Abstract
Cytochrome c unfolds locally and reversibly upon heating at pH 3. UV resonance Raman (UVRR) spectra reveal that instead of producing unordered structure, unfolding converts turns and some helical elements to β-sheet. It also disrupts the Met80-heme bond, and was earlier shown to induce peroxidase activity. Aromatic residues that are H-bonded to a heme propionate (Trp59 and Tyr48) alter their orientation, indicating heme displacement. T-jump/UVRR measurements give time constants of 0.2, 3.9 and 67 µs for successive phases of β-sheet formation and concomitant reorientation of Trp59. UVRR spectra reveal protonation of histidines, and specifically of His26, whose H-bond to Pro44 anchors the 40s Ω loop; this loop is known to be the least stable ‘foldon’ in the protein. His26 protonation is proposed to disrupt its H-bond with Pro44, triggering the extension of a short β-sheet segment at the ‘neck’ of the 40s Ω loop into the loop itself and back into the 60’s and 70’s helices. The secondary structure change displaces the heme via H-bonds from residues in the growing β-sheet, thereby exposing it to exogenous ligands, and inducing peroxidase activity. This unfolding mechanism may play a role in cardiolipin peroxidation by cyt c during apoptosis.
1. INTRODUCTION
Cytochrome c (cyt c), a much-studied electron carrier in the mitochondrial respiratory chain, has emerged as a renewed focus of biological interest because of its role as a mediator of apoptosis. Apoptotic cell death is triggered by migration of cyt c from the mitochondria into the cytoplasm, where it induces a cascade of biochemical events.1–7 Cyt c release into the cytoplasm results from its association with cardiolipin, a component of the mitochondrial membrane, accompanied by membrane pore formation.8 Cardiolipin forms a specific association with cyt c by inserting its hydrocarbon chains into hydrophobic channels in the protein, resulting in a conformation change that induces peroxidase activity.9–13 Peroxidation of cardiolipin induces the release of apoptotic pro-factors.14,15
We have shown that peroxidase activity is also induced by heating horse heart cyt c at pH 3, in concert with a protein conformation change involving β-sheet formation, and with conversion of the heme to a high-spin state.16 This conformation change was proposed to be initiated by protonation of His26,17,18 which H-bonds to Pro44, a residue at the tip of a long omega loop (40’s Ω loop) (Figure 1). This loop was characterized by Englander and coworkers19,20 as the least stable cooperative unfolding unit (‘foldon’) in cyt c. It is anchored at its other end by a short β-sheet ‘neck’, which we suggested could extend into the loop itself, when the His26-Pro44 H-bond is broken. Since the heme propionate substituents accept H-bonds from residues in the loop, this conformation change would be expected to perturb the heme, inducing dissociation of the Met80 ligand and allowing acesss of peroxide to the vacated coordination site.
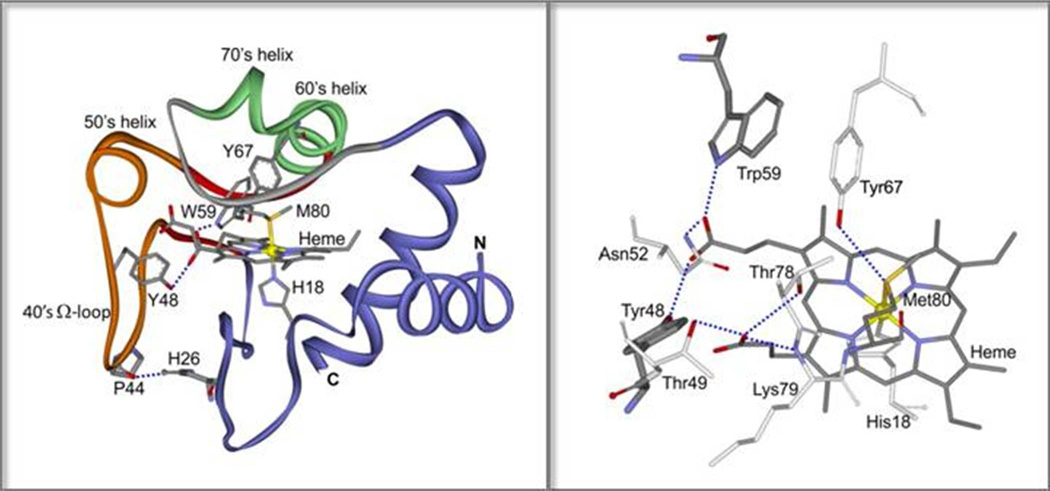
Figure 1.
Left: Ribbon diagram of hh cyt c (pdb id: 1hrc). The 40’s Ω loop is orange, its β-sheet ‘neck’ is red, the 60’s & 70’s helices are green and the Met80 loop is gray. H-bonds are shown in dashed blue lines for P44/H26, and for W59 and Y48 with a heme propionate. Right: The full H-bond pattern of the heme propionates with residues in the 40’s and Met80 loops, and the Tyr67/Met80 H-bond.
In the present work we characterize this conformation change more fully, using several UV resonance Raman spectroscopic probes. The spectra reveal that heating cyt c at pH 3 does indeed induce protonation of His26. The accompanying shift to β-sheet structure also induces changes in signals from tryptophan and tyrosine residues, which are consistent with the 40’s loop being the locus of protein conformation change, along with the adjacent 60’s and 70’s helices (Figure 1). These changes are reversible, and T-jump/UVRR experiments reveal that they occur on the microsecond time scale.
Since antibody to the Pro44 region was earlier shown to bind to apoptotic cells, as well as to cyt c when bound to lipid vesicles21, the proposed unfolding mechanism may be involved in cardiolipin peroxidation and apoptosis.
2. METHODS
2.1 Materials
Horse heart ferricytochrome c (hh cyt c) was purchased from Sigma Aldrich and pretreated as described below. Deuterated water (D2O), NaOD and DCl of greater than 99 % purity were purchased from Cambridge Isotope Laboratories Inc. and used without further purification. All other chemicals are of analytical grade and used as received. The protein was dissolved in 50 mM sodium phosphate buffer in H2O or D2O at appropriate pH or pD. Stock solutions of 1 mM protein were pretreated with 10 mM potassium ferricyanide to oxdize the residual reduced forms and desalted through PD10 column. Samples included 50 mM sodium perchlorate as a Raman intensity standard. The protein concentration was 50 – 100 µM for static RR measurements, 200µM for T-jump experiments, 50 FM for measurements of the 695 nm absorption band, and 20 µM for CD measurements.
2.2 Visible Absorption Spectroscopy
UV-Vis absorption spectra were recorded on an Agilent 8453 UV-Vis Spectrophotometer with a 1nm slit width. Temperature-dependent measurements were recorded on a thermostatted Cary 300 Bio UV-Visible spectrophotometer; samples were equilibrated for 10 min. 10 mm path length cells were used for absorption measurements. The baseline of the absorption spectra was modeled and subtracted using a spline function.
2.3 Circular Dichroism
Variable temperature far-UV circular dichroism wavelength scans were collected on an AVIV 62DS spectropolarimeter, equipped with a temperature-controlled cuvette holder, using a 1 mm quartz cell. The data were recorded from 260 to 190 nm with 1 nm spacing, and every 5 °C from 5 to 95 °C. 222 nm temperature scans were carried out from 20°C to 100°C at a rate of 2°C/0.5min. A bandwidth of 1.5 nm and average time of 5s were used. The data were converted to mean residue ellipticity [θ] in deg*cm2/dmol using the measured protein concentration.
2.4 Equilibrium UVRR Measurements
The experimental setup and the spectral acquisition scheme have previously been described.22 UV probe pulses (20 ns, 1 kHz) at 197 nm23 (~1 µJ/pulse) or 229 nm22,24 (~1 µJ/pulse) were obtained by frequency quadrupling the output of a Ti:sapphire laser, which was pumped (527 nm, ~10 mJ/pulse, 70 ns, 1 kHz) by an intracavity frequency-doubled Nd:YLF laser (GM30, Photonics International, Inc.). Raman scattered light was collected at 135° geometry and focused into a 1.26 m spectrograph (Spex 1269), which was equipped with a holographic grating (3600 groove/mm) and an intensified photodiode array detector (IPAD) (Roper Scientific). The sample solution was pumped through a wire-guided free-flowing cell to minimize the background. Temperature control of the sample was achieved with a water bath (RTE-100, Neslab), which regulates the temperature of the sample cell and reservoir. UVRR spectra were collected with 15 minutes accumulation time. The measured spectra were calibrated using the Raman spectrum of cyclohexane. The water background was subtracted from protein Raman spectra using the pure water Raman spectrum.
2.5 Time-resolved T-jump UVRR Measurements
UV probe pulses for time-resolved UVRR measurements were generated by the UV laser described above. T-jump pulses were obtained from an intracavity diode laser pumped Nd:YLF laser optical parametric oscillator (OPO), which can generate tunable near-infrared (NIR) 1.6–2.0 µm, 0.8–1.0 mJ/pulse, 20 ns, 1 kHz pulses (Photonics International, Inc.). The final temperature of the sample after a T-jump was determined to ±2 °C from the ~3400 cm−1 water Raman band intensity change.25 The wavelength of the T-jump pump pulse is within the near-IR water band centered at 1.9 µm. The final temperature of the sample within the laser interaction volume is achieved instantaneously after a T-jump pulse (complete thermalization is reached within 10 ~ 20 ps26). The probe and pump pulse sequence was alternated to minimize artifacts from spectrometer drift and photoproduct accumulation, which is critical for this study, since no internal standard was added. The timing between T-jump and probe pulses was adjusted with a computer-controlled pulse generator (DG 535, Stanford Research Systems). The final pump-probe (with T-jump) and probe-only (without T-jump) spectra were averages over 15 minutes of accumulation.
