by costaining with an anti-glucagon antibody (red). The images are single confocal frames. (Scale bar: 10 µm.) (C and D) Effects of GRA II on (prepro)glucagon mRNA levels in mouse and human islets (C) and clonal αTC1-9 cells (D). Islets and αTC1-9 cells were cultured at 16.7 mM glucose and stimulated for 15 min with 200 nM glucagon or left unstimulated. GRA II (400 nM) was added 30 min before the start of stimulation and maintained throughout the stimulation. (Prepro)glucagon mRNA levels represent the percentage of mRNA levels of nonstimulated control (given as 100%); n = 3. Data are expressed as mean ± SEM. Significance vs. stimulated expression without GRA II, *P < 0.05. (E and F) Effect of GRA II on glucagon promoter-driven DsRed2 expression in transfected αTC1-9 cells stimulated with either exogenously added glucagon (200 nM at 16.7 mM glucose for 15 min) (E) or 1 mM glucose for 15 min (F) to trigger glucagon secretion. Significance vs. stimulated expression without GRA II, *P < 0.05; #P < 0.01. GRA II (400 nM) was added 30 min before the start of stimulation and maintained throughout the stimulation. (G) Effect of [des-His1, Glu9]glucagon ([ΔH1,E9]glucagon) on glucagon-stimulated glucagon promoter-driven DsRed2 expression. Transfected αTC1-9 cells were preincubated for 30 min with 1 µM [ΔH1,E9]glucagon and then stimulated with 200 nM glucagon for 15 min at 16.7 mM glucose or left unstimulated. Significance vs. stimulated expression without [ΔH1,E9]glucagon, *P < 0.05; #P < 0.05. Data in E–G represent the ratio of DsRed2 fluorescence values obtained at 240 min and 60 min and are expressed as mean ± SEM for at least 15 monitored cells.
Signaling cascades that can be triggered by the glucagon receptor activate PKA via the adenylate cyclase system and activate PKC via the phospholipase C system (18, 19). We next analyzed whether exogenously applied or secreted glucagon activates either PKA or PKC. Stimulation with either exogenous glucagon (200 nM for up to 15 min) or secreted glucagon at low glucose concentration (1 mM glucose for up to 15 min) led to a more than twofold elevation in PKA activity (Fig. 3A) and to a more than threefold increase in PKC activity (Fig. 3B). Of note, the increased activity of both kinases was transient, and activity returned to basal levels by 30 min after the start of stimulation. To test whether signaling through either PKA or PKC is involved in glucagon-stimulated (prepro)glucagon gene transcription, we used Rp-cAMPS and bisindolylmaleimide-I (BIM) as pharmacologic inhibitors of PKA and PKC, respectively. Treatment of αTC1-9 cells with either 100 µM Rp-cAMPS or 150 nM BIM abolished the glucagon-stimulated increase in (prepro)glucagon mRNA levels (Fig. 3C), as well as glucagon promoter activity (Fig. 3D). Finally, treatment of mouse and human pancreatic islets with either inhibitor drastically reduced the stimulatory effect of exogenous glucagon on the transcription of its own gene (Fig. 3E).
Fig. 3.
Glucagon stimulates its own expression by signaling via PKA and PKC. (A and B) Effect of exogenously added (200 nM for up to 15 min) or secreted glucagon (in response to 1 mM glucose, for up to 15 min) on the activity for PKA (A) or PKC (B). Protein kinase activities are presented as percentage of activity in nonstimulated control (given as 100%); n = 6. Data are expressed as mean ± SEM. *P < 0.05 in A; *P < 0.01 in B. (C–E) Effect of inhibitors of PKA (Rp-cAMP) and PKC (BIM) on glucagon-stimulated (prepro)glucagon mRNA levels (C and E) and glucagon promoter-driven DsRed2 expression (D). Transfected αTC1-9 cells or islets were preincubated with either 100 µM Rp-cAMP or 150 nM BIM or vehicle (control) for 30 min and then stimulated with exogenous glucagon (200 nM at 16.7 mM glucose) or left unstimulated. (Prepro)glucagon mRNA levels at 60 min after the start of stimulation are presented as percentage of mRNA levels of nonstimulated control (given as 100%); n = 3. Data are expressed as mean ± SEM. Significance vs. stimulated expression without inhibitor, *P < 0.05. Data in D represent the ratio of DsRed2 fluorescence values obtained at 240 min and 60 min and are expressed as mean ± SEM for at least 15 monitored cells.
