Abstract
The biological function of Tripartite Motif 39 (TRIM39) remains largely unknown. In this study, we report that TRIM39 regulates the steady-state levels of p21 and is a pivotal determinant of cell fate. Ablation of TRIM39 leads to destabilization of p21 and increased G1/S transition in unperturbed cells. Furthermore, DNA damage-induced p21 accumulation is completely abolished in cells with depleted TRIM39. As a result, silencing of TRIM39 abrogates the G2 checkpoint induced by genotoxic stress, leading to increased mitotic entry and, ultimately, apoptosis. Importantly, we show p21 is a crucial downstream effector of TRIM39 mediating G1/S transition and DNA damage-induced G2 arrest. Mechanistically, TRIM39 interacts with p21, which subsequently prevents Cdt2 from binding to p21, therefore blocking ubiquitylation and proteasomal degradation of p21 mediated by CRL4Cdt2 E3 ligase. Strikingly, we found a significant correlation between p21 abundance and TRIM39 expression levels in human hepatocellular carcinoma samples. Our findings identify a causal role for TRIM39 in regulating cell cycle progression and the balance between cytostasis and apoptosis after DNA damage via stabilizing p21.
Tripartite motif 39 (TRIM39), also known as Ring finger protein 23 (RNF23), belongs to a family of proteins characterized by a TRIM consisting of RING domain, B-box, and coiled-coil domain. Recently, TRIM39 has been shown to stabilize modulator of apoptosis 1 (MOAP-1) by suppressing its polyubiquitylation process mediated by the anaphase promoting complex (APC/C)Cdh1 (1, 2). In agreement with the proapoptotic role of MOAP-1, TRIM39 was found to enhance apoptosis in response to high-dose etoposide treatment in HEK293T cells. Because p53 signaling is deficient in HEK293T cells, it remains to be elucidated what other mechanisms may contribute to the DNA damage responses regulated by TRIM39. In addition to this, the physiological functions of TRIM39 remain completely unknown.
The cyclin-dependent kinase inhibitor p21CIP1/WAF1 promotes cell cycle arrest in response to cellular stresses induced by chemotherapeutics, UV irradiation, or γ-irradiation. Levels of p21 increase in response to DNA damage primarily due to its transcriptional up-regulation by the tumor suppressor p53 (3). Although p21 accumulates after genotoxic stresses, it is degraded in response to DNA damage induced by low-dose UV irradiation (4, 5). Three E3 ubiquitin ligase complexes, SCFSkp2, CRL4Cdt2, and APC/CCdc20, have been shown to trigger p21 ubiquitylation and degradation during an unperturbed cell cycle (6–8). Following UV irradiation, the CRL4Cdt2 E3 ligase [composed of Cul4A/B, DNA damage-binding protein 1 (DDB1), and Cdt2] promotes the ubiquitylation and degradation of p21 in cooperation with proliferating cell nuclear antigen (PCNA). Positive regulators of p21 stability remain largely unknown. Phosphorylation of p21 by p38 and JNK has been reported to stabilize p21 (9). WISp39 and hSSB1 were found to stabilize p21 via protein–protein interactions (10, 11). Ras can stabilize p21 by promoting the formation of p21–cyclin D1 complexes that prevent ubiquitin-independent p21 degradation (12).
Here, we identified p21 as a unique TRIM39 interacting protein. TRIM39 positively regulates p21 stability by interfering with the formation of the Cdt2–p21 complex, therefore attenuating CRL4Cdt2-mediated ubiquitylation and degradation of p21. We demonstrated a crucial role of TRIM39 in G1/S transition under physiological conditions, as well as in regulating the balance between cytostasis and apoptosis after DNA damage via stabilizing p21.
Results
Splice Variants of Human TRIM39.
It was reported that the human TRIM39 protein is 98% identical to the mouse protein. The mouse TRIM39 protein lacked 30 amino acids (amino acids 269–298) due to an alternative splicing (13). Primers designed to amplify the reported human TRIM39 ORF (accession no. NM_021253; 1,557 bp, 518 aa) from HCT116 cells, amplified a second isoform (1,467 bp) encoding a protein of 488 aa that is missing amino acids 269–298 within TRIM39 Exon6. Sequencing results revealed this additional splice variant, TRIM39β (accession no. NM_172016), is 97.5% identical to mouse TRIM39 (Fig. S1A). Ectopically expressed TRIM39β corresponded to the faster migrating and most abundant form of TRIM39, whereas TRIM39α corresponded to the slower migrating form (Fig. S1B). PCR assay of cDNA from a variety of human cancer cells confirms that TRIM39β is relatively more abundant than TRIM39α in these cells (Fig. S1C).
TRIM39 Stabilizes p21.
