Abstract
We demonstrated previously that parathyroid hormone-related protein (PTHrP) 1-141 mRNA is the least stable of three isoforms and is the only isoform that is stabilized by TGF-β. In order to understand how PTHrP mRNA is stabilized by TGF-β, we first sought to elucidate the mechanism(s) that are responsible for the instability of PTHrP isoform 1-141 mRNA. The 3′-UTR of isoform 1-141 contains four AU-rich elements (AREs), which are known to mediate mRNA degradation. We utilized a luciferase reporter system to test whether these four AREs are responsible for the short half-life of PTHrP 1-141 mRNA. Our results demonstrated that ARE elements in the 3′-UTR of PTHrP 1-141 mRNA play a significant role in regulation of the stability of the mRNA. It is known that AREs mediate their effects on mRNA stability through a number of ARE-binding proteins that recruit the exosome, a complex of exonucleases that degrades the mRNA. We identified tristetraproline (TTP) as an RNA-binding protein that may be involved in ARE-mediated degradation of PTHrP 1-141 mRNA.
Keywords: Parathyroid hormone-related protein, mRNA stability, 3′-untranslated region, AU-rich elements, Tristetraproline
1. Introduction
PTHrP is a multifunctional protein that was originally identified as a major cause of humoral hypercalcemia of malignancy (HHM) (Mundy and Guise, 1997; Nissenson, 2000; Wysolmerski and Stewart, 1998). PTHrP is normally expressed in many human tissues, with its functions ranging from regulation of bone development to cell growth and differentiation (Nissenson, 2000; Wysolmerski and Stewart, 1998). However, overproduction of PTHrP in patients with breast, lung and other cancers can result in HHM which is characterized by increased blood calcium concentration due to increased bone resorption and reabsorption of calcium by the kidney. HHM results in increased patient morbidity due to muscular weakness and eventual renal failure (Mundy and Guise, 1997; Wysolmerski and Broadus, 1994). Elucidating the mechanisms of regulation of PTHrP will aid in developing novel treatments for HHM that control the production of PTHrP.
Alternative splicing of the 3′-end of PTHrP gene results in the synthesis of three distinct mRNA isoforms, which in turn are translated into three proteins that are 139, 141 and 173 amino acids long (Gillespie and Martin, 1994; Wysolmerski and Stewart, 1998) (Figure 1). The 1-173 mRNA isoform is stable with a half-life of more than 4 hours, while the 1-139 and 1-141 mRNA isoforms are relatively unstable with half-lives ranging from 30 to 90 minutes depending on cell type (Sellers et al., 2004). The mechanisms that determine the short half-lives of PTHrP 1-139 and 1-141 mRNA isoforms remain to be elucidated. It is known that short half-lives of mRNAs are often determined by AU-rich elements (AREs) present within 3′-untranslated regions (UTRs) (Chen et al., 1994; Chen and Shyu, 1995; Grzybowska et al., 2001). AREs mediate their mRNA-destabilizing effect through a number of proteins that bind to these elements and recruit the exosome, which is a complex of nucleases that degrades mRNA (Chen et al., 2001; Garneau et al., 2007; Mukherjee et al., 2002; van Hoof and Parker, 1999). The 3′-UTR of the 1-141 PTHrP isoform contains four such elements.
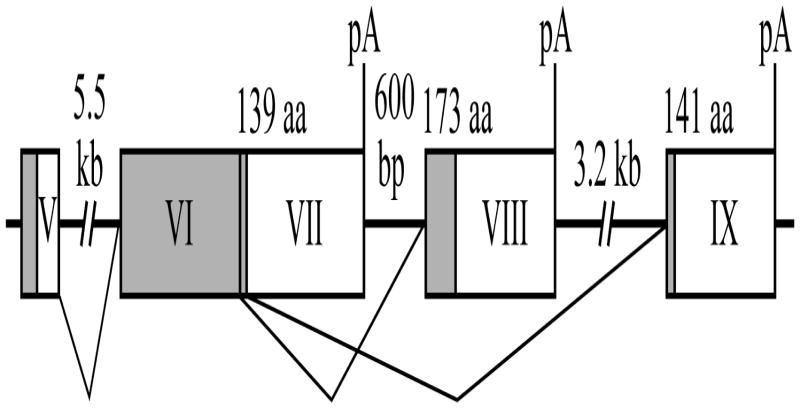
Fig. 1.
Alternative splicing of the 3′-end of the human PTHrP pre-mRNA. Exons 5 and 6 (coding exons) are alternatively spliced to exons 7, 8 or 9 at the 3′ end resulting in three mRNA variants that encode proteins of 139, 173 and 141 amino acids. Coding regions of mRNA are shaded. pA refers to the polyadenylation sites.
In this study, we examined the role of 3′-UTR AREs in the regulation of mRNA stability of the PTHrP 1-141 isoform. We demonstrated that the 1-141 isoform had reduced mRNA stability due to the presence of four functional AREs in its 3′-UTR. In addition, we showed that several proteins bind to the ARE elements in the 3′-UTR of the 1-141 isoform. The major binding protein has an electrophoretic mobility of 30-35 kDa, which is consistent with the molecular weight of tristetraprolin (TTP, 34 kDa), one of the best-known ARE-binding proteins (Blackshear, 2002; Hau et al., 2007; Lykke-Andersen and Wagner, 2005). Over-expression of exogenous TTP protein resulted in further decrease in the expression of the reporter gene containing the 3′-UTR of PTHrP 1-141 mRNA. We also showed direct binding of over-expressed TTP to AREs in the 3′-UTR of the 1-141 isoform. We concluded that TTP may play a role in ARE-mediated degradation of PTHrP 1-141 mRNA.
2. Materials and methods
2.1. HARA cell culture
Human lung squamous carcinoma cell line (HARA) was cultured as previously described (Sellers et al., 2004). The HARA cells express and secrete PTHrP and were kindly provided by Dr. Haruo Iguchi, Shikoku Cancer Center, Japan. The cell line was isolated from a patient with lung squamous cell carcinoma and established from a bone metastasis in a nude mouse.
2.2. Plasmid constructs
pGL3c-141UTR, pGL3c-141rUTR, pGL3c-ARE were constructed by subcloning the 3′UTR of PTHrP 1-141 mRNA in direct (pGL3c-141UTR) or reverse (pGL3c-141UTR) orientation or a smaller region (see Figure 2A) of the 1-141 3′-UTR containing the four ARE elements (pGL3c-141UTR) into the XbaI site of pGL3control luciferase reporter vector (Promega). 1-141 3′-UTR fragments for subcloning were generated by PCR using oligonucleotide primers listed in Table 1. pcDNA3FLAG and pcDNA3FLAG-TTP were kindly provided by Dr. Lukke-Andersen (University of Colorado).
