Abstract
The mammalian brain poses a formidable challenge to the study and treatment of neuropsychiatric diseases – owing to the complex interaction of genetic, epigenetic, and circuit-level mechanisms underlying pathogenesis. Technologies that facilitate functional dissection of distinct brain circuits are necessary for systematic identification of disease origin and therapy. Recent developments in the optogenetics technology have begun to address this challenge by enabling precise perturbation of distinct cell types based on molecular signatures, functional projections, and intracellular biochemical signaling pathways. With high temporal precision and reversible neuromodulation, optogenetics promises to improve existing disease models and advance our understanding of psychiatric conditions. In this review, we will describe the current state of molecular optogenetic tools and future directions of development.
Keywords: optogenetics, channelrhodopsin, neuropsychiatric disease, neuromodulation, gene therapy, brain machine interface
The brain is a particularly complicated organ to study due to its sheer diversity and heterogeneity of molecules, cells, and connections. Dysfunctions in specific cell types and circuits can lead to severe neuropsychiatric conditions [1]. To dissect the mechanisms of psychiatric conditions, it is essential to investigate the pathogenic mechanisms at every level of brain function: from subcellular molecular signaling and cellular physiology to circuit-level functions. Based on clinical observations and insights to neuropsychiatric diseases, it is possible to generate cellular and animal models that recapitulate the multilevel symptoms of human patients [2]. Systematic perturbation of the implicated circuit components in these disease models will help identify effective therapeutic targets and strategies. However, precise manipulation of the brain has been a difficult task to achieve. To fully recapitulate biological events, an ideal approach requires highly precise manipulations, including fast temporal control, cell-type specificity, and applicability within awake, behaving animals.
The development of optogenetic technologies enables such precise manipulations [3-6]. It combines the temporal and spatial precision of light pulses with cellular specificity of genetic targeting; by coupling cellular behavior with light, it provides a fast light-controlled approach to manipulate neural activities in intact brain circuits. The general strategy of optogenetics involves introducing a light-sensitive protein to a specific cell type, illuminating the targeted cells with defined parameters, and obtaining reliable readout of the cellular behavior. Compared to electrical microstimulation, optogenetic activation specifically targets the opsin-expressing neurons, whereas electrical stimulation may simultaneously affect diverse local, afferent, and passing axonal fibers [7, 8] in addition to cellular somata near the electrode insertion site. Currently, the molecular toolbox of optogenetics [4, 6] involves many different light-sensitive constructs capable of controlling a diverse range of neuronal functions, from electrical to biochemical signaling. Through a combination of molecular engineering and genomic discoveries of new light-sensitive proteins, the toolkit of neuron modulators is rapidly improving in specificity and versatility. At its present stage, optogenetics affords millisecond precision in controlling neuron activity, genetically defined cell type targeting, and the ability to modulate neural circuit function in awake-behaving animals. With these features, optogenetics can be used to causally probe circuitries underlying complex behavior, dissect signaling pathways and construct models of psychiatric disease through loss- and gain-of-function experiments. The present review will describe the optogenetic tools available for investigating neural circuit function and future directions in the development of this technology.
Modulating Neural Activity Using Light
The heterogeneous nature of brain tissue presents major challenges for selectively controlling subsets of well-defined neuron types in intact circuits. Most current neuromodulation techniques (i.e. electrical stimulation or pharmacological intervention) either simultaneously affect surrounding cells and processes, in addition to the target population, or have slow kinetics and reversibility. The lack of modulation specificity frequently limits the strength of conclusions drawn from conventional neuromodulation experiments. To overcome the spatial and temporal limitations of electrical, pharmacological and genetic neuromodulation approaches, various microbial and engineered opsins (Fig. 1a) have been developed recently to control electrical and biochemical activities of neurons with cell-type selectivity, high temporal precision, and rapid reversibility. Since most neurons in the brain are not naturally light-sensitive, selective expression of the opsin genes in targeted neural populations makes it possible to specifically control their activity. Additionally, the fast on-off kinetics of microbial opsins make it possible to evoke or inhibit neural activities within milliseconds, on a time scale relevant to physiological brain functions.
Figure 1.
