Abstract
The transcription factor B-lymphocyte-induced maturation protein-1 (Blimp-1) plays important roles in embryonic development and immunity. Blimp-1 is required for the differentiation of plasma cells, and mice with T-cell-specific deletion of Blimp-1 (Blimp-1 CKO mice) develop a fatal inflammatory response in the colon. Previous work demonstrated that lack of Blimp-1 in CD4+ and CD8+ T cells leads to intrinsic functional defects, but little is known about the functional role of Blimp-1 in regulating differentiation of T helper (h) cells in vivo and their contribution to the chronic intestinal inflammation observed in the Blimp1CKO mice.
Here we show that Blimp-1 is required to restrain the production of the inflammatory cytokine IL17 by Th cells in vivo. Blimp-1CKO mice have greater numbers of IL17-producing TCRβ+CD4+cells in lymphoid organs and in the intestinal mucosa. The increase in IL17-producing cells was not restored to normal levels in wild type and Blimp-1CKO-mixed bone marrow chimeric mice, suggesting an intrinsic role for Blimp-1 in constraining the production of IL17 in vivo. The observation that Blimp-1-deficient CD4+ T cells are more prone to differentiate into IL17+/IFNγ+ cells and cause severe colitis when transferred to Rag1-deficient mice provides further evidence that Blimp-1 represses IL17 production. Analysis of Blimp-1 expression at the single cell level during Th differentiation reveals that Blimp-1 expression is induced in Th1 and Th2 but repressed by TGFβ in Th17 cells. Collectively, the results described here establish a new role for Blimp-1 in regulating IL17 production in vivo.
Introduction
IL17 is an inflammatory cytokine produced by T helper (h) 17 cells (1), a Th subset that develops independently (2, 3) of the transcription factors T-bet and GATA-3, which are required for the development of Th1 and Th2 cells respectively (4). Th17 cells are associated with protection against bacterial and fungal infections encountered at mucosal surfaces (5) but also with several auto-inflammatory disorders, including rheumatoid arthritis, psoriasis, multiple sclerosis, corticosteroid-resistant asthma and inflammatory bowel disease (6, 7).
Differentiation of Th17 cells is induced by the cytokines TGFβ, IL6 and IL1β (8–10), and it is dependent upon the expression of the transcription factors RORγt and RORα (11, 12), which induce transcription of the Il17a gene (11). ROR-γt acts in cooperation with RORα and other transcription factors, including STAT3, IRF-4, BATF and Runx1, to induce full commitment of precursors to the Th17 subset (13–15). Activation of ROR-γt also promotes expression of the receptor for interleukin-23 (11), which is thought to maintain the expansion and pathogenesis of mature Th17 cells (16).
Although Th17 cells are clearly a distinct Th subset, recent data demonstrate that similar to other Th subpopulations, Th17 cells are considerably plastic and can acquire features and carry on effector functions characteristics of other Th subsets (17, 18). Th17 cells can express the regulatory T cell (Treg) specifying factor Foxp3 under certain conditions, and in the context of infectious and autoimmune inflammation, Th17 cells can also produce IFNγ and IL17 simultaneously (17, 19). IL17/IFNγ double-positive (IL17+/IFNγ+) T cells are found in elevated numbers in inflamed tissues of both humans and mice (20–22), and similar populations are observed in Th17 cells differentiated in vitro from human and murine naive CD4+ T cells (21–23). Recent studies indicate that IL23 signaling is important for the differentiation of IFNγ+/IL17+ CD4+ T cells both in vivo and in vitro (23, 24), but the transcriptional mechanisms underlying the differentiation and/or conversion of these cells from conventional (IFNγ negative) Th17 cells are poorly understood.
Blimp-1 (also called PRDI-BF1 in humans) is a transcription factor (encoded by the PRDM-1 gene) that plays crucial roles in regulating B and T lymphocyte function. Blimp-1 is expressed in both Treg and conventional T cells (25–27). Previous studies show that in Foxp3+ Treg cells Blimp-1 interacts with IRF-4 to induce IL-10 production (26). However, little is known about Blimp-1’s role in non Treg cell function and how lack of Blimp-1 contributes to the development of chronic mucosal inflammation. Despite their documented deficiency in producing IL-10, Blimp-1-deficient Tegs can suppress T cell-mediated colitis (25, 28) suggesting that defects in other T cell subsets may underlie the chronic intestinal inflammation developed in the Blimp-1CKO mice. The studies described herein address this possibility and identify a new, Treg-independent role for Blimp-1 in controlling intestinal mucosal homeostasis by limiting the numbers of IL17-producing Th cells in vivo.
Materials and Methods
Mice
C57BL/6 Prdm1flox/floxCD4-Cre+ (Blimp-1 CKO) and Prdm1+/+ CD4-Cre (Control) and mice were generated as previously described (25, 29). Previously described (30) C57BL/6 Prdm1flox/floxCD19-Cre+ mice were kindly donated by Dr. M. McHeyzer-Williams (Department of Immunology and Microbial Science, The Scripps Research Institute, La Jolla, California, USA). Mice bearing a BAC transgene encoding yellow fluorescent protein (YFP) under the control of Blimp-1 regulatory elements (Blimp-1 YFP reporter mice) (31) were also described previously (32). In these mice, YFP expression closely recapitulates Blimp-1 mRNA expression(32). Foxp3-IRES-GFP knock-in) (33) (Foxp3 reporter) mice were donated by Vijay Kuchroo (Brigham and Women’s Hospital Harvard Medical School) and crossed to C57BL/6 Prdm1flox/floxCD4-Cre+ mice to generate Ctrl and Blimp-1CKO Foxp3-reporter mice. All mice were maintained in a specific pathogen-free animal facility at the Cedars-Sinai Medical Center (CSMC) and handled in accordance with the institutional guidelines.
Large intestines (LI) lamina propria lymphocyte isolation
Large intestines (cecum, colon and rectum) were removed, opened longitudinally, cleaned and cut into strips 1 cm in length. Tissues were washed in ice-cold PBS and subjected to enzymatic digestion as previously described (34). Lamina propria mononuclear cells were purified on a 45%/72% Percoll gradient by centrifugation for 20 min at 25 °C and 600g. with no brake.
Antibodies and flow cytometry
Antibodies used for cell surface staining were Alexa-700 -conjugated TCRβ, Pacific Blue-conjugated anti-CD4, Phycoerythrin-conjugated anti-CD25, Allophycocyanin (APC)-conjugated anti-CD44 (all from Biolegend), PE-conjugated anti- IL17A, Alexa-647-conjugated anti-IL17F, PE-conjugated anti- IL4 and APC-conjugated IFNγ-(eBioscience). Cell surface staining was performed as previously described (25). Samples were analyzed on a LSRII analyzer (BD, San Jose, CA).
Determination of cytokine production
Total cell suspensions obtained from the LI-LP, spleen (SP), and mesenteric lymph nodes (mLN) were stimulated with plate-bound anti-CD3 (5 μg/ml) plus anti-CD28 (0.5 μg/ml) for 24 hours with addition of Brefeldin-A (BFA, 1mg/ml) for the last 6 hours. The production of IL17 and IFNγ was determined by ELISA on supernatants collected at 18 hours (before BFA addition) using eBioscience ELISA kits, following the manufacturer’s instructions. Where indicated, IL17, IL4 and IFNγ were also detected by intracellular staining, as previously described (29). Cells were analyzed on a LSRII analyzer. Cytokine staining was evaluated in live, CD4+ TCRβ+gated lymphocytes.
Quantitative real-time PCR (qRT-PCR)
Total mRNA was isolated using RNAeasy kits (Quiagen) according to the manufacturer’s instructions. Reverse transcription was performed on equal amounts of RNA (as determined by Nanodrop measurements) for each sample using Superscript III (Invitrogen). SYBR Green incorporation quantitative real-time PCR was performed using a FastStart SYBR Green Master mix (Roche) in the Realplex2 Mastercycler ep gradient S (Eppendorf). Primers used are described on Supplemental Table I.
Mixed Bone marrow chimera
Recipient mice (CD45.1+ wild-type) were lethally irradiated (10 Gy) for depletion of hematopoietic cells. Donor Ctrl (CD45.1+) or Blimp-1CKO (CD45.2+) Lin bone marrow cells, enriched for hematopoietic progenitors by magnetic bead depletion (StemCell Technologies), were injected intravenously and chimeras were analyzed 6–8 weeks later.
T Cell Transfer model of colitis
Induction of colitis by transfer of naïve CD4+ T cells was performed as previously described (24, 35), with the following modifications: C57BL/6 Rag1−/− mice were injected i.p. (4x105 cells in 200 ul of PBS) with naïve CD4+CD25−CD45RBHigh or CD4+ CD25− CD44low sorted from SP and LN from 4–6 week-old Ctrl or Blimp-1 CKO mice or CD4+CD25−CD44low Foxp3−cells sorted from Ctrl or Blimp-1CKO-Foxp3reporter mice. Mice were weighed weekly and inspected for clinical signs of disease (including weight loss, hunched appearance, pilo-erection of fur coat, and loose stool). Mice presenting clinically severe disease were sacrificed according to the CSMC Animal Care and Use Committee guidelines. At 7–12 weeks post-transfer, recipient mice were sacrificed and colons were removed, cleaned in ice-cold PBS, and pieces of ~0.5 cm in length were obtained from the proximal, middle and distal portion of the colon and fixed immediately in 10% formalin. Fixed tissue was later embedded in paraffin and 3 μm sections were cut and stained with Hematoxylin-Eosin (H&E). Samples were coded and scored by a pathologist in a blinded fashion as previously described (35).