All UVRR spectra were processed and analyzed using Grams/AI 7.00 (Galactic Industries Corp.) and plotted using Origin 6.0 software.
3. RESULTS
3.1 Avoidance of aggregation
Under conditions of thermal denaturation, cyt c is known to be susceptible to aggregation, through inter-molecular β-sheet association.27–31 We therefore determined the concentration range in which aggregation of horse heart cyt c at pH 3 could be avoided under our experimental conditions. Turbidity was monitored at 80°C 450–480 nm, in a trough of the cyt c absorption spectrum, where the optical density reflects diffuse scattering. At pH 3 and 80 °C, the solution became turbid when the concentration was higher than 150 µM. The apparent time constant for turbidity growth ranged from 2 to 4 hours for concentrations between 600 and 160 µM. Turbidity was not observed below 100 µM. Subsequent experiments were carried out at a concentration of 50 µM, except for the T-jump measurements. For these experiments, the reservoir temperature was held at 40°C, and only heated transiently. Consequently a higher concentration, 200µM, could be used without aggregation, in order to generate acceptable signal/noise in the pump-probe difference spectra.
3.2 Thermally induced β-sheet formation
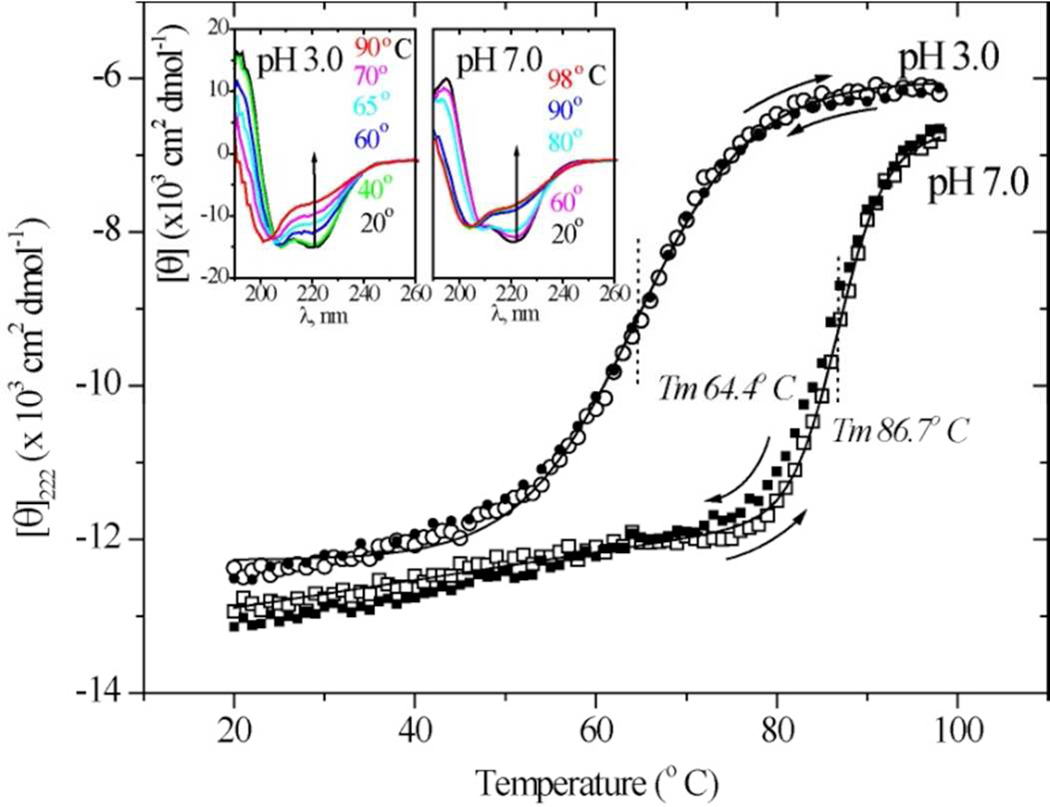
Thermally induced conformation change was initially monitored via UV circular dichroism (Figure 2). Cyt c spectra were recorded at a series of temperatures at pH 7.0 and 3.0. Progressive loss of helicity can be seen in the loss of the 222 nm trough. At pH 3.0, spectra at intermediate temperatures resemble those characteristic of β-sheet.32
Figure 2.
UVCD spectra (insets) for hh cyt c at pH 3.0 and 7.0, and the temperature profiles of the ellipticity at 222 nm. Reversibility can be seen from the agreement of [Θ]222 during heating (open symbols) and cooling (closed symbols).
The ellipticity at 222 nm gave well-behaved melting curves (Figure 2), with mid-point temperatures of 87 and 64°C at pH 7 and 3. Importantly, the temperature dependence was reversible, the same ellipticity being obtained on heating and cooling (arrows).
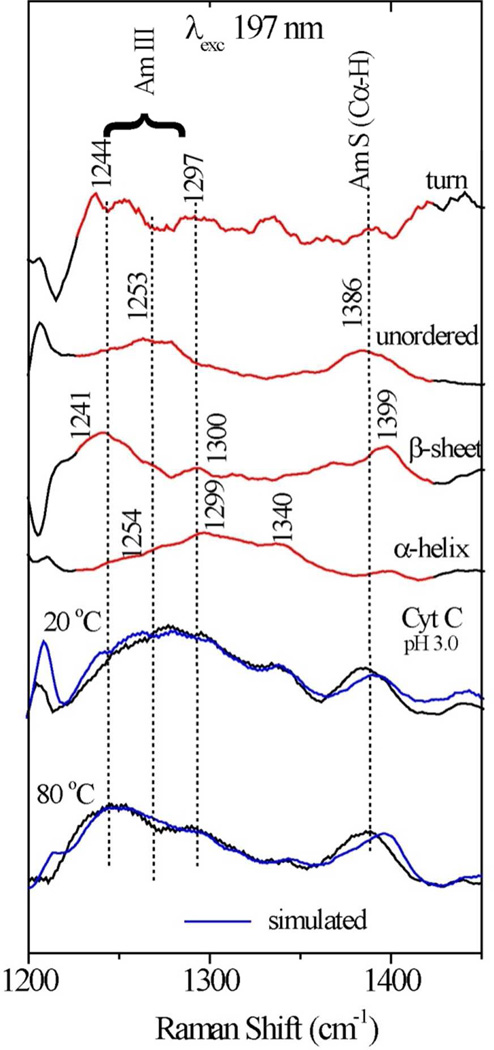
While the UVCD spectra can be analyzed for secondary structure composition,32–38 the considerable overlap of component spectra limits the reliability. We turned instead to UVRR spectra excited at 197 nm (Figure S1). Our procedure for secondary structure analysis39 is to subtract contributions from phenylalanine and tyrosine sidechains (Figure S2) and then to resolve the backbone scattering regions into contributions from α-helix, β-sheet, β-turn, and coil (unordered) contributions, using component spectra from a training set of proteins, via least squares fitting. The greatest discrimination is found in the 1200 – 1500 cm−1 region, where complex bands arising from the amide III and amide S vibrations39–43 give well-structured component spectra, as illustrated in Figure 3. The spectral fitting gave an average goodness-of-fit parameter, R2= 0.962, and relative uncertainties (one sigma) in the population estimates of 2% for helix and coil, 9% for β-sheet and 11% for β-turn (the latter two being somewhat correlated).
Figure 3.
Illustration of secondary structure component analysis of the hh cyt c 197 nm-excited UVRR spectra at pH 3.0 in the amide III and S region. Experimental and simulated spectra are compared at 20° and 80° C.
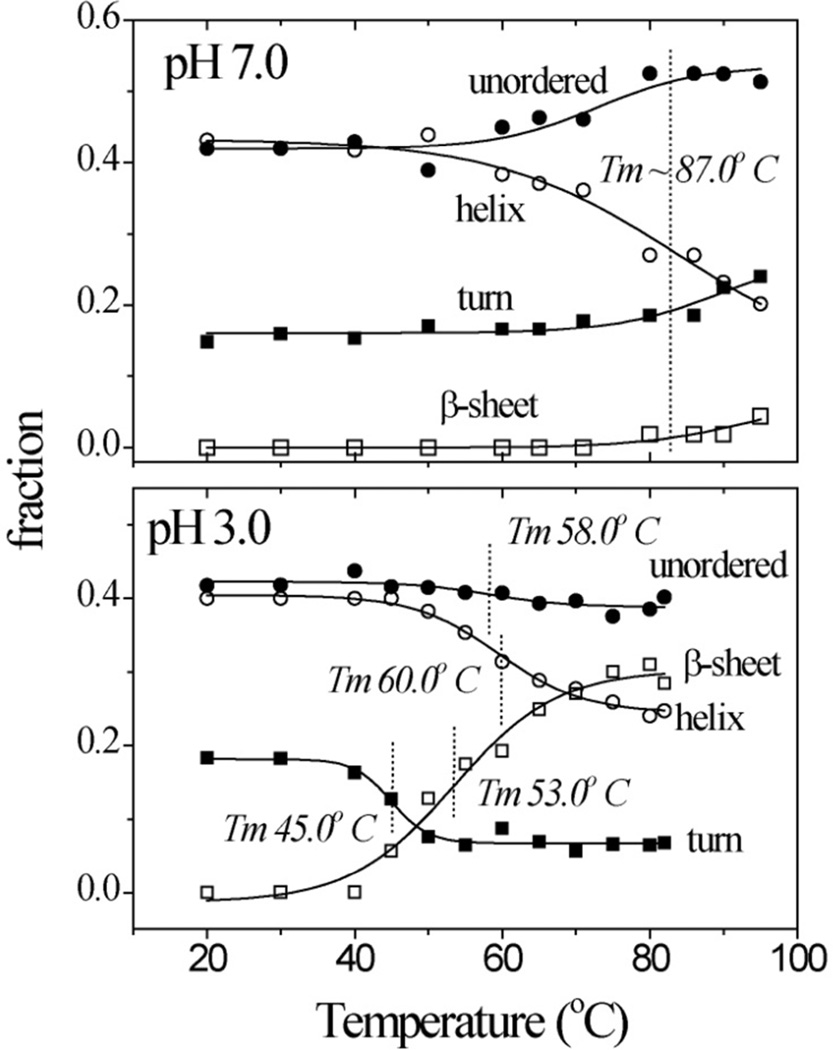
Figure 4 shows the results of this procedure. At 20°C, the spectra are the same at pH 7 and 3, and indicate the presence of 3% β-sheet, 16% β-turn, 41% α-helix, and 40% unordered structure. This composition is consistent with the crystal structure of native cyt c.44 At pH 7, unordered structure increases at high temperature, at the expense of helix melting, consistent with the 87°C Tm determined from UVCD at pH 7. (The temperature range was somewhat lower for the UVRR measurements, precluding an accurate Tm determination).