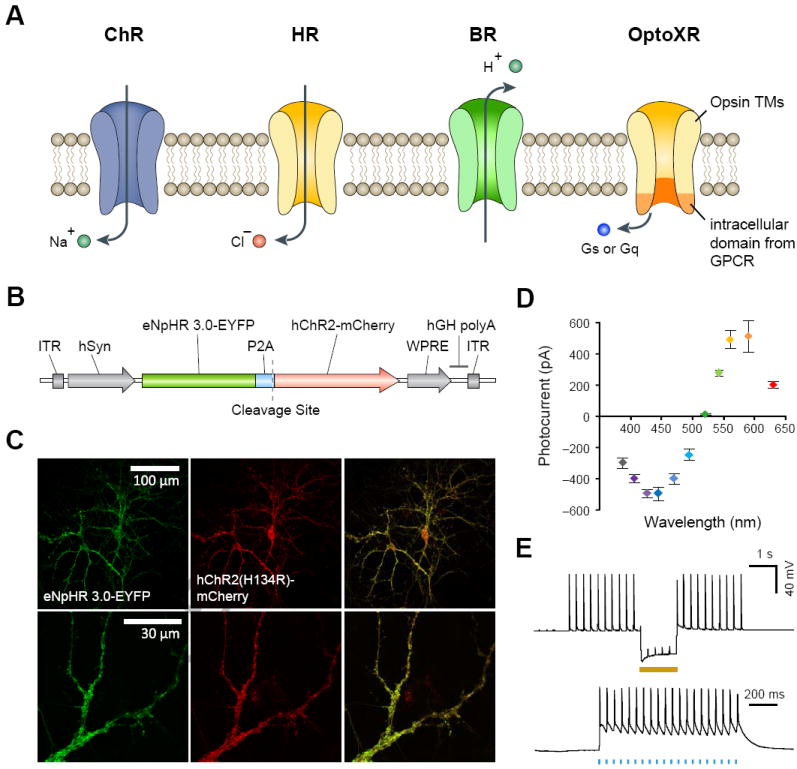
Microbial opsins as optogenetic tools. a. A variety of natural and engineered opsins can be used to control the membrane potential and intracellular biochemical signaling of neurons. Channelrhodopsins (ChRs) are light-activatable cation-conducting channels capable of activating neurons by depolarizing the membrane potential. Halorhodopsins (HRs) and bacteriorhodopsins (BRs) are light-activatable chloride and proton pumps capable of inhibiting neurons by hyperpolarizing the membrane potential. OptoXRs are engineered rhodopsin-GPCR chimeras useful for controlling intracellular G-protein coupled signaling cascades. OptoXRs are engineered by replacing the intracellular domain of vertebrate rhodopsin with the intracellular loops from GPCRs. b. A bi-cistronic vector used to express channelrhodopsin-2 (ChR2) and halorhodopsin (eNpHR3.0) in the same neuron. The two opsin genes are linked using a 2A self-cleavage linker. c. Images of neurons co-expressing ChR2 and eNpHR3.0. d. Activation spectra of neurons co-expressing ChR2 and eNpHR3.0. e. Bi-directional control of neurons co-expressing ChR2 and eNpHR3.0. Blue light (470nm) flashes induced action potential firing and yellow light (583nm) facilitated spike inhibition. Panel a is adapted with permission from ref [4] and panels b-e are adapted with permission from ref [22].
Activating Neurons Using Light
The discovery and introduction of algal light-sensitive cation conducting channelrhodopsins into neurons has enabled fast optical control of genetically defined neuronal populations in intact circuits, both in vitro and in vivo [9]. So far, channelrhodopsin genes from two algal species (Chlamydmonas reinhardtii and Volvox carteri) have been cloned and characterized in neurons (Fig. 1a). Most notably, channelrhodopsin-2 from Chlamydomonas reinhardtii (ChR2) [10-12] and channelrhodopsin-1 from Volvox carteri (VChR1) [13] have proven to be a powerful pair of ChRs for controlling intact neural circuits using light. ChR2 is maximally activated by blue light around 480 nm whereas VChR1 remains significantly light sensitive even at 589 nm, a wavelength at which ChR2 is no longer responsive. Therefore ChR2 and VChR1 can be integrated in combinatorial neural control experiments both in vitro and in vivo to test the necessity and sufficiency of a variety of neural circuit functions. Both ChR2 and VChR1 have fast onset kinetics with time constants on the order of milliseconds, and are therefore able to transduce high frequencies of millisecond-lasting light flashes into reliably evoked action potentials. However, because wildtype VChR1 has lower levels of photocurrent compared to ChR2, a chimera between Chlamydamonas ChR1 and Volvox ChR1 has been generated called C1V1. In conjunction with ChR2, C1V1 forms a powerful pair of opsins to simultaneously control the activity of distinct groups of neurons in vivo [14].
Mutagenesis studies could expand the function of channelrhodopsins by altering the spectral properties, conductance, or kinetics of the channels. Indeed, one mutant ChR2(H134R) has been shown to exhibit enhanced photocurrent[15], and three point-mutants of humanized ChR2 convert a brief pulse of light into a period of stable current influx that can last for many minutes[16]. These latter mutant channels were generated by substitution of the C128 position in the original ChR2 sequence. All three mutants are activated by blue light (470nm). Photocurrents generated by the opening of ChR2(C128A) and ChR2(C128S) can be effectively terminated by a pulse of green light (542nm). Other slow mutants were generated by mutating residue D156, which have been shown to result in similar stabilization of the conducting state [14, 17].