T cell isolation and in vitro T helper differentiation
Naïve CD4+ (CD25− CD44low) or effector (CD25− CD44low) T cells were sorted from spleen and lymph nodes cell suspensions, using a FACSAria III fluorescent cell sorter (BD Biosciences, San Jose, CA). Purity of sorted cells preparations were > 98%. For in vitro T helper differentiation, sorted naïve CD4+ T cells were stimulated in Iscove’s DMEM media (Cellgro, Manassas, VA) supplemented with 10% Fetal Bovine Serum (Omega scientific, Inc. Tarzana, CA) and penicillin/streptomycin (Cellgro) with plate-bound anti-CD3 (5 μg/ml), anti-CD28 (2.5 μg/ml) (both from BioXcell, West Lebanon, NH), and rHuIL-2 (25 U/ml; Roche) (neutral conditions). Experimental Th1 conditions also included rMuIL12 (5 ng/ml) plus anti-IL4 (10 μg/ml) in addition to IL2. Th2 conditions included rMuIL-4 (10 ng/ml) and anti-IFNγ (10ng/ml). Th17 conditions excluded IL2 and included IL1β (20ng/ml), IL23 (50 ng/ml), IL6 (10 ng/ml), TGFβ (5ng/ml) (Promega), anti-IL4 and anti-IFNγ. With the exception of TGFβ, all recombinant cytokines were obtained from eBioscience, Inc. Neutralizing antibodies anti- IFNγ and anti-IL4 were obtained from Bio X Cell. Cells were split every 3 days and fresh medium (with the appropriate recombinant cytokines) added each time cells were split. For experiments in which TGFβ was used to repress Blimp-1 expression, recombinant TGFβ was only added at the beginning of the cultures and not replaced upon cell splitting of the cells. Re-stimulation was performed with Phorbol Myristate Acetate (PMA, 50 ng/ml) and Ionomycin (500 ng/mL) (both from Sigma Aldrich) for 4 hours; BFA was added in the last 2 hours or with plate-bound anti-CD3 for 6 hours (BFA added in the last 2 hours).
Western Blots
Whole cell lysates (30 ng/sample) were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) transferred to Invitrolon™ PVDF membranes (Invitrogen) and immunoblotted with monoclonal α-mouse Blimp-1 (3H2E8), or monoclonal α-mouse β-actin (Sigma).
Chromatin Immunoprecipitation
ChIP assays were performed as previously described (29). Briefly, naïve CD4+ T cells were sort-purified from cell suspensions of lymph nodes and spleens from wild type mice and stimulated with plate bound anti-CD3, plus anti-CD28 under Th2 polarizing conditions. After seven to eight days cells were re-stimulated with PMA and ionomycin for 4 hours before crosslinking by fixation with 1.1 % paraformaldehyde for 10 min at room temperature. Sonicated chromatin from 4–5x107 cells was immunoprecipitated with 25 ul of either rabbit anti-Blimp-1 polyclonal antibody (clone 267, recognizing the C terminal of Blimp-1) (36) or pre-immune serum as a control. Quantitative real-time PCR using SYBR Green incorporation was performed in DNA recovered from IP and input samples (See table II for primers sequences). Fold enrichment for each sample was calculated by dividing the percentage of input values obtained with anti-Blimp by the values obtained with control antibody followed by normalization to the values obtained from the input chromatin. Analysis of sequence homology and identification of putative Blimp-1 consensus sites were performed using ECR browser (http://ecrbrowser.dcode.org) and rVista 2.0 software. All genomic sequences were obtained from Ensembl.
Retroviral gene transduction
Recombinant retroviruses encoding Blimp-1 (in the MigR1 vector, containing an internal ribosomal entry site–GFP cassette) were produced from transfected Plat-E packaging cells (37) and were used to transduce naïve CD4+ T cells stimulated under Th17-polarizing conditions or CD3−B220+ cells sorted from the spleens of Prdm1Flox/Flox CD19-CRE+/− and Prdm1Flox/Flox CD19 cre−/− mice and stimulated for 24hrs with 2 μg/ml of LPS (Sigma). Transduction was performed by mixing the B cells (24 hours after stimulation) or the T cells (48–56 hours after stimulation) with supernatant-containing virus and Polybrene (8 μg/ml, Sigma) and then centrifuged at RT for 90 minutes (10,000xg). After transduction B cells were re-plated in regular media and T cells re-plated under Th17-polarizing conditions. B cells were re-stimulated with LPS (10 μg/ml) two days after transduction and analyzed 36 hours after LPS re-stimulation. T cells were analyzed three and four days after transduction and upon re-stimulation with PMA and Ionomycin.
Statistics
Student’s t-test with two-tailed distribution of equal variances was used to calculate probability (p) values, using the JMP software (SAS Institute Inc., NC).
Results
Lack of Blimp-1 is associated with accumulation of CD4+ T cells in the colon and increased production of IL17 in vivo
In order to identify the cellular mechanisms leading to the inflammatory response observed in the intestinal mucosa of the Blimp-1CKO mice, we first investigated the presence of T cells in the inflammatory infiltrate observed in 8–16 week-old mice with signs of colitis development. We found that the absolute numbers of mononuclear cells recovered from the large intestines (LI; cecum, colon and rectum) lamina propria (LI-LP) were significantly higher in the Blimp-1 CKO mice (Fig. 1A). Both the percentages and absolute numbers of TCRβ+CD4+ were elevated in the LI-LP of Blimp-1CKO mice; CD4+ T cells percentages were on average twofold higher in the Blimp-1CKO mice (Fig. 1B), while the absolute numbers were three to four-fold higher in the Blimp-1 CKO mice (Fig. 1B). Thus, Blimp-1-deficient CD4+ T cells accumulate in the colonic lamina propria.
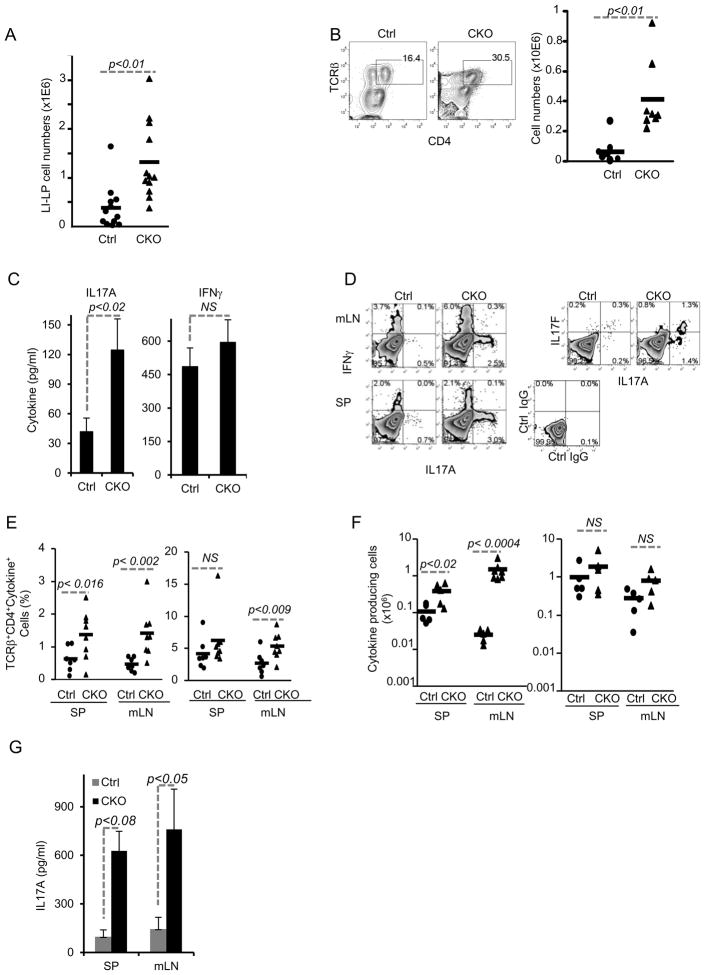
Figure 1. Accumulation of CD4+ TCRβ+ cells in the colon and increased amounts of IFNγ and IL17-producing cells in Blimp-1 CKO mice.
