Abstract
Here we develop a minimal model of the cell actomyosin cortex by forming a quasi-2D cross-linked filamentous actin (F-actin) network adhered to a model cell membrane and contracted by myosin thick filaments. Myosin motors generate both compressive and tensile stresses on F-actin and consequently induce large bending fluctuations, which reduces their effective persistence length to <1 μm. Over a large range of conditions, we show the extent of network contraction corresponds exactly to the extent of individual F-actin shortening via buckling. This demonstrates an essential role of buckling in breaking the symmetry between tensile and compressive stresses to facilitate mesoscale network contraction of up to 80% strain. Portions of buckled F-actin with a radius of curvature ∼300 nm are prone to severing and thus compressive stresses mechanically coordinate contractility with F-actin severing, the initial step of F-actin turnover. Finally, the F-actin curvature acquired by myosin-induced stresses can be further constrained by adhesion of the network to a membrane, accelerating filament severing but inhibiting the long-range transmission of the stresses necessary for network contractility. Thus, the extent of membrane adhesion can regulate the coupling between network contraction and F-actin severing. These data demonstrate the essential role of the nonlinear response of F-actin to compressive stresses in potentiating both myosin-mediated contractility and filament severing. This may serve as a general mechanism to mechanically coordinate contractility and cortical dynamics across diverse actomyosin assemblies in smooth muscle and nonmuscle cells.
Keywords: cytoskeleton, active gels, nonequilibrium, myosin II
Changes in cell shape, as required for migration and division, are mediated by the cell cortex, a thin shell of cross-linked actin filaments (F-actin) bound to the inner leaflet of the plasma membrane. The actin cortex is an apolar, disordered network of F-actin decorated with myosin II motors. Myosin II motors drive contractility of the cortical actin network, enabling shape change and cytoplasmic flows underlying diverse physiological processes ranging from cell division and migration to tissue morphogenesis (1–6). Variable coupling of these actomyosin forces to the plasma membrane regulates transmission of stresses to cell–matrix or cell–cell adhesions to modulate cell shape change and force transmission (7–9). Actomyosin stresses are also implicated in driving cortical F-actin filament turnover in vivo to maintain a highly dynamic actin cytoskeleton (2–4). Thus, actin polymerization dynamics and contractile force generation are intimately coupled in the actomyosin cortex.
The actin filaments within the cortex are highly dynamic and constantly turning over, renewing themselves within minutes (10). The initial step of actin cortex turnover involves the severing of F-actin, which both destabilizes existing filaments and releases monomers for de novo polymerization. To date, the predominant mechanism identified for destabilizing F-actin both in vivo and in vitro is severing by ADF/cofilin. Recent data have indicated that the mechanism for severing by cofilin is mechanical in nature, making it prone to spontaneous fragmentation (11). There is mounting evidence that myosin II also plays a role in the turnover of cortical actin (2–4), and myosin II-mediated F-actin disassembly is observed in vitro (12). Given the known role of myosin II in the generation of contractile stresses and its association with F-actin disassembly, these mechanisms are likely related, although no such mechanism has been directly shown.
Myosin II motors generate local stresses at the molecular level within the actin cytoskeleton to drive contractility of the cortex at 10- to 100-μm length and nano-Newton force scales. The mechanisms of force transmission within a disordered actomyosin cortex are not well understood. In the sarcomeric organization of actomyosin found in striated muscle, the relative sliding of myosin against polarity-organized F-actin preferentially generates tensile stresses to drive contraction (13). However, actomyosin networks in nonmuscle cells lack sarcomeric organization (14) and a mechanism to break the symmetry between tensile and compressive stresses is necessary (15). Theoretical work has demonstrated that such symmetry-breaking mechanisms could arise in 2D and 3D networks due to inherent mechanical stability of tensile stresses (16), by myosin II “sorting” of F-actin polarity (17) or spatial regulation of F-actin polymerization (18). The role of the nonlinear elastic response of F-actin in facilitating contraction of bundles (19) and networks (20–22) has also been explored in theory and simulation, and F-actin buckling is observed during contraction of actomyosin bundles (19). However, it is not clear to what extent each of these microscopic mechanisms contributes to contractile deformation of a quasi-2D network at larger length scales. Moreover, mechanisms to mechanically coordinate network contractility with F-actin turnover are not well understood.