Substituting ChR2 residue E123 with T or A was found to accelerate channel closure kinetics and significantly increase the fidelity of fast optogenetic control [18]. These E123 mutations can be combined with other gain-of-function modifications such as the H134R and T159C mutations [18, 19], or membrane trafficking signals [14, 20-22]. The E123 mutations appear unique thus far as they eliminate the sensitivity of channel kinetics to membrane potential[19]. Opsins of this class (E123 mutations alone or in combination with other modifications [18] are termed ChETAs (ChR E123T/A). ChETA tools have been shown to deliver improved performance (up to 200Hz) within intact mammalian brain tissue[18]. A caveat is that faster deactivation is accompanied by reduced light sensitivity for long pulses of light.
Inhibiting Neurons Using Light
A number of light-activated anion pumps have been explored for neural activity silencing. In particular, the chloride-pumping halorhodopsin from Natronomonas pharaonis (NpHR)[23] has been shown to hyperpolarize neurons upon illumination with green or yellow light (Fig. 1a). Also, NpHR and ChR2 have sufficient spectral separation and can be simultaneously expressed in the same neurons to facilitate bidirectional control of neural activity, to test both the necessity and sufficiency of a circuit component in neural function (Fig. 1b-e). eNpHR3.0, an enhanced version of NpHR, was used along with bilateral optical fiber devices to inhibit the cholinergic neurons of the nucleus accumbens and elucidate a causal role for these rare cells in implementing cocaine conditioning in freely moving mice, which appears to operate via enhancing inhibition of inhibitory striatal medium spiny neurons [24].
Ecological prospecting has led to the discovery of additional neural silencers to eNpHR3.0, such as archaerhodopsin-3 (Arch), a green light (~550nm) activated proton pump isolated from the archaebacteria Halorubrum sodomense [25, 26]. Arch is well expressed in neurons and can drive large inhibitory currents. Other prominent channels for silencing neurons include bacteriorhopsin (BR) and Guillardia theta rhodopsin-3 (GtR3). While the peak absorbance of BR is ~560 nm[27], GtR3 is maximally excited by ~470 nm light[22]. The diversity in their spectral properties renders them useful in selective silencing of neurons using multiple wavelengths.
Neuron Activity Sensors
Optical neural activity actuators can be combined with genetically encoded neural activity reporters to greatly enhance our ability to probe complex circuit-level mechanisms underlying neuropsychiatric diseases. By genetically targeting activity actuators and reporters into specific cell types, one can selectively perturb a specific cell population and simultaneously observe its effect on downstream circuit components. Two major classes of genetically encoded activity reporters have been developed to detect changes in intracellular calcium or membrane potential.
Calcium Sensors
Due to the involvement of voltage-gated calcium channels in propagating action potentials, much effort has been spent in developing fast and accurate calcium sensors to report neural activity. Traditional synthetic calcium dyes cannot be targeted to specific cell types and exhibit fast photobleaching. To overcome these limitations, genetically encoded calcium indicators (GECI) have been developed [28]. One class of GECI, called GCaMP, consists of a circularly permutated GFP that fluoresces in response to calcium influx[29, 30]. Modifications of the original GCaMP led to new variants, the best of which is GCaMP3, which contains amino acid mutations T116V, M66K, and N363D[28].
Voltage Sensors
Another route to measure neural activity is by monitoring the membrane potential changes in neurons. Genetically encoded voltage-sensitive fluorescent proteins (VSFPs) have been constructed by fusing fluorescent reporters with voltage-sensing domains. Upon voltage fluctuations, the voltage-sensing domain undergoes conformational change, thereby shifting the spectral response of the reporter protein[31].
More recently, Kralj et al showed that Archaerhodopsin 3 (Arch) isolated from Halorubrum sodomense can also be used as a genetically encoded voltage sensor[32]. After virally delivering Arch to rat hippocampal neurons cultures, they demonstrated that Arch exhibits high sensitivity and speed, both of which exceed that of existing VSFPs by a factor of ten. These properties allow Arch to detect single action potentials in vitro, which is a major step forward in achieving precise optical detection of neural circuit dynamics [32].
Modulation of Biochemical Signaling
Optogenetic control of well-defined biochemical signaling events can be achieved by constructing chimeras [33] between vertebrate rhodopsin and conventional ligand-gated GPCRs [34, 35]. Many of the membrane receptors involved in neurotransmission and neuromodulation, such as dopaminergic, serotonergic, and adrenergic receptors belong to the GPCR family. When used as optogenetic tools these vertebrate opsin chimera are referred to as optoXRs (Fig. 1a), which allow for optically controlled intracellular signaling with temporal resolution suitable for modulating behavior in freely behaving animals [34]. OptoXRs provide speed and cellular precision that are not achievable with pharmacological and genetic tools.
Targeting Optogenetic Modulators to Specific Neural Circuits
Neuropsychiatric diseases affect complex neural circuits. To precisely pinpoint the circuit-level mechanism of pathogenesis, we must be able to target brain tissue with cellular- and circuit-level specificity. Opsins can be selectively introduced into subsets of neurons based on their molecular signature, projection pattern, anatomical organization, and functional activity.