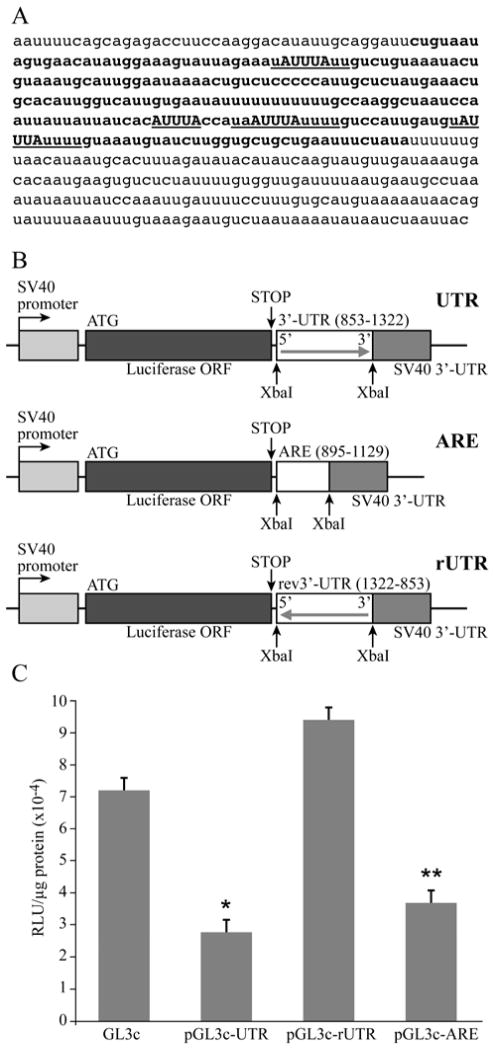
Fig. 2.
3′-UTR of the 1-141 isoform of PTHrP mRNA downregulated luciferase expression in HARA cells. A. Nucleotide sequence of the 3′-UTR of PTHrP 1-141 mRNA. AREs are underlined, core ARE sequences are capitalized. Small region used in transient transfections is indicated in bold face. B. Diagram of luciferase-3′-UTR constructs used in transient transfections. The 3′-UTR of PTHrP 1-141 mRNA in both orientations or the small region of the 3′-UTR containing AREs were subcloned into the XbaI site of pGL3 control vector between the luciferase open reading frame (ORF) and SV-40 3′-UTR sequence. C. Transient transfections of luciferase-3′-UTR reporter constructs into HARA cells. HARA cells were transfected with 2 μg of either pGL3control vector or pGL3control vector containing either the 1-141 isoform 3′-UTR in either direct (pGL3c-UTR) or reverse orientation (pGL3c-rUTR) or the smaller 3′-UTR region (pGL3c-ARE). Luciferase activity was assayed, normalized to protein concentration and plotted. pGL3c-UTR was significantly lower than pGL3c (*, p-value = 0.0002), pGL3c-ARE was significantly lower than pGL3c (**, p-value = 0.001) and pGL3c-ARE was not statistically different from pGL3c-UTR (p-value = 0.40).
Table 1.
Primers used for PCR-amplification of fragments 1-141 PTHrP mRNA isoform. XbaI restriction site sequence is underlined
| Construct | Forward primer, 5′ to 3′ | Reverse primer, 5′ to 3′ |
|---|---|---|
| pGL3c-141UTR, pGL3c-141rUTR | TGCTCTAGAAATTTTCAGCAGAGACCTTCC | TGCTCTAGAGTAATTAGATTATATTTTATTAGAC |
| pGL3c-ARE | TGCTCTAGACTGTAATAGTGAACA TATGG | TGCTCTAGAATATAGAAATTCAG CAGCACC |
2.3. Site-directed mutagenesis
Site-directed mutagenesis was performed using the Quick Change XL mutagenesis kit (Stratagene). Oligonucleotide primers and DNA templates for each mutagenesis reaction are listed in Table 2.
Table 2.
Oligonucleotides used for site-directed mutagenesis. Mismatched nucleotides in each primer are underlined
| Template Construct | Resulting construct | Forward primer, 5′ to 3′ | Reverse primer, 5′ to 3′ |
|---|---|---|---|
| pGL3c-141UTR | pGL3c-MutA1 | GTAATAGTGAACATATGGAAAGTATTAGAAATAGGCCTTGTCTGTAAATACTGTAAATGCATTGGAATAA | TTATTCCAATGCATTTACAGTATTTACAGACAAGGCCTATTTCTAATACTTTCCATATGTTCAC TATTAC |
| pGL3c-MutA1 | pGL3c-MutA12 | TTTTTTTTGCCAAGGCTAATCCAATTATTATTATCATCGCGACCATAATTTATTTTGTCCATTGATGTATTTATTTTGTA | TACAAAATAAATACATCAATGGACAAAATAAATTATGGTCGCGATGATAATAATAATTGGATTAGCCTTGGCAAAAAAAA |
| pGL3c-MutA12 | pGL3c-MutA123 | CTAATCCAATTATTATTATCATCGCGACCATAACGCGTTTTGTCCATTGATGTATTTATTTTGTAAATGTAT | ATACATTTACAAAATAAATACATCAATGGACAAAACGCGTTATGGTCGCGATGATAATAATAATTGGATTAG |
| pGL3c-MutA123 | pGL3c-MutA1234 | CATAACGCGTTTTGTCCATTGATGTGCGCATTTTGTAAATGTATCTTGGTGCTG | CAGCACCAAGATACATTTACAAAATGCGCACATCAATGGACAAAACGCGTTATG |
| pGL3c-141UTR | pGL3c-MutA4 | TTTACCATAATTTATTTTGTCCATTGATGTGCGCATTTTGTAAATGTATCTTGGTGCTGCTG | CAGCAGCACCAAGATACATTTACAAAATGCGCACATCAATGGACAAAATAAATTATGGTAAA |
| pGL3c-MutA4 | pGL3c-MutA34 | CTAATCCAATTATTATTATCACATTTACCATAACGCGTTTTGTCCATTGATGTGCGCATTTTGTAAAT | ATTTACAAAATGCGCACATCAATGGACAAAACGCGTTATGGTAAATGTGATAATAATAATTGG ATTAG |
| pGL3c-MutA34 | pGL3c-MutA234 | GCCAAGGCTAATCCAATTATTATTATCATCGCGACCATAACGCGTTTTGTCCATTGATGT | ACATCAATGGACAAAACGCGTTATGGTCGCGATGATAATAATAATTGGATTAGCCTTGGC |
2.4. Transient transfections of HARA cells
One confluent 10 cm plate of HARA cells was split into a 6-well plate the day before transfections. Cells were transfected using the Superfect Transfection Reagent (QIAgen). Cells were harvested using Reporter Lysis Buffer (Promega) 16–24 hours after transfections. Luciferase activity was assayed on a LumiCount luminometer (Packard) using the Luciferase Assay System (Promega). Protein concentration in cell lysates was measured on a UVmax Kinetic Multiplate Reader (Molecular Devices) using the Coomassie Protein Assay Reagent (Pierce).