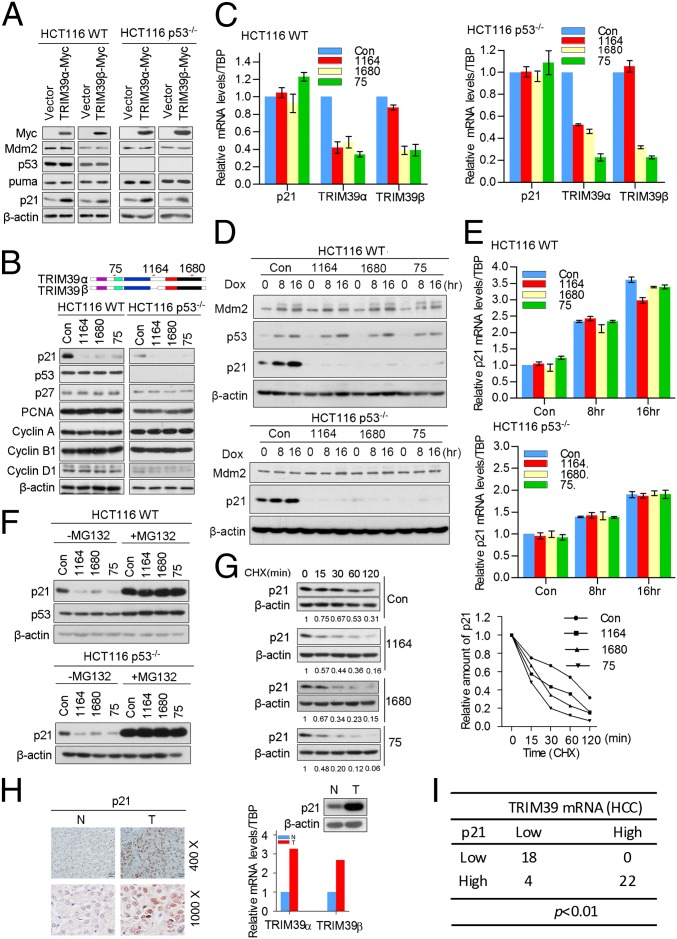
TRIM39 has been shown to promote apoptosis through stabilization of MOAP-1 (2). To test if regulation of apoptosis by TRIM39 has any cross-talk with the p53 signaling pathway, we introduced TRIM39 into HCT116 WT cells as well as HCT116 p53−/− cells, followed by examining the expression levels of p53 and its downstream target genes. Surprisingly, cells expressing ectopic TRIM39α or TRIM39β resulted in a significant accumulation of p21 in both cell lines (Fig. 1A), suggesting TRIM39-mediated up-regulation of p21 was p53-independent. By contrast, the levels of p53, Mdm2, and Puma were not affected by TRIM39 overexpression (Fig. 1A). Notably, p21 mRNA levels remained unchanged in the presence of ectopic TRIM39 (Fig. S2A). Up-regulation of exogenously expressed p21 was also observed in HEK293T cells expressing ectopic TRIM39 (Fig. S2B). To clarify further the role of TRIM39 in regulating p21 expression, we performed a knockdown experiment using two independent shRNAs targeting both TRIM39α and TRIM39β (75 shRNA and 1,680 shRNA) and one shRNA targeting TRIM39α alone (1,164 shRNA) (Fig. 1B). Because the TRIM39 antibodies generated by us (as well as one commercially available TRIM39 antibody) were not working well with endogenous TRIM39, we conducted a quantitative RT-PCR (qRT-PCR) assay using Taqman probes to evaluate the knockdown efficiency. Cells stably expressing 75 shRNA or 1,680 shRNA profoundly reduced both TRIM39α and TRIM39β mRNA levels, whereas cells expressing 1,164 shRNA only abolished TRIM39α mRNA expression (Fig. 1C). To verify the knockdown specificity further, we transiently transfected TRIM39α or TRIM39β along with GFP expression vector into HEK293T cells, followed by delivering lentiviral shRNAs against different TRIM39 isoforms. As shown in Fig. S2C, 75 shRNA or 1,680 shRNA largely attenuated both TRIM39α and TRIM39β protein levels, whereas 1,164 shRNA only reduced TRIM39α expression. We next determined the effect of TRIM39 ablation on p21 expression. Strikingly, TRIM39α/β compound knockdown or depleting TRIM39α alone in HCT116 cells abolished p21 protein levels (Fig. 1B). The reduction of p21 protein after TRIM39 depletion was not a consequence of transcriptional repression, as shown by the lack of a decrease in p21 mRNA levels on TRIM39 knockdown (Fig. 1C). Notably, depleting TRIM39 failed to affect the protein levels of other selected cell cycle regulators (Fig. 1B). On TRIM39 knockdown, p21 degradation was not limited to HCT116 cells, because p21 proteolysis was also observed in additional tumor cell lines, including HeLa, Saos2, and U2OS cells (Fig. S2D). DNA damage is known to induce p21 via both p53-dependent and p53-independent pathways. To assess whether TRIM39 regulates p21 expression under DNA damage conditions, we exposed TRIM39-depleted HCT116 cells with genotoxic agents. In HCT116 WT and HCT116 p53−/− cells, doxorubicin treatment led to up-regulation of p21 protein levels. Strikingly, TRIM39 knockdown completely abolished p21 elevation triggered by doxorubicin treatment (Fig. 1D). Likewise, etoposide-induced p21 accumulation was also profoundly attenuated in TRIM39-depleting cells (Fig. S2 E and F). Notably, depletion of TRIM39 did not abolish p21 mRNA induction on genotoxic treatment (Fig. 1E). We also determined the mRNA levels of TRIM39 during the cell cycle and under DNA damage conditions. TRIM39 mRNA abundance remained unchanged throughout the cell cycle or on DNA damage treatment (Fig. S2 G and H). These data suggest TRIM39 family members are essential for p21 expression under physiological conditions as well as in response to DNA damage treatment.
Fig. 1.
TRIM39 stabilizes p21. (A) HCT116 WT and p53−/− cells were infected with the indicated lentiviral constructs. Cell extracts were then prepared and analyzed by Western blotting with the indicated antibodies. (B, Upper) Schematic representation of the indicated shRNAs targeting TRIM39. (B, Lower) Protein extracts from cells infected with indicated lentiviral shRNA constructs were analyzed by Western blotting. Total RNA was isolated, and qRT-PCR was performed. (C) Error bars represent the SD of triplicate measurements. Con, control. (D and E) Cells expressing indicated shRNAs were treated with 0.2 μM doxorubicin (Dox) for 8 or 16 h. (D) Cell lysates were then extracted and subjected to Western blotting. Total RNA from cells was isolated and subjected to qRT-PCR. (E) Error bars represent the SD of triplicate measurements. (F) Cells infected with lentivirus encoding the indicated shRNAs were treated with DMSO or MG132 (20 μM) for 4 h. Cell extracts were analyzed by Western blotting using the indicated antibodies. (G) HCT116 WT cells expressing the indicated shRNAs were treated with 25 μg/mL cycloheximide (CHX) for the indicated time. (Left) Cell lysates were harvested and analyzed by Western blotting. (Right) Semiquantification with β-actin as a loading control and relative p21 levels at time 0 set as 1. (H) Immunohistochemical staining of p21 in HCC samples and adjacent hepatic tissues. N, normal hepatic tissue adjacent to tumor region; T, tumor region. (Scale bar, 400×, 20 μm; 1,000×, 8 μm). (Upper Right) Expression levels of p21 were measured by Western blotting. (Lower Right) Expression levels of TRIM39 were determined by qRT-PCR assay. Representative results are shown. (I) Abundance of p21 is positively correlated with TRIM39 mRNA levels in HCC samples. The correlation between TRIM39 and p21 expression in HCC samples was analyzed by Fisher’s exact test.