Targeting Specific Cell Types Using Molecular Signatures
The cellular substrate of pathology for many neuropsychiatric diseases tends to involve molecularly defined cell types. For example, parvalbumin, dopaminergic, and orexinergic neurons are affected in schizophrenia, Parkinson’s disease, and narcolepsy respectively. Many cell types can be targeted to express microbial opsins using the promoters of their signature genes – such as parvalbumin [36], tyrosine hydroxylase[37], and hypocretin [38] for the cell types described above. Depending on the genomic structure, some promoters are compact enough to be packaged into viral vectors so that specific cell types can be used to target across a wide range of animal models. Cell types whose promoters are too large to be packaged into viral vectors can be targeted using transgenic technologies, although this practice is mostly limited to mice and rats.
Viral Gene Delivery
Viral vectors are versatile tools for delivering genetic constructs into neurons. Both lentivirus and adeno-associated virus (AAV) vectors have been used to deliver optogenetic constructs into genetically defined cell populations. By fusing microbial opsin genes to cell-type specific promoter one can obtain restricted gene expression in the cell type of interest. For in vivo applications stereotaxic injection can be used to deliver concentrated viral vectors into target brain regions. The advantages of using lentiviral and AAV vectors include stable long-term expression and high transgene levels [39]. While lentiviral vectors are permanently integrated into the genome of target cells, AAV vectors are mostly maintained extra-chromosomally. One major constraint of the viral delivery method is the limited packaging capacity: AAV can accommodate transgene constructs up to 5 kb while lentiviral vectors are limited to 10 kb for the complete packaged genome. Due to the package length constraint, AAV and lentiviruses can only be used to deliver relatively small promoters (~2kb for AAV and ~5kb for lentiviral vectors; microbial opsin gene plus a fluorescent tag can take up to 2kb of sequence length). This exclusion of long promoters limits the expression specificity, as some cell type-specific promoters have much longer regulatory elements. Another caveat is that the spatial precision of stereotaxic injection is limited, especially when targeting small brain nuclei, and multiple injections are required to achieve efficient expression in large regions of brain tissue [4].
Transgenic Technology
For promoters that exceed the packaging limit of viral vectors, transgenic animal lines can be generated to express optogenetic constructs in a cell-type specific fashion. Transgenic constructs can be introduced into transgenic mice either through bacterial artificial chromosomes (BAC) or using short transgene cassettes containing recombinant cell-type specific promoters [40]. Several transgenic mouse lines expressing channelrhodopsin and/or halorhodopsin under the pan-neuronal Thy-1 promoter have been developed [41, 42]. Compared to viral delivery, one disadvantage is the amount of time needed to generate and breed transgenic animal lines. In addition, due to the untargeted nature of the transgene insertion, each transgenic animal line needs to be carefully characterized to avoid undesirable gene interruptions.
Conditional Expression
A major challenge with generating transgenic animals constitutively expressing microbial opsins is that some cell-type specific promoters do not drive strong expression of the downstream gene. Since many of the opsins have low conductance or pumping properties[6], high levels of opsin expression are required for achieving reliable neural activation or silencing. To amplify the expression level, inducible AAV vectors carrying double floxed microbial opsin genes (DIO AAV vectors, [36, 37]) can be delivered into the brain of transgenic animals expressing Cre recombinase under the control of cell type-specific promoters. Cre recombinase will recognize the double floxed opsin construct and enable transgene expression after recombination [36, 37]. The DIO AAV system allows the gene of interest to be expressed using strong ubiquitous promoters, while deriving cell type-specificity from Cre recombinase expression. The availability of numerous constitutive and inducible Cre transgenic mouse (and rats in the near future) lines from the Allen Brain Institute for Brain Science, Jackson Laboratory, GENSAT, and other transgenic animal repositories provides a convenient solution for targeting a wide variety of cell types in the brain.
Circuit-Specific Targeting
Even cell populations identified via a common molecular signature can represent a heterogenous mixture of cell types. For example, the tyrosine hydroxylase-positive dopaminergic neurons in the midbrain project to multiple downstream brain regions, including the striatum, nucleus accumbens, prefrontal cortex, habenula, and many other areas. While dopaminergic neurons projecting to the dorsal striatum play a critical role in motor function, dopaminergic neurons projecting to the nucleus accumbens are involved in reward conditioning[37, 43]. The fact that a single molecularly defined cell population can project to multiple downstream brain regions and affect diverse brain processes warrants the need to improve cell type-specific targeting based on their wiring. Circuit-specific targeting also holds the potential for uncovering the therapeutic mechanisms of deep brain stimulation (DBS), which has demonstrated therapeutic efficacy for neuropsychiatric diseases such as Parkinson’s disease [44] and major depressive disorder[45, 46]. However, as implanted electrodes non-selectively modulate all the cells as well as processes in the vicinity of implantation site, optogenetic stimulation can be used to identify the specific circuit components involved in the symptomatic rescue and facilitate more efficacious therapies.