2.5. Synthesis of radiolabeled RNA
32P-radiolabeled RNA for UV-cross-linking assay and RNA EMSA was synthesized using T7 Maxi-Script Kit (Ambion) and purified from a 5% polyacrylamide-8M urea gel according to manufacturer’s instructions. DNA templates for in vitro transcription reactions were synthesized by PCR using pGL3c-141UTR or pGL3c-141UTRmutA1234 as templates (Table 2) and the following oligonucleotide primers: 5′-TAATACGACTCACTATAGGGACTGTAATAGTGAACATATGG-3′ (forward) and 5′-ATATA GAAATTCAGCAGCACC-3′ (reverse).
2.6. RNA electrophoretic mobility shift assay (EMSA)
20 μg of HeLa S100 extract was incubated in 40 μg/ml tRNA, 0.2 U/ml RNAse inhibitor (Invitrogen) and 10% glycerol (final concentrations) for 10 min at 30°C. 32P-labeled RNA probe (200,000 cpm) was added and the reaction mix was incubated for 20 min at 4°C. Reactions were resolved on a 4% native polyacrylamide gel, pre-run for 30 min, at 4°C.
2.7. UV-cross-linking assay
The RNA-protein UV-cross-linking assay was performed as previously described by our laboratory (Sellers et al., 2004). Briefly, S100 extracts (10 mg) were combined with 200,000 c.p.m. of in vitro transcribed RNA, incubated on ice for 20 min, and cross-linked with U.V. light (0.25 J total energy, 254 nm wavelength, 8 cm from source, Stratalinker, Stratagene) in 0.75 ml Eppendorf tubes on ice. Samples were treated with 1 μg/μl RNase A (Sigma) at 37°C for 30 min, mixed with loading buffer, heated to 95°C for 3 min, and separated using a 12% SDS–PAGE gel. The gel was run at 4°C for 11 h at 30 mA.
2.8. Preparation of S100 extracts
S100 extracts from HARA cells were prepared as previously described by our laboratory (Sellers et al., 2004). S100 extracts from HeLa cells were purchased from Biovest (Worcester, MA). Briefly, cells were washed with ice-cold PBS, pelleted at 500 g at 4°C, re-suspended in homogenization buffer, and allowed to swell on ice for 20 min. Samples were homogenized and nuclei were pelleted at 12 000 g for 2 min at 4°C. Supernatants were transferred to an ultrafuge tube with 0.11 volumes of 10X extraction buffer and centrifuged at 100 000 g at 4°C for 1 h and the supernatant (S100) frozen in liquid nitrogen.
2.9. TTP RT-PCR
RNA for reverse transcription reaction was isolated from HARA cells using Trizol reagent (Invitrogen). The reverse transcription reaction was performed with 1 μg of total RNA isolated from HARA cells using the Superscript RT kit (Invitrogen) with oligo dT as the primer. 2 μl of the RT reaction were used as a template in the PCR reaction. TTP cDNA was amplified under the following cycling conditions: 94°C for 1 min, 55°C for 30 sec and 72°C for 1 min for 30 cycles using the following oligonucleotide primers: 5′-TGTCCTCCAGCTCCTTC-3′ (forward) and 5′-TGAGATCCAGCTGATCTGACC-3′ (reverse).
2.10. Data analysis
Generalized linear models were used to determine if transfection with the luciferase-3′-UTR reporter constructs had an effect on luciferase activity (Figs. 2 and 3). Adjustments for multiple comparisons were made using Tukey methods. Adjusted p-values are reported in the figure legends.
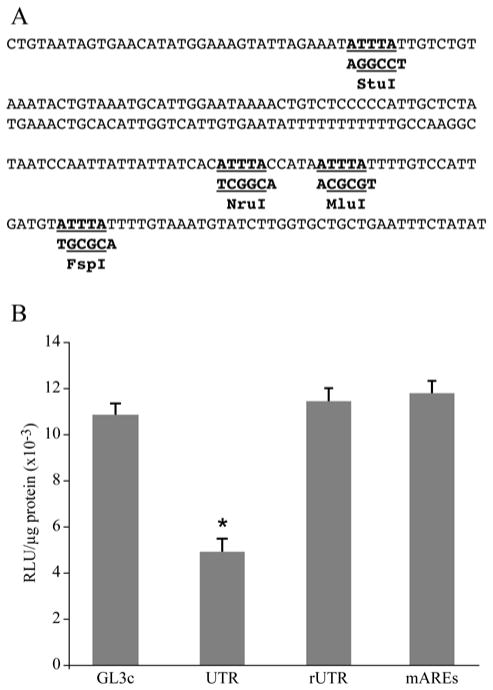
Fig. 3.
ARE elements were responsible for downregulation of luciferase activity by the 3′-UTR of the PTHrP 1-141 mRNA. A. Sequence of the 3′-UTR of PTHrP 1-141 mRNA containing four AREs. Core ARE sequences are underlined. The mutated nucleotides are underlined and the restriction sites generated are shown below the sequence. B. Transient transfections of HARA cells with mutated 3′-UTR-luciferase reporter construct. HARA cells were transfected with 2 μg of either empty pGL3control vector or pGL3vector containing either the wild-type 3′-UTR in direct (pGL3c-UTR) or reverse (pGL3c-rUTR) orientations or the 3′-UTR with mutated AREs (pGL3c-mutA1234). UTR was significantly lower than pGL3c (*, p-value = 0.0002), but both rUTR and mAREs were not significantly different from pGL3c (p-values = 0.84 and 0.59, respectively).
Experimental effects were taken into account, since two sets of experiments were carried out separately (Fig. 4). ANOVA models (using SAS PROC GLM procedure) with covariates ‘experiment’ and ‘treatment,’ as well as the interaction term between them were used to test paired difference of interests. Holm’s method was used to adjust for multiple comparisons for paired differences.