It is known that p21 is degraded by proteasome-mediated degradation. Interestingly, proteasome inhibitor MG132 failed to stabilize p21 protein levels further in the presence of ectopically expressed TRIM39α or TRIM39β (Fig. S2I), arguing that TRIM39 may prevent proteasome-mediated p21 destruction. Furthermore, down-regulation of p21 protein levels mediated by TRIM39 knockdown can be blocked by MG132 treatment (Fig. 1F), thereby suggesting that TRIM39 maintains the steady-state levels of p21 by blocking its proteasomal degradation. To prove that TRIM39 affects p21 stability per se, we treated control cells or cells stably expressing TRIM39 shRNA with cycloheximide and examined the half-life of p21. A significant decrease in p21 half-life was observed on TRIM39 knockdown (Fig. 1G). By contrast, ectopically expressed TRIM39 profoundly extended the half-life of p21 protein (Fig. S2 J and K). Thus, TRIM39 is essential for the maintenance of steady-state levels of p21.
Levels of p21 are often misregulated in human cancers (14, 15). To assess the correlation between p21 and TRIM39 abundance in hepatocellular carcinoma (HCC), we first determined by qRT-PCR the expression status of TRIM39 and p21 using 62 pairs of HCC tissues. Notably, we excluded 18 HCC samples that showed concordant changes in p21 mRNA and protein levels compared with their normal controls. TRIM39α or TRIM39β mRNA levels were reduced in 22 of 62 samples (35.5%) and were elevated in 22 of 62 samples (35.5%). We then measured its cellular localization by immunohistochemistry (IHC) staining. In HCC samples, p21 mainly localized in the nuclei of the cancer cells (Fig. 1H). Strikingly, a significant correlation between p21 protein levels and TRIM39 mRNA abundance was observed (Fig. 1I), further supporting a possible link between p21 and TRIM39.
TRIM39 Binds p21.
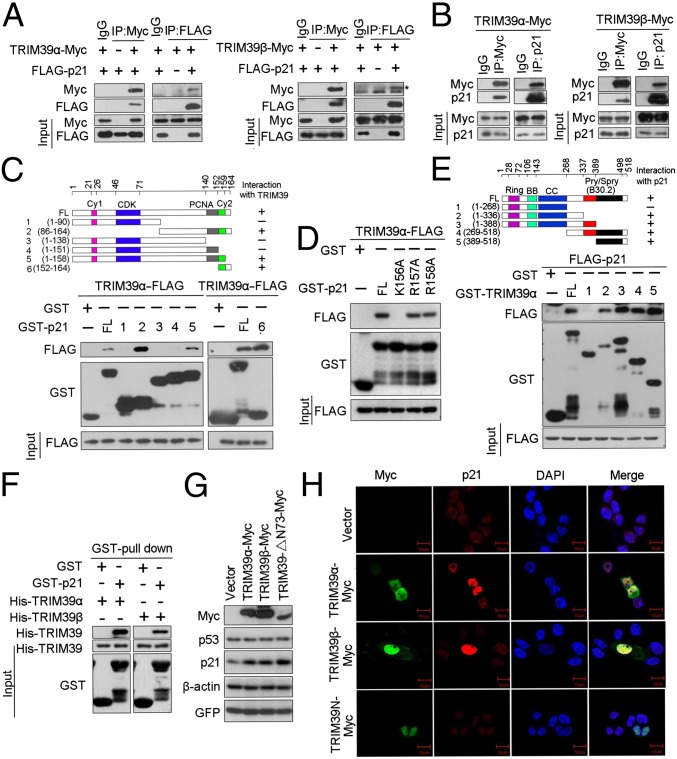
Protein–protein interactions play essential roles in regulating p21 stability. We therefore asked if TRIM39 physically associates with p21. As shown in Fig. 2A, exogenously expressed TRIM39α or TRIM39β was coimmunoprecipitated with ectopically expressed p21. Reciprocal immunoprecipitation with exogenous p21 also brought down overexpressed TRIM39α or TRIM39β. To test if TRIM39 associates with endogenous p21 protein, we introduced TRIM39α-Myc or TRIM39β-Myc into HCT116 WT cells and then performed immunoprecipitation with anti-Myc antibody. The association between p21 and TRIM39α or TRIM39β was readily detected (Fig. 2B). These results suggest a specific interaction between TRIM39 and p21 in vivo. Data from GST pull-down experiments suggest that the C-terminal region (amino acids 152–158) of p21 mediates TRIM39-p21 association (Fig. 2C). Furthermore, GST-p21 (amino acids 152–164) itself could interact with TRIM39, suggesting that amino acids 152–164 of p21 are both required and sufficient for TRIM39 interaction. To identify key amino acid residues of p21 required for TRIM39 interaction, we constructed mutant p21 expression vectors bearing a single point mutation within the region of amino acids 156–158. Lysine-to-alanine substitution at K156 completely disrupted p21–TRIM39 complex formation both in vitro and in vivo (Fig. 2D and Fig. S2L), indicating K156 residue of p21 is essential for TRIM39 interaction. Mapping the region of TRIM39 required for p21 binding revealed that amino acids 268–337 and amino acids 389–518 were critical regions for the interaction between TRIM39 and p21 (Fig. 2E). By contrast, TRIM39 N-terminal domain (TRIM39-N; amino acids 1–268) failed to bind p21 both in vitro and in vivo (Fig. 2E and Fig. S2M). To determine whether the interaction of TRIM39 and p21 is direct, we generated and purified recombinant TRIM39 and p21. Purified His-TRIM39α or His-TRIM39β was able to interact with GST-p21 under cell-free conditions, suggesting a direct interaction between TRIM39 and p21 (Fig. 2F). Several TRIM family members are known to demonstrate E3 ubiquitin ligase activity. We next investigated whether the RING domain of TRIM39 is required for stabilizing p21. To this end, full-length TRIM39 or TRIM39-ΔN73 (RING domain deletion mutant) was transfected into HCT116 WT and U2OS cells, respectively. Endogenous p21 levels were then assessed by Western blotting. Similar to full-length TRIM39, TRIM39-ΔN73 exhibited a very robust effect on stabilizing p21 (Fig. 2G and Fig. S2N), suggesting that the E3 ligase activity of TRIM39 is dispensable for regulating the steady-state levels of p21.