Figure 4.
Temperature profiles of UVRR-determined secondary structure content in hh cyt c at pH 7.0 and 3.0.
A very different pattern emerged at pH 3 (Figure 4). Helix melting was still observed, and its Tm, 60°C, was consistent with the UVCD determination (Figure 2). However, β-sheet, rather than unordered structure, increased with temperature, and the Tm for this increase was distinctly lower, 53°C. The β-turn population decreased perceptibly, with an even lower Tm, 45°C.
3.3 Associated changes in aromatic residues
229 nm excitation selectively enhances UVRR bands of tyrosine and tryptophan (Figure S3). These signals lose intensity as the temperature increases (Figure 5). This effect is associated with exposure to solvent of buried sidechains.45–47
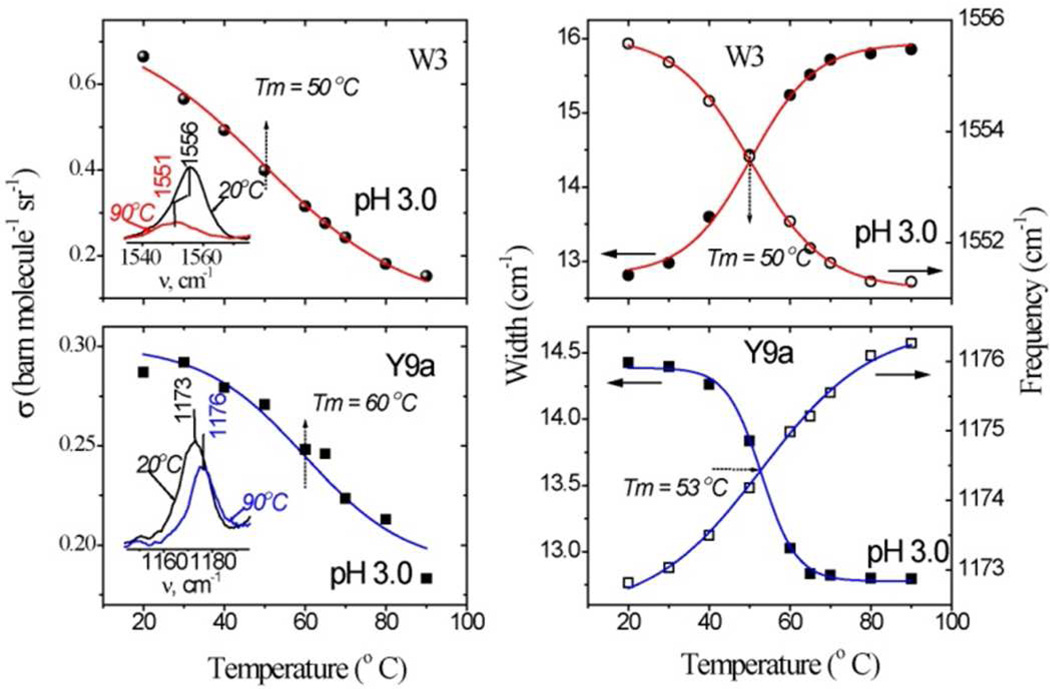
Figure 5.
Temperature profiles of the Trp W3 and Tyr 9a band intensities (left), frequencies (open symbols) and bandwidths (filled symbols) (right) in hh cyt c at pH 3.0. Inset to the intensity profiles shows the corresponding experimental band measured at the indicated lowest and highest temperatures. Sigmoid fits to the experimental data are shown as solid lines.
In addition, the Trp W3 band shifts down in frequency and also become broader. The W3 frequency reflects the dihedral angle of the indole sidechain,48–50 and the observed broadening indicates a wider distribution of this angle. The sole Trp residue is Trp 59, and the W3 broadening indicates an increased distribution of its sidechain angles. The Trp59 indole is H-bonded to one of the heme propionate substituents (Figure 1), and its increased mobility implies weakening of the H-bond. At pH 3, the Tm for this process, 50°C (Figure 5), matches the Tm for β-sheet formation, as does that for the W3 intensity loss.
Essentially the same Tm, 53°C describes an upshift of the Tyr 9a band (Figure 5) at pH 3, but in this case the band narrows. The Y9a band has been found to reflect the dihedral angle of the tyrosine OH substituent with respect to the phenyl ring of tyrosine, more in-plane orientations giving higher frequencies.51 The upshift and narrowing indicated that one or more of the Tyr OH groups shift from a largely out-of-plane to a more in-plane orientation, and thereby narrow the range of orientations among the four residues. A higher Tm, 60°C, is found for the Y9a intensity loss, indicating that one or more of the Tyr residues retains a hydrophobic environment beyond the temperature where others change their OH orientation.
There are four Tyr residues in cyt c, three of which donate OH H-bonds. Jordan et al. calculated the OH dihedral angles from crystallographic coordinates by assuming standard C-O-H distances and angle, and minimizing the H-bond distances.52 The computed dihedral angles are 37°, 22°, and 3° for Tyr48, 67, and 74. The fourth residue, Tyr97, is at the protein surface, and likely has a large dihedral angle (steric hindrance is minimized at 90°), which, however, is unlikely to change much with temperature. Since Tyr48 has the largest angle of the H-bonded residues, it can most readily account for the observed Y9a upshift and narrowing, if its H-bond partner shifts position in a way that decreases the OH dihedral angle, bringing it to a more in-plane orientation. Also supporting the Tyr48 assignment for the Y9a shift is the coincidence of its Tm with that of the Trp59 W3 band, since both residues are H-bonded to the same heme propionate (Figure 1, see Discussion).
From the behavior of the Tyr48 and Trp59 UVRR signals, it can be inferred that the heme is displaced during the temperature-induced protein conformation change, shifting the propionate and decreasing the Tyr48 OH angle, while freeing up the Trp59 indole. The Tyr67 OH orientation may also be perturbed, since it is H-bonded to the S atom of the Met80 heme ligand, which is broken upon heating at pH 3 (see below). However, Tyr67 is unlikely to contribute much to the Y9a upshift, since its starting OH orientation is more in-plane than that of Tyr48. Tyr74, which is H-bonded to an ordered water molecule near the protein surface cannot contribute since its starting OH orientation is in-plane.
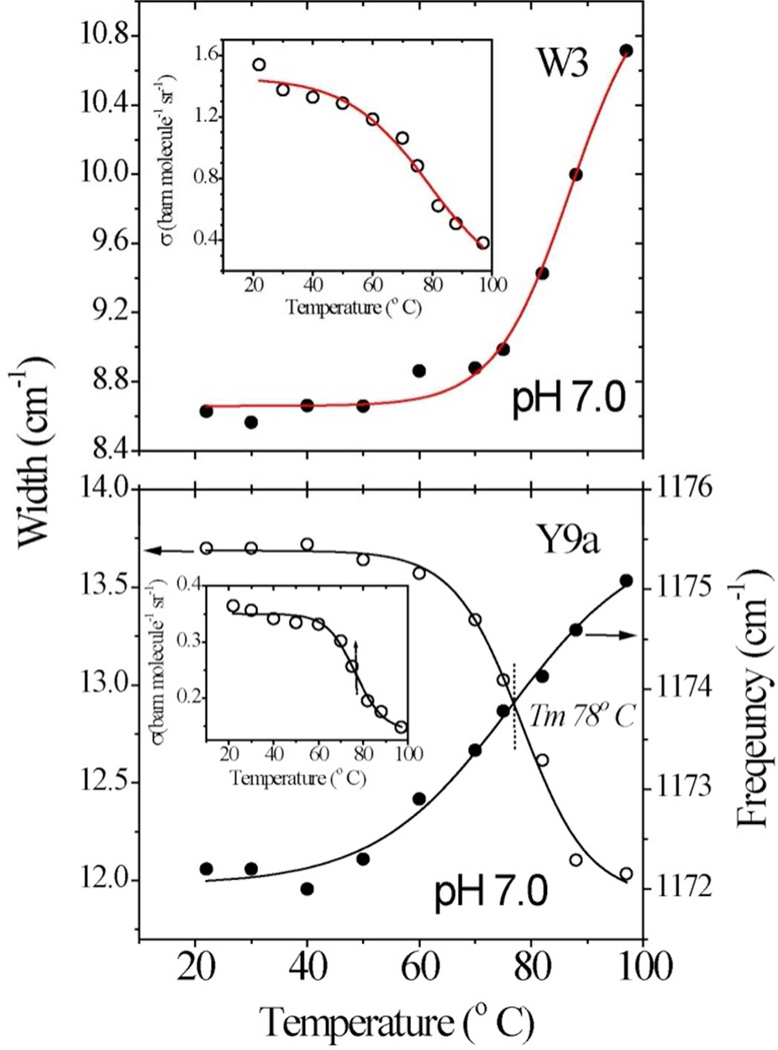
Similar changes in the aromatic residue UVRR signals are seen at pH 7 only near the global melting temperature (Figure 6).