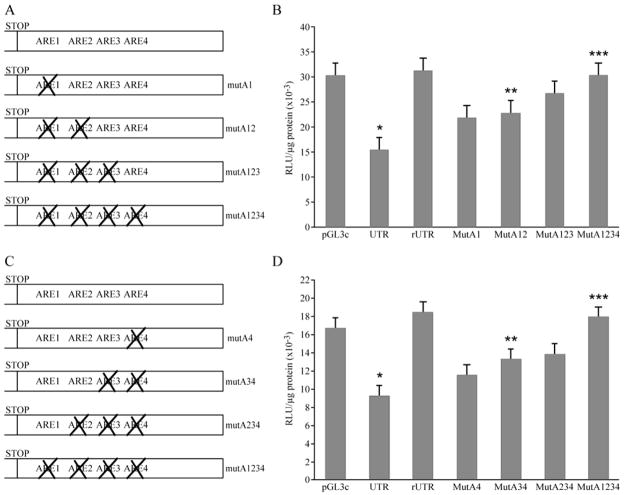
Fig. 4.
Mapping of the ARE elements responsible for the regulation of PTHrP 1-141 mRNA stability. A. Diagram of mutations introduced into the 3′-UTR to sequentially disrupt AREs in a 5′ to 3′direction. B. Transient transfections of HARA cells with mutated 3′-UTR-luciferase reporter constructs. HARA cells were transfected with 2 μg of either empty pGL3control vector or pGL3vector containing either the wild-type 3′-UTR in direct (pGL3c-UTR) or reverse (pGL3c-rUTR) orientations or the 3′-UTR with mutated AREs (pGL3c-MutA1, pGL3c-MutA12, pGL3c-MutA123 and pGL3c-MutA1234). After adjusting for multiple comparisons, UTR was significantly lower than GL3 (*, p-value < 0.0004), mutA12 was significantly greater than UTR (**, p-value = 0.009), mutA1234 was significantly greater than mutA12 (***, p-value = 0.0123) and mutA1234 was not significantly different from GL3 (p-value = 0.9958). C. Diagram of mutations introduced into 3′-UTR of 1-141 isoform to sequentially disrupt AREs in a 3′ to 5′ direction. D. Transient transfections of HARA cells with mutated 3′-UTR-luciferase reporter constructs. HARA cells were transfected with 2 μg of either empty pGL3control vector or pGL3vector containing either the wild-type 3′-UTR in direct (pGL3c-UTR) or reverse (pGL3c-rUTR) orientations or the 3′-UTR with mutated AREs (pGL3c-MutA4, pGL3c-MutA34, pGL3c-MutA234 and pGL3c-MutA1234). After adjusting for multiple comparisons, UTR was significantly lower than GL3 (*, p-value < 0.0004), mutA34 was significantly greater than UTR (**, p-value = 0.002), mutA1234 was significantly greater than mutA34 (***, p-value = 0.0006) and mutA1234 was not significantly different from GL3 (p-value = 0.29).
Generalized linear models were used to determine whether treatment (TTP or V) had any effect on counts within each of the two groups (UTR or MutA1234; Fig. 6). Experiment number was included in the model as a block effect.
Fig. 6.
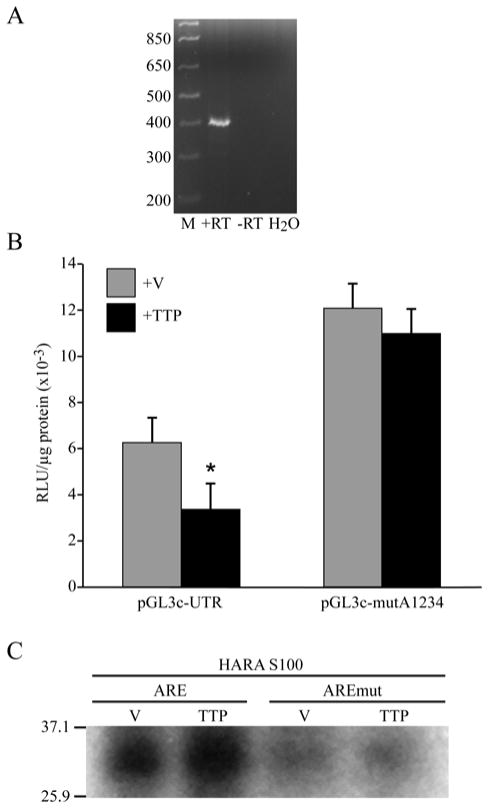
ARE-dependent downregulation of luciferase activity was mediated by TTP. A. TTP was expressed in HARA cells. RT-PCR was performed using RNA isolated from HARA cells (+RT). Reactions without addition of reverse transcriptase enzyme (−RT) or using water as a template (H2O) were used as negative controls. B. Overexpression of TTP reduced the activity of luciferase-3′-UTR reporter in HARA cells. HARA cells were transfected with 1 μg of pGL3control vector containing either the intact 3′-UTR (pGL3c-UTR) or 3′-UTR with four disrupted AREs (pGL3c-mutA1234) together with 1 μg of either pcDNA3FLAG (+V) or pcDNA3FLAG-TTP (+TTP). Within ‘UTR’group, mean counts in the ‘V’ treatment group were greater compared to those in ‘TTP’ treatment group (*, p-value = 0.0003). Within ‘MUT’group, mean counts in the ‘V’ treatment group were not significantly different from those in ‘TTP’ treatment group (p-value = 0.5543). C. Binding of TTP to ARE elements in the 3′-UTR of PTHrP 1-141 mRNA. 5 μg of S100 extract prepared from HARA cells transfected either with pcDNA3FLAG (V) or pcDNA3FLAG-TTP (TTP) were UV-crosslinked to a 32P-labeled RNA fragment of the 1-141 isoform 3′-UTR containing either four intact AU-rich elements (ARE) or four mutated elements (AREmut). Cross-linked products were separated on 10% SDS-PAGE gels and visualized by autoradiography.
3. Results
3.1. 3′-UTR of PTHrP 1-141 mRNA downregulated expression of the luciferase reporter
The PTHrP 1-141 mRNA is the least stable of the three PTHrP mRNA isoforms (Sellers et al., 2004). The mechanisms that determine the instability of this isoform remain to be elucidated. Unstable mRNAs often contain AU-rich elements (AREs) in their 3′-UTRs (Chen et al., 1994; Chen and Shyu, 1995; Grzybowska et al., 2001). AREs consist of AUUUA motifs imbedded in a larger U-rich context. The 3′-UTR of PTHrP 1-141 mRNA contains four AREs (Figure 2A). We investigated whether these elements contribute to the instability of the PTHrP 1-141 isoform by utilizing a luciferase reporter system to study the effects of various 3′UTR sequences on mRNA stability (Wang et al., 2006). The 3′UTR of PTHrP 1-141 mRNA, either in the direct or reverse orientation (to serve as a negative control), or a smaller region of the 3′-UTR containing all four ARE elements were sub-cloned into the 3′-UTR of luciferase cDNA in the pGL3-Control vector (Figure 2B). The resulting plasmid constructs were transiently transfected into HARA human squamous carcinoma cells. Our results demonstrated that insertion of the 3′-UTR of the 1-141 isoform in the direct orientation, as well as the smaller ARE-containing region of the 3′-UTR, resulted in decreased activity of luciferase reporter compared to the original pGL3-Control vector and the vector containing the 1-141 3′-UTR sub-cloned in the reverse orientation (Figure 2C). These results indicated that the 3′-UTR of the 1-141 isoform, and specifically the 235-nucleotide region containing the four AREs, negatively regulated mRNA expression, possibly by decreasing mRNA stability.