Fig. 2.
TRIM39 interacts with p21. (A) HEK293T cells were transiently transfected with plasmid DNA expressing Flag-p21 and/or TRIM39-Myc. Total cell lysates were immunoprecipitated with the indicated antibodies. The asterisk indicates the specific TRIM39β-Myc band. IP, immunoprecipitate. (B) HCT116 WT cells infected with the indicated lentiviral constructs were collected, and the cell lysates were immunoprecipitated with anti-Myc and anti-p21 antibodies. The immunoprecipitates were subjected to Western blotting with the indicated antibodies. (C and D) Extracts from HEK293T cells transfected with TRIM39α-FLAG were incubated with recombinant full-length (FL) GST-p21 or GST-p21 mutant coupled to GSH-Sepharose. Proteins retained on Sepharose were then blotted with the indicated antibodies. (E) Extracts from HEK293T cells transfected with FLAG-p21 were incubated with recombinant full-length GST-TRIM39α or GST-TRIM39α deletion mutant protein coupled to GSH-Sepharose. Proteins retained on Sepharose were then blotted with the indicated antibodies. (F) Purified His-TRIM39 proteins were incubated with GST-p21. Proteins retained on Sepharose were then blotted with the indicated antibodies. (G) HCT116 WT cells were transfected with the indicated expression constructs for 48 h. Cell lysates were then harvested and analyzed by Western blotting with the indicated antibodies. (H) HCT116 WT cells were transiently transfected with the indicated constructs for 48 h. Cells were then fixed and stained as indicated. (Scale bars, 10 μm.)
To determine the subcellular localization of the TRIM39–p21 complex, we transfected HCT116 WT cells with vectors expressing full-length TRIM39-Myc or TRIM39-N–Myc. Immunofluorescent staining revealed that TRIM39α, TRIM39β, or TRIM39-N predominantly localized in the nucleus. Furthermore, exogenous TRIM39α or TRIM39β colocalized with endogenous p21 in the nucleus (Fig. 2H). Notably, cells introduced with full-length TRIM39α or TRIM39β exhibited a profound increase of nucleus p21. By contrast, overexpression of TRIM39-N failed to up-regulate p21 protein levels (Fig. 2H and Fig. S2I). Taken together, these data indicate that TRIM39 exerts a positive effect on p21 stability through interacting with p21 in the nucleus.
TRIM39α Interacts with TRIM39β.
During the course of examining the half-life of p21 protein in cells expressing exogenous TRIM39α or TRIM39β, we found that TRIM39α is an unstable protein with a short half-life of around 40 min in asynchronous cells. TRIM39β showed a relatively longer protein half-life, which is about 4 h (Fig. S2 J and K). Importantly, both TRIM39α and TRIM39β could be stabilized by the addition of MG132 (Fig. S2I), arguing that the steady-state levels of TRIM39α or TRIM39β might be regulated in a proteasome-dependent manner.
The fact that cells depleted of both TRIM39α and TRIM39β showed a very similar effect on p21 destabilization compared with cells depleted of TRIM39α alone raised two possibilities: (i) TRIM39α is a dominant regulator of p21, or (ii) TRIM39α and TRIM39β are both required for maintaining p21 stability. To distinguish these possibilities, we first performed a rescue experiment by introducing a shRNA-resistant form of TRIM39α or/and TRIM39β into HCT116 WT cells stably depleted of both α and β isoforms of TRIM39 (Fig. S3 A and B). The stability of p21 was decreased in cells stably expressing 75 shRNA or 1,680 shRNA. Surprisingly, reconstitution with shRNA-resistant TRIM39α (TRIM39α-75R or TRIM39α-1,680R) or TRIM39β (TRIM39β-75R or TRIM39β-1,680R) in these cells failed to restore p21 expression levels, suggesting TRIM39α or TRIM39β alone is insufficient for maintaining p21 stability. By contrast, coexpression of both shRNA-resistant α and β isoforms of TRIM39 significantly restored p21 protein levels in cells expressing 75 shRNA or 1,680 shRNA (Fig. S3C). Likewise, delivering shRNA-resistant TRIM39α (TRIM39α-1,164R) into cells expressing 1,164 shRNA also restored p21 protein levels (Fig. S3D). These data further indicate that both TRIM39α and TRIM39β may be required for maintaining the steady-state levels of p21.
We hypothesized that TRIM39α and TRIM39β may form a protein complex to regulate p21. To test this possibility, we transiently transfected HEK293T cells with TRIM39α or TRIM39β individually or with both plasmids and then performed coimmunoprecipitation with antibodies against different tag proteins. As shown in Fig. S3 E and F, ectopically expressed TRIM39α and TRIM39β physically interacted with each other. We next showed that purified recombinant His-TRIM39β was able to interact with GST-TRIM39α under cell-free conditions (Fig. S3G), suggesting a direct interaction between TRIM39α and TRIM39β. To assess the biochemical consequence of this interaction, we examined the expression levels of exogenously expressed TRIM39α (with a shorter half-life) in the absence or presence of ectopic TRIM39β. Our results revealed that overexpression of TRIM39β led to up-regulation of TRIM39α protein levels (Fig. S3H). Indeed, in cells depleted of TRIM39β alone (the TRIM39α-resistant form was introduced into cells stably expressing 75 shRNA or 1,680 shRNA), the half-life of TRIM39α was significantly decreased (Fig. S3I). Taken together, TRIM39β is essential for maintaining the steady-state levels of TRIM39α, likely through protein–protein interactions.
TRIM39 Protects p21 from Ubiquitin-Mediated Degradation via Competing with Cdt2 for p21 Interaction.