Results and Discussion
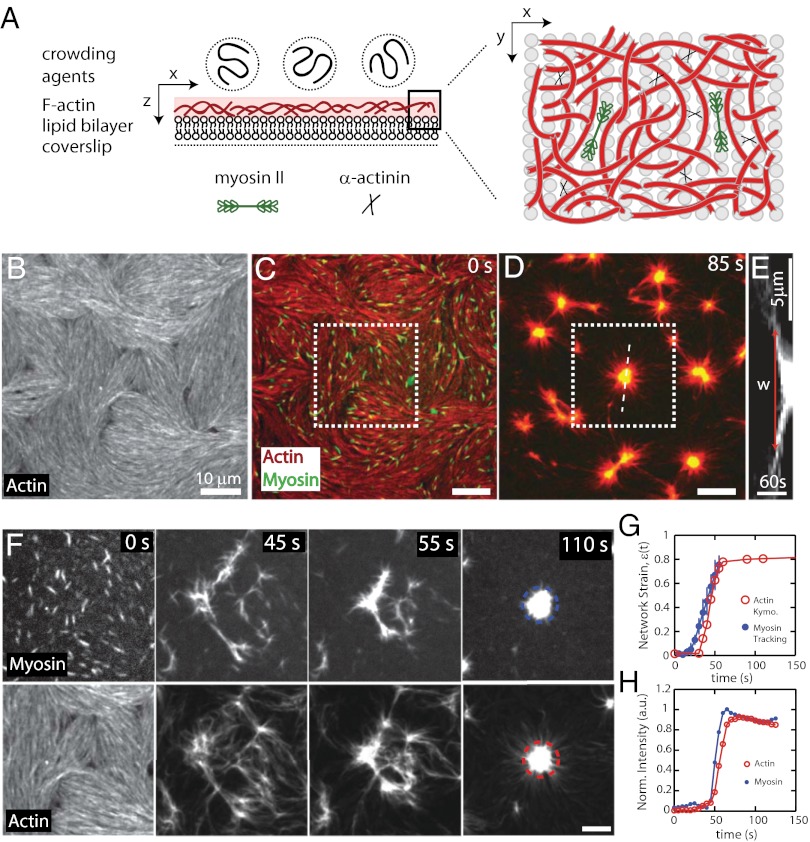
To identify mechanisms of contractility in disordered actin networks, we developed an in vitro model of the actomyosin cortex reconstituted from a minimal set of components that is amenable to high-resolution imaging of fluorescently tagged constituents. In this model, the crowding agent methylcellulose (MC) is used to localize F-actin to the surface of a supported lipid bilayer (Fig. 1 A and B and Fig. S1). We use a MC concentration (0.25%) that is sufficient to crowd phalloidin-stabilized F-actin near the surface of the lipid bilayer (Fig. 1 A and B, Figs. S1 and S2, and Movies S1 and S2), but low enough to prevent significant F-actin bundling (Fig. S3). Nevertheless, MC can induce interactions between F-actin filaments (23) and spatial variation of filament density is observed. The alignment of F-actin we observe during accumulation onto the membrane can be altered by perturbing formation conditions (Movie S3); changes to filament alignment do not impact any of the conclusions of our work, as myosin activity rapidly reorganizes F-actin. To cross-link F-actin into space-spanning networks, we use the flexible cross-linking protein α-actinin (24), which at low concentrations (Rxlink = 0.003) forms a network of isotropically cross-linked filaments and at high concentrations (Rxlink = 0.03) forms a network of dense bundles (Fig. S3). Specific adhesion of F-actin to the model membrane is mediated by the incorporation of nickel-tagged (NTA) lipids in the bilayer and the addition of a histidine-tagged actin-binding domain of fimbrin (FimA2) (25) (Fig. S1). Membrane adhesion via FimA2 immobilizes F-actin and prevents filament alignment (Fig. S1, Movie S4, and Fig. 4A). Thus, we can create a thin (∼200 nm) and dense quasi-2D network of F-actin (Fig. S1) whose cross-linking density (Rxlink = [α-actinin]/[G-actin]) and membrane adhesion (RAdh = [FimA2]/[G-actin]) can be controlled precisely.
Fig. 1.
Reconstitution of a model contractile actomyosin cortex. (A) Schematic illustration of reconstituted system. F-actin is crowded to the surface of a supported lipid bilayer (SLB) after which cross-linking proteins and skeletal muscle myosin II filaments are added. Data in B–H are from 1 μM F-actin (100% labeled) crowded to the surface of 91% EPC/9% nickel-tagged lipid (NTA) (mol/mol) SLB with Rxlink = 0 and RAdh = 0. (B) Alexa-568–labeled F-actin at the SLB surface. (C and D) F-actin (red) and myosin II (green) (C) immediately (0 s) after myosin thick filament formation and (D) 85 s after myosin filaments assembled (t = 0 s). (E) Kymograph of the actin over time taken at the white dotted line in D with the width of the contractile zone w indicated. (F) Magnified images of myosin and F-actin from square region indicated in C during contraction. Myosin filaments appear at 0 s. (G) Contractile strain of the actin network from the kymograph in E. (H) Average normalized myosin and actin fluorescence intensity within the center of contraction, indicated in F.
Fig. 4.
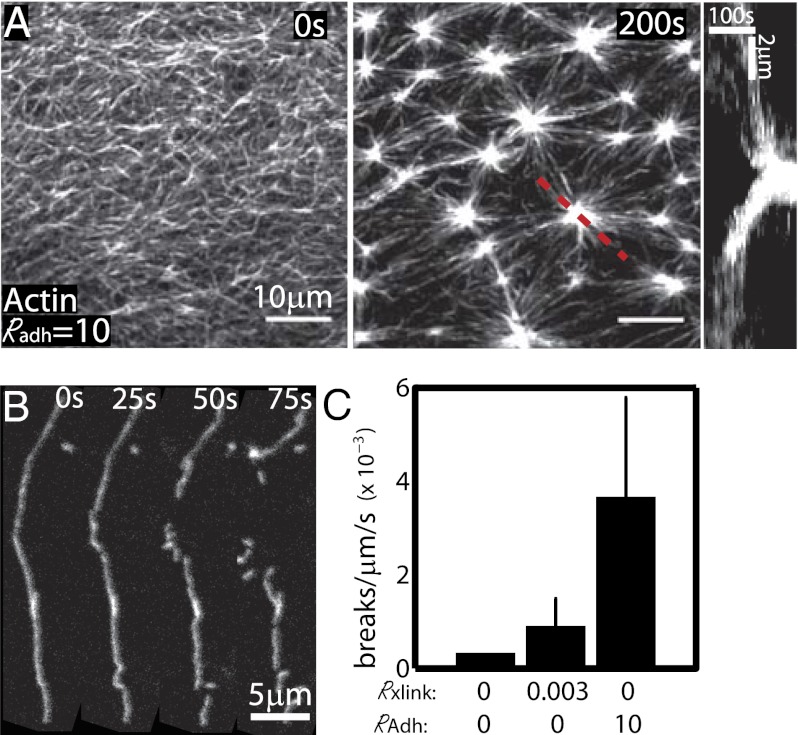
Membrane adhesion modulates mechanically induced severing. (A) F-actin images in RAdh = 10, Rxlink = 0. (Left) Precontraction, (Middle) postcontraction, and (Right) a kymograph of actin intensity for red dashed line. (B) Individual F-actin during contraction of a sparsely labeled network with RAdh = 10, Rxlink = 0. (C) Severing rates for Rxlink = 0/Radh = 0 (Nfil = 48, Nexpt = 1), Rxlink = 0.003 (Nfil = 204, Nexpt = 4), and Radh = 10 (Nfil = 133, Nexpt = 3). Nfil is the total number of filaments observed through a total of Nexp independent experiments.