Another approach to target specific neural projections can be achieved using trans-synaptic viral vectors or proteins. A number of trans-synaptic proteins and viral vectors with unique anterograde- or retrograde-transporting properties [47-49] may be engineered with recombinases to activate gene expression in subpopulations of neurons with cell type- and circuit-specificity. For example, fusion proteins containing Cre and either wheat germ agglutinin (WGA) or tetanus toxin fragment C (TTC) can be expressed in the cell bodies of one brain region and allow the recombinase to be trans-synpatically delivered to the postsynaptic or presynaptic neurons in another brain region[22]. Similarly, retrograde- and anterograde-transporting viral vectors [50, 51] such as rabies virus (RV), herpes simplex virus 1 (HSV-1), and vesicular stomatitis virus (VSV) can also be used to deliver recombinases or transgene cassettes in a projection specific fashion. When combined with conditional expression systems, either Cre-dependent transgenic mice or viral vectors, these projection-specific strategies will allow circuit-specific gene expression in a variety of animal models.
Future Developments
Optogenetics provides a basis for linking behavioral functions with cellular and circuit-level activity patterns. Already, systematic optogenetic perturbation of distinct cell types and brain projections has begun to shed light on the cellular substrates of neurological and psychiatric disorders including Parkinson’s disease, anxiety, retinal degeneration, cocaine conditioning, social dysfunction, and depression [14, 24, 52-56]. Tantalizing possibilities for the future of optogenetics lie in several directions, including: the extension of optogenetic tools with distinct wavelengths of sensitivity and novel modes of control beyond electrical signaling; the emergence of gene expression technologies capable of targeting optogenetic tools to functionally defined cell populations and circuit elements; and the continued intersection of optogenetics with a variety of readout modalities and especially with high-throughput system-wide electrical and genome-wide molecular profiling technologies.
The optogenetics toolbox of the future will enable multiplexed control of several populations of neurons within the same experimental setup and will enable the combinatorial testing of signal processing roles of distinct neural circuit elements in complex behavior. Genome prospecting and molecular engineering of opsin genes from diverse microbial species have already expanded the optogenetic toolbox with a diverse palette of neuronal activators and inhibitors with distinct wavelengths of excitation, ion selectivity, and kinetics [18, 26, 57-59]. The search continues for proteins sensitive to far-red wavelengths which will not only enable probing of deeper brain regions, but will also facilitate the simultaneous investigation of multiple neural circuits when used in conjunction with other opsins. Recent successes in obtaining an atomic resolution structure of channelrhodopsin will catalyze opsin engineering efforts [60, 61]. In addition, proteins responsive to wavelengths from the ultrasound or magnetic frequency ranges will possibly enable the remote control of neural tissue without the necessity of any surgical implants. New opsins with selective conductance for potassium and calcium, ions important in synaptic events and intraneuronal signaling pathways, will result in precise dissection of the role of specific signaling processes in neural function.
Optogenetic control will also expand beyond electrical signaling by enabling precise modulation of endogenous transcription within specific neural populations. The development of light sensing protein domains from plant and microbial species [62-65] will enable the design of photo-activatable enzymes and transcription factors. These light-sensing domains can be coupled with programmable DNA binding domains, including designer zinc finger proteins (ZFPs) [66] and transcription activator-like effectors (TALEs) [67-70], to achieve optically-controlled targeted modulation of endogenous transgene and RNA expression in mammalian genomes. The ability to regulate transcription within specific circuits in awake behaving animals will be important for dissecting the molecular and epigenetic basis of neuropsychiatric diseases.
In order to fully understand the circuit-level mechanisms underlying neuropsychiatric disease, it is important to pinpoint the specific circuits underlying behavioral dysfunctions. The targeting of optogenetic tools into specific cells based on their activity during behavioral testing allows the identification of functional circuits. This can be achieved using molecular markers of neural activity [71] including immediate early genes (IEG) such as c-Fos, Zif268, Arc, and Npas4. Leveraging the activity-dependent transcription regulatory elements of IEGs [72, 73], it is possible to target the expression of opsin genes to distinct groups of neurons that are active during behavioral testing. For example, in an animal model of posttraumatic stress disorder (PTSD), activity-dependent targeting of opsins will enable the identification of cells involved in the phenotype. Subsequent activation of the targeted cells in the absence of trauma-inducing stimuli can be used to determine the necessity or sufficiency of specific cell populations in the manifestation of the PTSD phenotype. The epigenetic profile of these cells can be further analyzed to identify molecular signatures of PTSD.