3.2. Four ARE elements in the 3′-UTR of PTHrP 1-141 mRNA were responsible for the downregulation of luciferase reporter expression
We demonstrated that a 235-nucleotide region of the 3′-UTR of PTHrP 1-141 mRNA containing four ARE elements decreased the expression of the luciferase reporter, possibly by decreasing the stability of luciferase mRNA. However, other sequences present within this region might be responsible for the observed effect. To confirm that the four ARE elements in the 3′-UTR of the PTHrP 1-141 mRNA were responsible for the observed effect, we disrupted all four AU-rich elements in the 3′-UTR by introducing point mutations (Figure 3A). HARA cells were transiently transfected with the resulting plasmid constructs. These results demonstrated that mutating all four AREs abolished the 3′-UTR-mediated downregulation of the luciferase reporter activity (Figure 3B). We concluded that the effects of the 3′-UTR of PTHrP 1-141 mRNA on the expression of the luciferase reporter can be attributed to the four AREs, likely through decreased mRNA stability. Therefore, the instability of PTHrP 1-141 mRNA was due to the presence of the AU-rich elements in its 3′-UTR.
3.3. The four ARE elements in the 3′-UTR contributed to the instability of PTHrP 1-141 mRNA
To determine which of the four AU-rich elements in the 3′-UTR of PTHrP 1-141 mRNA contributed to the regulation of mRNA stability, we constructed a series of luciferase reporter/3′-UTR plasmid constructs containing point mutations of AREs that sequentially disrupted one, two, three or all four of the ARE elements in a 5′ to 3′ direction (Figure 4A). We tested these constructs in the transient transfection assays with HARA cells. As observed previously, insertion of the 3′-UTR of PTHrP 1-141 mRNA into luciferase cDNA resulted in downregulation of luciferase activity when compared to the two control vectors. Sequential mutation of individual AREs showed a stepwise increase in luciferase activity, though the increases observed with each individual step did not reach statistical significance. However, when the first two AREs from the 5′end of the 3′-UTR were mutated, we observed a significant increase in luciferase activity compared to the intact 3′-UTR (Figure 4B). When the mutations in the third and the forth AREs were added, we observed a further significant increase in luciferase activity. We concluded that all 4 ARE elements (Figure 4A) likely contributed to the ARE-mediated regulation of PTHrP 1-141 mRNA stability.
To confirm our observation, we created another series of point mutations, this time mutating the AREs sequentially in a 3′ to 5′ direction (Figure 4C). Again, sequential mutation of individual AREs showed a stepwise increase in luciferase activity that did not reach statistical significance. However, when both AREs 3 and 4 were mutated, we observed a significant increase in the reporter activity compared to the intact 3′-UTR (Figure 4D). Mutation of the last 2 elements (AREs 1 and 2) resulted in a further significant increase in reporter activity (Figure 4D). These results confirmed our conclusion that all 4 ARE elements were important for the regulation of PTHrP 1-141 mRNA stability.
3.4. Protein binding to the AU-rich elements in the 3′-UTR of PTHrP 1-141 isoform
We demonstrated that the AU-rich elements in the 3′-UTR of PTHrP 1-141 mRNA regulate mRNA expression, most likely through decreased mRNA stability. AU-rich elements are known to regulate mRNA stability by recruiting ARE-binding proteins, such as tristetraprolin (TTP) or A+U-Rich Element Binding Factor 1 (AUF1), which in turn recruit the exosome, a complex of exonucleases that degrades the mRNA (Blackshear, 2002; Chen et al., 2001; Garneau et al., 2007; Guhaniyogi and Brewer, 2001; Hau et al., 2007; Lapucci et al., 2002; Lykke-Andersen and Wagner, 2005; Mukherjee et al., 2002; van Hoof and Parker, 1999). Therefore, to elucidate the mechanism(s) of ARE-mediated regulation of PTHrP 1-141 mRNA stability, we examined the proteins that bind to the AREs in the PTHrP 3′-UTR.
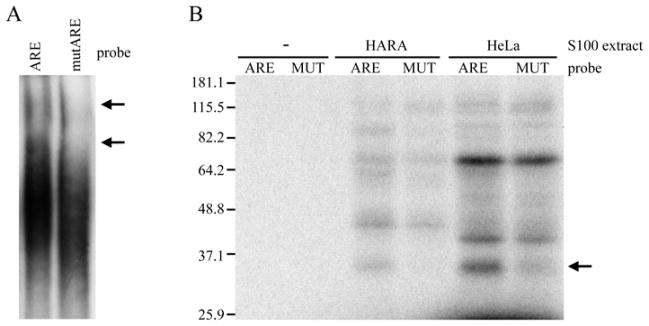
First, we performed an RNA electrophoretic mobility shift assay (EMSA). Addition of a 32P-labeled 235-nucleotide RNA fragment of the PTHrP 1-141 3′-UTR containing all four ARE elements to a HeLa S100 cytoplasmic extract resulted in a mobility shift of the probe (Figure 5A, left lane). However, when the probe with the mutated AREs (Figure 3A) was tested there was a decrease in the amount of shifted probe (Figure 5A, right lane). This indicated that binding of the protein complex to the PTHrP 1-141 3′-UTR depended on the presence of intact AREs. The protein complex is likely to consist of multiple proteins bound to AU-rich elements both directly and indirectly. EMSA assays do not reveal how many proteins bind to RNA directly, nor their individual molecular weights. To answer these questions, we performed an RNA-protein UV cross-linking assay. This assay demonstrated, in both HARA and HeLa S100 cytoplasmic extracts, the binding of a protein with the mobility of approximately 30–35 kDa to the 32P-labeled 235-nucleotide RNA fragment of the PTHrP 1-141 3′-UTR containing the AU-rich elements (Figure 5B). When the probe with 4 mutated AREs was used, the binding of this protein was significantly reduced (Figure 5B). We concluded that this protein specifically binds to the AREs in the PTHrP 1-141 3′-UTR and plays a role in the ARE-mediated regulation of PTHrP 1-141 mRNA stability.