Glucagon-Stimulated Glucagon Gene Transcription Is Mediated via cAMP-Element Binding Protein.
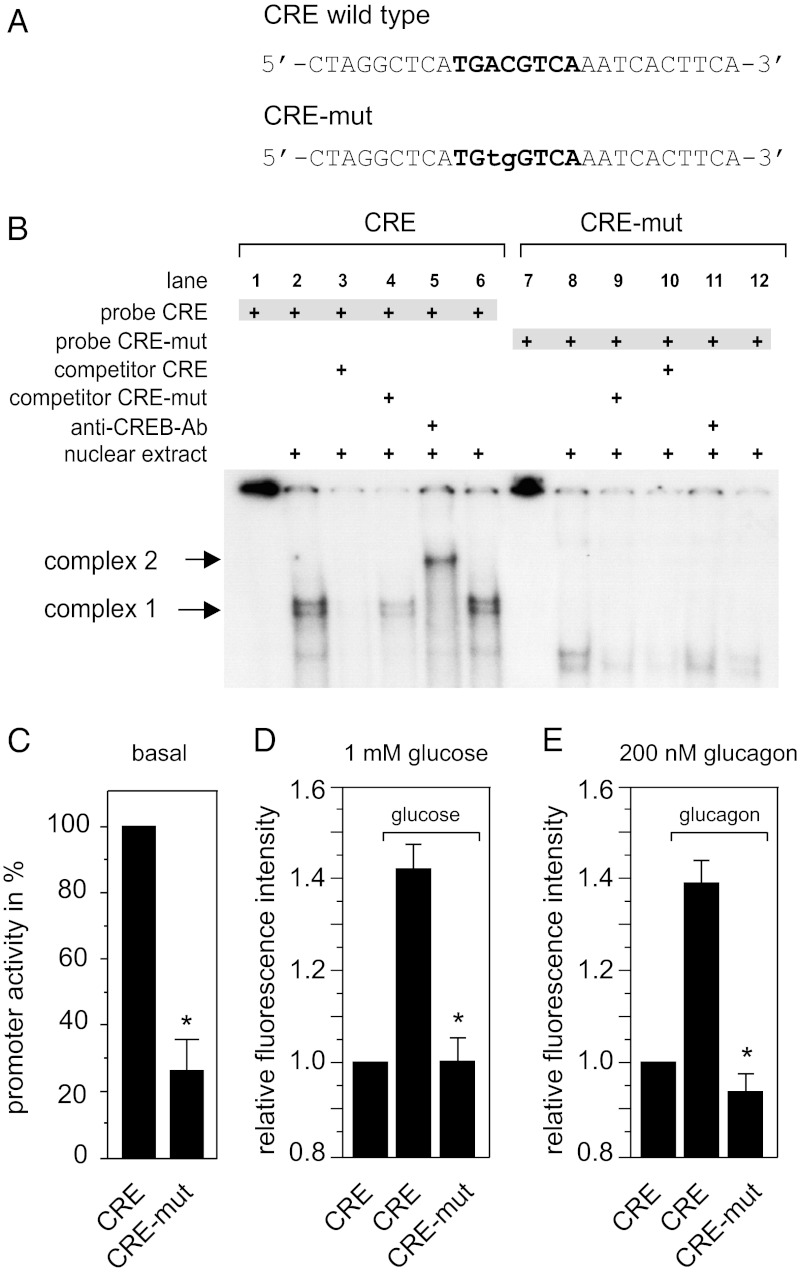
The promoter of the (prepro)glucagon gene is well characterized, and cis- and transacting factors that contribute to transcriptional control have been identified (20). Among these transcription factors, the prime candidates for glucagon-stimulated transcriptional control are the cAMP-response element (CRE) (21) and the respective CRE-binding protein (CREB), which generally can be activated by PKA and PKC (22, 23). CREB has been shown to be activated in response to glucagon stimulation in hepatocytes and pancreatic β cells (24, 25). To test whether the positive autocrine feedback action by glucagon on the transcription of its own gene involves CREB, we mutated the CREB binding site in the glucagon promoter of our reporter construct. Mutating the CRE motif from TGACGTCA to TGTGGTCA (CRE-mut; Fig. 4A) abolished binding of CREB to respective ds oligonucleotide probes in electrophoretic mobility shift assays (Fig. 4B). The CREB/ds oligonucleotide complex (complex 1) was identified by “supershifting” this complex in response to incubation with an anti-CREB antibody, which resulted in the formation of complex 2 (Fig. 4B). Introduction of the same mutation into the CRE motif of our reporter construct reduced basal glucagon promoter activity (Fig. 4C), and also abolished the stimulatory effect of secreted glucagon (Fig. 4D) and exogenously applied glucagon (Fig. 4E) on glucagon promoter-driven DsRed2 expression/fluorescence.