Upon the addition of skeletal muscle myosin dimers to the sample chamber, myosin thick filaments ∼1.5 μm in length and containing ∼400 motor domains assemble rapidly (∼1–2 min) on the F-actin network (Fig. S4). After assembly, myosin II thick filaments drive rapid motions of F-actin and myosin to form a dynamic and contractile network (Fig. 1 C–E and Movies S5 and S6). Myosin activity drives the contraction of the F-actin network over a certain length scale into dense foci at rates of 150 nm/s (from kymograph) (Fig. 1 D–F). Once contracted, these aggregates can undergo further coalescence with each other (22). We quantify compressive strain within the contractile domain by tracking the position of myosin thick filaments and measuring the separation distance between select pairs during contraction, w(t) (Fig. S5). The contractile strain of the network, ɛ, is then determined by ɛ(t) = 1 − w(t)/w0, where w0 is the initial separation distance. Over the first 60–90 s, the network strain increases from 0 to 80% (Fig. 1G) and coincides with a rapid increase in local density of actomyosin into foci (Fig. 1H and Fig. S5). After this time, no significant network strain is measured although movement of actomyosin continues to persist for >15 min.
As the network architecture is perturbed by changes to F-actin length, cross-linking density, and strength of membrane adhesion, contraction is qualitatively similar, but the length scale over which it occurs is quantitatively distinct. In the absence of α-actinin, the contractile domain size is similar to the F-actin length and indicates that myosin or putative cross-linking by MC is insufficient to facilitate cross-linking over large distances (Figs. S6 and S7). When a low concentration of α-actinin (Rxlink = 0.003) is added, the contractile domain size increases and is consistent with enhanced network connectivity (Movie S7 and Fig. S6). When Rxlink = 0.03, the network of bundled actin filaments contracts as a single domain over millimeter length scales (Movie S8 and Fig. S8). To permit quantification of contraction in these networks of α-actinin/F-actin bundles, we locally activate myosin activity within the field of view to spatially restrict contraction (Movie S9 and Fig. S8). By contrast, adhesion to the membrane via His-FimA2 significantly reduces the contractile domain size to <3 μm (Fig. S6), reflecting an adhesive drag impeding contraction. Thus, changes to F-actin length, cross-link density, and strength of membrane adhesion strongly modulate the contractile domain size.
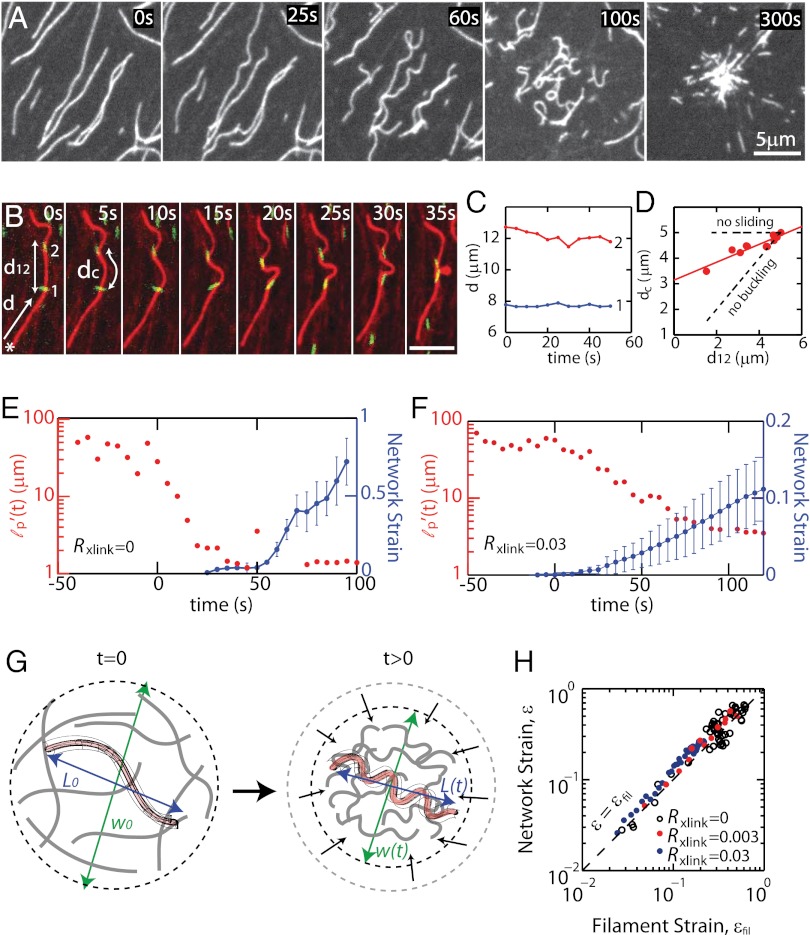
To identify microscopic origins of contraction across all these conditions, we image individual F-actin by labeling on 1–2% of the filaments within the quasi-2D network (Fig. 2A). Immediately upon the addition of myosin thick filaments, F-actin bending and buckling are observed, with the amplitude and frequency of bends increasing during contraction (Movie S10). F-actin buckling results from compressive stress on F-actin segments and could arise from several mechanisms, including from differences in the rate of myosin sliding along filaments (Fig. 2 B–D and Movie S11) or the nature of stress fluctuations within the surrounding network, as has been previously observed for microtubules in actomyosin networks (26). The minimum forces required to drive these curvatures in the absence of adhesion and bundling are determined by the Euler buckling threshold for F-actin and are ∼3–4 pN for a radius of curvature rc = 300 nm (27), well within the range of forces generated by myosin II filaments. Filament bending is observed over all conditions, including networks of thick α-actinin–mediated bundles (Fig. S8 and Movie S9) and high membrane adhesion (Fig. 4B and Movie S12), although the mean curvatures of bends in these conditions differ. In bundles and high membrane adhesion, large radii of curvature are rarely observed, reflecting constraints of the surrounding environment.