Fig. 5.
Binding of proteins to AREs within the 3′-UTR of PTHrP 1-141 mRNA. A. EMSA using ARE-containing region of the 3′-UTR and S100 extract from HeLa cells. 20 μg of HeLa S100 cytoplasmic extract were incubated with a 32P-labeled RNA fragment of the 1-141 isoform 3′-UTR containing either four intact AU-rich elements (ARE) or four mutated elements (mutARE). Protein-RNA complexes were separated on 4% native polyacrylamide gels and visualized by autoradiography. Mobility shifts are indicated by arrows. B. UV-cross-linking of HeLa and HARA S100 extracts to ARE-containing region of the PTHrP 1-141 mRNA 3′-UTR. 20 g of S100 extract from HeLa cells (HeLa) or 5 g of S100 extract from HARA cells (HARA) were incubated with a 32P-labeled RNA fragment of the 1-141 isoform 3′-UTR containing either four intact AU-rich elements (ARE) or four mutated elements (MUT). Cross-linked products were separated on 10% SDS-PAGE gels and visualized by autoradiography. Labeled RNA incubated with reaction buffer alone (−) was used as a negative control. The ARE-dependent binding of the 30-35 kDa protein is indicated by an arrow.
3.5. ARE-binding protein TTP regulated expression of the 3′-UTR PTHrP 1-141 luciferase reporter
We demonstrated that a protein with a molecular weight of 30-35 kDa directly binds to AU-rich elements in the 3′-UTR of PTHrP 1-141 mRNA. This protein may play a role in the regulation of ARE-mediated decay of PTHrP 1-141 mRNA. Next, we determined the identity of the 30-35 kDa PTHrP 1-141 mRNA binding protein.
A number of proteins that bind to ARE elements and recruit the exosome have been described (Chen et al., 2001; Mukherjee et al., 2002; van Hoof and Parker, 1999). These proteins include AUF1, TTP, BRF1 and 2, KSRP, and HuR (Blackshear, 2002; Brennan and Steitz, 2001; Guhaniyogi and Brewer, 2001; Hau et al., 2007; Lapucci et al., 2002; Lykke-Andersen and Wagner, 2005; Stoecklin et al., 2002). All of these proteins promote degradation of mRNA, with the exception of HuR, which stabilizes mRNA (Brennan and Steitz, 2001; Lopez de Silanes et al., 2005; Srikantan and Gorospe, 2012). The ARE-binding protein TTP has a molecular weight of 34 kDa, which fits best with the mobility of the ARE-binding protein that we detected in our UV cross-linking assay. In order to determine whether TTP had any effect on PTHrP 1-141 mRNA stability in our model system, we first determined whether TTP was expressed in HARA cells. Indeed, using RT-PCR, we detected expression of TTP mRNA in this cell line (Figure 6A). Quantitative real-time RT-PCR demonstrated that the TTP mRNA was expressed at 12.2 copies/1,000 copies of β2-microglubulin mRNA. Next, we transiently transfected a TTP expression vector in combination with a luciferase reporter construct containing either the intact 3′-UTR of PTHrP 141 mRNA or with mutated AREs (Figure 3A). Co-transfection of the TTP expression vector with the PTHrP 1-141 3′-UTR-luciferase construct containing intact AREs resulted in a reduction in luciferase reporter activity when compared to the empty expression vector (Figure 6B), indicating that endogeneous levels of TTP are not saturating and TTP can be over-expressed. However, co-transfection of the TTP expression vector with the PTHrP 1-141 3′-UTR- luciferase reporter construct containing mutated AREs had no effect on reporter activity (Figure 6B). We concluded that TTP downregulated the expression of the luciferase reporter containing the 3′-UTR of PTHrP 1-141 mRNA and did so by activating ARE-mediated decay through binding to AREs in the 3′-UTR. Thus, we concluded that TTP may play a role in the ARE-mediated decay of PTHrP 1-141 mRNA by binding to these elements and recruiting the exosome that degrades the mRNA.
To further verify that TTP mediated ARE-dependent degradation of PTHrP 1-141 mRNA, we transiently transfected HARA cells with the TTP mammalian expression vector or with an empty vector as a control. S100 cytoplasmic extracts were prepared from transfected cells and analyzed using an RNA-protein UV cross-linking assay (Figure 6C). The intensity of the previously observed 30-35 kDa band increased in S100 extracts prepared from cells transfected with the TTP expression vector when compared to the empty vector. No significant protein binding in the 30-35 kDa range was observed when the RNA probe with mutated AREs was used. Therefore, we concluded that TTP is at least one of the proteins that plays a role in ARE-mediated degradation of PTHrP 1-141 mRNA.
4. Discussion
Alternative splicing of the 3′-end of PTHrP pre-mRNA results in the production of three mRNA isoforms, namely 1-141, 1-173 and 1-139 (Wysolmerski and Stewart, 1998). These isoforms display different half-lives, with isoform 1-173 being the most stable (half-life >4 hrs) and isoform 1-141 being the least stable (half life=30–45 min) (Sellers et al., 2004). In this study, we proposed to identify the mechanism(s) that determines the short half-life of PTHrP 1-141 mRNA.
The rapid turnover rate of mRNAs is often determined by the presence of AU-rich elements (AREs) in the 3′-UTR (Chen et al., 1994; Chen and Shyu, 1995; Grzybowska et al., 2001). mRNAs that possess short half-lives, such as transcripts encoding immediately early genes, often contain multiple AREs in their 3′-UTRs (Chen et al., 1994; Chen and Shyu, 1995; Grzybowska et al., 2001). Similar to 3′-untranslated regions of immediately-early genes, the 3′-UTR of PTHrP isoform 1-141 contains four AU-rich elements.
We demonstrated that inserting either the complete 3′-UTR of PTHrP 1-141 mRNA or a smaller 235-nucleotide region of the 3′-UTR containing all four ARE elements into the 3′-UTR of luciferase cDNA resulted in 2–3 fold decrease in luciferase activity when compared to control vectors (Figure 1). We also demonstrated that mutating all four AU-rich elements in the 3′-UTR of PTHrP 1-141 isoform completely abolished the effect of the 3′-UTR on luciferase activity. It is evident, therefore, that AREs in the 3′-UTR of PTHrP 1-141 mRNA mediated the decreased luciferase reporter activity.