Fig. 4.
Glucagon-stimulated (prepro)glucagon gene transcription is mediated via CREB and the CRE motif in the (prepro)glucagon promoter. (A and B) CREB binds in electrophoretic mobility shift assays to ds oligonucleotides containing an intact CRE motif (CRE), but not to ds oligonucleotides containing a mutated CRE (CRE-mut). The normal ds oligonucleotide/CREB complex 1 (B, lanes 2, 4, and 6) is “supershifted” and forms complex 2 after incubation with an anti-CREB antibody (B, lane 5). (C–E) Effect of CRE-mut on basal (C) or stimulated (prepro)glucagon promoter activity by secreted (D) or exogenous (E) glucagon. In C, αTC1-9 cells were cotransfected with pGL4.CMV.hRlucCP and either WT pGlcg1.luc2neo (CRE) or mutant pGlcg1.CRE-mut.luc2neo (CRE-mut). Basal promoter activity was analyzed as described in Materials and Methods and calculated by dividing Glcg1.CRE or Glcg1.CRE-mut promoter-controlled firefly luciferase luminescence by CMV promoter-controlled Renilla luciferase luminescence. Glucagon promoter activity is presented as percentage of WT promoter activity (given as 100%); n = 3. Data are expressed as mean ± SEM; *P < 0.05. In D and E, αTC1-9 cells were transfected with either WT pGlcg1.DsRed2 (CRE) or mutant pGlcg1.mut.DsRed2 (CRE-mut). Cells were left unstimulated or were stimulated with either 1 mM glucose (D) or 200 nM glucagon (at 16.7 mM glucose) (E) for 15 min. Data represent the ratio of DsRed2 fluorescence values obtained at 240 min and 60 min and are expressed as mean ± SEM for at least 15 monitored cells. Significance vs. stimulated expression of CRE, *P < 0.05.
Conclusion
Positive autocrine feedback leading to a secretion-biosynthesis coupling, which allows replenishment of the peptide hormone stores of the hormone-producing cell, is not a widely accepted concept. Such a feedback mechanism has been reported for the peptide hormones insulin (1–11) and gastrin (26), as well as for the cytokines IL-1 (27), IL-21 (28), and erythropoietin (29); growth factors HGF, KGF, and kit (30); and the vasoconstrictor endothelin (31, 32).
Our data demonstrate that secreted glucagon stimulates its own resynthesis via a positive autocrine feedback mechanism. The increase in (prepro)glucagon mRNA levels is only transient, however, and the feedback loop is turned off by a lack of glucagon stimulus, leading to transient activation of protein kinases PKA and PKC in response to rising blood glucose levels, as well as to inhibition of glucagon gene transcription by insulin (33). Interestingly, GLP-1, another product of the (prepro)glucagon gene that is synthesized and secreted by intestinal l cells, is also able to activate the glucagon promoter in αTC1-9 cells (Fig. S2).