Fig. 2.
Myosin-induced F-actin buckling occurs concomitantly with contraction. (A) Images of individual F-actin (1–2% labeled) during contraction of sample with Rxlink = 0 and RAdh = 0 (Movie S10). Myosin thick filaments form at 0 s. Data in B–D are from sample with Rxlink = 0.003 and RAdh = 0 (Movie S11). (B) Myosin (green) translocating along F-actin (red) where d is the distance of myosin from F-actin barbed end (*), dc is length of F-actin between myosin punctae, and d12 is the Euclidean distance between myosin thick filaments. (C) Distance d of myosin punctae from the barbed end of F-actin over time. (D) dc as a function of d12, with dashed lines indicating the results for changes expected due to buckling, with no actomyosin sliding (no sliding), and those for relative sliding, with no F-actin buckling (no buckling). (E and F) Persistence length, lp′ (red) and network strain (blue) for (E) Rxlink = 0 (RAdh = 0) (Movie S10) and (F) Rxlink = 0.03 (RAdh = 0) (Movie S9). The lp′ data are for a single F-actin, and the network strain reflects the average of four myosin pairs in E and five in F, each within a single experiment. (G) Schematic indicating measurement of filament compressive strain, determined by tracking changes in end-to-end length of single filaments, and network contractile strain, determined by changes in the size of the network. (H) Network strain ɛ as a function of filament strain ɛfil during contraction for Rxlink = 0, 0.003, and 0.03 (RAdh = 0). Dashed line indicates ɛ = ɛfil. The data are the average of four myosin pairs and nine filaments for Rxlink = 0, seven myosin pairs and six filaments for Rxlink = 0.003, and five myosin pairs and seven filaments for Rxlink = 0.03, each for a single experiment.
The change in filament curvature during contraction can be quantified by measuring F-actin’s effective persistence length over time, 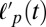 . At times before and up to myosin addition, the persistence length
. At times before and up to myosin addition, the persistence length 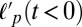 remains constant and is >10 μm, consistent with the known persistence length of F-actin (17 μm) with the increased value presumed to reflect the quasi-2D and crowded environment. Upon myosin addition,
remains constant and is >10 μm, consistent with the known persistence length of F-actin (17 μm) with the increased value presumed to reflect the quasi-2D and crowded environment. Upon myosin addition, 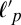 drops rapidly to ∼1 μm for both entangled (Rxlink = 0) and highly bundled (Rxlink = 0.03) networks as contraction occurs (Fig. 2 E and F and Fig. S9). The persistence length continues to decrease further during contraction up to strains of 80%. Thus, the generation of large contractile strains in a disordered actin network is associated with compressive stresses that bend and buckle actin filaments.
drops rapidly to ∼1 μm for both entangled (Rxlink = 0) and highly bundled (Rxlink = 0.03) networks as contraction occurs (Fig. 2 E and F and Fig. S9). The persistence length continues to decrease further during contraction up to strains of 80%. Thus, the generation of large contractile strains in a disordered actin network is associated with compressive stresses that bend and buckle actin filaments.
The origin of network contraction could arise from numerous mechanisms. A sarcomere-like mechanism could arise from tensile stresses driving actomyosin sliding to increase overlap of actin and myosin filaments and, thus, shorten the overall length. Filament buckling is another means to shorten the end-to-end length of F-actin and, in the presence of tensile forces to drive coalescence, could also result in contraction. We measure the filament strain by measuring the fraction change in end-to-end filament length L(t) during contraction, ɛfil = 1 − L(t)/L0, where L0 is the initial distance between filament ends (Fig. 2G), and find that the extent of network contraction corresponds exactly to the extent of filament shortening via buckling such that ɛ = ɛfil (Fig. 2H). Moreover, this exact matching of filament compressive strain and network contraction is observed over a large range of cross-link concentrations (Fig. 2H). If F-actin buckling did not play a significant role in network contraction, we would expect a lack of correlation between these parameters and ɛ < ɛfil. By contrast, a sarcomere-like sliding contraction mechanism would enable contraction without any changes to filament length, such that ɛ > ɛfil. Instead, we observe that network contraction occurs concomitantly with filament buckling, suggesting an important role in its regulation.