Figure 6.
Same as Fig. 5 but for hh cyt c at pH 7.0
3.4 Met80-Fe bond breaking
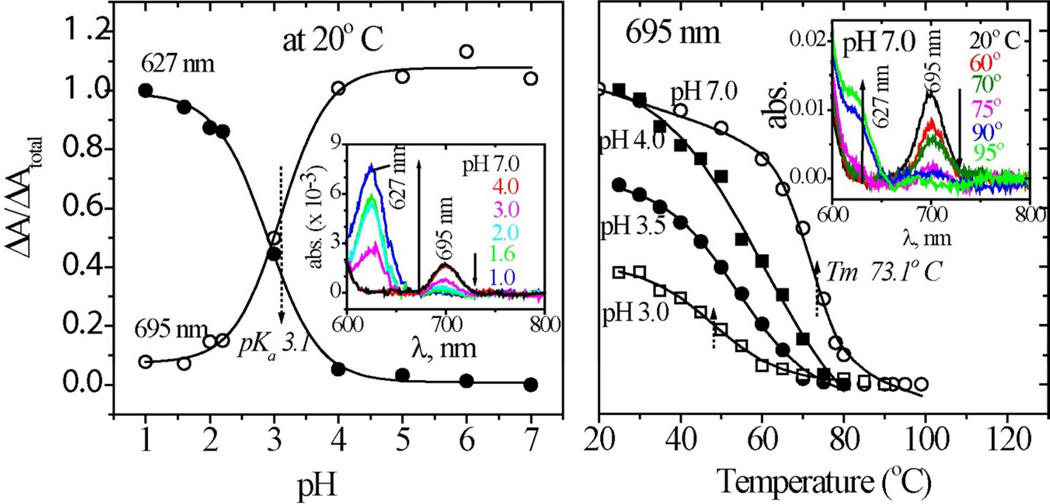
Heme ligation was monitored via the absorption band at 695 nm associated with the Fe-methionine bond53,54 (Figure 7). This band diminishes as the pH is decreased, reflecting the well-known acid-induced dissociation of the Met80 ligand.55 The 695 nm band is replaced by one at 627 nm, assigned to a charge transfer transition of the high-spin heme that results from displacement of Met80 by a water molecule.56 At 20° C, both absorbances follow pH titration curves with a pKa of 3.1. The 695 nm band is also replaced by the 627 nm band when the temperature is increased, describing a series of thermal transitions that depend on pH. At pH 7, Tm for the 695 nm band is 73°C, slightly below the global melting temperature, while at pH 3, Tm= 48°C, near the Tm for the first phase of β-sheet formation. Consistent with the pH titration curve, the fraction of methionine-bound heme is 40% at 20°C and pH 3.
Figure 7.
Left: pH titration curve for the 695 (heme-Met80, open circles) and 627 nm (heme-H2O, filled circles) absorption bands of hh cyt c. Right: Temperature profiles of the 695 nm absorbance at pH = 7.0 (open circles), 4.0 (filled squares), 3.5 (filled circles), and 3.0 (open squares). Inset show the baseline-corrected absorption spectra, as a function of pH and temperature (various colors).
3.5 Histidine protonation
To explore the coupling of pH with protein structure change, we monitored the protonation status of histidine residues, using the intensity of the 220 nm-excited UVRR band57,58 of HisD2+ at 1408 cm−1. The histidine UVRR spectrum is complex and weak, but it is simplified upon protonation, and further simplified upon H/D exchange, which eliminates mixing of the N-H bending coordinate into the imidazole ring modes.59,60 A single prominent band remains, which, although weaker than other protein bands, is sufficiently isolated to permit measuring the number of protonated histidine residues57,58 (Figure S4).
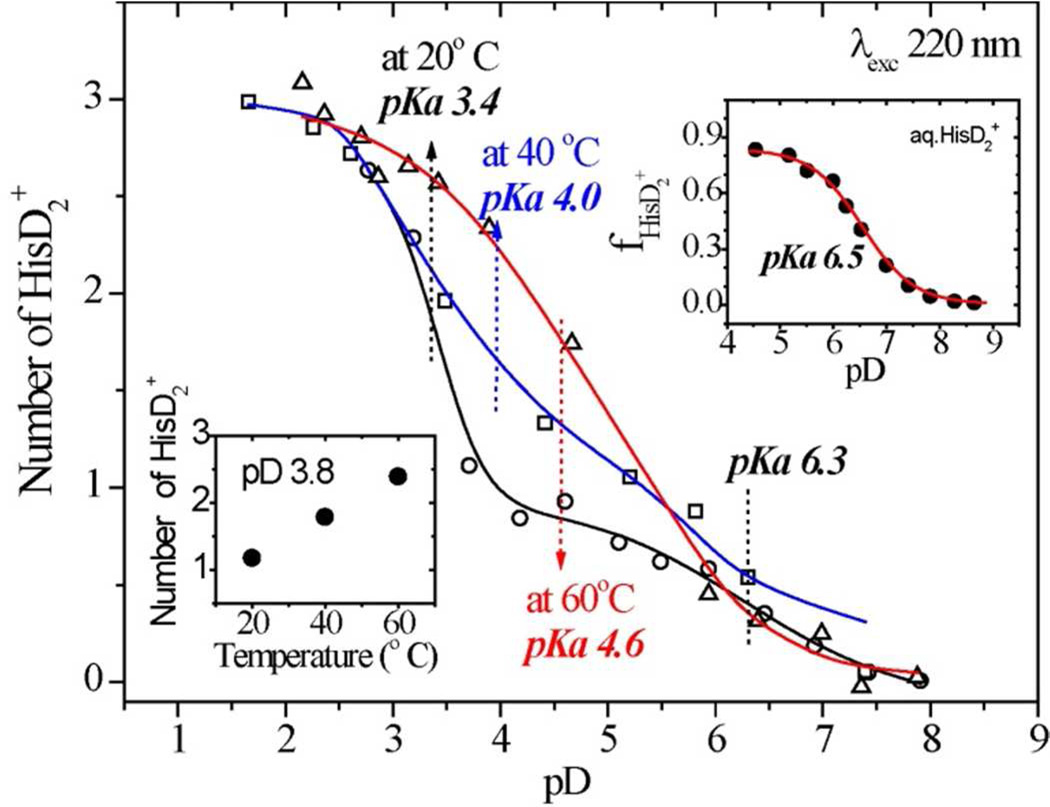
The pD values obtained after correction (pD=0.929×pH*+0.42, where pH* is the pH meter reading in D2O)61 shown in Figure 8 reveals a marked dependence of histidine protonation on protein unfolding. Cyt c contains three His residues, 18, 26 and 33. His18 is one of the heme ligands, His26 forms a H-bond with Pro44, and His33 is exposed to solvent. At 20°C, the HisD2+ titration curve reveals protonation of one His residue with an apparent pKa of 6.3, and two His residues with an apparent pKa of 3.4. The first pKa is close to that of aqueous HisD2+ (Fig. 8 inset), and is assigned62 to the exposed His33. The second pKa is assigned to both His18 and His26. This pKa is lowered to about the same extent from the free HisD2+ value by coordination to the heme Fe and by the H-bond from Pro44. Previous studies had indicated a pKa of <3.5 for His2662 and 2.5 for His18.63 The differences between the latter estimate and ours is likely not significant.
Figure 8.
pD titration curves for the 1408 cm−1 HisD2+ of hh cyt c at 20° C (circles), 40° C (squares) and 60° C (triangles). The insets show the aqueous histidine titration curve (upper) and the intensity-derived average protonation number as function of temperature(bottom). [The pH meter readings (pH*, in D2O) were converted to pD = 0.929×pH*+0.42 as described in ref. 61]
As the temperature is raised, the titration curve shifts markedly to higher pD values. The first His residue still titrates with an apparent pKa ~ 6.3, as expected for the exposed His33, but the pKa shifts steadily higher for the second and third residues at 40 and 60°C. At pH 3.8, the proton number rises from one at 20°C toward three at 60°C. This behavior is consistent with an increase in the His26 pKa as the H-bond from Pro44 is broken due to temperature-induced protein conformation change; the His18-Fe bond is also weakened in this process, resulting in His18 protonation at high temperatures.