Although the presence of all involved players in pancreatic α cells has been documented in earlier studies—that is, the presence in α cells of glucagon receptor (15–17) and the roles of CRE and CREB (21, 34, 35) and of cAMP, PKC, and PKA (35–38) in (prepro)glucagon gene expression—their involvement in glucagon-stimulated (prepro)glucagon gene expression had not been studied previously. There may be two major reasons for this. First, a mouse model with a general knockout of the glucagon receptor showed an increase in circulation glucagon levels as well as increased pancreatic glucagon content (39). However, it should be stressed that under conditions of “global glucagon resistance,” the increased demand for glucagon is compensated for by an increase in α cell mass, as well as glucagon biosynthesis. The involved signaling mechanisms “override” regulatory circuits that function under physiological conditions. The fact that signaling and function of α cells under these conditions is not physiological is underscored by the finding that the α cells in this mouse model show an immature phenotype expressing genes normally repressed in mature α cells, such as genes encoding GLUT2 or Pdx1 (40). A second reason for the lack of previous studies may be the “textbook wisdom” suggesting that the existence of a positive autocrine feedback mechanism for peptide hormone biosynthesis is counterintuitive, because elevated/continuous exposure to a peptide hormone leads to desensitization to the hormone at the effector cell level, and, consequently, cells that produce and secrete a peptide hormone should be desensitized to the hormone because of its continuous presence. However, it should be kept in mind that a broad spectrum of fuel-regulating hormones, including insulin and glucagon, are secreted in a pulsatile manner (41, 42). Along with being sensed more potently by peripheral target cells, pulsatility also provides an off state of ligand presence for the hormone-secreting cells themselves, thereby allowing hormone sensing.
Although our data indicate the contribution of CREB to the glucagon autocrine feedback mechanism, we do not exclude the possible involvement of other transcription factors that can be activated by PKA and PKC. Moreover, given that synthesis of peptide hormones must be coupled to the highly complex regulated biosynthesis of secretory granules, as for insulin (43), we do not exclude the possible existence of further retrograde signaling mechanisms.
In conclusion, together with previously published data on insulin and gastrin feedback, the present data on glucagon support the concept of positive autocrine feedback action as a more general mechanism involved in the secretion–biosynthesis coupling of peptide hormones.
Materials and Methods
Pancreatic islets were prepared from 4-mo-old BALB/C mice using collagenase digestion as described previously (44). Human islets were provided by O. Korsgren (Department of Oncology, Radiology, and Clinical Immunology, Uppsala University Hospital, Uppsala, Sweden) via the Nordic Islet Network. The glucagon promoter (−773/+35 bp) was amplified by PCR from genomic rat DNA using 5′-TCCTTCTGTTGAATGGCCAG-3′ as the upstream primer and 5′-TTTGAGTGTGTTCTGCGCC-3′ as the downstream primer. Total RNA was purified from 20 islets using the RNeasy Micro Kit (Qiagen) according to the manufacturer’s instructions. Reverse-transcription reactions were performed using the SuperScriptIII Kit (Invitrogen).
(Prepro)glucagon mRNA levels were determined by real-time PCR. Taqman reactions were carried out following the manufacturer’s instructions for Taqman gene expression assays (Applied Biosystems). Relative quantification of glucagon mRNA expression was calculated by the comparative Ct method (45) using peptidylpropyl isomerase A as an internal reference. Glucagon secretion was measured using the LINCO glucagon RIA kit (Millipore). Total protein content of the cell lysate, determined by the Bradford assay, was used to normalize for between well variations in cell density. Glucagon biosynthesis was analyzed by using l-[4,5- 3H] leucine (PerkinElmer) for labeling and a sheep polyclonal antibody directed toward the C terminus of glucagon (ab36215; Abcam) for immunoprecipitation. Nuclear extracts from αTC1-9 cells for EMSA were prepared as described previously (46). Supershift assays were performed using a CREB-antibody (Abcam). The following inhibitors were used: PKA inhibitor Rp-cAMPS, PKC inhibitor BIM I, glucagon receptor antagonist II (all Calbiochem/Merck), and the glucagon receptor inhibitory peptide [Des-His1,Glu9] glucagon amide (sequence: Ser-Gln-Gly-Thr-Phe-Thr-Ser-Glu-Tyr-Ser-Lys-Tyr-Leu-Asp-Ser-Arg-Arg-Ala-Gln-Asp-Phe-Val-Gln-Trp-Leu-Met-Asn-Thr-NH2) (American Peptide). The experiments are described in more detail in SI Materials and Methods.
Supplementary Material
Acknowledgments
This study was supported by the Swedish Research Council; Novo Nordisk Foundation; Karolinska Institutet; Swedish Diabetes Association; Knut and Alice Wallenberg Foundation; European Foundation for the Study of Diabetes; Diabetes Research and Wellness Foundation; Bert von Kantzow Foundation; Grant FP7-228933-2 [in vivo imaging of beta-cell receptors by applied nanotechnology (VIBRANT)]; Skandia Insurance Company, Ltd.; World Class University program through the National Research Foundation of Korea (funded by Ministry of Education, Science and Technology Grant CR31-2008-000-10105-0); Strategic Research Programme in Diabetes at Karolinska Institutet; Stichting af Jochnick Foundation; and Erling-Persson Family Foundation. Human islets were provided through the European Consortium of Islet Transplantation (ECIT) Islets for Basic Research program (Juvenile Diabetes Research Foundation Award 31-2008-413).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1212870110/-/DCSupplemental.