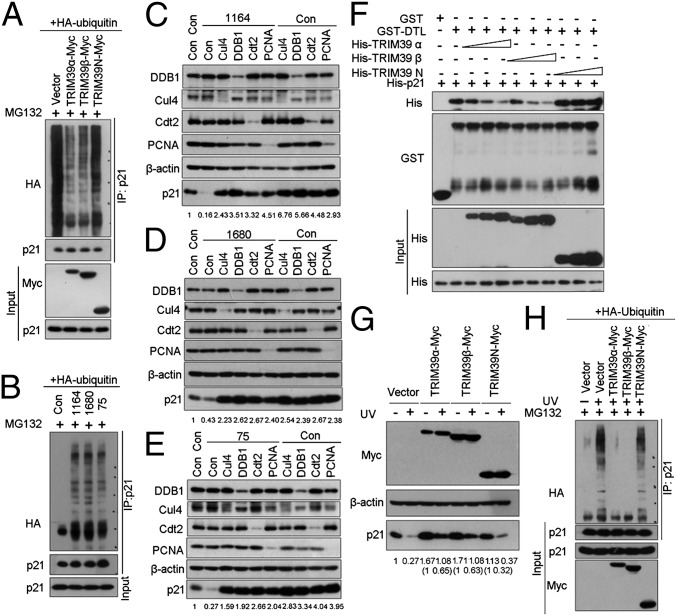
Ubiquitylation is crucial for proteasome-mediated destruction of p21. To test if TRIM39 affects p21 ubiquitylation in vivo, we measured polyubiquitylated p21 levels in cells expressing full-length TRIM39 or TRIM39-N. As shown in Fig. 3A, endogenous p21 was efficiently ubiquitylated in the presence of proteasome inhibitor MG132. Interestingly, both TRIM39α and TRIM39β largely abolished the polyubiquitylation of endogenous p21. By contrast, TRIM39-N failed to protect p21 from ubiquitylation, suggesting that the direct binding of TRIM39 with p21 is important for blocking p21 polyubiquitylation. Conversely, ablation of endogenous TRIM39 by 75, 1,164, or 1,680 shRNA resulted in a significant increase of p21 polyubiquitylation (Fig. 3B).
Fig. 3.
TRIM39 protects p21 from CRL4Cdt2-mediated ubiquitylation and degradation. (A) HCT116 WT cells expressing HA-ubiquitin were infected with the indicated lentiviral plasmids for 48 h. Endogenous p21 protein was immunoprecipitated and immunoblotted with the indicated antibodies. (B) HCT116 WT cells expressing HA-ubiquitin were infected with the indicated lentiviral shRNA. The immunoprecipitates were subjected to Western blotting with the indicated antibodies. Con, control. (C–E) HCT116 WT cells infected with lentivirus encoding the indicated shRNAs were lysed, and cell extracts were subjected to Western blotting with the indicated antibodies. The ratio of p21 protein normalized to β-actin relative to the control (marked as 1) was indicated below each lane. (F) Bacterially expressed GST or GST-Cdt2 was bound to glutathione-Sepharose beads and incubated with purified His-p21 (5 μg) in the presence or absence of increasing amounts of His-TRIM39α, His-TRIM39β, or His–TRIM39-N. Proteins retained on Sepharose were blotted with the indicated antibodies. (G) HCT116 p53−/− cells infected with lentiviral vectors expressing the indicated TRIM39 constructs were treated with UV irradiation (20 J/m2) for 6 h. Cell lysates were then extracted and subjected to Western blot analysis. The ratio of p21 protein normalized to β-actin relative to the control (marked as 1) was indicated below each lane. (H) HCT116 p53−/− cells expressing HA-ubiquitin were infected with the indicated shRNAs. Cells were then exposed to UV irradiation (20 J/m2) in the presence of MG132 (20 μM) for 6 h before harvesting. The p21 was immunoprecipitated with anti-p21 polyclonal antibody. The immunoprecipitates (IP) were subjected to Western blotting with the indicated antibodies.
It has been proposed that on association with PCNA, p21 is targeted for ubiquitylation by CRL4Cdt2 ubiquitin ligase complex in S phase and in response to low-dose UV treatment. Because p21 associates with Cdt2 via residues 156–161, which is overlapping with the region (amino acids 152–158) required for TRIM39 interaction, we speculate that the mechanisms underlying TRIM39-dependent p21 stabilization might be associated with the CRL4Cdt2 ubiquitin ligase complex. To clarify this, we first examined whether p21 degradation on TRIM39 depletion is dependent on Cul4A/B, DDB1, Cdt2, and PCNA. Down-regulation of TRIM39 by shRNA significantly decreased the basal levels of p21 without affecting the steady-state levels of Cul4, Cdt2, PCNA, and DDB1. Silencing each one of the components of CRL4Cdt2 resulted in significant protection of TRIM39 depletion-induced p21 destabilization (Fig. 3 C–E). By contrast, knocking down Skp2 or Cdh1 (TRIM39 was recently reported to inhibit APC/CCdh1) failed to prevent p21 degradation in cells depleted of TRIM39 (Fig. S4 A–C), suggesting TRIM39 stabilizes p21 via disrupting CRL4Cdt2-mediated proteolytic degradation of p21.
Cdt2 was proposed as the substrate factor recruiting p21 to the rest of the CRL4 ubiquitin ligase complex (16). We hypothesized that TRIM39 may compete with Cdt2 for p21 interaction. To address this, we conducted an in vitro competition assay to assess the interaction between GST-Cdt2 and His-p21 in the absence or presence of an increasing amount of full-length TRIM39 or TRIM39-N. Strikingly, only full-length TRIM39α or TRIM39β, but not TRIM39-N, disrupted Cdt2–p21 complex formation in a dose-dependent manner (Fig. 3F), suggesting that association between TRIM39 and p21 is critical for preventing Cdt2 from binding to p21. Notably, no in vitro interaction between GST-Cdt2 and His-TRIM39 was detected (Fig. S4D). Furthermore, WT p21 and p21PCNA (PCNA binding-deficient mutant of p21) bind to TRIM39 with equal efficiency (Fig. S4E), arguing that the interaction between TRIM39 and p21 is independent of the PCNA-interacting peptide (PIP) box. It is known that the destruction of p21 by CRL4Cdt2 requires p21 binding to PCNA. Consistent with this notion, p21PCNA exhibited increased protein stability compared with WT p21 (Fig. S4F, lanes 1 and 4). Importantly, exogenously expressed TRIM39 failed to stabilize p21PCNA further as it did WT p21 (Fig. S4F), suggesting inhibition of CRL4Cdt2-mediated p21 degradation is the key mechanism underlying TRIM39-dependent stabilization of p21 protein.