A) Total numbers of mononuclear cells in the large intestines (LI: cecum, colon and rectum) lamina propria (LP) in 8–16 week-old Prdm1+/+CD4-CRE+/−(Ctrl) and Prdm1F/F CD4-CRE+/− (CKO) mice. Each symbol represents one animal (circles, Ctrl; triangles, CKO) and bars represent average for each group. Results shown are representative of 5 independent experiments. B) Percentages of TCRβ+CD4+ T cells (FACS plots) and total numbers of TCRβ+CD4+ T cells (chart) in LI-LP from Ctrl and Blimp-1CKO mice. C) Production of and IL17A (left) and IFNγ (right) measured by ELISA on supernatants of LI-LP cells stimulated with plate-bound anti-CD3 plus anti-CD28 for 24 hours. D) Intracellular staining (ICC) for lL17A and IFNγ (D, left plots) or IL17A and IL17F (D, right plots) and FACS analysis of CD4+TCRβ+ cells in mesenteric lymph nodes (mLN, top row) and spleen (SP, bottom row) from Ctrl and CKO mice. SP and mLN total cell suspensions were stimulated as in C) before analysis (BFA was added in the last 6 hours). Data shown are from gated TCRβ+CD4+ cells. E–F) combined results of the percentage (E) or absolute numbers (F) of IL17A or IFNγ+ CD4+ T cells in SP or mLN from 5–6 independent experiments for a total of 6–8 mice per group. G) Production of IL17A (ELISA) by cells from SP or mLN after 18 hours stimulation w/plate-bound anti-CD3 plus anti-CD28. Data show is the average and SEM from three different experiments with three to seven mice per group.
To characterize the CD4+ T cells that accumulated in the intestinal mucosa of Blimp-1CKO mice, we next investigated the production of the cytokines IFNγ and IL17A by these cells, as these two cytokine have been shown to play effectors roles in intestinal inflammation. LI-LP were isolated from Ctrl and Blimp-1 CKO mice and stimulated with plate-bound anti-CD3 plus anti-CD28 for 24 hours and IFNγ and IL17A production were measured in the supernatants (Fig. 1C). The amount of IFNγ detected in the supernatant of these cultures was similar in Ctrl and Blimp-1 CKO mice (Fig. 1C, right), but LI-LP from Blimp-1 CKO mice produced significantly more IL17A than cells from Ctrl mice (Fig. 1C, left).
To determine if the increased production of IL17A and IFN-γ was restricted to the LI mucosa, we measured production of these cytokines in SP and mLN of Blimp-1 CKO mice after in vitro TCR stimulation. Intracellular cytokine staining (ICC) and FACS analysis showed increased percentages (Fig. 1D–E) and absolute numbers (Fig. 1F) of IL17A- producing cells in both SP and mLN of Blimp-1 CKO mice. In addition, we also observed an increase in the percentage of IL17F+ cells in the SP of Blimp-1 CKO mice (Fig. 1D). In both Ctrl and Blimp-1 CKO mice, approximately 50% of the IL17A+ cells were also IL17F+, but CKO mice had two to three- fold more IL17-producing cells (Fig. 1D). Different from the observed for IL17 production, the percentages of IFNγ-producing cells was elevated in the mLNs but not in the spleen of Blimp-1CKO mice (Fig. 1D–E) and no significant differences were observed in the absolute numbers of IFNγ-producing cells in Ctrl and Blimp-1CKO mice (Fig. 1F). Secretion of IL17A was also significantly increased in the SP and mLNs of the CKO mice (Fig. 1G). Thus, lack of Blimp-1 is associated with increased production of IL17, and to a lesser extent, IFNγ in vivo.
Blimp-1-deficient Effector/Memory CD4+ T cells have increased expression of genes associated with the Th17 differentiation program
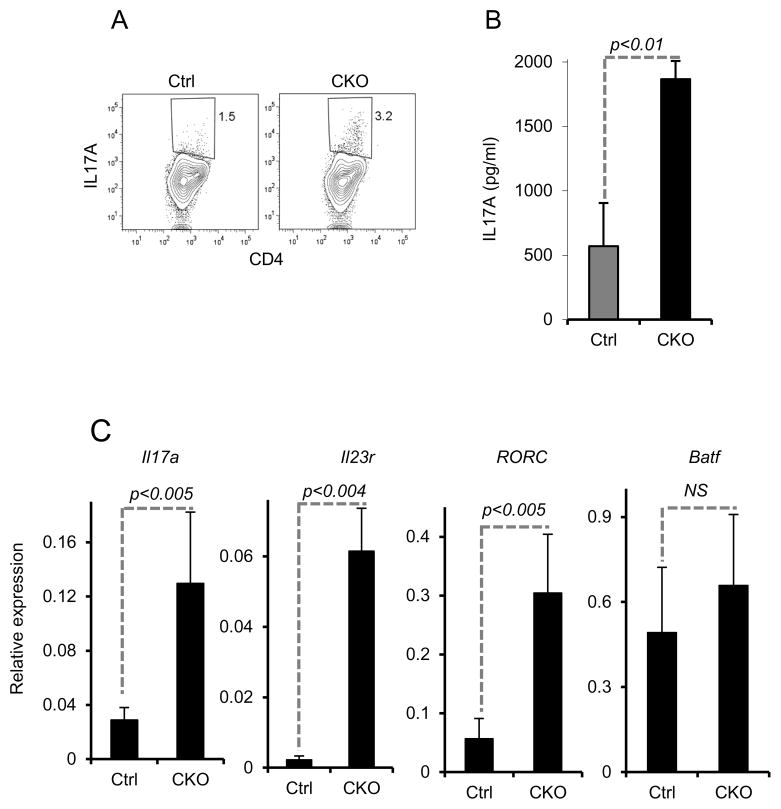
Characterization of the IL17-producing cells present in the Blimp-1CKO mice revealed that they were all antigen-experienced cells, which expressed high levels of CD44 (data not shown). Sort-purified CD44high CD25− CD4+ cells from pooled mLN and SP from Blimp-1CKO had higher frequency of IL17 producers and secreted significantly more IL17A protein than Ctrl cells upon in vitro re-stimulation (Fig. 2A–B). They also expressed significantly higher amounts of Il17a, Rorc, Il23r but not Batf mRNA than Ctrl cells when stimulated in vitro (Fig. 2C). Thus, the expression of Th17 signature genes is up-regulated in Blimp-1-deficient antigen-experience CD4+ T cells, suggesting that Blimp-1 is required to control the differentiation and/or accumulation of Th17 cells in vivo.
Figure 2. Increased expression of IL17 and Th17-signature genes in Blimp-1-deficient effector/memory cells.
CD4+ antigen-experienced (CD44High) T cells were sorted from pooled SP and mLN from Ctrl or Blimp-1 CKO mice and stimulated in vitro with plate-bound anti-CD3 plus anti-CD28 and IL17 production was measured by ICC staining and FACS analysis at 6 hours (A) or in the supernatants by ELISA at 48 hours (B). Il17a, Il23r, RORC, and BATF steady state mRNA levels (C) were measured by qRT PCR in Ctrl and Blimp-1 CKO stimulated as in (A). Results shown in A-C are representative of two to three independent experiments with three to five mice/group. Bars show average and error bars represent SEM. Results in C are presented relative to 18S expression.
Regulation of IL17 production by Blimp-1 is intrinsic to CD4+ effector T cells
Increased numbers of IL17-producing CD4+ T cells in Blimp-1 CKO mice could result from intrinsic effects of Blimp-1 in regulating the differentiation/accumulation of Th17-cells in vivo or could result from the previously described impaired Treg cell function in these mice (25, 26), as defective Treg cell responses can facilitate priming and development of IL17 and IFNγ-producing cells (38–40). Alternatively, the increased numbers of IL-17 producing cells could be a secondary effect of the ongoing inflammation in the colon of these mice (25). In order to distinguish these possibilities, we next analyzed the expression of IL17 in CD4+ T cells from chimeric mice generated with a mixture of wild type and Blimp-1CKO bone marrow cells. In order to delay the development of severe inflammation in the chimeric mice, we designed our experiments such that the hematopoietic compartment in the reconstituted mice would contain significantly more Ctrl than Blimp-1-deficient cells. Thus, in all sites analyzed, the majority of TCRβ+CD4+ cells were Blimp-1-sufficient (Fig. 3A and data not shown).
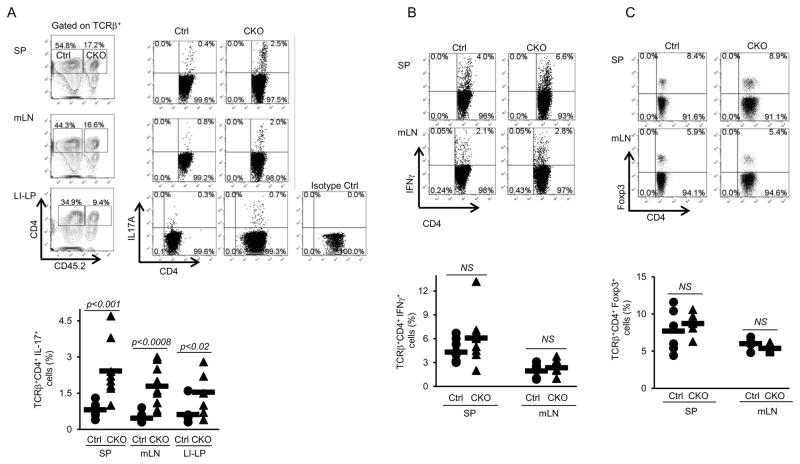
Figure 3. Increased production of IL17 by Blimp-1-deficient CD4+ T cells in mixed-bone marrow chimeric mice.