A third major avenue for the future will be to integrate the use of optogenetics with other system-level technologies to facilitate global analysis of brain functions. A combination of optogenetics and fMRI [74] or large-scale multi-electrode arrays [75] will allow us to identify target brain regions and activity patterns corresponding to specific behavioral functions. Subsequent genome-wide transcriptome [75] and epigenome [76] analysis of the identified circuits will bridge molecular signatures with circuit dynamics and behavior. The integration of optogenetics with a diverse range of readout modalities will further bridge our understanding of nervous system functions and disease processes from molecules and cells to circuits and behavior.
As the optogenetics technology matures, it will continue to augment our ability to dissect the mammalian brain with increasing precision and expanding modes of control. The ability to specifically modulate the electrical, biochemical, and transcriptional activity of specific neuronal circuits will bring us one step closer to understanding the mechanism of normal and pathological nervous system function. The intersection of optogenetics with other disciplines of biology and engineering will lead to novel therapeutic targets and innovative therapeutic interventions for neuropsychiatric diseases.
Acknowledgments
Y.M. is supported by NIGMS training grant, T32 GM007484. F.Z. is supported by a NIH Transformative R01, the McKnight, Simons, Gates, Damon-Runyon Foundations, Robert Metcalfe, and Michael Boylan. Additional sequence information and plasmid maps can be found at the Optogenetics Resources Website (http://www.optogenetics.org).
Footnotes
Competing Financial Interests
The authors report no biomedical financial interest or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ressler KJ, Mayberg HS. Targeting abnormal neural circuits in mood and anxiety disorders: from the laboratory to the clinic. Nature neuroscience. 2007;10(9):1116–24. doi: 10.1038/nn1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nestler EJ, Hyman SE. Animal models of neuropsychiatric disorders. Nature neuroscience. 2010;13(10):1161–9. doi: 10.1038/nn.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deisseroth K. Optogenetics. Nature methods. 2011;8(1):26–9. doi: 10.1038/nmeth.f.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang F, et al. Optogenetic interrogation of neural circuits: technology for probing mammalian brain structures. Nature protocols. 2010;5(3):439–56. doi: 10.1038/nprot.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang F, et al. Circuit-breakers: optical technologies for probing neural signals and systems. Nature reviews Neuroscience. 2007;8(8):577–81. doi: 10.1038/nrn2192. [DOI] [PubMed] [Google Scholar]
- 6.Yizhar O, et al. Optogenetics in neural systems. Neuron. 2011;71(1):9–34. doi: 10.1016/j.neuron.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Llewellyn ME, et al. Orderly recruitment of motor units under optical control in vivo. Nature medicine. 2010;16(10):1161–5. doi: 10.1038/nm.2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Histed MH, Bonin V, Reid RC. Direct activation of sparse, distributed populations of cortical neurons by electrical microstimulation. Neuron. 2009;63(4):508–22. doi: 10.1016/j.neuron.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang F, et al. The microbial opsin family of optogenetic tools. Cell. 2011;147(7):1446–57. doi: 10.1016/j.cell.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagel G, et al. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci U S A. 2003;100(24):13940–5. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyden ES, et al. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8(9):1263–8. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 12.Zhang F, et al. Channelrhodopsin-2 and optical control of excitable cells. Nat Methods. 2006;3(10):785–92. doi: 10.1038/nmeth936. [DOI] [PubMed] [Google Scholar]
- 13.Zhang F, et al. Red-shifted optogenetic excitation: a tool for fast neural control derived from Volvox carteri. Nat Neurosci. 2008;11(6):631–3. doi: 10.1038/nn.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yizhar O, et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 2011;477(7363):171–8. doi: 10.1038/nature10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagel G, et al. Light activation of channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Current biology : CB. 2005;15(24):2279–84. doi: 10.1016/j.cub.2005.11.032. [DOI] [PubMed] [Google Scholar]
- 16.Berndt A, et al. Bi-stable neural state switches. Nat Neurosci. 2009;12(2):229–34. doi: 10.1038/nn.2247. [DOI] [PubMed] [Google Scholar]
- 17.Bamann C, et al. Structural guidance of the photocycle of channelrhodopsin-2 by an interhelical hydrogen bond. Biochemistry. 2009;49(2):267–278. doi: 10.1021/bi901634p. [DOI] [PubMed] [Google Scholar]
- 18.Gunaydin LA, et al. Ultrafast optogenetic control. Nature neuroscience. 2010;13(3):387–92. doi: 10.1038/nn.2495. [DOI] [PubMed] [Google Scholar]