3.6 T-jump/UVRR spectroscopy
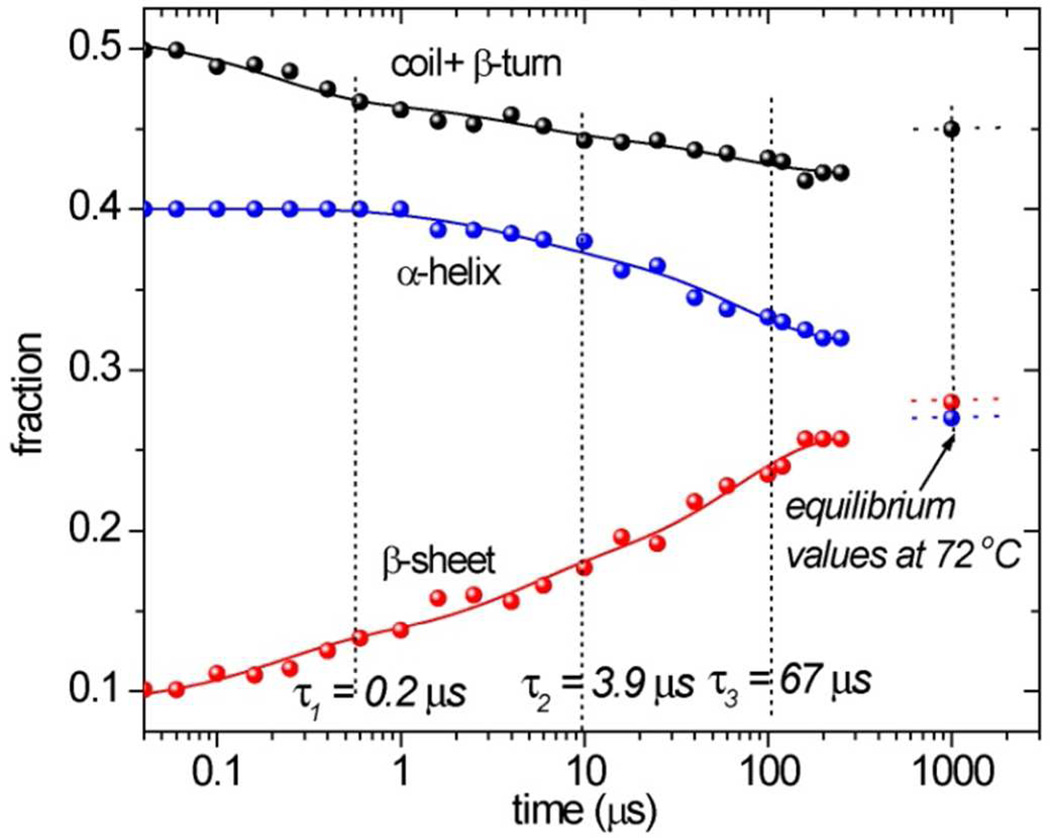
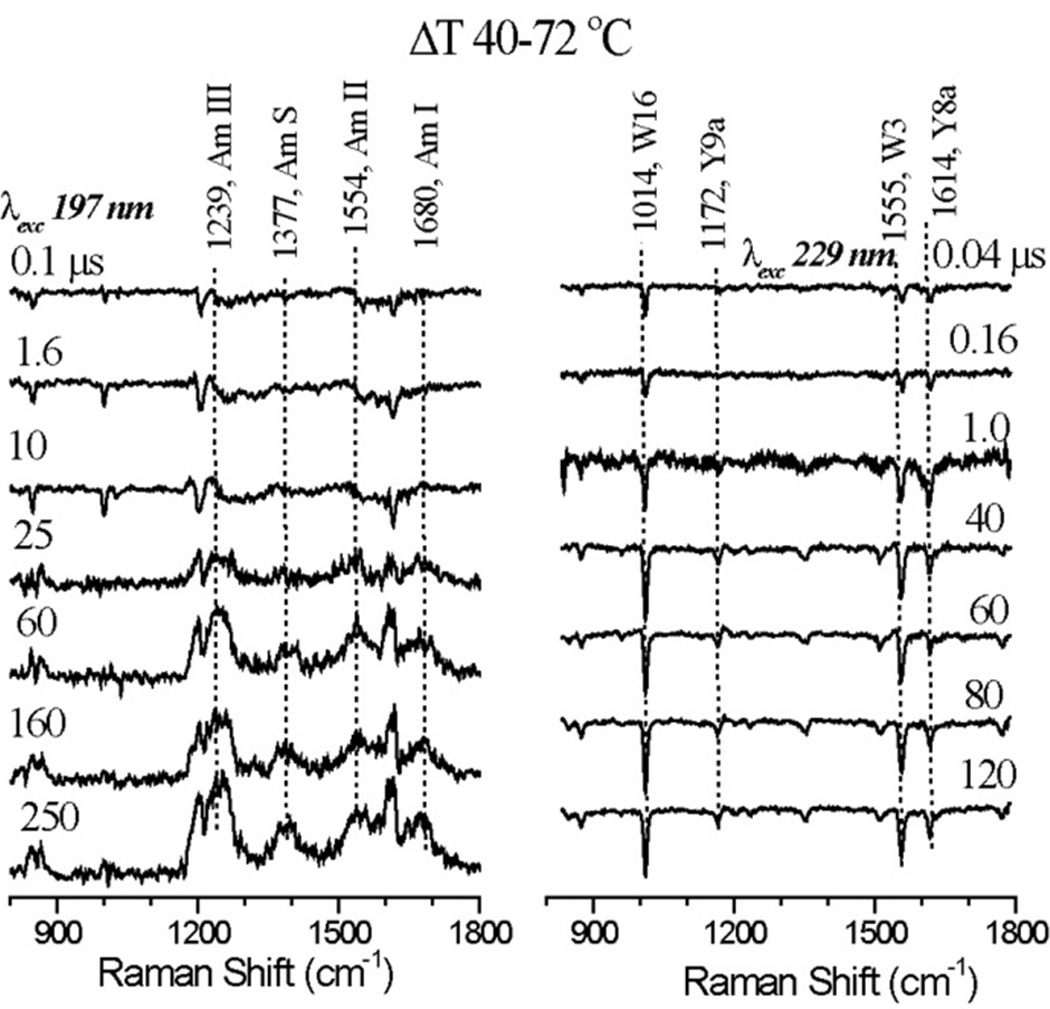
Firgures 9 display UVRR difference spectra induced by a 40° to 72°C temperature jump in cyt c, at 197 and 229 nm probe wavelengths. Figure 10 displays the time course of secondary structure change, obtained via the same analysis of the 197 nm UVRR spectra as was used to determine temperature profiles from the static spectra. However, because of a higher noise level induced by the T-jump, the procedure failed to reliably separate coil and β-turn contributions, which are therefore summed in Figure 10. Also indicated in Figure 10 axes are the secondary structure fractions obtained from the equilibrium spectrum at the final temperature 72°C. These values show that β-sheet formation is essentially completed in the 200 µs time window of the T-jump measurements, but some conversion of α-helix to unordered structure (~5%) continues after 200 µs
Figure 10.
Time evolution of UVRR-determined secondary structure content in hh cyt c at pH 3.0 following a 40–72° C T-jump. Solid curves are fits to successive exponentials, with the indicated time constants.
The growth of β-sheet structure, at the expense of the other secondary structures, occurs in a multi-stage fashion, over four decades in time. The dotted lines indicate best-fit time constants, 0.2, 3.9 and 67 µs, for three sequential first-order processes. About half of the β sheet amplitude is encompassed by the first two processes.
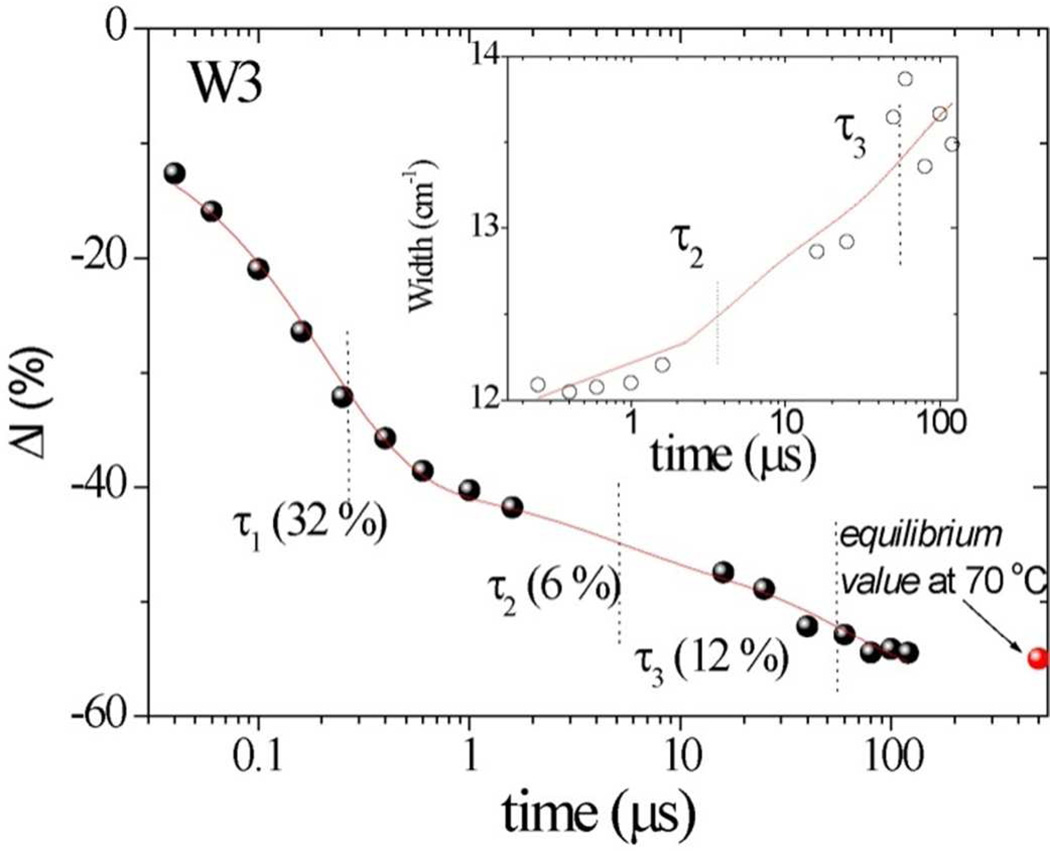
Similar multi-exponential evolution is seen for the TrpW3 band intensity and broadening, (Figure 11); at 200 µs the data approach the 72°C equilibrium values. The curves drawn through the points are computed with the time constants obtained from the secondary structure fits. The fastest process (0.2 µs) accounts for most of the W3 intensity loss, but produces little W3 broadening and only a small change in the secondary structure (Figure 11). We attribute this process to solvent penetration into the interior of the protein, exposing Trp59 to a hydrophillic environment. Similar sub-microsecond decays of aromatic residue intensity have been seen in other protein T-jump studies.64,65 The second (3.9 µs) and third (67 µs) processes account for most of the β-sheet formation, as well as of the W3 broadening, consistent with coupling of the protein conformation change and the Trp59 mobility.