Wild type CD45.1-irradiated mice received a total of ~4 × 106 HSC-enriched mixed BM cells from Ctrl (CD451.1) and Blimp-1CKO (CD451.2) mice. Eleven weeks after bone marrow cells transfer, mLN, SP and LI-LP cells were isolated from chimeric mice, stimulated as described in Fig. 1D and IL17 (A) production was analyzed in TCRβ+CD4+ cells in the CD45.2− (Ctrl) or CD45.2+ (Blimp1CKO) gates. B, Frequency of IFNγ-producing TCRβ+CD4+ cells from the MLN and SP from chimeric mice (stimulation and analysis done as in A). C) Frequency of Foxp3-expressing cells in the Ctrl and Blimp-1CKO compartments in the SP and mLN from chimeric mice (gated as shown in A). Plots (A–C) show compiled results from four to nine different chimeric mice.
In both compartments, Blimp-1-suficient and Blimp-1-deficient, the frequency of Foxp3+ Treg cells was similar (Fig. 3C). In the Blimp-1-deficient compartment however, the frequency of IL17-producing (but not of IFNγ-producing) cells was significantly increased in comparison to the wild type compartment in all sites evaluated, including the LI-LP (Fig. 3). Thus, Blimp-1-deficient CD4+ T cells are intrinsically more prone to differentiate into IL17-producing cells.
Blimp-1-deficient CD4+ T cells differentiate into IL-17+/IFNγ+ cells and cause severe colitis in Rag1-deficient mice
To determine if the absence of Blimp-1 can lead to the preferential differentiation of IL17 and/or IFNγ producing cells in vivo, we transferred CD4+ naïve (CD44low/CD25−) Ctrl or Blimp-1 CKO cells into Rag1-deficient (Rag1−/−) mice and monitored colitis development in the recipient mice. Rag1−/− mice receiving Ctrl cells developed colitis and wasting disease as indicated by histological analysis and total body weight loss. Rag1−/− mice receiving Blimp-1-deficient cells showed signs of disease earlier than mice receiving Blimp-1sufficient cells (Fig. 4A) and also developed more severe colitis, with more prominent inflammation in the colon (Fig. 4B) and significantly increased colitis scores (average score=3±0.9) than in those receiving Ctrl cells (average score=1±0.7) (Fig. 4C).
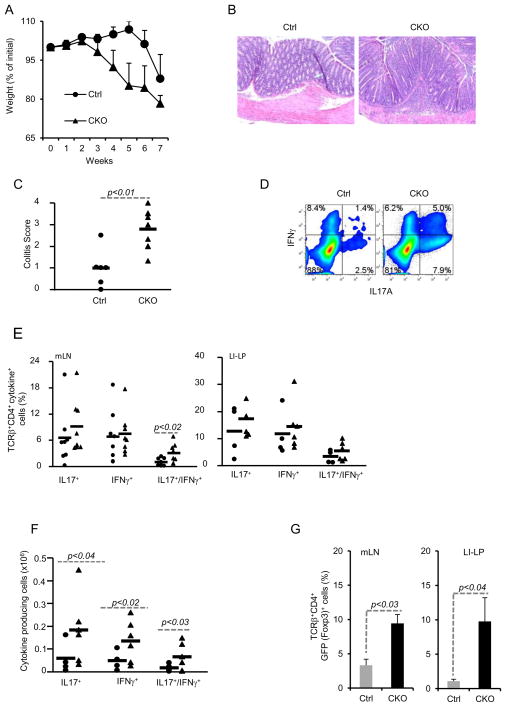
Figure 4. Blimp-1 CKO CD4+ T cells differentiate into IL17 and IFNγ-double producer cells in vivo and cause severe colitis in Rag1−/− mice.
Rag1−/− mice were injected i.p. with 4x105 naïve (CD44lowCD25−) CD4+ T cells sorted from Ctrl or Blimp-1 CKO mice and monitored for symptoms of colitis development and wasting disease, including body weight loss daily (A). Six to seven weeks after T cell transfer, recipient mice were euthanized and colons were collected and processed for H&E staining (B, 20 X magnifications) and histological analysis. Slides were analyzed blindly. Colitis score (C) was graded semi-quantitatively from 0–4, as previously described (35) each symbol represent one animal (the circles and triangles represent Ctrl and CKO, respectively). D) ICC staining and FACS analysis of cells from mLN from recipient mice 7 weeks after adoptive T cell transfer of Ctrl (circles) or Blimp1CKO (triangles) cells. mLN cells were stimulated in vitro for 24 hours with plate-bound αCD3 plus αCD28 before staining. FACS plots show cells in the TCRβ+CD4+ gate. E) Combined results (each symbol represent one animal, and bars represent average of five to six different experiments) of the percentage of IL17A+, IFNγ+, or IL17A+/IFNγ+ CD4+ T cells in the mLN (left panel), or LI-LP (right panel) for a total of six to seven mice per group. F) Absolute numbers of IL17A+, IFNγ+, or IL17A+/IFNγ+ CD4+ T cells in the mLN based on the percentages shown in E. G) Percentages of TCRβ+ CD4+Foxp3+ cells in the mLN (left) or LI-LP (right) of Rag−/− mice injected with Ctrl or Blimp-1 CKO 4x105 naïve [CD44lowCD25−GFP− (Foxp3−)] CD4+ T cells and analyzed six to seven weeks later. Data are representative of five (A–C) three (E–F) different experiments, with a total of four to seven mice per group.
Evaluation of cytokine production upon ex vivo TCR stimulation of cells from mLN and LI-LP revealed that while the percentages of IFNγ or IL17-single producer cells were only marginally increased in the Blimp-1CKO mice, the percentage of IFNγ and IL17-double producer cells were significantly increased in the mLN of mice transferred with Blimp-1-deficient cells (Fig. 4D–E). However, the absolute numbers of IL17+, IFN-γ+, and IL17+/IFN-γ+ cells were all significantly increased in the LI-LP of mice injected with Blimp-1-deficient cells in comparison to mice injected with Ctrl cells (Fig. 4F). To determine if the differences we observed between Ctrl and Blimp-1 CKO cells-mediated colitis was due to differential induction Foxp3+ Treg cells from the injected naive cells, we repeated these experiments using naïve CD4+ T cells isolated from Ctrl or Blimp-1-CKO mice that had been previously bred with Foxp3-reporter mice. This strategy allowed us to exclude Foxp3+ cells from the pool of naïve cells adoptively transferred to the Rag1−/− mice and subsequently monitor the development of Foxp3-expressing cells in the recipient mice. Rag1−/− mice injected with CD4+ naïve [CD44lowCD25−GFP−(Foxp3−) cells] from Blimp-1CKO mice developed more severe colitis than mice injected with Blimp-1-suficent cells (data shown), similarly to that described above, indicating that the increased severity of colitis caused by Blimp-1deficient cells was not due to differential contamination of the transferred cells with Foxp3+ Treg cells. Moreover, analysis of GFP expression in cells from the recipient mice showed that Foxp3+ cells developed in greater numbers from Blimp-1-deficient than from Ctrl cells (Fig. 4G), indicating that increased inflammation in mice injected with Blimp-1-deficient was not due to impaired induction of Foxp3+ cells.
Blimp-1 expression is selectively regulated during Th differentiation
The results described above indicated that expression of Blimp-1 suppresses the differentiation of Th17 cells in vivo, and that in the absence of Blimp-1 more cells tend to differentiate into IL17+ or IL17+/IFNγ+ cells. We reasoned that if Blimp-1 antagonizes Th17 differentiation, its expression should be down regulated in cells differentiated under Th17 conditions, as opposed to cells that usually do not produce IL17, such as Th1 and Th2 cells. Previous studies have shown that Blimp-1 is expressed in CD4+ T cells differentiating into either Th1 or Th2 conditions, but while one group showed increased expression of Blimp-1 in Th2 cells (41) another showed similar expression in Th1 and Th2 cells (28). Neither of these studies investigated Blimp-1 expression in Th17 cells. In order to clarify whether Blimp-1 expression is, in fact, differentially regulated during Th differentiation, we used cells from previously described Blimp-1-YFP reporter mice (29, 32) to analyze Blimp-1 expression at the single cell level during Th differentiation in vitro.