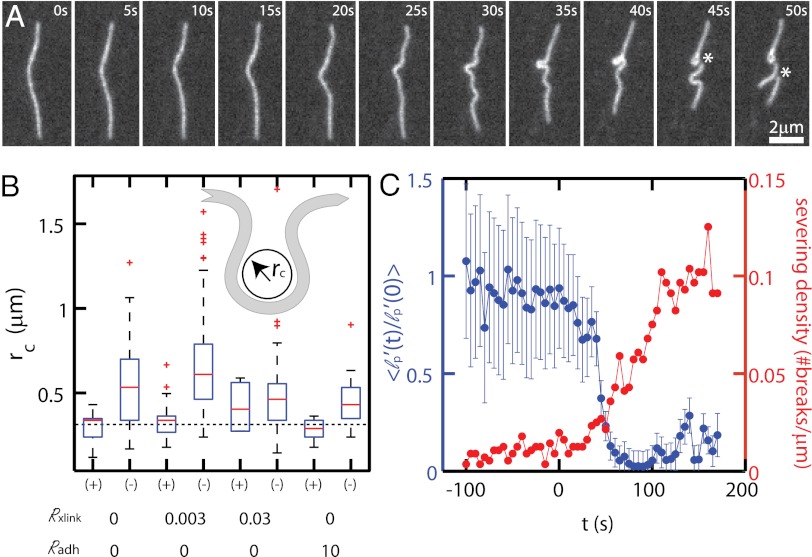
Filament severing occurs during myosin-mediated bending fluctuations and motions. Severing events predominately localize to sites of high F-actin curvature (>99%) (Fig. 3A) with remaining events occurring at apparently taut and straight filament portions (Fig. S10). To explore the connection between F-actin curvature and severing, we measure the radius of curvature, rc, of F-actin immediately before a severing event (Fig. 3B). We find that severing occurs predominately at or below an rc of ∼300 nm, irrespective of the cross-linking density (Fig. 3B), and is not a product of photodamage (Movie S13). Moreover, filament severing occurs throughout contraction as myosin-generated stresses continually drive bending fluctuations of filaments (Fig. 3C). At the initial stages of contraction, the severing rate is low and large radii of curvature bends facilitate contractility. Severing increases at the later stages of contraction, where small radii of curvature are apparent, and severing events do not contribute significantly to further contraction within the next 10–20 s (Fig. 3A). Thus, the compressive stresses arising from myosin motors result in mechanically mediated F-actin severing.
Fig. 3.
Filament buckling at high curvature induces severing. (A) F-actin images during contraction of a sparsely labeled network (Rxlink = 0, RAdh = 0). (B) Box plot of the filament radii of curvature rc measured preceding a severing event (+) or not (−). Dashed line indicates 300 nm. The sample sizes for the different conditions are as follows: Rxlink = 0/Radh = 0 (Nsever = 14, Nstable = 58), Rxlink = 0.003/Radh = 0 (Nsever = 22, Nstable = 123), Rxlink = 0.03/Radh = 0 (Nsever = 4, Nstable = 58), and Rxlink = 0/RAdh = 10 (Nsever = 10, Nstable = 21). Nsever is the number of measurements of rc taken that sever in the following time point. Nstable is the number of rc measurements, selected at random, that do not sever in the next time point. Data reflect eight independent experiments. (Inset) Schematic of radius of curvature. (C) Persistence length normalized to initial value before contraction (blue) and mean severing density as a function of time (red) for Rxlink = 0.003/RAdh = 0. The data reflect the curvature of 5 filaments (lp′) and the severing (Nsever) of 102 filaments from a single experiment.
In the cell, the actomyosin cortex is coupled to the plasma membrane by cross-linking and regulatory proteins (28). To explore the consequence of membrane adhesion on actomyosin contraction, we mimic this condition by coupling the actin network to the lipid bilayer with a histidine-purified actin-binding domain of fimbrin (FimA2) (Fig. 4A). At low FimA2 concentrations (Radh = [fimA2]/[G-actin] = 10), the contraction is qualitatively similar to that observed with no adhesion but the length scale of contraction is reduced (Movie S14). As the adhesion to the membrane is increased (Radh = 100–1,000), contraction is impeded further, evidenced by the formation of smaller aggregates that are spaced closer together (Fig. S6). Using sparse labeling of F-actin (1–2% fluorescent), we readily observe that the consequence of high adhesion is to dramatically constrain the transverse motions of F-actin and prevent buckling with large rc (Fig. 4B and Movie S12). The rc at which severing occurs is nearly identical to those observed in the absence of adhesion (Fig. 3B) but occurs with greater frequency due to adhesion constraints. As such, the rate of F-actin severing is dramatically enhanced in these networks (Fig. 4C). The resistance provided by membrane adhesion reduces filament translocation (Figs. S6 and S10 and Movies S12 and S14) and constrains bending to smaller radii of curvature, thereby impeding contraction but enhancing mechanically mediated severing.