Figure 11.
Time evolution of Trp W3 intensity in hh cyt c at pH 3.0 following a 40–72 ° C T-jump. Solid curve is obtained using the time constants obtained from 197 nm-excited UVRR spectra with the indicated time constants. Inset shows the time-dependent bandwidth change for W3 band.
4. DISCUSSION
4.1 His26 protonation triggers 40s Ω loop conversion to β-sheet
The β-sheet is an important structural element in many proteins, and has become a focus of interest because of its role in protein aggregation, including fibril formation in amyloid diseases.66,67 The interstrand H-bond motif of the β-sheet lends itself to inter-protein association. β-sheet peptides and proteins are therefore prone to aggregation.68,69
The present work reveals a novel instance of intra-protein β-sheet formation as a stage in protein unfolding. The UVRR analysis of secondary structure shows melting of the cyt c helices and turns to unordered structure at neutral pH, but at pH 3, heating produces β-sheet at the expense of turns and helices. This conformation change is reversible, provided the solution is dilute. As the cyt c concentration is raised above 150 µM, heating leads to protein aggregation, probably due to intermolecular association of the thermally induced β-sheet strands. The reversibility of the transition in dilute solution, where the protein molecules are isolated, implies that β-sheet interstrand H-bond requirements are satisfied intramolecularly. T-jump/UVRR spectra show this process to occur in microseconds, consistent with unfolding rates in other small proteins.70–77
The UVRR analysis of histidine protonation reveals that one of the three His residues in cyt c titrates with a normal pKa, 6.3. This pKa is assigned to His33,62 whose sidechain is exposed to solvent in the native cyt c structure. The remaining two His residues have greatly depressed apparent pKa’s, both around 3.4 at 20° C. One of these, His18, is ligated to the heme Fe, and the Fe-His bond must be broken to permit its protonation at pKa of 2.5.63 The remaining residue, His26, is H-bonded to the backbone carbonyl of Pro44. Evidently this interaction is strong enough to depress the His26 pKa by ~3 units.62 As the temperature is raised both of the depressed pKa’s are also elevated, reaching an average value of 4.6 at 60° C.
We infer that protonation of His26 disrupts its interaction with Pro44 and triggers the conformation change that induces β-sheet formation. Pro44 sits at the tip of a large Ω loop (40s Ω loop) containing residues 40–57 (Figure 1). Using H-exchange NMR measurements Englander and coworkers19 have identified this loop, along with another loop containing the Met80 ligand, as being the least stable ‘foldons’ of cyt c. The 40s Ω loop was found to be strongly destabilized at low pH, an effect attributed to protonation of His26, and possibly one of the heme propionates20. Our observed shift of ~3 units in the pKa of His26 supports this view, as does the finding by Sinibaldi et al.17 that mutation of the His26 significantly destabilizes the cyt c structure.
Disruption of the His26/Pro44 H-bond thus leaves the 40s Ω loop only marginally stable. Heating the solution then overcomes the remaining stabilization. Instead of melting to unordered structure, however, the 40s Ω loop converts to a β-sheet structure. In the native cyt c structure (Figure 1), the ‘neck’ of the 40s Ω loop consists of a short antiparallel β-sheet, consisting of residues 37–39 and 58–61. We envision that this β-sheet segment extends itself into the remainder of the now-labile 40s Ω loop, at the expense of the three β-turns and the short 50s helix contained in the native loop.
The 40s Ω loop, including its β-neck (residues 37–61) contains 20 % of the cyt c residues, whereas the extent of thermally induced β-sheet structure is ~30 % (Figure 4). Moreover the 50s helix (residues 49–55) contains only 5 % of the cyt c residues, while heating induces a loss of helix from 40 % to 25 %. The remaining 10 % loss could result from melting of the short 60s and 70s helices. These are immediately upstream of the β-neck, and we envision the β-sheet formation extending from the 40s Ω loop on through the 60s and 70s helices. It is notable that the Tm for helix melting, 60°C, is higher than the Tm for β-sheet formation, 53°C, whereas the β-turn Tm, 45°C is lower (Figure 4). These data indicate that the 40s Ω loop conformation change is initiated at 45°C, and that additional thermal energy is needed to extend the sheet from the 40s Ω loop into the 60s and 70s region. The 53°C Tm for β-sheet formation represents an average for these two processes.
4.2 Aromatic signals reveal heme displacement
Tm’s of 50 and 53°C are observed for frequency shifts in the Trp W3 and Tyr 9a UVRR bands (Figure 5), coincident with the β-sheet Tm. The W3 signal arises from the sole Trp residue, Trp59. Its shift and broadening indicates increased mobility of the indole sidechain.48,49 The Y9a band arises from the four Tyr residues; its upshift and narrowing implicate a more in-plane average orientation of the OH substituents, and a narrowing of its distribution.52 This effect is likely associated mainly with Tyr48, which has the most out-of-plane OH orientation in the native structure.52
Both Trp59 and Tyr48 are H-bonded to the same heme propionate substituent. Trp59 sits in the middle of the β-neck of the 40s Ω loop, while Tyr48 is in the loop, just downstream of the 50s helix (Figure 1). The same propionate also forms H-bonds with Asn52, in the 50s helix, and with a backbone amide adjacent to Gly41, further into the loop. As well, the residue adjacent to Tyr48, Thr49, is H-bonded to the second propionate substituent (Figure 1). This extensive set of H-bonds with residues in the 40s Ω loop, provide a mechanism for heme displacement when the loop converts to β-sheet, thereby altering the sidechain positions. The Y9a upshift indicates that the displacement forces the Tyr48 OH into a more in-plane orientation, while the W3 broadening indicates an increase in the Trp59 mobility, due to weakening of its H-bond.
The second propionate also form H-bonds with Lys79 and Thr78 (Figure 1), which are in the loop containing the Met80 ligand. Thus, the heme displacement resulting from the 40s Ω loop/β-sheet conversion provides a mechanism for breaking the Met80-Fe bond. The Tm for Met80-Fe bond breaking, 48°C, corresponds to the first stage of β-sheet formation.
We note as well the likely involvement of Tyr67, which H-bonds to Met80. Breaking the Met80-Fe bond would destabilize the 60’s helix, contributing to its melting, proposed to occur at a slightly higher temperature (Tm = 60°C).
4.3 Microsecond conformational rates
The T-jump/UVRR analysis reveals a rapid process, 0.2 µs, during which the W3 signal from Trp59 loses significant intensity (Figure 12); other Trp and Tyr bands lose intensity as well (Figure 9). This phase is attributed to thermally-driven solvent penetration, which exposes the aromatic sidechains to an aqueous environment.64,65
Figure 9.
Pump-probe UVRR difference spectra with 197 (left) and 229 (right) nm excitation, induced by the indicated T-jump, at the indicated delay intervals, for hh cyt c at pH 3.0.
During this early phase there is little change in secondary structure (Figure 11), whose subsequent alteration occurs with two time constants, 3.9 and 67 µs. The microsecond time scale is consistent with intramolecular unfolding processes that have been reported for small proteins.70–77 The early phases account for about half of the β-sheet formation, while the 67 µs phase accounts for the remainder. These amplitudes are roughly congruent with the two successive thermal equilibrium phases (Figure 4), which we assign to the 40s Ω loop and the 60s and 70s helices. The Trp59 W3 band broadens during these two phases (Figure 12), consistent with increased Trp59 mobility as a result of the protein conformation change.
4.4 Possible mechanism of cardiolipin-induced apoptosis
We suggest that the 40s Ω loop β-sheet conversion trigered by loss of the Pro44-His26 H-bond may also initiate apoptotic events associate with cardiolipin binding to cyt c. Jemmerson et al.21 found that cyt c in apoptotic cells, or in association with lipid vesicles, binds to an antibody that recognizes a region around the Pro44 residue in non-native forms of cyt c, indicating exposure of Pro 44. Moreover, cyt c bound to cardiolipin vesicles has been reported to contain substantial fractions of β-sheet, on the basis of UVCD measurements.78
Cardiolipin induces its own peroxidation via its interactions with cyt c, and the resulting hydroperoxides induce the release of pro-apoptotic factors. Peroxidase activity would result from His26-triggered β-sheet conversion, which displaces the heme, breaking the Met80-Fe bond, and allowing access of peroxide to the heme Fe. We previously showed that heating cyt c at pH 3 induces peroxidase activity with the same Tm as is observed for β-sheet formation.16
In its complex with cyt c, cardiolipin has been shown to insert its acyl chains into hydrophobic regions of the protein. Two hydrophobic sites have been suggested, one in the region of the Met80 loop,9 and the other a channel extending into the interior from Asn52, an invariant residue.10 Sinibadi et al.13 have proposed that both sites are occupied. Rytomaa et al.79 proposed that Asn52 forms an H-bond with the phosphate head group at the end of the acyl chain that is inserted in the Asn52 channel. Asn52 is on the 50s helix, which is part of the 40s Ω loop. We suggest that the geometry of the complex with cardiolipin brings Pro44, at the end of the 40s Ω loop, into contact with the membrane surface, and that its H-bond with His26 is broken because His26 H-bonds instead with a phosphate head group of the membrane. Alternatively His26 may actually become protonated by proximity to the surface, given the lowered pH at the surface of anionic membranes.80
His26-triggered β-sheet conversion, and the associated melting of the 60’s helix may also loosen the C-terminal helix; one of its residues, the invariant Arg91, which has a significant effect on cardiolipin-induced redox modulation,81,82 is H-bonded to Met65, on the 60’s helix. FRET measurements by Hanske et al.5 indicate that cardiolipin induces unfolding of the C-terminal helix. Groves and coworkers have proposed insertion of this helix into the lipid membrane to explain their important discovery of cardiolipin-promoted membrane pore formation by cyt c,83 permitting its own penetration through the membrane. This finding can explain the phenomenon of cyt c migration from the mitochondrion to the cytosol, where it binds and activates the apoptosis protease activation factor, APAf-1.8,14,15 The conformation change induced by breaking the Pro44-His26 H-bond, by destabilizing the 60’s helix, may lower the barrier for unmooring the C-terminal helix, allowing it to penetrate the lipid membrane.