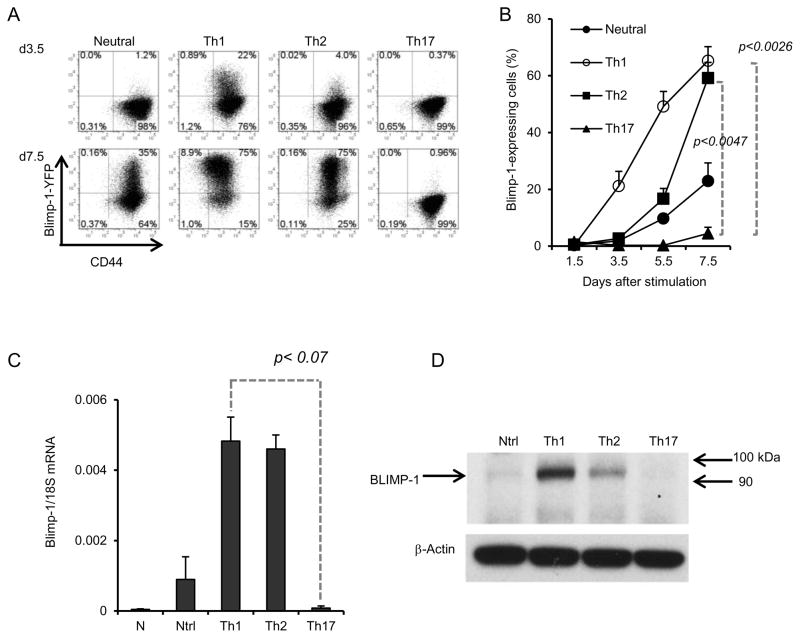
Sorted naïve (CD44lowCD25−) CD4+T cells from Blimp-1-YFP reporter mice were stimulated under neutral, Th1, Th2 or Th17 conditions (Supplementary Fig. 1) and Blimp-1 mRNA expression (as reported by YFP) was analyzed at different time points. As expected from previous reports (41, 42), stimulation of naïve CD4+ T with plate-bound anti-CD3 plus anti-CD28 induced expression of Blimp-1 in a small percentage of cells; addition of IL2 to these cultures (neutral conditions) increased expression, especially at later time points (Fig. 5A). Cells cultured under Th1 or Th2 conditions began to express Blimp-1 sooner than cells cultured in neutral conditions, but Th1 cells expressed significantly more than Th2 cells at earlier time points (Fig. 5A). However, at day 7.5, more than 70% of Th1 and Th2 cells expressed high levels of Blimp-1 (Fig. 5A–B). In contrast to cells stimulated under neutral, Th1 or Th2 conditions, cells cultured under Th17 conditions did not up-regulate Blimp-1 significantly throughout the experiment, and less than 5% of the cells expressed Blimp-1 at day 7.5 post stimulation (Fig. 5B). Analysis of Blimp-1 steady-state mRNA (Fig. 5C) and protein levels (Fig. 5D) in these cells confirmed the differential expression of Blimp-1 during Th differentiation. Thus, Blimp-1 is expressed in Th1 and Th2 cells but not in Th17 cells.
Figure 5. Blimp-1 is expressed in Th1 and Th2 but not Th17 cells.
A) Blimp-1 expression as reported by YFP in Naïve CD4+ T cells sorted from YFP transgenic Blimp-1 reporter mice and stimulated under neutral (with IL2), Th1,Th2 or Th17 conditions for 3.5 (top row) or 7.5 (bottom row) days. FACS Plots shown are from the gated live, CD4+ cells in the well. B) Average percentages (and standard deviation) of YFP positive cells shown in A for five independent experiments, for a total of seven different mice. C) Steady state Blimp-1 mRNA at d7.5 in cells from experiment shown in A). D) Immunoblotting of total cell lysates obtained from cells stimulated as in A) for 5.5 days (Ntrl=Neutral), using antibodies to Blimp-1 or β-actin.
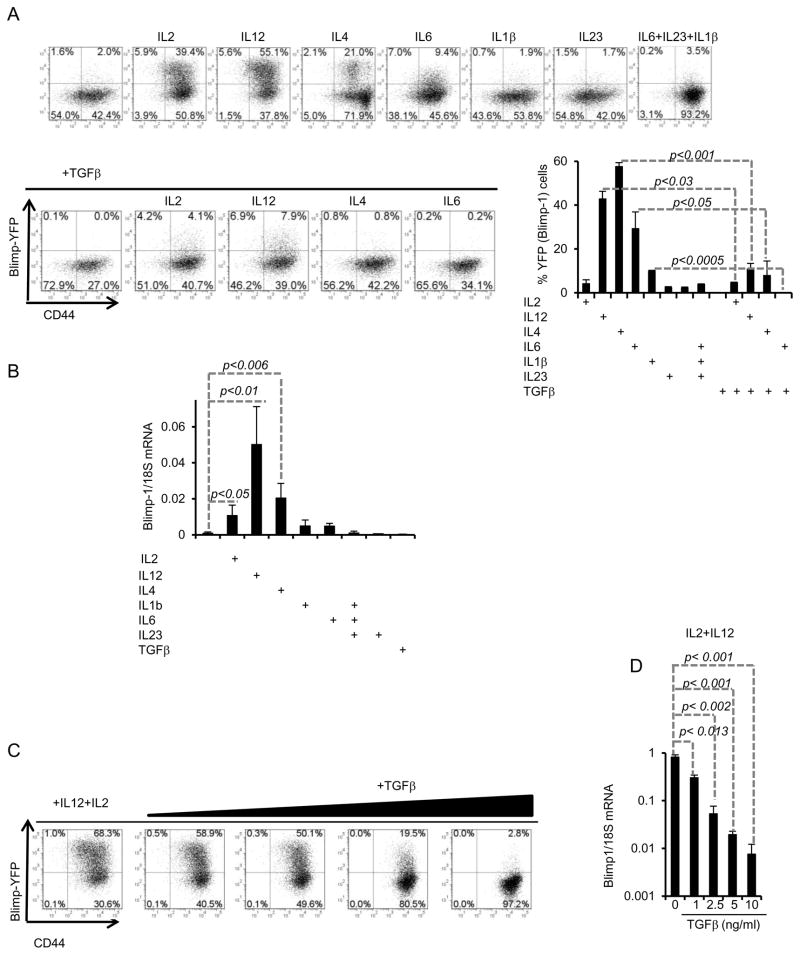
We next investigated the regulation of Blimp-1 expression by different Th polarizing cytokines. Previous studies (28, 41, 42) have reported contradictory results on the induction of Blimp-1 by Th1 and Th2-inducing cytokines. We found that IL2, IL4, IL6 and IL12 were each capable of enhancing expression of Blimp-1 by anti-CD3 plus anti-CD28 stimulation (which alone induced little expression of Blimp-1) (Fig. 6A), but at different magnitudes, with IL12 being the best inducer, followed by IL2 and then IL4. Amongst Th17 inducing cytokines, IL6 was the best inducer of Blimp-1 expression, although in comparison to IL2, IL4 and IL12, IL6 was a poor inducer. Neither IL1β nor IL23 induced Blimp-1 expression; in fact these cytokines partially inhibited Blimp-1 expression induced by IL6 (Fig. 6A). Since the expression of Blimp-1 in cells stimulated with a combination of IL6, IL23 and IL1β was higher than in cells stimulated under Th17 conditions (Fig. 6A) (which included TGFβ in addition to IL6, IL23 and IL1β),we suspected that TGFβ could have an inhibitory effect on Blimp-1 expression. We, therefore, compared Blimp-1 expression induced by the combination of TCR (and co-stimulation) and IL2, IL12, or IL4 in the absence or presence of TGFβ. We found that TGFβ led to significant inhibition of Blimp-1 expression in these cultures (Fig. 6A). Evaluation of Blimp-1 steady-state mRNA levels confirmed these results (Fig. 6B).
Figure 6. Blimp-1 expression is induced by Th1 and Th2-inducing cytokines but repressed by TGFβ in Th17-developing cells.
A) Naïve CD4+ T cells from Blimp-1 reporter mice were stimulated with plate-bound anti-CD3 plus anti-CD28 alone or in the presence of the indicted cytokines and YFP (Blimp-1) expression was analyzed by FACS at d5. B) qRT-PCR analysis of Blimp-1 mRNA expression in the same cells from experiment shown in (A). C) Dose-dependent inhibition of Blimp-1 expression by TGFβ in Blimp-1 reporter naïve CD4+ T cells stimulated as indicated in the presence of different amounts of TGFβ for 5 days and then analyzed by FACS. D) qRT-PCR analysis of Blimp-1 mRNA expression in the same cells from experiment shown in (C). Data shown are from one experiment with pooled cells from three mice; similar results were obtained in 2 independent experiments.
We next sought to determine if TGFβ could inhibit expression of Blimp-1 induced by the combination of IL2 and IL12. TGFβ inhibited Blimp-1 expression induced by TCR and co-stimulation in combination with IL2 and IL12 in a dose-dependent manner (Fig. 6C). Even at concentrations lower than the required amounts used to induce Th17 differentiation in vitro, TGFβ led to significant inhibition Blimp-1 expression in cells cultured simultaneously with IL12 and IL2. These results were also confirmed by mRNA expression measured by qRT-PCR (Fig. 6D). Together, these results establish TGFβ as a potent suppressor of TCR and cytokine-induced Blimp-1 expression during Th17 differentiation.