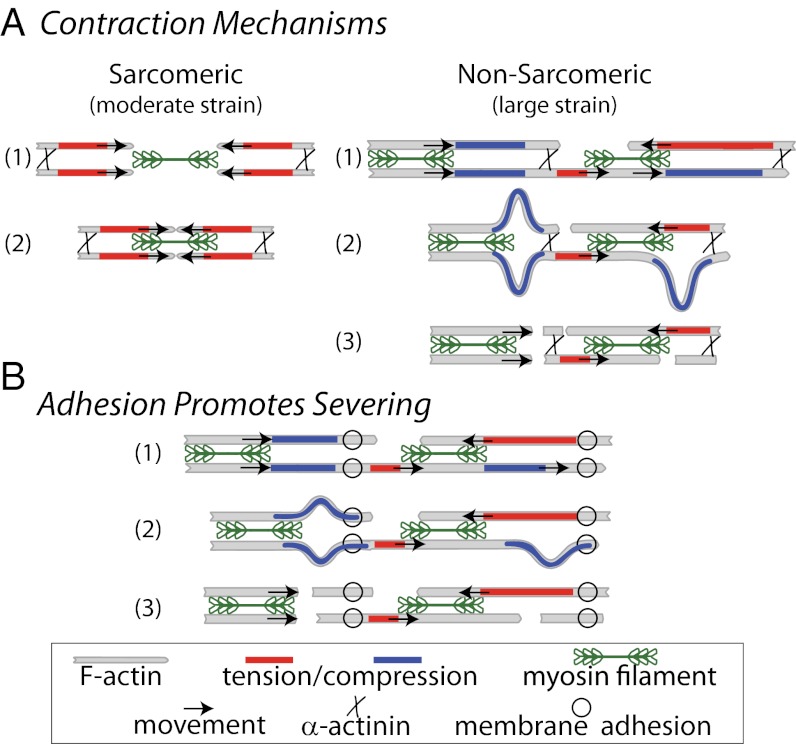
Our data demonstrate a prominent role of F-actin bending and buckling in the contraction of nonsarcomeric actomyosin networks found in nonmuscle and smooth muscle cells. In striated muscle, contraction is facilitated by enhanced actomyosin overlap arising from tensile forces applied to F-actin with well-defined polarity (Fig. 5A). The extent of contraction is limited by the extent of increased overlap of myosin thick filaments and F-actin thin filaments and is ∼30% for striated muscle (29). In actomyosin networks and bundles that lack such organization, myosin generates both compressive and tensile stresses on F-actin (Fig. 5A) (19). These stresses can arise either locally by the stochastic interactions of myosin motors directly on individual F-actin (15, 19) or from myosin-generated fluctuations of the surrounding network in which the filament is embedded (26). Filament portions under compressive stresses yield readily via buckling, thereby breaking the symmetry between compressive and tensile stresses that myosin motors generate in disordered actin networks. The remaining tensile stresses drive shortening of the filament segments to result in network contraction. Contraction persists over large strains as filament portions change from being under tension to compression as the network architecture dynamically changes during contraction. Thus, this contractility mechanism is robust to disorder and accommodates a large extent of contraction, as is observed in smooth and nonmuscle cells. Our data demonstrate that buckling can account for the contractile strains up to 60–80% at the initial stages of contraction; at later times, filament severing and reorganization may start to dominate further contraction and myosin-mediated remodeling but these later stages are not amenable to our analysis.
Fig. 5.
Schematics of contraction mechanisms. (A) In sarcomeres, myosin filaments (green) are segregated toward pointed ends of F-actin and cross-links (black “x”) are at the barbed end. This configuration permits solely tensile forces (red) and, thus, translocation of actin filaments (black arrows), resulting in contraction. In disordered, nonsarcomeric actomyosin bundles and networks that lack segregation of motors and cross-links, actomyosin interactions result in both tension and compression (blue). Compressive stresses are relieved through filament buckling and severing, keeping only tensile forces and, thus, driving contraction. (B) Cross-links to the membrane (open circles) spatially constrain stresses generated by motors and thus promote severing and prevent long-range transmission of tensile stresses. Numbers delineate sequential steps in each process.
Here we demonstrate that local contractions can occur in quasi-2D networks containing myosin and actin alone. In these conditions, the length scale over which contraction occurs within the network is proportional to the F-actin length, consistent with poor network connectivity by myosin motors (30–33) or putative cross-linking by MC. Through the addition of the passive cross-linker α-actinin, we are able to increase the length scale of contraction to macroscopic length scales. Previous work in semidilute, 3D networks has shown the necessity of cross-linkers for contractility (30–33) whereas a sufficiently high density of myosin motors in the absence of cross-linkers is sufficient to facilitate contractility in 1D bundles (34). We attribute the differences in efficiency of myosin cross-linking observed under these diverse experimental conditions to changes in local concentration of actin and myosin. Our current findings appear to be consistent with a local concentration of actomyosin in a 2D network being between that of a semidilute 3D network and a highly dense 1D bundle.
Our results suggest that control of filament bending is an important regulator of contractility. In weakly cross-linked networks (Rxlink = 0–0.003), large radii of curvature are observed and facilitate a large extent of local network compression. As the cross-link concentration is increased and bundles are formed (Rxlink = 0.03), the filaments are constrained, thus reducing the mean radii of curvature and limiting the local compression. We speculate that filament bends and, thus, the regime of buckling-mediated contractility could be eliminated in the regime where myosin-generated stresses are reduced (e.g., by reduced myosin concentration or thick filament size) or have increased bundle bending rigidity. In this regime, other mechanisms of contractility arising from filament reorganization may occur (16).
We find that portions of F-actin that are bent to high curvature are prone to severing. This finding is consistent with previous measurements of mechanically mediated F-actin severing induced by externally applied or thermal forces (11, 35), which found that severing occurred at a radius of curvature of <0.4 μm (35) or an angle of 56° (11). In the absence of external mechanical constraints, such severing occurs with forces of 3–4 pN. By contrast, F-actin breakage under tension occurs at forces of ∼400 pN (36). This discrepancy likely underlies the predominance of filament severing via buckling that we observe. Thus, F-actin severing enhances the asymmetric response to facilitate contractility of disordered actomyosin. In addition to relieving compressive stress, severing creates F-actin barbed and pointed ends that change F-actin density and length. Whereas previous measurements have demonstrated a myosin-II–mediated destabilization of F-actin networks and bundles both in vitro (12, 37) and in vivo (4), our results demonstrate that these likely arise from compressive stresses generated within disordered networks. F-actin severing changes the local density of barbed and pointed filament ends, which, depending on the biochemical environment, can result in changes of polymer density. In the presence of capping protein or the absence of G-actin, F-actin would disassemble whereas, in the presence of formins or high concentrations of G-actin, actin polymerization would be stimulated (38). This demonstrates the importance of compressive stresses generated by actomyosin interactions in disordered networks as a mechanism to couple contractility and F-actin turnover, as has been reported in vivo (2–4). Such mechanically regulated F-actin polymerization dynamics may enhance or complement biochemical regulation by severing proteins such as ADF/cofilin.