- 19.Berndt A, et al. High-efficiency channelrhodopsins for fast neuronal stimulation at low light levels. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(18):7595–600. doi: 10.1073/pnas.1017210108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao S, et al. Improved expression of halorhodopsin for light-induced silencing of neuronal activity. Brain cell biology. 2008;36(1-4):141–54. doi: 10.1007/s11068-008-9034-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gradinaru V, Thompson KR, Deisseroth K. eNpHR: a Natronomonas halorhodopsin enhanced for optogenetic applications. Brain cell biology. 2008;36(1-4):129–39. doi: 10.1007/s11068-008-9027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gradinaru V, et al. Molecular and cellular approaches for diversifying and extending optogenetics. Cell. 2010;141(1):154–65. doi: 10.1016/j.cell.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang F, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446(7136):633–9. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- 24.Witten IB, et al. Cholinergic interneurons control local circuit activity and cocaine conditioning. Science. 2010;330(6011):1677–81. doi: 10.1126/science.1193771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Idnurm A, Howlett BJ. Characterization of an opsin gene from the ascomycete Leptosphaeria maculans. Genome / National Research Council Canada = Genome / Conseil national de recherches Canada. 2001;44(2):167–71. doi: 10.1139/g00-113. [DOI] [PubMed] [Google Scholar]
- 26.Chow BY, et al. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature. 2010;463(7277):98–102. doi: 10.1038/nature08652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park DG, et al. Effect of high pressure on the light-induced structural change of bacteriorhodopsin reconstituted in liposome. Biochimica et Biophysica Acta (BBA)-Bioenergetics. 1989;973(1):19–22. [Google Scholar]
- 28.Tian L, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nature methods. 2009;6(12):875–81. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akerboom J, et al. Crystal structures of the GCaMP calcium sensor reveal the mechanism of fluorescence signal change and aid rational design. Journal of Biological Chemistry. 2009;284(10):6455. doi: 10.1074/jbc.M807657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagai T, et al. Circularly permuted green fluorescent proteins engineered to sense Ca2+ Proceedings of the National Academy of Sciences. 2001;98(6):3197. doi: 10.1073/pnas.051636098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akemann W, et al. Imaging brain electric signals with genetically targeted voltage-sensitive fluorescent proteins. Nature methods. 7(8):643–649. doi: 10.1038/nmeth.1479. [DOI] [PubMed] [Google Scholar]
- 32.Kralj JM, et al. Optical recording of action potentials in mammalian neurons using a microbial rhodopsin. Nature methods. 9(1):90–95. doi: 10.1038/nmeth.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim JM, et al. Light-driven activation of beta 2-adrenergic receptor signaling by a chimeric rhodopsin containing the beta 2-adrenergic receptor cytoplasmic loops. Biochemistry. 2005;44(7):2284–92. doi: 10.1021/bi048328i. [DOI] [PubMed] [Google Scholar]
- 34.Airan RD, et al. Temporally precise in vivo control of intracellular signalling. Nature. 2009;458(7241):1025–9. doi: 10.1038/nature07926. [DOI] [PubMed] [Google Scholar]
- 35.Oh E, et al. Substitution of 5-HT1A receptor signaling by a light-activated G protein-coupled receptor. The Journal of biological chemistry. 2010;285(40):30825–36. doi: 10.1074/jbc.M110.147298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sohal VS, et al. Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature. 2009;459(7247):698–702. doi: 10.1038/nature07991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsai HC, et al. Phasic firing in dopaminergic neurons is sufficient for behavioral conditioning. Science. 2009;324(5930):1080–4. doi: 10.1126/science.1168878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adamantidis AR, et al. Neural substrates of awakening probed with optogenetic control of hypocretin neurons. Nature. 2007;450(7168):420–4. doi: 10.1038/nature06310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davidson BL, Breakefield XO. Viral vectors for gene delivery to the nervous system. Nature reviews Neuroscience. 2003;4(5):353–64. doi: 10.1038/nrn1104. [DOI] [PubMed] [Google Scholar]
- 40.Gong S, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425(6961):917–25. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 41.Arenkiel BR, et al. In vivo light-induced activation of neural circuitry in transgenic mice expressing channelrhodopsin-2. Neuron. 2007;54(2):205–18. doi: 10.1016/j.neuron.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao S, et al. Cell type-specific channelrhodopsin-2 transgenic mice for optogenetic dissection of neural circuitry function. Nature methods. 2011;8(9):745–52. doi: 10.1038/nmeth.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adamantidis AR, et al. Optogenetic interrogation of dopaminergic modulation of the multiple phases of reward-seeking behavior. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31(30):10829–35. doi: 10.1523/JNEUROSCI.2246-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Limousin P, et al. Effect of parkinsonian signs and symptoms of bilateral subthalamic nucleus stimulation. Lancet. 1995;345(8942):91–5. doi: 10.1016/s0140-6736(95)90062-4. [DOI] [PubMed] [Google Scholar]
- 45.Mayberg HS, et al. Deep brain stimulation for treatment-resistant depression. Neuron. 2005;45(5):651–60. doi: 10.1016/j.neuron.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 46.Schlaepfer TE, et al. Deep brain stimulation to reward circuitry alleviates anhedonia in refractory major depression. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2008;33(2):368–77. doi: 10.1038/sj.npp.1301408. [DOI] [PubMed] [Google Scholar]