Blimp-1 binds to the Il17a gene in vivo but is not sufficient to repress Th17 differentiation
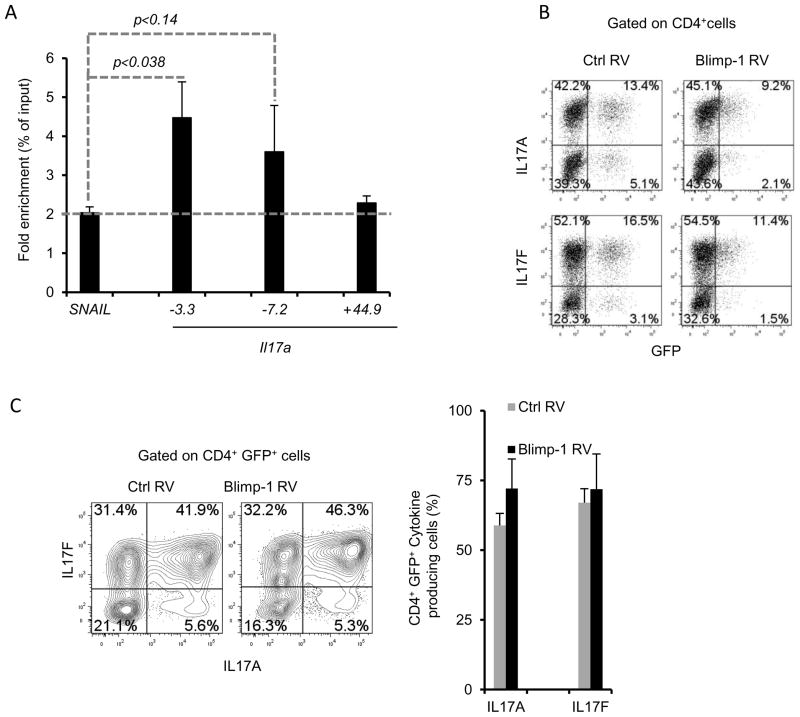
The observation that lack of Blimp-1 is associated with increased expression of Il17a mRNA and protein as well as several other Th17 genes (Fig. 2) and that Blimp-1 expression is selectively down regulated during Th17 differentiation (Fig. 5) suggested that Blimp-1 could function as a repressor of the Th17 program. Blimp-1 has potent transcriptional repression capabilities and directly regulates the transcription of several cytokine genes (26, 29, 41). Thus, we first sought to determine if Blimp-1 response elements could be identified in the Il17a gene. We identified many putative Blimp-1 binding sites at the Il17a promoter region as well as in regions downstream of the transcription start site (data not shown). We then tested if Blimp-1 could bind to three of the sites we identified. Using wild type Th2 polarized cells, we found significant enrichment for Blimp-1 at at least one of these sites (located at approximately 3.3 Kb upstream of the transcription start site, TSS). We also detected some enrichment for Blimp-1 binding on a site located 7.2 Kb upstream of the TSS, but in this case there was no significant difference in comparison to nonspecific binding on a control gene (Fig. 7A). The third site investigated is located further downstream (approximately 44.9 Kb from the TSS) showed no enrichment for Blimp-1 binding (Fig. 7A). Thus, in Th2 cells Blimp-1 can bind to at least one site in the Il17a gene.
Figure 7. Blimp-1 binds to the murine Il17a gene in Th2 cells but it is not sufficient to repress IL17 production in Th17 cells.
A) Chromatin Immunoprecipitation (ChIP) analysis of wild-type Th2 polarized cells re-stimulated with PMA and ionomycin, followed by immunoprecipitation of chromatin with anti-Blimp-1 or control antibody and quantitative PCR analysis of binding of Blimp-1 at different regions upstream (−3.3, −7.2 Kb ) or downstream (+44.9Kb) of the transcription start site of the murine Il17a gene or a negative control region in the SNAIL3 gene; results were normalized to those of the input chromatin, followed by the ratio of results obtained with anti-Blimp-1 antibody and control antibody (nonspecific background). Data are representative of three to four independent experiments (mean and SEM). B–C) Production of IL17 upon enforced expression of Blimp-1 in Th17 cells. Naïve CD4+ T cells were stimulated under Th17 conditions and transduced with retrovirus expressing GFP only (Ctrl RV) or GFP and Blimp-1 as a bi-cistronic message (Blimp-1 RV); cells were re-stimulated 3 (B) or 4 (C) days after transduction and analyzed for IL17A (top plots) or IL17F (bottom plots) production. Plots shown are from CD4+ (B) or CD4+GFP+(C) cells. Chart in C shows the average and SEM of three independent experiments.
To determine if Blimp-1 could suppress Il17a activity in Th17 cells, we used retrovirus transduction to enforce Blimp-1 expression in cells stimulated under Th17 polarizing conditions and measured IL17 production. Expression of Blimp-1 (as indicated by a GFP reporter), and confirmed by qRTPCR (data not shown) did not result in significant decrease of IL17A or IL17F production (Fig. 7B–C). However, transduction with the same retroviral construct was able to partially restore plasma cell formation in Blimp-1-deficient B cells stimulated in vitro (Supplemental Fig. 2), indicating that functional Blimp-1 protein is expressed from this construct. Therefore, although Blimp-1 is bound to the Il17a gene in Th2 cells, it is not sufficient to repress Il17a transcription in cells stimulated under Th17 conditions.
Discussion
The results of this study provide evidence for a non-redundant role for the transcriptional regulator Blimp-1 in constraining the numbers of IL17-producing Th cells in vivo. We showed that spontaneous development of chronic intestinal inflammation in mice with T cell-specific deletion of Blimp-1 was associated with accumulation of IL17-producing Th cells in vivo. Furthermore, Blimp-1-deficient naïve CD4+ T cells preferentially differentiated into IL17 and IFNγ-double-producing cells and caused severe colitis when transferred to Rag1−/− mice. Consistent with a role for Blimp-1 in keeping the numbers of Th17-producing cells under control, we demonstrated that Blimp-1 was selectively expressed in Th1 and Th2 but not in Th17 cells.
Our observation that IL17-producing cells selectively accumulate in the Blimp-1-deficient CD4+ T cell compartment of Ctrl and Blimp-1CKO-mixed-bone marrow chimeric mice (Fig. 3) indicate a Treg-independent, effector T cell-intrinsic role for Blimp-1 in repressing the production of IL17 in vivo. This is further supported by our findings that Blimp1-deficient naïve CD4+ T cells generated more IL17-producing cells than control cells and caused severe colitis upon adoptive transfer to Rag1−/−mice. These results cannot be attributed to a defective Treg response in the absence of Blimp-1, because Blimp-1 deficient CD4+ T cells led to the generation of Foxp3+ Treg cells in greater numbers than control cells. Although IL10 production is defective in a small subset of Foxp3+ Blimp-1-deficient Tregs (25, 28, 41), IL10-deficient Tregs can block adoptive T cell transfer–induced colitis (43, 44) and similar findings have been reported for Blimp-1-deficent Tregs (28).
The mechanisms underlying repression of IL17 production/Th17 differentiation by Blimp-1 remains to be fully elucidated. Our observation that Blimp-1-deficient CD4+ T cells had increased amounts of Il17a mRNA, suggested that Blimp-1 could regulate IL17 production at the transcriptional level. In addition, Blimp-1 deficient CD4+ effector T cells had increased amounts of Il23r and RORC mRNA, suggesting that Blimp-1 might directly repress these genes. Consistent with this possibility Blimp-1 consensus binding sites can be found in conserved, putatively regulatory regions in both loci (data not shown). Alternatively, since expression of Il23r and Il17a are both positively regulated by Rorc (encoding RORγt), Blimp-1 might also regulate Il23r and Il17a indirectly by acting as a repressor of Rorc.
Our observation that Blimp-1 can bind to at least one site on the Il17a promoter in Th2 cells suggested that Blimp-1 might function to directly repress the Il17a gene during Th2 differentiation. In fact, Il17a site -3.2, which shown significant enrichment for Blimp-1 binding is located approximately 2 Kb downstream of conserved non coding sequence 2 (CNS2, also called CNS5), an important regulatory region in the il17a promoter (45, 46). Consistent with the idea that Blimp-1 could repress Il17a in Th2 cells, deficiency of the histone lysine methyltransferase G9a, which can function as a co-factor for Blimp-1-mediated repression (47) leads to increased production of IL17A in Th2 polarized cells (48). However, forced expression of Blimp-1 in cells differentiating under Th17 conditions did not result in a significant decrease of IL17A or IL17F production (Fig. 7B–C). Expression of Prdm1 mRNA in transduced Th17 cells that expressed Blimp-1 was similar to that of developing Th2 cells (data not shown). Nevertheless, it is possible that repression of Il17a in Th17 cells requires higher amounts of Blimp-1 protein than achieved in our experiments. Alternatively, repression of the Il17a by Blimp-1 might require co-repressors that are selectively expressed in Th2 (and potentially Th1) but not in Th17 cells. Such a mechanism could play a role in preserving Il17a activity in Th17 cells that are exposed to conditions where Blimp-1 expression can be induced.
Our results presented here show that TGFβ signaling is a potent repressor of Blimp-1 expression and might function to repress Blimp-1 expression during Th17 differentiation. Interestingly, a recent study using in vitro generated Th17 cells to induce colitis upon transfer into Rag1−/− mice demonstrate that the extinction of IL17 production in Th17 cells that become IFNγ-producers in the presence of IL12 and IL23 is contingent upon limited amounts or complete absence of TGFβ (23). The presence of TGFβ resulted in maintenance of IL17 production in these cells (23). Using Experimental Autoimmune Encephalomyelitis (EAE) as a model of inflammation, Ghoreschi et al (49) made similar observations, showing that Th17 cells generated in the presence of IL23 and absence of TGFβ retained the capacity to make IL17 while simultaneously turning on IFNγ production (49). Based on these observations, our results showing that Blimp-1-deficient T cells generate more IL17+/IFNγ+ cells under inflammatory conditions suggest a model whereby induction of Blimp-1 expression could be required to repress IL17 production in Th17 cells converting into IFNγ-producing cells. According to this model, under highly inflammatory conditions where IL12 is abundant and TGFβ scarce, IL12 would lead to induction of Blimp-1 expression in Th17 cells, in addition to inducing expression of T-bet and IFNγ. Blimp-1 expression under these conditions would result in the repression of both, IL17 and IL23R, thus restraining the formation and/or maintenance of the IL17+/IFNγ+ cells. In the absence of Blimp-1, IL12 would lead to IFNγ production, but would fail to repress IL17 and favor the generation/maintenance of the IL17+/IFNγ+ cells.