In the presence of adhesion to the lipid bilayer, F-actins are subject to large compressive stresses that promote severing, but force transmission through the network is not permitted due to enhanced adhesive drag (Fig. 5B). Thus, adhesion of the cortical network to the membrane modulates the coordination between actomyosin contractility and filament severing, which may be a regulatory mechanism to coordinate cortex contractility and turnover in vivo. These results may explain the reduced contractility with enhanced cortical adhesion observed in vivo (39). Moreover, our results suggest that membrane adhesion could be a means to use actomyosin stresses to generate a high density of F-actin barbed ends, as is found at focal adhesion plaques in adherent cells (40). Contractility in our simplified, reconstituted system demonstrates the importance of the nonlinear mechanical response of individual actin filaments to compressive stresses, as their buckling is crucial for early symmetry breaking events to drive cortical actomyosin contractility and polymerization dynamics, which are necessary in cell physiology.
Methods
A supported lipid bilayer from egg phosphatidyl choline (91%) and nickel lipid (9%) is formed on a Piranha-treated coverslip glass surface. In conditions with membrane adhesion, a His-tagged actin-binding domain of fimbrin is added and allowed to bind to the membrane. In all experiments, 1.3 μM phalloidin-stabilized actin filaments (F-actin) are crowded to the lipid bilayer surface with a 0.25% methylcellulose solution for >15 min, after which α-actinin and myosin II are sequentially added and gently mixed. To prevent shear of the F-actin network proximal to the lipid bilayer, the α-actinin and myosin are mixed with a small-volume pipette in the immediate vicinity of the positioned objective. At elevated concentrations of α-actinin (Rxlink = 0.03) contraction is initiated through the inactivation of blebbistatin. All imaging is performed using spinning-disk confocal microscopy. Details of all experimental methods can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Martin Lenz (Centre National de la Recherche Scientifique, Orsay, France) for his advice on calculations. We also thank Tobias Falzone and Yujie Li for providing purified proteins. M.L.G. acknowledges support from the Burroughs Wellcome Fund, the Packard Foundation, and National Institutes of Health Grant DP10D00354. This work was supported by the Materials Research Science and Engineering Center at University of Chicago. M.P.M. is funded by a postdoctoral fellowship from the National Science Foundation Institute for Complex Adaptive Matter (ICAM).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1214753109/-/DCSupplemental.
References
- 1.Bray D, White JG. Cortical flow in animal cells. Science. 1988;239(4842):883–888. doi: 10.1126/science.3277283. [DOI] [PubMed] [Google Scholar]
- 2.Guha M, Zhou M, Wang YL. Cortical actin turnover during cytokinesis requires myosin II. Curr Biol. 2005;15(8):732–736. doi: 10.1016/j.cub.2005.03.042. [DOI] [PubMed] [Google Scholar]
- 3.Medeiros NA, Burnette DT, Forscher P. Myosin II functions in actin-bundle turnover in neuronal growth cones. Nat Cell Biol. 2006;8(3):215–226. doi: 10.1038/ncb1367. [DOI] [PubMed] [Google Scholar]
- 4.Wilson CA, et al. Myosin II contributes to cell-scale actin network treadmilling through network disassembly. Nature. 2010;465(7296):373–377. doi: 10.1038/nature08994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol. 2009;10(11):778–790. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pollard TD. Mechanics of cytokinesis in eukaryotes. Curr Opin Cell Biol. 2010;22(1):50–56. doi: 10.1016/j.ceb.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wozniak MA, Chen CS. Mechanotransduction in development: A growing role for contractility. Nat Rev Mol Cell Biol. 2009;10(1):34–43. doi: 10.1038/nrm2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardel ML, Schneider IC, Aratyn-Schaus Y, Waterman CM. 2010. Mechanical integration of actin and adhesion dynamics in cell migration Annu Rev Cell Dev Biol 26:3.1–3.19.