To verify further that competing with Cdt2 for p21 binding is central to TRIM39-mediated p21 stabilization, we next asked if TRIM39 could protect p21 ubiquitylation and subsequent degradation triggered by UV irradiation. To this end, we infected HCT116 p53−/− cells with lentiviral vector expressing full-length TRIM39 or TRIM39-N and then detected p21 levels in the presence or absence of UV damage. In agreement with previous reports, irradiation of HCT116 p53−/− cells with low-dose UV exposure caused the degradation of p21. Intriguingly, both TRIM39α and TRIM39β, but not TRIM39-N, significantly reversed UV-mediated degradation of p21 (Fig. 3G). Consistent with the protection of p21 from UV-induced degradation, polyubiquitylation of p21 on UV exposure was profoundly abolished in cells expressing TRIM39α or TRIM39β (Fig. 3H). These data demonstrate that TRIM39α and TRIM39β are essential for stabilizing p21 in unperturbed cells as well as in UV-irradiated cells. Taken together, our data demonstrate that TRIM39 regulates p21 stability via competing with Cdt2 for p21 binding, therefore disrupting CRL4Cdt2 ubiquitin ligase-mediated p21 ubiquitylation and degradation.
Regulation of Cell Cycle Progression and DNA Damage Responses by TRIM39 Is p21-Dependent.
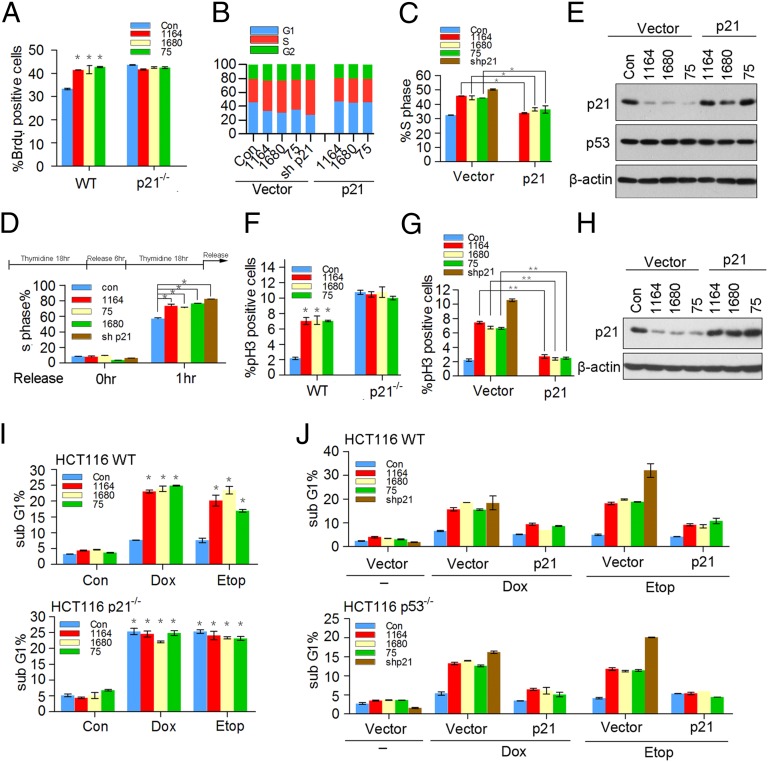
We reasoned that if TRIM39 is crucial for maintaining the basal levels of p21 protein, TRIM39 should affect cell cycle progression from G1 to S phase. Indeed, the fraction of cells in S phase, as determined by incorporation of BrdU into DNA and measurement of DNA content, as well as by double thymidine block and release, was substantially increased by TRIM39 silencing, similar to what was observed in p21-null or p21-depleting HCT116 cells (Fig. 4 A–D). In contrast, cells were arrested in G1 phase when TRIM39 was overexpressed (Fig. S5). Notably, ablation of TRIM39 in HCT116 p21−/− cells showed no additional effect on G1/S transition (Fig. 4A), ruling out the possibility that TRIM39 may additionally regulate S-phase entry by mechanisms independent of p21. Importantly, complementation of TRIM39-depleting cells with exogenous p21 fully blocked accelerated G1/S transition induced by TRIM39 ablation (Fig. 4 B, C, and E). Taken together, these data indicate p21 is a critical downstream effector of TRIM39 in mediating G1/S transition.
Fig. 4.
TRIM39 regulates the G1/S transition and DNA damage-induced G2 checkpoint in a p21-dependent manner. (A) HCT116 WT and HCT116 p21−/− cells infected with the indicated shRNAs were pulse labeled with BrdU for 30 min before harvesting and then analyzed by flow cytometry. Bar graph showing the percentages for S phase cells. The error bars represent the mean ± SD of three independent experiments. *P < 0.05. (B) HCT116 WT cells stably expressing vector or p21 were infected with the indicated lentiviral shRNAs for 72 h. Propidium iodide (PI)-stained cells were then subjected to flow cytometry analysis for their DNA content. The bar graph shows the percentages for G1, S, and G2/M cells. Results are one trial representative of three independent experiments. (C) S-phase percentage of cells in B is graphed. The error bars represent the mean ± SD of three independent experiments. *P < 0.05. (D) HCT116 WT cells stably expressing the indicated shRNAs were synchronized by a double thymidine block. The released cells were then harvested and analyzed by flow cytometry. The error bars represent the mean ± SD of three independent experiments. *P < 0.05. (E) Cell lysates in B were harvested and analyzed by Western blotting. (F) HCT116 WT and HCT116 p21−/− cells stably expressing the indicated shRNAs were pretreated with 0.2 μM doxorubicin for 2 h, followed by synchronization with nocodazole (100 ng/mL) for 16 h. The mitotic index was determined by mitotic marker pH3 staining. The error bars represent the mean ± SD of three independent experiments. *P < 0.05. (G) HCT116 p53−/− cells stably expressing vector or p21 were infected with the indicated lentiviral shRNAs for 72 h. Cells were then treated as described in F. The error bars represent the mean ± SD of three independent experiments. **P < 0.01. (H) Cell lysates in G were harvested and analyzed by Western blotting. (I) HCT116 WT cells (Upper) and HCT116 p21−/− cells (Lower) were infected with the indicated lentiviral shRNAs for 72 h. Cells were then treated with 0.2 μM doxorubicin (Dox) or 5 μM etoposide (Etop) for 24 h, followed by FACS analysis of the sub-G1 fraction. The error bars show the mean ± SD of three independent experiments. *P < 0.05. (J) HCT116 WT cells (Upper) and HCT116 p53−/− cells (Lower) stably expressing vector or p21 were infected with the indicated lentiviral shRNAs for 72 h. Cells were then treated as described in I. The error bars show the mean ± SD of three independent experiments.