References
- 1.Leibiger IB, Leibiger B, Moede T, Berggren PO. Exocytosis of insulin promotes insulin gene transcription via the insulin receptor/PI-3 kinase/p70 s6 kinase and CaM kinase pathways. Mol Cell. 1998;1(6):933–938. doi: 10.1016/s1097-2765(00)80093-3. [DOI] [PubMed] [Google Scholar]
- 2.Xu GG, Rothenberg PL. Insulin receptor signaling in the β-cell influences insulin gene expression and insulin content: Evidence for autocrine β-cell regulation. Diabetes. 1998;47(8):1243–1252. doi: 10.2337/diab.47.8.1243. [DOI] [PubMed] [Google Scholar]
- 3.Wu H, et al. Insulin stimulates pancreatic-duodenal homoeobox factor-1 (PDX1) DNA-binding activity and insulin promoter activity in pancreatic β cells. Biochem J. 1999;344(Pt 3):813–818. [PMC free article] [PubMed] [Google Scholar]
- 4.Leibiger B, Wåhlander K, Berggren PO, Leibiger IB. Glucose-stimulated insulin biosynthesis depends on insulin-stimulated insulin gene transcription. J Biol Chem. 2000;275(39):30153–30156. doi: 10.1074/jbc.M005216200. [DOI] [PubMed] [Google Scholar]
- 5.da Silva Xavier G, Varadi A, Ainscow EK, Rutter GA. Regulation of gene expression by glucose in pancreatic β cells (MIN6) via insulin secretion and activation of phosphatidylinositol 3′-kinase. J Biol Chem. 2000;275(46):36269–36277. doi: 10.1074/jbc.M006597200. [DOI] [PubMed] [Google Scholar]
- 6.Andreozzi F, et al. Activation of the hexosamine pathway leads to phosphorylation of insulin receptor substrate-1 on Ser307 and Ser612 and impairs the phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin insulin biosynthetic pathway in RIN pancreatic β-cells. Endocrinology. 2004;145(6):2845–2857. doi: 10.1210/en.2003-0939. [DOI] [PubMed] [Google Scholar]
- 7.Luciani DS, Johnson JD. Acute effects of insulin on beta-cells from transplantable human islets. Mol Cell Endocrinol. 2005;241(1-2):88–98. doi: 10.1016/j.mce.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 8.Muller D, Huang GC, Amiel S, Jones PM, Persaud SJ. Identification of insulin signaling elements in human beta-cells: Autocrine regulation of insulin gene expression. Diabetes. 2006;55(10):2835–2842. doi: 10.2337/db06-0532. [DOI] [PubMed] [Google Scholar]
- 9.Solinas G, Naugler W, Galimi F, Lee MS, Karin M. Saturated fatty acids inhibit induction of insulin gene transcription by JNK-mediated phosphorylation of insulin-receptor substrates. Proc Natl Acad Sci USA. 2006;103(44):16454–16459. doi: 10.1073/pnas.0607626103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leibiger IB, Leibiger B, Berggren PO. Insulin signaling in the pancreatic β-cell. Annu Rev Nutr. 2008;28:233–251. doi: 10.1146/annurev.nutr.28.061807.155530. [DOI] [PubMed] [Google Scholar]
- 11.Leibiger B, et al. Selective insulin signaling through A and B insulin receptors regulates transcription of insulin and glucokinase genes in pancreatic β cells. Mol Cell. 2001;7(3):559–570. doi: 10.1016/s1097-2765(01)00203-9. [DOI] [PubMed] [Google Scholar]
- 12.Uhles S, Moede T, Leibiger B, Berggren PO, Leibiger IB. Selective gene activation by spatial segregation of insulin receptor B signaling. FASEB J. 2007;21(7):1609–1621. doi: 10.1096/fj.06-7589com. [DOI] [PubMed] [Google Scholar]
- 13.Moede T, Leibiger B, Berggren PO, Leibiger IB. Online monitoring of stimulus-induced gene expression in pancreatic β-cells. Diabetes. 2001;50(Suppl 1):S15–S19. doi: 10.2337/diabetes.50.2007.s15. [DOI] [PubMed] [Google Scholar]
- 14.Hamaguchi K, Leiter EH. Comparison of cytokine effects on mouse pancreatic alpha-cell and beta-cell lines: Viability, secretory function, and MHC antigen expression. Diabetes. 1990;39(4):415–425. doi: 10.2337/diab.39.4.415. [DOI] [PubMed] [Google Scholar]