- 47.Wickersham IR, et al. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007;53(5):639–47. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maskos U, et al. Retrograde trans-synaptic transfer of green fluorescent protein allows the genetic mapping of neuronal circuits in transgenic mice. Proc Natl Acad Sci U S A. 2002;99(15):10120–5. doi: 10.1073/pnas.152266799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sugita M, Shiba Y. Genetic tracing shows segregation of taste neuronal circuitries for bitter and sweet. Science. 2005;309(5735):781–5. doi: 10.1126/science.1110787. [DOI] [PubMed] [Google Scholar]
- 50.Callaway EM. Transneuronal circuit tracing with neurotropic viruses. Curr Opin Neurobiol. 2008;18(6):617–23. doi: 10.1016/j.conb.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beier KT, et al. Anterograde or retrograde transsynaptic labeling of CNS neurons with vesicular stomatitis virus vectors. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(37):15414–9. doi: 10.1073/pnas.1110854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Busskamp V, et al. Genetic reactivation of cone photoreceptors restores visual responses in retinitis pigmentosa. Science. 2010;329(5990):413–7. doi: 10.1126/science.1190897. [DOI] [PubMed] [Google Scholar]
- 53.Covington HE, 3rd, et al. Antidepressant effect of optogenetic stimulation of the medial prefrontal cortex. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30(48):16082–90. doi: 10.1523/JNEUROSCI.1731-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gradinaru V, et al. Optical deconstruction of parkinsonian neural circuitry. Science. 2009;324(5925):354–9. doi: 10.1126/science.1167093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kravitz AV, et al. Regulation of parkinsonian motor behaviours by optogenetic control of basal ganglia circuitry. Nature. 2010;466(7306):622–6. doi: 10.1038/nature09159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tye KM, et al. Amygdala circuitry mediating reversible and bidirectional control of anxiety. Nature. 2011;471(7338):358–62. doi: 10.1038/nature09820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang F, et al. Red-shifted optogenetic excitation: a tool for fast neural control derived from Volvox carteri. Nature neuroscience. 2008;11(6):631–3. doi: 10.1038/nn.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Berndt A, et al. Bi-stable neural state switches. Nature neuroscience. 2009;12(2):229–34. doi: 10.1038/nn.2247. [DOI] [PubMed] [Google Scholar]
- 59.Kleinlogel S, et al. Ultra light-sensitive and fast neuronal activation with the Ca(2)+-permeable channelrhodopsin CatCh. Nature neuroscience. 2011;14(4):513–8. doi: 10.1038/nn.2776. [DOI] [PubMed] [Google Scholar]
- 60.Watanabe HC, et al. Structural model of channelrhodopsin. Journal of Biological Chemistry. doi: 10.1074/jbc.M111.320309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kato HE, et al. Crystal structure of the channelrhodopsin light-gated cation channel. Nature. doi: 10.1038/nature10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Levskaya A, et al. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461(7266):997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shimizu-Sato S, et al. A light-switchable gene promoter system. Nature biotechnology. 2002;20(10):1041–4. doi: 10.1038/nbt734. [DOI] [PubMed] [Google Scholar]
- 64.Yazawa M, et al. Induction of protein-protein interactions in live cells using light. Nature biotechnology. 2009;27(10):941–5. doi: 10.1038/nbt.1569. [DOI] [PubMed] [Google Scholar]
- 65.Kennedy MJ, et al. Rapid blue-light-mediated induction of protein interactions in living cells. Nature methods. 2010;7(12):973–5. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sander JD, et al. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA) Nature methods. 2011;8(1):67–9. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang F, et al. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nature biotechnology. 2011;29(2):149–53. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boch J, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326(5959):1509–12. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 69.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326(5959):1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 70.Scholze H, Boch J. TAL effectors are remote controls for gene activation. Current opinion in microbiology. 2011;14(1):47–53. doi: 10.1016/j.mib.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 71.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465(7295):182–7. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reijmers LG, et al. Localization of a stable neural correlate of associative memory. Science. 2007;317(5842):1230–3. doi: 10.1126/science.1143839. [DOI] [PubMed] [Google Scholar]
- 73.Barth AL, Gerkin RC, Dean KL. Alteration of neuronal firing properties after in vivo experience in a FosGFP transgenic mouse. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2004;24(29):6466–75. doi: 10.1523/JNEUROSCI.4737-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee JH, et al. Global and local fMRI signals driven by neurons defined optogenetically by type and wiring. Nature. 2010;465(7299):788–92. doi: 10.1038/nature09108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang J, et al. Integrated device for optical stimulation and spatiotemporal electrical recording of neural activity in light-sensitized brain tissue. Journal of neural engineering. 2009;6(5):055007. doi: 10.1088/1741-2560/6/5/055007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guo JU, et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nature neuroscience. 2011;14(10):1345–51. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]