Consistent with the notion that p21 is essential to sustain the G2 checkpoint after DNA damage in human cells (17, 18), we found that after low-dose doxorubicin treatment, HCT116 WT and HCT116 p53−/− cells arrested in G2 phase, whereas p21-depleting or HCT116 p21−/− cells entered mitosis more readily (Fig. 4 F and G), as judged by phosphohistone H3 staining. Intriguingly, a large fraction of TRIM39-depleting cells entered mitosis on doxorubicin exposure, which can be blocked by introducing exogenously expressed p21 (Fig. 4 G and H). Notably, on doxorubicin treatment, silencing TRIM39 had no additional effect on mitotic entry in cells lacking p21 (Fig. 4F). These data strongly indicate p21 is a key downstream mediator of TRIM39 in sustaining the DNA damage-induced G2 checkpoint. To assess the effect of TRIM39 depletion on apoptosis elicited by DNA-damaging agents, we treated HCT116 WT cells or HCT116 p53−/− cells with doxorubicin or etoposide and then performed propidium iodide staining. Flow cytometry analysis revealed that a significant portion of the TRIM39-depleting cells became apoptotic by 24 h of genotoxic treatment, in contrast to a marginal induction of apoptosis in control shRNA-expressing cells (Fig. 4I). Silencing p21 exerted similar effects on DNA damage-induced apoptosis (Fig. 4J). Notably, knockdown TRIM39 had no additional effect on DNA damage-induced apoptosis in cells lacking p21 (Fig. 4I). Importantly, reintroducing p21 into TRIM39-depleting cells profoundly protected cells from doxorubicin- or etoposide-triggered apoptotic death (Fig. 4J). These data suggest that TRIM39 ablation may sensitize cells to DNA damage-induced apoptosis via blunting p21 accumulation. Taken together, our findings uncover the physiological significance of the TRIM39-p21 axis in regulating G1/S transition. Moreover, our study identifies a causal role of TRIM39 in determining the outcome of DNA damage response and reveals TRIM39 as a crucial regulator in triggering cytostasis.
Discussion
In this study, we identified TRIM39 as a binding partner of p21 in the nucleus. Exogenous TRIM39 stabilized p21 by preventing its ubiquitylation, whereas TRIM39 depletion resulted in destabilization of p21, which was accompanied by increased ubiquitylation. Mechanistically, we found that TRIM39 competed with Cdt2 for p21 interaction, which subsequently abrogated p21 proteolysis mediated by CRL4Cdt2 E3 ligase. Indeed, destruction of p21 on TRIM39 knockdown was significantly blocked by silencing of Cul4, Cdt2, DDB1, or PCNA. Furthermore, UV-induced ubiquitylation and degradation of p21 were profoundly abolished by ectopically expressed TRIM39. Therefore, disrupting CRL4Cdt2 E3 ligase-mediated p21 ubiquitylation and degradation serves as a crucial mechanism underlying TRIM39-dependent stabilization of p21. Strikingly, TRIM39 ablation prevented p21 elevation induced by doxorubicin or etoposide, indicating TRIM39-mediated posttranslational stabilization of p21 is essential for p21 accumulation on genotoxic treatment.
Given the crucial role of TRIM39 in maintaining the steady-state levels of p21, we hypothesized that TRIM39 might affect cell cycle progression via acting on p21. In unperturbed cells, TRIM39 depletion resulted in an accelerated G1/S transition, which can be reversed by reintroduction of p21. In response to DNA damage, p21 is essential for G2 arrest (18). Consistent with impaired DNA damage-induced p21 elevation in the absence of TRIM39, down-regulation of TRIM39 attenuated G2 arrest in cells exposed to genotoxic treatment, leading to increased mitotic entry and apoptosis, which can be rescued by introduction of ectopic p21. More importantly, silencing TRIM39 in HCT116 p21−/− cells did not produce additional effects on S-phase entry and the DNA damage-induced G2 checkpoint. Taken together, these data indicate that the TRIM39-p21 axis plays a crucial role in controlling cell cycle progression and DNA damage-induced G2 arrest. Furthermore, the biological function of TRIM39 in cell fate determination is largely dependent on its ability to stabilize p21.
How TRIM39 is regulated remains largely unknown. We found that both α and β can be stabilized on proteasome inhibitor treatment, suggesting TRIM39 proteins may undergo proteasome-mediated degradation. Because DNA replication, as well as DNA damage repair, after UV irradiation requires p21 to be degraded, it is conceivable that the short half-life of TRIM39 (especially TRIM39α) operates in concert with other signaling pathways to ensure the rapid turnover of p21 during the G1/S transition or after low-dose UV irradiation. The regulation of TRIM39 family members during cell cycle progression or after DNA damage needs further investigation. Surprisingly, proteolytic destruction of p21 on TRIM39α and TRIM39β compound knockdown can only be rescued by introduction of both α and β shRNA-resistant constructs, suggesting the posttranslational regulation of p21 by TRIM39 is dependent on the presence of both α and β isoforms. How exactly α and β isoforms work as a complex to regulate p21 remains to be elucidated. Notably, MOAP-1 was recently reported as a substrate of the APC/CCdh1 ubiquitin ligase. TRIM39 was proposed to regulate MOAP-1 via acting on APC/CCdh1 (1). Unfortunately, the molecular mechanisms underlying TRIM39-dependent regulation of APC/CCdh1 are obscure. The biological function of TRIM39 has been proposed to enhance apoptosis because MOAP-1 is a proapoptotic factor in response to genotoxic insults (2). However, these data were solely based on exposing HEK293T cells to high-dose etoposide (100 μM) treatment. In HEK293T cells, p21 protein levels are undetectable. Hence, it is conceivable that in the absence of p21, TRIM39 functions as a proapoptotic factor via distinct binding partners like MOAP-1.