- 15.Ma X, et al. Glucagon stimulates exocytosis in mouse and rat pancreatic α-cells by binding to glucagon receptors. Mol Endocrinol. 2005;19(1):198–212. doi: 10.1210/me.2004-0059. [DOI] [PubMed] [Google Scholar]
- 16.Tian G, Sandler S, Gylfe E, Tengholm A. Glucose- and hormone-induced cAMP oscillations in α- and β-cells within intact pancreatic islets. Diabetes. 2011;60(5):1535–1543. doi: 10.2337/db10-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Z, et al. Insulin and glucagon regulate pancreatic α-cell proliferation. PLoS ONE. 2011;6(1):e16096. doi: 10.1371/journal.pone.0016096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Christophe J. Glucagon and its receptor in various tissues. Ann N Y Acad Sci. 1996;805:31–42. doi: 10.1111/j.1749-6632.1996.tb17471.x. [DOI] [PubMed] [Google Scholar]
- 19.Unson CG. Molecular determinants of gluca/gon receptor signaling. Biopolymers. 2002;66(4):218–235. doi: 10.1002/bip.10259. [DOI] [PubMed] [Google Scholar]
- 20.Jin T. Mechanisms underlying proglucagon gene expression. J Endocrinol. 2008;198(1):17–28. doi: 10.1677/JOE-08-0085. [DOI] [PubMed] [Google Scholar]
- 21.Miller CP, Lin JC, Habener JF. Transcription of the rat glucagon gene by the cyclic AMP response element-binding protein CREB is modulated by adjacent CREB-associated proteins. Mol Cell Biol. 1993;13(11):7080–7090. doi: 10.1128/mcb.13.11.7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez GA, Montminy MR. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59(4):675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- 23.Yamamoto KK, Gonzalez GA, Biggs WH, 3rd, Montminy MR. Phosphorylation-induced binding and transcriptional efficacy of nuclear factor CREB. Nature. 1988;334(6182):494–498. doi: 10.1038/334494a0. [DOI] [PubMed] [Google Scholar]
- 24.Herzig S, et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413(6852):179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- 25.Dalle S, et al. Glucagon promotes cAMP-response element-binding protein phosphorylation via activation of ERK1/2 in MIN6 cell line and isolated islets of Langerhans. J Biol Chem. 2004;279(19):20345–20355. doi: 10.1074/jbc.M312483200. [DOI] [PubMed] [Google Scholar]
- 26.Kovac S, Xiao L, Shulkes A, Patel O, Baldwin GS. Gastrin increases its own synthesis in gastrointestinal cancer cells via the CCK2 receptor. FEBS Lett. 2010;584(21):4413–4418. doi: 10.1016/j.febslet.2010.09.046. [DOI] [PubMed] [Google Scholar]
- 27.Zoja C, Bettoni S, Morigi M, Remuzzi G, Rambaldi A. Interleukin-1 regulates cytokine gene expression in human mesangial cells through the interleukin-1 receptor type 1. J Am Soc Nephrol. 1992;2(12):1709–1715. doi: 10.1681/ASN.V2121709. [DOI] [PubMed] [Google Scholar]
- 28.Caprioli F, et al. Autocrine regulation of IL-21 production in human T lymphocytes. J Immunol. 2008;180(3):1800–1807. doi: 10.4049/jimmunol.180.3.1800. [DOI] [PubMed] [Google Scholar]
- 29.Kim I, Kim CH, Yim YS, Ahn YS. Autocrine function of erythropoietin in IGF-1–induced erythropoietin biosynthesis. Neuroreport. 2008;19(17):1699–1703. doi: 10.1097/WNR.0b013e32831743fb. [DOI] [PubMed] [Google Scholar]
- 30.Parrott JA, Mosher R, Kim G, Skinner MK. Autocrine interactions of keratinocyte growth factor, hepatocyte growth factor, and kit-ligand in the regulation of normal ovarian surface epithelial cells. Endocrinology. 2000;141(7):2532–2539. doi: 10.1210/endo.141.7.7581. [DOI] [PubMed] [Google Scholar]