One caveat of this model is the assumption that IL12 could induce Blimp-1 and IFNγ simultaneously, which is at odds with our previous observation that in CD4+ T cells stimulated under neutral conditions, Blimp-1 binds to and suppresses transcription of both Tbx21 and Ifng (41). It is possible, however, that the Ifng gene regions bound by Blimp-1 are not equally accessible in T cells stimulated under neutral conditions and in cells responding to IL12 and transitioning into Th1 cells (or “ex Th17 cells”) developing during mucosal inflammation. In addition to differential accessibility of Blimp-1 binding regions at the Ifng locus, these different Th subpopulations may also diverge in the expression of co-activators and co-repressors which could interact with Blimp-1 to counteract or favor repression of Ifng. The same arguments can be made to explain the fact that Th1 cells express high levels of Blimp-1 while making high amounts of IFNγ. Evaluation of Blimp-1-binding sites occupancy in cells stimulated under neutral conditions side by side with cell transitioning from Th17 to Th1 will be required to clarify this.
In line with our observation that IL17-producing cells accumulate in the Blimp-1CKO mice and that Blimp-1-deficient T cells fail to turn off the Il17 gene under inflammatory conditions, recent genome-wide association studies (GWAS) reveal strong association between the gene encoding Blimp-1 (PRDM1) and Inflammatory (50, 51)Bowel Disease (IBD). One of the forms of IBD, Crohn’s disease (CD) is also associated with the accumulation of IL17+/IFNγ+ cells in the intestinal lesions) (21, 22). Although the functional implications of the PRDM1 polymorphisms associated with IBD remain unknown, one intriguing possibility is that these polymorphisms cause decreased expression or partial loss of function of Blimp-1 resulting in the accumulation of IL17+/IFNγ+ cells in the intestinal lesions of CD patients.
Plasticity of the CD4+ T cells is now a recognized trait of immune responses, especially under conditions associated with inflammatory disorders. Understanding the mechanisms regulating T helper cell plasticity in these conditions will be essential in considering new therapeutic approaches to treat these diseases. The involvement of Blimp-1 in counteracting the differentiation of the highly inflammatory IL17+/IFNγ+ producing cells in vivo as well as its potential role in repressing Il17a expression in Th1 and Th2 cells that we report here might represent initial steps towards the understanding the mechanisms regulating effector T cell response under chronic inflammatory conditions.
Supplementary Material
Acknowledgments
We thank Dr. Kathryn Calame for her invaluable support of these studies and Dr. Jonathan Kaye for helpful discussions and for critical reading of the manuscript. We are also grateful to Brian de la Torre and to Asha Kadavallore for helping with mice irradiation and retrovirus supernatant production respectively, and to the CSMC Flow Cytometry core, especially Gillian Hultin for sorting. We also thank Carol Landers and Richard Deem for technical help and Loren Karp for editorial assistance.
Footnotes
This work was supported by NIH-NIAID grant # AI083948-01 (to G. Martins), by the F. Widjaja Foundation Inflammatory Bowel and Immunobiology Institute (IBIRI) Funds, and a pre-doctoral scholarship from Fundacao de Amparo a Pesquisa e ao Ensino do Estado de Sao Paulo (Brazil) FAPESP#2008/04606-2 (to L. Benevides).
The authors have no conflicting financial interests.
References
- 1.Weaver CT, Hatton RD, Mangan PR, Harrington LE. IL-17 family cytokines and the expanding diversity of effector T cell lineages. Annu Rev Immunol. 2007;25:821–852. doi: 10.1146/annurev.immunol.25.022106.141557. [DOI] [PubMed] [Google Scholar]
- 2.Park H, Li Z, Yang XO, Chang SH, Nurieva R, Wang YH, Wang Y, Hood L, Zhu Z, Tian Q, Dong C. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol. 2005;6:1133–1141. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, Weaver CT. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6:1123–1132. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 4.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*) Annu Rev Immunol. 28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaschitz C, Raffatellu M. Th17 cytokines and the gut mucosal barrier. J Clin Immunol. 30:196–203. doi: 10.1007/s10875-010-9368-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miossec P. IL-17 and Th17 cells in human inflammatory diseases. Microbes Infect. 2009;11:625–630. doi: 10.1016/j.micinf.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 7.Sallusto F, Lanzavecchia A. Human Th17 cells in infection and autoimmunity. Microbes Infect. 2009;11:620–624. doi: 10.1016/j.micinf.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–189. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 9.Manel N, Unutmaz D, Littman DR. The differentiation of human T(H)-17 cells requires transforming growth factor-beta and induction of the nuclear receptor RORgammat. Nat Immunol. 2008;9:641–649. doi: 10.1038/ni.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou L, Ivanov II, Spolski R, Min R, Shenderov K, Egawa T, Levy DE, Leonard WJ, Littman R. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol. 2007;8:967–974. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]
- 11.Ivanov I, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 12.Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y, Ma L, Shah B, Panopoulos AD, Schluns KS, Watowich SS, Tian Q, Jetten AM, Dong C. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou L, Littman DR. Transcriptional regulatory networks in Th17 cell differentiation. Curr Opin Immunol. 2009;21:146–152. doi: 10.1016/j.coi.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bettelli E, Korn T, Oukka M, Kuchroo VK. Induction and effector functions of T(H)17 cells. Nature. 2008;453:1051–1057. doi: 10.1038/nature07036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schraml BU, Hildner K, Ise W, Lee WL, Smith WA, Solomon B, Sahota G, Sim J, Mukasa R, Cemerski S, Hatton RD, Stormo GD, Weaver CT, Russell JH, Murphy TL, Murphy KM. The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature. 2009;460:405–409. doi: 10.1038/nature08114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Shea JJ, Steward-Tharp SM, Laurence A, Watford WT, Wei L, Adamson AS, Fan S. Signal transduction and Th17 cell differentiation. Microbes Infect. 2009;11:599–611. doi: 10.1016/j.micinf.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bluestone JA, Mackay CR, O’Shea JJ, Stockinger B. The functional plasticity of T cell subsets. Nat Rev Immunol. 2009;9:811–816. doi: 10.1038/nri2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou L, Chong MM, Littman DR. Plasticity of CD4+ T cell lineage differentiation. Immunity. 2009;30:646–655. doi: 10.1016/j.immuni.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 19.Murphy KM, Stockinger B. Effector T cell plasticity: flexibility in the face of changing circumstances. Nat Immunol. 11:674–680. doi: 10.1038/ni.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bending D, De la Pena H, Veldhoen M, Phillips JM, Uyttenhove C, Stockinger B, Cooke A. Highly purified Th17 cells from BDC2.5NOD mice convert into Th1-like cells in NOD/SCID recipient mice. J Clin Invest. 2009;119:565–572. doi: 10.1172/JCI37865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Annunziato F, Cosmi L, Santarlasci V, Maggi L, Liotta F, Mazzinghi B, Parente E, Fili L, Ferri S, Frosali F, Giudici F, Romagnani P, Parronchi P, Tonelli F, Maggi E, Romagnani S. Phenotypic and functional features of human Th17 cells. J Exp Med. 2007;204:1849–1861. doi: 10.1084/jem.20070663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cosmi L, De Palma R, Santarlasci V, Maggi L, Capone M, Frosali F, Rodolico G, Querci V, Abbate G, Angeli R, Berrino L, Fambrini M, Caproni M, Tonelli F, Lazzeri E, Parronchi P, Liotta F, Maggi E, Romagnani S, Annunziato F. Human interleukin 17-producing cells originate from a CD161+CD4+ T cell precursor. J Exp Med. 2008;205:1903–1916. doi: 10.1084/jem.20080397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee YK, Turner H, Maynard CL, Oliver JR, Chen D, Elson CO, Weaver CT. Late developmental plasticity in the T helper 17 lineage. Immunity. 2009;30:92–107. doi: 10.1016/j.immuni.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahern PP, Schiering C, Buonocore S, McGeachy MJ, Cua DJ, Maloy KJ, Powrie F. Interleukin-23 drives intestinal inflammation through direct activity on T cells. Immunity. 33:279–288. doi: 10.1016/j.immuni.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martins GA, Cimmino L, Shapiro-Shelef M, Szabolcs M, Herron A, Magnusdottir E, Calame K. Transcriptional repressor Blimp-1 regulates T cell homeostasis and function. Nat Immunol. 2006;7:457–465. doi: 10.1038/ni1320. [DOI] [PubMed] [Google Scholar]