- 9.Roh-Johnson M, et al. Triggering a cell shape change by exploiting preexisting actomyosin contractions. Science. 2012;335(6073):1232–1235. doi: 10.1126/science.1217869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ponti A, Machacek M, Gupton SL, Waterman-Storer CM, Danuser G. Two distinct actin networks drive the protrusion of migrating cells. Science. 2004;305(5691):1782–1786. doi: 10.1126/science.1100533. [DOI] [PubMed] [Google Scholar]
- 11.McCullough BR, et al. Cofilin-linked changes in actin filament flexibility promote severing. Biophys J. 2011;101(1):151–159. doi: 10.1016/j.bpj.2011.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haviv L, Gillo D, Backouche F, Bernheim-Groswasser A. A cytoskeletal demolition worker: Myosin II acts as an actin depolymerization agent. J Mol Biol. 2008;375(2):325–330. doi: 10.1016/j.jmb.2007.09.066. [DOI] [PubMed] [Google Scholar]
- 13.Huxley HE. The mechanism of muscular contraction. Science. 1969;164(3886):1356–1365. doi: 10.1126/science.164.3886.1356. [DOI] [PubMed] [Google Scholar]
- 14.Svitkina TM, Verkhovsky AB, McQuade KM, Borisy GG. Analysis of the actin-myosin II system in fish epidermal keratocytes: Mechanism of cell body translocation. J Cell Biol. 1997;139(2):397–415. doi: 10.1083/jcb.139.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lenz M, Gardel ML, Dinner AR. Requirements for contractility in disordered cytoskeletal bundles. New J Phys. 2012;14:033037. doi: 10.1088/1367-2630/14/3/033037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dasanayake NL, Michalski PJ, Carlsson AE. General mechanism of actomyosin contractility. Phys Rev Lett. 2011;107(11):118101. doi: 10.1103/PhysRevLett.107.118101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kruse K, Joanny JF, Jülicher F, Prost J, Sekimoto K. Asters, vortices, and rotating spirals in active gels of polar filaments. Phys Rev Lett. 2004;92(7):078101. doi: 10.1103/PhysRevLett.92.078101. [DOI] [PubMed] [Google Scholar]
- 18.Vavylonis D, Wu JQ, Hao S, O’Shaughnessy B, Pollard TD. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 2008;319(5859):97–100. doi: 10.1126/science.1151086. [DOI] [PubMed] [Google Scholar]
- 19.Lenz M, Thoresen T, Gardel ML, Dinner AR. Contractile units in disordered actomyosin bundles arise from f-actin buckling. Phys Rev Lett. 2012;108(23):238107. doi: 10.1103/PhysRevLett.108.238107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang S, Wolynes PG. Active contractility in actomyosin networks. Proc Natl Acad Sci USA. 2012;109(17):6446–6451. doi: 10.1073/pnas.1204205109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liverpool TB, Marchetti MC, Joanny JF, Prost J. Mechanical response of active gels. Europhys Lett. 2009;85:18007. [Google Scholar]
- 22.Soares e Silva M, et al. Active multistage coarsening of actin networks driven by myosin motors. Proc Natl Acad Sci USA. 2011;108(23):9408–9413. doi: 10.1073/pnas.1016616108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Köhler S, Lieleg O, Bausch AR. Rheological characterization of the bundling transition in F-actin solutions induced by methylcellulose. PLoS ONE. 2008;3(7):e2736. doi: 10.1371/journal.pone.0002736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Courson DS, Rock RS. Actin cross-link assembly and disassembly mechanics for alpha-Actinin and fascin. J Biol Chem. 2010;285(34):26350–26357. doi: 10.1074/jbc.M110.123117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skau CT, et al. Actin filament bundling by fimbrin is important for endocytosis, cytokinesis, and polarization in fission yeast. J Biol Chem. 2011;286(30):26964–26977. doi: 10.1074/jbc.M111.239004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brangwynne CP, Koenderink GH, Mackintosh FC, Weitz DA. Nonequilibrium microtubule fluctuations in a model cytoskeleton. Phys Rev Lett. 2008;100(11):118104. doi: 10.1103/PhysRevLett.100.118104. [DOI] [PubMed] [Google Scholar]
- 27.Howard J. Mechanics of Motor Proteins and the Cytoskeleton. Sunderland, MA: Sinauer; 2001. [Google Scholar]
- 28.Saarikangas J, Zhao H, Lappalainen P. Regulation of the actin cytoskeleton-plasma membrane interplay by phosphoinositides. Physiol Rev. 2010;90(1):259–289. doi: 10.1152/physrev.00036.2009. [DOI] [PubMed] [Google Scholar]
- 29.Gordon AM, Homsher E, Regnier M. Regulation of contraction in striated muscle. Physiol Rev. 2000;80(2):853–924. doi: 10.1152/physrev.2000.80.2.853. [DOI] [PubMed] [Google Scholar]
- 30.Janson LW, Kolega J, Taylor DL. Modulation of contraction by gelation/solation in a reconstituted motile model. J Cell Biol. 1991;114(5):1005–1015. doi: 10.1083/jcb.114.5.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bendix PM, et al. A quantitative analysis of contractility in active cytoskeletal protein networks. Biophys J. 2008;94(8):3126–3136. doi: 10.1529/biophysj.107.117960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Köhler S, Schaller V, Bausch AR. Structure formation in active networks. Nat Mater. 2011;10(6):462–468. doi: 10.1038/nmat3009. [DOI] [PubMed] [Google Scholar]
- 33.Backouche F, Haviv L, Groswasser D, Bernheim-Groswasser A. Active gels: Dynamics of patterning and self-organization. Phys Biol. 2006;3(4):264–273. doi: 10.1088/1478-3975/3/4/004. [DOI] [PubMed] [Google Scholar]
- 34.Thoresen T, Lenz M, Gardel ML. Reconstitution of contractile actomyosin bundles. Biophys J. 2011;100(11):2698–2705. doi: 10.1016/j.bpj.2011.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arai Y, et al. Tying a molecular knot with optical tweezers. Nature. 1999;399(6735):446–448. doi: 10.1038/20894. [DOI] [PubMed] [Google Scholar]
- 36.Tsuda Y, Yasutake H, Ishijima A, Yanagida T. Torsional rigidity of single actin filaments and actin-actin bond breaking force under torsion measured directly by in vitro micromanipulation. Proc Natl Acad Sci USA. 1996;93(23):12937–12942. doi: 10.1073/pnas.93.23.12937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmoller KM, Semmrich C, Bausch AR. Slow down of actin depolymerization by cross-linking molecules. J Struct Biol. 2011;173(2):350–357. doi: 10.1016/j.jsb.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 38.Campellone KG, Welch MD. A nucleator arms race: Cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11(4):237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Charras GT, Hu CK, Coughlin M, Mitchison TJ. Reassembly of contractile actin cortex in cell blebs. J Cell Biol. 2006;175(3):477–490. doi: 10.1083/jcb.200602085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gupton SL, Eisenmann K, Alberts AS, Waterman-Storer CM. mDia2 regulates actin and focal adhesion dynamics and organization in the lamella for efficient epithelial cell migration. J Cell Sci. 2007;120(Pt 19):3475–3487. doi: 10.1242/jcs.006049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.