It was found that p21 is overexpressed in a variety of human cancers. We found a strong correlation between TRIM39 mRNA abundance and p21 protein levels in HCC samples. Surprisingly, both up-regulation and down-regulation of TRIM39 levels were found in HCC samples. How TRIM39 acts to regulate tumorigenesis, as well as the antitumor response, remains to be elucidated. Furthermore, because p21 also plays a role in DNA repair, senescence, and reprogramming of induced pluripotent stem cells, examining the biological functions of TRIM39 under these physiological and physiopathological conditions could provide a clue as to how to explore TRIM39 for potential therapeutic applications. In summary, our study identifies TRIM39 as a key regulator in modulating p21 stability and establishes a clear functional role of the TRIM39-p21 axis in cell cycle progression and cellular responses to genotoxic insults.
Materials and Methods
Results are reported as the mean ± SD of three or more independent experiments. Unless stated otherwise, comparisons were performed with a two-tailed Student t test.
A description of cell culture and reagents, plasmids and antibodies, real-time PCR, RNAi, in vivo ubiquitylation, flow cytometry, immunofluorescence, IHC, and in vitro binding is provided in SI Material and Methods.
Supplementary Material
Acknowledgments
We thank Drs. Hongrui Wang, Aidong Han, Guanghui Jin, and Hongbing Zhang for reagents and discussions. V.C.Y. is supported by grants from Singapore Ministry of Health's National Medical Research Council under its IRG (NMRC/1317/2011) and National University of Singapore (R148-000-121-133). This study was supported by National Natural Science Foundation of China Grant 31170718 (to H.Y.), National Basic Research Program of China 973 Program Grant 2009CB522202 (to H.Y.), Fundamental Research Funds for the Central Universities Grant 2010121086 (to H.Y.), Science Planning Program of Fujian Province Grants 2009J1010 and 2010J1008, and Ministry of Education of China 111 Project B06016 and B12001.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1214156110/-/DCSupplemental.
References
- 1.Huang NJ, et al. The Trim39 ubiquitin ligase inhibits APC/CCdh1-mediated degradation of the Bax activator MOAP-1. J Cell Biol. 2012;197(3):361–367. doi: 10.1083/jcb.201111141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee SS, et al. TRIM39 is a MOAP-1-binding protein that stabilizes MOAP-1 through inhibition of its poly-ubiquitination process. Exp Cell Res. 2009;315(7):1313–1325. doi: 10.1016/j.yexcr.2008.11.021. [DOI] [PubMed] [Google Scholar]
- 3.el-Deiry WS, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75(4):817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 4.Bendjennat M, et al. UV irradiation triggers ubiquitin-dependent degradation of p21(WAF1) to promote DNA repair. Cell. 2003;114(5):599–610. doi: 10.1016/j.cell.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Lee H, Zeng SX, Lu H. UV Induces p21 rapid turnover independently of ubiquitin and Skp2. J Biol Chem. 2006;281(37):26876–26883. doi: 10.1074/jbc.M605366200. [DOI] [PubMed] [Google Scholar]
- 6.Amador V, Ge S, Santamaría PG, Guardavaccaro D, Pagano M. APC/C(Cdc20) controls the ubiquitin-mediated degradation of p21 in prometaphase. Mol Cell. 2007;27(3):462–473. doi: 10.1016/j.molcel.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bornstein G, et al. Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase. J Biol Chem. 2003;278(28):25752–25757. doi: 10.1074/jbc.M301774200. [DOI] [PubMed] [Google Scholar]
- 8.Kim Y, Starostina NG, Kipreos ET. The CRL4Cdt2 ubiquitin ligase targets the degradation of p21Cip1 to control replication licensing. Genes Dev. 2008;22(18):2507–2519. doi: 10.1101/gad.1703708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim GY, et al. The stress-activated protein kinases p38 alpha and JNK1 stabilize p21(Cip1) by phosphorylation. J Biol Chem. 2002;277(33):29792–29802. doi: 10.1074/jbc.M201299200. [DOI] [PubMed] [Google Scholar]
- 10.Xu S, et al. hSSB1 binds and protects p21 from ubiquitin-mediated degradation and positively correlates with p21 in human hepatocellular carcinomas. Oncogene. 2011;30(19):2219–2229. doi: 10.1038/onc.2010.596. [DOI] [PubMed] [Google Scholar]
- 11.Jascur T, et al. Regulation of p21(WAF1/CIP1) stability by WISp39, a Hsp90 binding TPR protein. Mol Cell. 2005;17(2):237–249. doi: 10.1016/j.molcel.2004.11.049. [DOI] [PubMed] [Google Scholar]
- 12.Coleman ML, Marshall CJ, Olson MF. Ras promotes p21(Waf1/Cip1) protein stability via a cyclin D1-imposed block in proteasome-mediated degradation. EMBO J. 2003;22(9):2036–2046. doi: 10.1093/emboj/cdg189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orimo A, et al. Molecular cloning of testis-abundant finger Protein/Ring finger protein 23 (RNF23), a novel RING-B box-coiled coil-B30.2 protein on the class I region of the human MHC. Biochem Biophys Res Commun. 2000;276(1):45–51. doi: 10.1006/bbrc.2000.3380. [DOI] [PubMed] [Google Scholar]
- 14.Abbas T, Dutta A. p21 in cancer: Intricate networks and multiple activities. Nat Rev Cancer. 2009;9(6):400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wagayama H, et al. High expression of p21WAF1/CIP1 is correlated with human hepatocellular carcinoma in patients with hepatitis C virus-associated chronic liver diseases. Hum Pathol. 2002;33(4):429–434. doi: 10.1053/hupa.2002.124724. [DOI] [PubMed] [Google Scholar]
- 16.Abbas T, et al. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 2008;22(18):2496–2506. doi: 10.1101/gad.1676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minemoto Y, et al. Characterization of adriamycin-induced G2 arrest and its abrogation by caffeine in FL-amnion cells with or without p53. Exp Cell Res. 2001;262(1):37–48. doi: 10.1006/excr.2000.5072. [DOI] [PubMed] [Google Scholar]
- 18.Bunz F, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282(5393):1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.