5. CONCLUSIONS
At pH 3, cyt c undergoes a reversible unfolding process, which differs from the thermal unfolding seen at neutral pH. UVRR analysis shows that instead of a simple loss of secondary structure, there is a conformation change to β-sheet. This change occurs in two phases, involving first the loss of β-turns, and then the loss of helix structure. T-jump studies indicate that these phases occur with 3.9 and 67 µs time constants, consistent with intramolecular unfolding dynamics in small proteins. UVRR analysis also shows reorientation of the Trp59 indole ring on this time scale, following a faster, solvent penetration phase.
UVRR/pH titrations reveal a depressed pKa for His26, reflecting its H-bond interaction with Pro44. Loss of this interaction labilizes the 40s Ω loop, one of two low-energy foldons in cyt c. It is proposed that in the first phase of thermally induced β-sheet formation a short native β-sheet segment at the 40s Ω loop neck is extended into the rest of the loop, while in the second phase the sheet is further extended into the adjacent 60s and 70s helices. The conformation change alters the orientation of the Trp59 and Tyr48 sidechains, which are H-bonded to a heme propionate substituent. This reorientation is suggested to reflect displacement of the heme as a result of the 40s Ω loop conformation change. Heme displacement accounts for the observed loss of the Met80-Fe bond, and the induction of peroxidase activity. Tm’s for these events coincide with that of β-sheet formation and of Trp59 and Tyr48 reorientation.
These observations point to a mechanism for the initiation of apoptosis by cardiolipin binding to cyt c, inasmuch as the binding event appears to labilize the 40s Ω loop, and induces β-sheet formation as well as peroxidase activity. Insertion of cardiolipin acyl chains into cyt c, is suggested to disrupt the Pro44/His26 H-bond, through interaction of His26 with phosphate head groups in the lipid bilayer.
Supplementary Material
ACKNOWLEDGEMENT
This work was supported by NIH grant GM 25158 (to T.G.S). We thank Dr. Yu-San Huang and Dr. Oyeyemi F. Oyerinde for helping with the pKa measurements of histidine and Dr. Swarnalatha Venkateshrao for assistance with the UVRR experiments at neutral pH.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Temperature dependent UVRR spectra of cyt c at pH 3, excited at 197 and 229 nm; illustration of the subtraction procedure used to enhance the amide bands at 197 nm excitation; and the spectrum of cyt c in D2O excited at 220 nm. This material is available free of charge via the internet at http://acs.pub.org.
REFERENCES
- 1.Kluck RM, BossyWetzel E, Green DR, Newmeyer DD. Science. 1997;275:1132–1136. doi: 10.1126/science.275.5303.1132. [DOI] [PubMed] [Google Scholar]
- 2.Cai JY, Yang J, Jones DP. BBA-Bioenergetics. 1998;1366:139–149. doi: 10.1016/s0005-2728(98)00109-1. [DOI] [PubMed] [Google Scholar]
- 3.Jiang XJ, Wang XD. Annu. Rev. Biochem. 2004;73:87–106. doi: 10.1146/annurev.biochem.73.011303.073706. [DOI] [PubMed] [Google Scholar]
- 4.Huttemann M, Pecina P, Rainbolt M, Sanderson TH, Kagan VE, Samavati L, Doan JW, Lee I. Mitochondrion. 2011;11:369–381. doi: 10.1016/j.mito.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanske J, Toffey JR, Morenz AM, Bonilla AJ, Schiavoni KH, Pletneva EV. Proc. Natl. Acad. Sci. USA. 2012;109:125–130. doi: 10.1073/pnas.1112312108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kulikov AV, Shilov ES, Mufazalov IA, Gogvadze V, Nedospasov SA, Zhivotovsky B. Cell. Mol. Life Sci. 2012;69:1787–1797. doi: 10.1007/s00018-011-0895-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patriarca A, Polticelli F, Piro MC, Sinibaldi F, Mei G, Bari M, Santucci R, Fiorucci L. Arch. Biochem. Biophys. 2012;522:62–69. doi: 10.1016/j.abb.2012.03.028. [DOI] [PubMed] [Google Scholar]
- 8.Hu YM, Benedict MA, Ding LY, Nunez G. EMBO J. 1999;18:3586–3595. doi: 10.1093/emboj/18.13.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rytomaa M, Kinnunen PKJ. J. Biol. Chem. 1994;269:1770–1774. [PubMed] [Google Scholar]
- 10.Tuominen EKJ, Wallace CJA, Kinnunen PKJ. J. Biol. Chem. 2002;277:8822–8826. doi: 10.1074/jbc.M200056200. [DOI] [PubMed] [Google Scholar]
- 11.Ascenzi P, Ciaccio C, Sinibaldi F, Santucci R, Coletta M. Biochem. Biophys. Res. Commun. 2011;404:190–194. doi: 10.1016/j.bbrc.2010.11.091. [DOI] [PubMed] [Google Scholar]
- 12.Belikova NA, Vladimirov YA, Osipov AN, Kapralov AA, Tyurin VA, Potapovich MV, Basova LV, Peterson J, Kurnikov IV, Kagan VE. Biochemistry. 2006;45:4998–5009. doi: 10.1021/bi0525573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sinibaldi F, Howes BD, Piro MC, Polticelli F, Bombelli C, Ferri T, Coletta M, Smulevich G, Santucci R. J. Biol. Inorg. Chem. 2010;15:689–700. doi: 10.1007/s00775-010-0636-z. [DOI] [PubMed] [Google Scholar]
- 14.Saleh A, Srinivasula SM, Acharya S, Fishel R, Alnemri ES. J. Biol. Chem. 1999;274:17941–17945. doi: 10.1074/jbc.274.25.17941. [DOI] [PubMed] [Google Scholar]
- 15.Zou H, Henzel WJ, Liu XS, Lutschg A, Wang XD. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- 16.Balakrishnan G, Hu Y, Oyerinde OF, Su J, Groves JT, Spiro TG. J. Am. Chem. Soc. 2007;129:504–505. doi: 10.1021/ja0678727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sinibaldi F, Piro MC, Howes BD, Smulevich G, Ascoli F, Santucci R. Biochemistry. 2003;42:7604–7610. doi: 10.1021/bi034132r. [DOI] [PubMed] [Google Scholar]
- 18.Fetrow JS, Dreher U, Wiland DJ, Schaak DL, Boose TL. Protein Sci. 1998;7:994–1005. doi: 10.1002/pro.5560070417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krishna MMG, Lin Y, Rumbley JN, Englander SW. J. Mol. Biol. 2003;331:29–36. doi: 10.1016/s0022-2836(03)00697-1. [DOI] [PubMed] [Google Scholar]
- 20.Krishna MMG, Maity H, Rumbley JN, Lin Y, Englander SW. J. Mol. Biol. 2006;359:1410–1419. doi: 10.1016/j.jmb.2006.04.035. [DOI] [PubMed] [Google Scholar]
- 21.Jemmerson R, Liu J, Hausauer D, Lam KP, Mondino A, Nelson RD. Biochemistry. 1999;38:3599–3609. doi: 10.1021/bi9809268. [DOI] [PubMed] [Google Scholar]
- 22.Zhao XJ, Tengroth C, Chen RP, Simpson WR, Spiro TG. J. Raman Spectrosc. 1999;30:773–776. [Google Scholar]
- 23.Balakrishnan G, Hu Y, Nielsen SB, Spiro TG. Appl. Spectrosc. 2005;59:776–781. doi: 10.1366/0003702054280702. [DOI] [PubMed] [Google Scholar]
- 24.Zhao XJ, Chen RP, Tengroth C, Spiro TG. Appl. Spectrosc. 1999;53:1200–1205. [Google Scholar]
- 25.Balakrishnan G, Hu Y, Spiro TG. Appl. Spectrosc. 2006;60:347–351. doi: 10.1366/000370206776593799. [DOI] [PubMed] [Google Scholar]
- 26.Anfinrud PA, Han C, Hochstrasser RM. Proc. Natl. Acad. Sci. USA. 1989;86:8387–8391. doi: 10.1073/pnas.86.21.8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alakoskela JM, Jutila A, Simonsen AC, Pirneskoski J, Pyhajoki S, Turunen R, Marttila S, Mouritsen OG, Goormaghtigh E, Kinnunen PKJ. Biochemistry. 2006;45:13447–13453. doi: 10.1021/bi0611107. [DOI] [PubMed] [Google Scholar]
- 28.de Groot NS, Ventura S. Spectrosc.-Iint. J. 2005;19:199–205. [Google Scholar]
- 29.Speare JO, Rush TS. Biopolymers. 2003;72:193–204. doi: 10.1002/bip.10337. [DOI] [PubMed] [Google Scholar]