In order to study the role of AREs in the 3′-UTR of 1-141 PTHrP mRNA, we utilized a luciferase reporter system that has been widely used to study such processes as transcription (Alam and Cook, 1990), translation (Harding et al., 2000) or mRNA stability (Wang et al., 2006). Protein expression is regulated on multiple levels and the decrease or increase in reporter activity might be attributed to effects on transcription, translation and/or mRNA stability. AU-rich elements sometimes affect gene expression at the level of translation, but most act through mRNA stability, resulting in increased degradation of ARE-containing mRNAs (Barreau et al., 2005; Chen et al., 1994; Chen and Shyu, 1995; Grzybowska et al., 2001). The effects of the AU-rich elements in the 3′-UTR of 1-141 isoform on luciferase activity may be mediated by TTP, which is involved in ARE-mediated degradation of mRNA (Anderson et al., 2004; Ross et al., 2012). Therefore, we concluded that ARE-mediated degradation is one of the mechanisms that determine the short half-life of PTHrP 1-141 mRNA.
Degradation of ARE-containing mRNAs is mediated by a number of ARE-binding proteins that bind to AU-rich elements and recruit the exosome, a complex of nucleases that degrades mRNA (Blackshear, 2002; Chen et al., 2001; Hau et al., 2007; Lykke-Andersen and Wagner, 2005; Mukherjee et al., 2002; van Hoof and Parker, 1999). We identified TTP, one of the known ARE-binding proteins, as a potential player of the ARE-mediated degradation of PTHrP 1-141 mRNA. First, in RNA-protein UV-cross linking experiments, we established that the major protein that binds specifically to the AU-rich elements in the 3′-UTR of PTHrP 1-141 mRNA had a mobility of 30-35 kDa (Figure 5B), which corresponds best to the mobility of TTP among all known ARE-binding proteins (Blackshear, 2002). Second, we demonstrated that overexpression of TTP further reduced the activity of the luciferase-PTHrP 3′-UTR reporter in an ARE-dependent manner (Figure 6B). Third, transfection of a TTP expression vector into HARA cells resulted in increased cross-linking of the 30-35 kDa protein to the PTHrP 3′-UTR when compared to cells transfected with an empty vector (Figure 6C). Therefore, we concluded that TTP may play an important role in the ARE-dependent degradation of PTHrP 1-141 mRNA, though we cannot rule out the possibility that additional ARE-binding proteins in the 30-35 kDa range are also involved.
In addition to the 35 kDa binding protein, proteins of approximately 60 and 80 kDa bound specifically to the AREs in HARA, but not HeLa, S100 extracts (Figure 5B). This is consistent with an earlier study showing that several proteins in HARA S100 extracts cross-linked to a smaller probe (probe no. 2) containing two-thirds of the ARE sequence, including proteins of approximately of 30-50 kDa, 60 and 80 kDa (Sellers et al., 2004). Perhaps these tissue-specific factors complement or modify the binding of the 35 kDa protein and play a role in the ARE-mediated regulation of PTHrP 1-141 mRNA stability.
It has been previously demonstrated that the half-life of the 1-141 mRNA of PTHrP is increased by treatment of various cell lines with such growth factors as TGF-β or EGF (Cho et al., 2004; Heath et al., 1995; Sellers et al., 2002). TGF-β and EGF are known to be produced by a number of malignant tumors (Kim et al., 2003; Yu and Stamenkovic, 2004). Upregulation of PTHrP expression by TGF-β and EGF on the level of both transcription and mRNA stability is thought to be the major factor contributing to overproduction of PTHrP in such pathological conditions as breast and prostate cancer (Wysolmerski and Stewart, 1998). One of the potential ways in which TGF-β and EGF may increase the stability of PTHrP 1-141 mRNA is by suppressing the ARE-mediated degradation of PTHrP 1-141 mRNA. It is possible that treatment of cells with TGF-β or EGF may result in an increased expression and/or increased binding of HuR, one of the few ARE-binding proteins whose activity is known to stabilize mRNAs (Brennan and Steitz, 2001; Lopez de Silanes et al., 2005; Srikantan and Gorospe, 2012), to AREs in the 3′-UTR of the PTHrP 1-141 mRNA, allowing HuR to compete with TTP for binding to the AREs and increase the stability of PTHrP 1-141 mRNA. This scenario is supported by recent experiments showing that PTHrP and TGF-β mRNAs are both stabilized by HuR in a von Hippel-Lindau (VHL) tumor suppressor gene-dependent manner in renal cell carcinoma (Danilin et al., 2009). Future experiments will determine whether TGF-β or EGF changes expression of TTP and/or HuR or the ability of TTP and/or HuR to bind to the AREs in the 3′-UTR of PTHrP 1-141 mRNA in HARA cells.
In summary, we demonstrated that the instability of PTHrP 1-141 mRNA is determined, at least in part, by the four AREs in its 3′-UTR. Effects of these AREs are potentially mediated by the TTP ARE-binding protein. It is possible that sequences in addition to the AU-rich elements in the 3′-UTR, such as instability elements in the coding region, are involved in determining the instability of the PTHrP 1-141 mRNA. It is also possible that other sequences are involved in the stabilization of PTHrP 1-141 mRNA by TGF-β and EGF, such as regulation through other regions of the 3′-UTR or through regulatory sequences in the coding region. The current findings significantly advance our understanding of the mechanisms that regulate the production of PTHrP in normal and cancerous tissues.
The instability of PTHrP 1-141 mRNA is determined by four AREs in its 3′-UTR.
Effects of these AREs are mediated by the tristetraproline (TTP) ARE-binding protein.
Additional proteins (60-80 kDa) bind specifically to the AREs in HARA cell extracts.
Acknowledgments
We would like to thank Dr. Lukke-Andersen (University of Colorado) for providing us with pcDNA3FLAG and pcDNA3FLAG-TTP vectors and Cristobal Valdebenito for performing the TTP mRNA real-time RT-PCR. We would like to thank Tim Vojt for assistance in making the figures. This work was supported by National Cancer Institute grants CA100730 and CA77911.