- 31.Saijonmaa O, Nyman T, Fyhrquist F. Endothelin-1 stimulates its own synthesis in human endothelial cells. Biochem Biophys Res Commun. 1992;188(1):286–291. doi: 10.1016/0006-291x(92)92382-8. [DOI] [PubMed] [Google Scholar]
- 32.Saito S, Hirata Y, Imai T, Marumo F. Autocrine regulation of the endothelin-1 gene in rat endothelial cells. J Cardiovasc Pharmacol. 1995;26(Suppl 3):S84–S87. [PubMed] [Google Scholar]
- 33.Philippe J. Glucagon gene transcription is negatively regulated by insulin in a hamster islet cell line. J Clin Invest. 1989;84(2):672–677. doi: 10.1172/JCI114214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Philippe J, et al. Alpha cell-specific expression of the glucagon gene is conferred to the glucagon promoter element by the interactions of DNA-binding proteins. Mol Cell Biol. 1988;8(11):4877–4888. doi: 10.1128/mcb.8.11.4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knepel W, Chafitz J, Habener JF. Transcriptional activation of the rat glucagon gene by the cyclic AMP-responsive element in pancreatic islet cells. Mol Cell Biol. 1990;10(12):6799–6804. doi: 10.1128/mcb.10.12.6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drucker DJ, Campos R, Reynolds R, Stobie K, Brubaker PL. The rat glucagon gene is regulated by a protein kinase A-dependent pathway in pancreatic islet cells. Endocrinology. 1991;128(1):394–400. doi: 10.1210/endo-128-1-394. [DOI] [PubMed] [Google Scholar]
- 37.Philippe J, Drucker DJ, Habener JF. Glucagon gene transcription in an islet cell line is regulated via a protein kinase C-activated pathway. J Biol Chem. 1987;262(4):1823–1828. [PubMed] [Google Scholar]
- 38.Fürstenau U, Schwaninger M, Blume R, Kennerknecht I, Knepel W. Characterization of a novel protein kinase C response element in the glucagon gene. Mol Cell Biol. 1997;17(4):1805–1816. doi: 10.1128/mcb.17.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gelling RW, et al. Lower blood glucose, hyperglucagonemia, and pancreatic alpha cell hyperplasia in glucagon receptor knockout mice. Proc Natl Acad Sci USA. 2003;100(3):1438–1443. doi: 10.1073/pnas.0237106100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vuguin PM, et al. Ablation of the glucagon receptor gene increases fetal lethality and produces alterations in islet development and maturation. Endocrinology. 2006;147(9):3995–4006. doi: 10.1210/en.2005-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weigle DS. Pulsatile secretion of fuel-regulatory hormones. Diabetes. 1987;36(6):764–775. doi: 10.2337/diab.36.6.764. [DOI] [PubMed] [Google Scholar]
- 42.Hellman B, Salehi A, Gylfe E, Dansk H, Grapengiesser E. Glucose generates coincident insulin and somatostatin pulses and antisynchronous glucagon pulses from human pancreatic islets. Endocrinology. 2009;150(12):5334–5340. doi: 10.1210/en.2009-0600. [DOI] [PubMed] [Google Scholar]
- 43.Liu Y, Mziaut H, Ivanova A, Solimena M. Beta cells at the crossroads: Choosing between insulin granule production and proliferation. Diabetes Obes Metab. 2009;11(Suppl 4):54–64. doi: 10.1111/j.1463-1326.2009.01107.x. [DOI] [PubMed] [Google Scholar]
- 44.Leibiger B, et al. Short-term regulation of insulin gene transcription by glucose. Proc Natl Acad Sci USA. 1998;95(16):9307–9312. doi: 10.1073/pnas.95.16.9307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-Delta Delta C(T) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 46.Dyer RB, Herzog NK. Isolation of intact nuclei for nuclear extract preparation from a fragile B-lymphocyte cell line. Biotechniques. 1995;19(2):192–195. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.