- 30.Paquet MJ, Laviolette M, Pezolet M, Auger M. Biophys. J. 2001;81:305–312. doi: 10.1016/S0006-3495(01)75700-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pertinhez TA, Bouchard ML, Tomlinson EJ, Wain R, Ferguson SJ, Dobson CM, Smith LJ. FEBS Lett. 2001;495:184–186. doi: 10.1016/s0014-5793(01)02384-5. [DOI] [PubMed] [Google Scholar]
- 32.Sreerama N, Woody RW. Anal. Biochem. 1993;209:32–44. doi: 10.1006/abio.1993.1079. [DOI] [PubMed] [Google Scholar]
- 33.Chen YH, Yang JT, Chau KH. Biochemistry. 1974;13:3350–3359. doi: 10.1021/bi00713a027. [DOI] [PubMed] [Google Scholar]
- 34.Vanstokkum IHM, Spoelder HJW, Bloemendal M, Vangrondelle R, Groen FCA. Anal. Biochem. 1990;191:110–118. doi: 10.1016/0003-2697(90)90396-q. [DOI] [PubMed] [Google Scholar]
- 35.Johnson WC. Proteins. 1999;35:307–312. [PubMed] [Google Scholar]
- 36.Sreerama N, Venyaminov SY, Woody RW. Anal. Biochem. 2000;287:243–251. doi: 10.1006/abio.2000.4879. [DOI] [PubMed] [Google Scholar]
- 37.Sreerama N, Woody RW. Anal. Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 38.Sreerama N, Woody RW. Numerical Computer Methods, Pt D. 2004;Vol. 383:318–351. doi: 10.1016/S0076-6879(04)83013-1. [DOI] [PubMed] [Google Scholar]
- 39.Huang CY, Balakrishnan G, Spiro TG. J. Raman Spectrosc. 2006;37:277–282. [Google Scholar]
- 40.Chi ZH, Chen XG, Holtz JSW, Asher SA. Biochemistry. 1998;37:2854–2864. doi: 10.1021/bi971160z. [DOI] [PubMed] [Google Scholar]
- 41.Williams RW. Methods Enzymol. 1986;130:311–331. doi: 10.1016/0076-6879(86)30016-8. [DOI] [PubMed] [Google Scholar]
- 42.Oladepo SA, Xiong K, Hong ZM, Asher SA, Handen J, Lednev IK. Chem. Rev. 2012;112:2604–2628. doi: 10.1021/cr200198a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Balakrishnan G, Weeks CL, Ibrahim M, Soldatova AV, Spiro TG. Curr. Opin. Struc. Biol. 2008;18:623–629. doi: 10.1016/j.sbi.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bushnell GW, Louie GV, Brayer GD. J. Mol. Biol. 1990;214:585–595. doi: 10.1016/0022-2836(90)90200-6. [DOI] [PubMed] [Google Scholar]
- 45.Asher SA. Annu. Rev. Phys. Chem. 1988;39:537–588. doi: 10.1146/annurev.pc.39.100188.002541. [DOI] [PubMed] [Google Scholar]
- 46.Kitagawa T. Prog. Biophys. Mol. Biol. 1992;58:1–18. doi: 10.1016/0079-6107(92)90009-u. [DOI] [PubMed] [Google Scholar]
- 47.Austin JC, Rodgers KR, Spiro TG. Metallobiochemistry, Part C. 1993;226:374–396. doi: 10.1016/0076-6879(93)26017-4. [DOI] [PubMed] [Google Scholar]
- 48.Takeuchi H. Biopolymers. 2003;72:305–317. doi: 10.1002/bip.10440. [DOI] [PubMed] [Google Scholar]
- 49.Miura T, Takeuchi H, Harada I. J. Raman Spectrosc. 1989;20:667–671. [Google Scholar]
- 50.Juszczak LJ, Desamero RZB. Biochemistry. 2009;48:2777–2787. doi: 10.1021/bi801293v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takeuchi H, Watanabe N, Satoh Y, Harada I. J. Raman Spectrosc. 1989;20:233–237. [Google Scholar]
- 52.Jordan T, Eads JC, Spiro TG. Protein Sci. 1995;4:716–728. doi: 10.1002/pro.5560040411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shechter E, Saludjia P. Biopolymers. 1967;5:788-&. doi: 10.1002/bip.1967.360050812. [DOI] [PubMed] [Google Scholar]
- 54.Schejter A, George P. Biochemistry. 1964;3:1045-&. doi: 10.1021/bi00896a006. [DOI] [PubMed] [Google Scholar]
- 55.Greenwoo C, Wilson MT. Eur. J. Biochem. 1971;22:5-&. doi: 10.1111/j.1432-1033.1971.tb01507.x. [DOI] [PubMed] [Google Scholar]
- 56.Makinen MW, Churg AK. In: Iron porphyrins. Lever ABP, Gray HB, editors. Vol. Part 1. Reading, Massachusetts: Addison-Wesley; 1983. pp. 141–325. [Google Scholar]
- 57.Wu Q, Li FB, Wang WX, Hecht MH, Spiro TG. J. Inorg. Biochem. 2002;88:381–387. doi: 10.1016/s0162-0134(01)00354-3. [DOI] [PubMed] [Google Scholar]
- 58.Zhao XJ, Wang DJ, Spiro TG. J. Am. Chem. Soc. 1998;120:8517–8518. [Google Scholar]
- 59.Balakrishnan G, Jarzecki AA, Wu Q, Kozlowski PM, Wang DJ, Spiro TG. J. Phys. Chem. B. 2012;116:9387–9395. doi: 10.1021/jp305083t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Markham LM, Mayne LC, Hudson BS, Zgierski MZ. J. Phys. Chem. 1993;97:10319–10325. [Google Scholar]
- 61.Krezel A, Bal W. J. Inorg. Biochem. 2004;98:161–166. doi: 10.1016/j.jinorgbio.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 62.Cohen JS, Hayes MB. J. Biol. Chem. 1974;249:5472–5477. [PubMed] [Google Scholar]
- 63.Babul J, Stellwag E. Biochemistry. 1972;11:1195–1200. doi: 10.1021/bi00757a013. [DOI] [PubMed] [Google Scholar]
- 64.Huang CY, Balakrishnan G, Spiro TG. Biochemistry. 2005;44:15734–15742. doi: 10.1021/bi051578u. [DOI] [PubMed] [Google Scholar]
- 65.Gilmanshin R, Gulotta M, Dyer RB, Callender RH. Biochemistry. 2001;40:5127–5136. doi: 10.1021/bi002255v. [DOI] [PubMed] [Google Scholar]
- 66.Chiti F, Dobson CM. Annu. Rev. Biochem. 2006;Vol. 75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 67.Fink AL. Fold. Des. 1998;3:R9–R23. doi: 10.1016/S1359-0278(98)00002-9. [DOI] [PubMed] [Google Scholar]
- 68.Sunde M, Blake CCF. Quarterly Reviews of Biophysics. 1998;31:1-+. doi: 10.1017/s0033583598003400. [DOI] [PubMed] [Google Scholar]
- 69.Ross CA, Poirier MA. Nature Medicine. 2004;10:S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 70.Blanco FJ, Rivas G, Serrano L. Nat. Struc. Biol. 1994;1:584–590. doi: 10.1038/nsb0994-584. [DOI] [PubMed] [Google Scholar]
- 71.Munoz V, Thompson PA, Hofrichter J, Eaton WA. Nature. 1997;390:196–199. doi: 10.1038/36626. [DOI] [PubMed] [Google Scholar]
- 72.Cochran AG, Skelton NJ, Starovasnik MA. Proc. Natl. Acad. Sci. USA. 2001;98:5578–5583. doi: 10.1073/pnas.091100898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu Y, Oyola R, Gai F. J. Am. Chem. Soc. 2003;125:15388–15394. doi: 10.1021/ja037053b. [DOI] [PubMed] [Google Scholar]
- 74.Dyer RB, Maness SJ, Peterson ES, Franzen S, Fesinmeyer RM, Andersen NH. Biochemistry. 2004;43:11560–11566. doi: 10.1021/bi049177m. [DOI] [PubMed] [Google Scholar]
- 75.Snow CD, Qiu LL, Du DG, Gai F, Hagen SJ, Pande VS. Proc. Natl. Acad. Sci. USA. 2004;101:4077–4082. doi: 10.1073/pnas.0305260101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chung HS, Khalil M, Smith AW, Ganim Z, Tokmakoff A. Proc. Natl. Acad. Sci. USA. 2005;102:612–617. doi: 10.1073/pnas.0408646102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Krejtschi C, Huang R, Keiderling TA, Hauser K. Vib. Spectrosc. 2008;48:1–7. [Google Scholar]
- 78.Su J. Princeton University; 2008. [Google Scholar]
- 79.Rytomaa M, Kinnunen PKJ. J. Biol. Chem. 1995;270:3197–3202. doi: 10.1074/jbc.270.7.3197. [DOI] [PubMed] [Google Scholar]
- 80.Prats M, Teissie J, Tocanne JF. Nature. 1986;322:756–758. [Google Scholar]
- 81.Rajagopal BS, Silkstone GG, Nicholls P, Wilson MT, Worrall JAR. BBA-Bioenergetics. 2012;1817:780–791. doi: 10.1016/j.bbabio.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 82.Bradley JM, Silkstone G, Wilson MT, Cheesman MR, Butt JN. J. Am. Chem. Soc. 2011;133:19676–19679. doi: 10.1021/ja209144h. [DOI] [PubMed] [Google Scholar]
- 83.Bergstrom CL, Beales PA, Lv Y, Kyle Vanderlick T, Groves JT. Proc. Natl. Acad. Sci. USA. 2012 doi: 10.1073/pnas.1303819110. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.