- 26.Cretney E, Xin A, Shi W, Minnich M, Masson F, Miasari M, Belz GT, Smyth GK, Busslinger M, Nutt SL, Kallies A. The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nat Immunol. 12:304–311. doi: 10.1038/ni.2006. [DOI] [PubMed] [Google Scholar]
- 27.Martins G, Calame K. Regulation and functions of blimp-1 in T and B lymphocytes. Annu Rev Immunol. 2008;26:133–169. doi: 10.1146/annurev.immunol.26.021607.090241. [DOI] [PubMed] [Google Scholar]
- 28.Kallies A, Hawkins ED, Belz GT, Metcalf D, Hommel M, Corcoran LM, Hodgkin PD, Nutt SL. Transcriptional repressor Blimp-1 is essential for T cell homeostasis and self-tolerance. Nat Immunol. 2006;7:466–474. doi: 10.1038/ni1321. [DOI] [PubMed] [Google Scholar]
- 29.Martins GA, Cimmino L, Liao J, Magnusdottir E, Calame K. Blimp-1 directly represses Il2 and the Il2 activator Fos, attenuating T cell proliferation and survival. J Exp Med. 2008;205:1959–1965. doi: 10.1084/jem.20080526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shapiro-Shelef M, Lin KI, McHeyzer-Williams LJ, Liao J, McHeyzer-Williams MG, Calame K. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19:607–620. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- 31.Misulovin Z, Yang XW, Yu W, Heintz N, Meffre E. A rapid method for targeted modification and screening of recombinant bacterial artificial chromosome. J Immunol Methods. 2001;257:99–105. doi: 10.1016/s0022-1759(01)00452-5. [DOI] [PubMed] [Google Scholar]
- 32.Rutishauser RL, Martins GA, Kalachikov S, Chandele A, Parish IA, Meffre E, Jacob J, Calame K, Kaech SM. Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. 2009;31:296–308. doi: 10.1016/j.immuni.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bettelli ECY, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, Kuchroo VK. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 34.Weigmann B, Tubbe I, Seidel D, Nicolaev A, Becker C, Neurath MF. Isolation and subsequent analysis of murine lamina propria mononuclear cells from colonic tissue. Nat Protoc. 2007;2:2307–2311. doi: 10.1038/nprot.2007.315. [DOI] [PubMed] [Google Scholar]
- 35.Read S, Powrie F. Induction of inflammatory bowel disease in immunodeficient mice by depletion of regulatory T cells. Curr Protoc Immunol Chapter. 2001;15(Unit 15):13. doi: 10.1002/0471142735.im1513s30. [DOI] [PubMed] [Google Scholar]
- 36.Kuo TC, Calame KL. B lymphocyte-induced maturation protein (Blimp)-1, IFN regulatory factor (IRF)-1, and IRF-2 can bind to the same regulatory sites. J Immunol. 2004;173:5556–5563. doi: 10.4049/jimmunol.173.9.5556. [DOI] [PubMed] [Google Scholar]
- 37.Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- 38.Chaudhry A, Rudra D, Treuting P, Samstein RM, Liang Y, Kas A, Rudensky AY. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326:986–991. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chaudhry A, Samstein RM, Treuting P, Liang Y, Pils MC, Heinrich JM, Jack RS, Wunderlich FT, Bruning JC, Muller W, Rudensky AY. Interleukin-10 signaling in regulatory T cells is required for suppression of Th17 cell-mediated inflammation. Immunity. 34:566–578. doi: 10.1016/j.immuni.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huber S, Gagliani N, Esplugues E, O’Connor W, Jr, Huber FJ, Chaudhry A, Kamanaka M, Kobayashi Y, Booth CJ, Rudensky AY, Roncarolo MG, Battaglia M, Flavell RA. Th17 cells express interleukin-10 receptor and are controlled by Foxp3 and Foxp3+ regulatory CD4+ T cells in an interleukin-10-dependent manner. Immunity. 34:554–565. doi: 10.1016/j.immuni.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cimmino L, Martins GA, Liao J, Magnusdottir E, Grunig G, Perez RK, Calame KL. Blimp-1 attenuates Th1 differentiation by repression of ifng, tbx21, and bcl6 gene expression. J Immunol. 2008;181:2338–2347. doi: 10.4049/jimmunol.181.4.2338. [DOI] [PubMed] [Google Scholar]
- 42.Gong D, Malek TR. Cytokine-dependent Blimp-1 expression in activated T cells inhibits IL-2 production. J Immunol. 2007;178:242–252. doi: 10.4049/jimmunol.178.1.242. [DOI] [PubMed] [Google Scholar]
- 43.Uhlig HH, Coombes J, Mottet C, Izcue A, Thompson C, Fanger A, Tannapfel A, Fontenot JD, Ramsdell F, Powrie F. Characterization of Foxp3+CD4+CD25+ and IL-10-secreting CD4+CD25+ T cells during cure of colitis. J Immunol. 2006;177:5852–5860. doi: 10.4049/jimmunol.177.9.5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murai MTO, Kim G, Madan R, Karp CL, Cheroutre H, Kronenberg M. Interleukin 10 acts on regulatory T cells to maintain expression of the transcription factor Foxp3 and suppressive function in mice with colitis. Nat Immunol. 2009;10:1178–1184. doi: 10.1038/ni.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang FMG, Strober W. Interactions among the transcription factors Runx1, RORgammat and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nat Immunol. 2008;11:1297–1306. doi: 10.1038/ni.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang XZY, Yang XO, Nurieva RI, Chang SH, Ojeda SS, Kang HS, Schluns KS, Gui J, Jetten AM, Dong C. Transcription of Il17 and Il17f is controlled by conserved noncoding sequence 2. Immunity. 2012;36:23–31. doi: 10.1016/j.immuni.2011.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gyory I, Wu J, Fejer G, Seto E, Wright KL. PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat Immunol. 2004;5:299–308. doi: 10.1038/ni1046. [DOI] [PubMed] [Google Scholar]
- 48.Lehnertz B, Northrop JP, Antignano F, Burrows K, Hadidi S, Mullaly SC, Rossi FM, Zaph C. Activating and inhibitory functions for the histone lysine methyltransferase G9a in T helper cell differentiation and function. J Exp Med. 207:915–922. doi: 10.1084/jem.20100363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ghoreschi K, Laurence A, Yang XP, Tato CM, McGeachy MJ, Konkel JE, Ramos HL, Wei L, Davidson TS, Bouladoux N, Grainger JR, Chen Q, Kanno Y, Watford WT, Sun HW, Eberl G, Shevach EM, Belkaid Y, Cua DJ, Chen W, O’Shea JJ. Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling. Nature. 467:967–971. doi: 10.1038/nature09447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Imielinski M, Baldassano RN, Griffiths A, Russell RK, Annese V, Dubinsky M, Kugathasan S, Bradfield JP, Walters TD, Sleiman P, Kim CE, Muise A, Wang K, Glessner JT, Saeed S, Zhang H, Frackelton EC, Hou C, Flory JH, Otieno G, Chiavacci RM, Grundmeier R, Castro M, Latiano A, Dallapiccola B, Stempak J, Abrams DJ, Taylor K, McGovern D, Silber G, Wrobel I, Quiros A, Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmuda MM, Bitton A, Dassopoulos T, Datta LW, Green T, Griffiths AM, Kistner EO, Murtha MT, Regueiro MD, Rotter JI, Schumm LP, Steinhart AH, Targan SR, Xavier RJ, Libioulle C, Sandor C, Lathrop M, Belaiche J, Dewit O, Gut I, Heath S, Laukens D, Mni M, Rutgeerts P, Van Gossum A, Zelenika D, Franchimont D, Hugot JP, de Vos M, Vermeire S, Louis E, Cardon LR, Anderson CA, Drummond H, Nimmo E, Ahmad T, Prescott NJ, Onnie CM, Fisher SA, Marchini J, Ghori J, Bumpstead S, Gwillam R, Tremelling M, Delukas P, Mansfield J, Jewell D, Satsangi J, Mathew CG, Parkes M, Georges M, Daly MJ, Heyman MB, Ferry GD, Kirschner B, Lee J, Essers J, Grand R, Stephens M, et al. Common variants at five new loci associated with early-onset inflammatory bowel disease. Nat Genet. 2009;41:1335–1340. doi: 10.1038/ng.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McGovern DP, Gardet A, Torkvist L, Goyette P, Essers J, Taylor KD, Neale BM, Ong RT, Lagace C, Li C, Green T, Stevens CR, Beauchamp C, Fleshner PR, Carlson M, D’Amato M, Halfvarson J, Hibberd ML, Lordal M, Padyukov L, Andriulli A, Colombo E, Latiano A, Palmieri O, Bernard EJ, Deslandres C, Hommes DW, de Jong DJ, Stokkers PC, Weersma RK, Sharma Y, Silverberg MS, Cho JH, Wu J, Roeder K, Brant SR, Schumm LP, Duerr RH, Dubinsky MC, Glazer NL, Haritunians T, Ippoliti A, Melmed GY, Siscovick DS, Vasiliauskas EA, Targan SR, Annese V, Wijmenga C, Pettersson S, Rotter JI, Xavier RJ, Daly MJ, Rioux JD, Seielstad M. Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat Genet. 42:332–337. doi: 10.1038/ng.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.