The abbreviations used are
- ARE
AU-rich element
- PTHrP
Parathyroid Hormone-Related Protein
- 3′-UTR
3′-Untranslated Region
- TGF-β
Transforming Growth Factor-beta
- HHM
Humoral Hypercalcemia of Malignancy
- TTP
Tristetraproline
- EMSA
Electrophoretic Mobility Shift Assay
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Alam J, Cook JL. Reporter genes: application to the study of mammalian gene transcription. Anal Biochem. 1990;188:245–254. doi: 10.1016/0003-2697(90)90601-5. [DOI] [PubMed] [Google Scholar]
- Anderson P, Phillips K, Stoecklin G, Kedersha N. Post-transcriptional regulation of proinflammatory proteins. J Leukoc Biol. 2004;76:42–47. doi: 10.1189/jlb.1103536. [DOI] [PubMed] [Google Scholar]
- Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackshear PJ. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem Soc Trans. 2002;30:945–952. doi: 10.1042/bst0300945. [DOI] [PubMed] [Google Scholar]
- Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Chen TM, Shyu AB. Interplay of two functionally and structurally distinct domains of the c-fos AU-rich element specifies its mRNA-destabilizing function. Mol Cell Biol. 1994;14:416–426. doi: 10.1128/mcb.14.1.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- Cho YM, Lewis DA, Koltz PF, Richard V, Gocken TA, Rosol TJ, Konger RL, Spandau DF, Foley J. Regulation of parathyroid hormone-related protein gene expression by epidermal growth factor-family ligands in primary human keratinocytes. J Endocrinol. 2004;181:179–190. doi: 10.1677/joe.0.1810179. [DOI] [PubMed] [Google Scholar]
- Danilin S, Sourbier C, Thomas L, Rothhut S, Lindner V, Helwig JJ, Jacqmin D, Lang H, Massfelder T. von Hippel-Lindau tumor suppressor gene-dependent mRNA stabilization of the survival factor parathyroid hormone-related protein in human renal cell carcinoma by the RNA-binding protein HuR. Carcinogenesis. 2009;30:387–396. doi: 10.1093/carcin/bgn275. [DOI] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- Gillespie MT, Martin TJ. The parathyroid hormone-related protein gene and its expression. Mol Cell Endocrinol. 1994;100:143–147. doi: 10.1016/0303-7207(94)90293-3. [DOI] [PubMed] [Google Scholar]
- Grzybowska EA, Wilczynska A, Siedlecki JA. Regulatory functions of 3′UTRs. Biochem Biophys Res Commun. 2001;288:291–295. doi: 10.1006/bbrc.2001.5738. [DOI] [PubMed] [Google Scholar]
- Guhaniyogi J, Brewer G. Regulation of mRNA stability in mammalian cells. Gene. 2001;265:11–23. doi: 10.1016/s0378-1119(01)00350-x. [DOI] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- Hau HH, Walsh RJ, Ogilvie RL, Williams DA, Reilly CS, Bohjanen PR. Tristetraprolin recruits functional mRNA decay complexes to ARE sequences. J Cell Biochem. 2007;100:1477–1492. doi: 10.1002/jcb.21130. [DOI] [PubMed] [Google Scholar]
- Heath JK, Southby J, Fukumoto S, O’Keeffe LM, Martin TJ, Gillespie MT. Epidermal growth factor-stimulated parathyroid hormone-related protein expression involves increased gene transcription and mRNA stability. Biochem J. 1995;307(Pt 1):159–167. doi: 10.1042/bj3070159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Uehara H, Karashima T, Shepherd DL, Killion JJ, Fidler IJ. Blockade of epidermal growth factor receptor signaling in tumor cells and tumor-associated endothelial cells for therapy of androgen-independent human prostate cancer growing in the bone of nude mice. Clin Cancer Res. 2003;9:1200–1210. [PubMed] [Google Scholar]
- Lapucci A, Donnini M, Papucci L, Witort E, Tempestini A, Bevilacqua A, Nicolin A, Brewer G, Schiavone N, Capaccioli S. AUF1 Is a bcl-2 A + U-rich element-binding protein involved in bcl-2 mRNA destabilization during apoptosis. J Biol Chem. 2002;277:16139–16146. doi: 10.1074/jbc.M201377200. [DOI] [PubMed] [Google Scholar]
- Lopez de Silanes I, Lal A, Gorospe M. HuR: post-transcriptional paths to malignancy. RNA Biol. 2005;2:11–13. doi: 10.4161/rna.2.1.1552. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 2005;19:351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee D, Gao M, O’Connor JP, Raijmakers R, Pruijn G, Lutz CS, Wilusz J. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 2002;21:165–174. doi: 10.1093/emboj/21.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundy GR, Guise TA. Hypercalcemia of malignancy. Am J Med. 1997;103:134–145. doi: 10.1016/s0002-9343(97)80047-2. [DOI] [PubMed] [Google Scholar]
- Nissenson RA. Parathyroid hormone-related protein. Rev Endocr Metab Disord. 2000;1:343–352. doi: 10.1023/a:1026578720736. [DOI] [PubMed] [Google Scholar]
- Ross CR, Brennan-Laun SE, Wilson GM. Tristetraprolin: Roles in cancer and senescence. Ageing Res Rev. 2012 doi: 10.1016/j.arr.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers RS, Capen CC, Rosol TJ. Messenger RNA stability of parathyroid hormone-related protein regulated by transforming growth factor-beta1. Mol Cell Endocrinol. 2002;188:37–46. doi: 10.1016/s0303-7207(01)00752-3. [DOI] [PubMed] [Google Scholar]
- Sellers RS, Luchin AI, Richard V, Brena RM, Lima D, Rosol TJ. Alternative splicing of parathyroid hormone-related protein mRNA: expression and stability. J Mol Endocrinol. 2004;33:227–241. doi: 10.1677/jme.0.0330227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikantan S, Gorospe M. HuR function in disease. Front Biosci. 2012;17:189–205. doi: 10.2741/3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G, Colombi M, Raineri I, Leuenberger S, Mallaun M, Schmidlin M, Gross B, Lu M, Kitamura T, Moroni C. Functional cloning of BRF1, a regulator of ARE-dependent mRNA turnover. EMBO J. 2002;21:4709–4718. doi: 10.1093/emboj/cdf444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Parker R. The exosome: a proteasome for RNA? Cell. 1999;99:347–350. doi: 10.1016/s0092-8674(00)81520-2. [DOI] [PubMed] [Google Scholar]
- Wang J, Pitarque M, Ingelman-Sundberg M. 3′-UTR polymorphism in the human CYP2A6 gene affects mRNA stability and enzyme expression. Biochem Biophys Res Commun. 2006;340:491–497. doi: 10.1016/j.bbrc.2005.12.035. [DOI] [PubMed] [Google Scholar]
- Wysolmerski JJ, Broadus AE. Hypercalcemia of malignancy: the central role of parathyroid hormone-related protein. Annu Rev Med. 1994;45:189–200. doi: 10.1146/annurev.med.45.1.189. [DOI] [PubMed] [Google Scholar]
- Wysolmerski JJ, Stewart AF. The physiology of parathyroid hormone-related protein: an emerging role as a developmental factor. Annu Rev Physiol. 1998;60:431–460. doi: 10.1146/annurev.physiol.60.1.431. [DOI] [PubMed] [Google Scholar]
- Yu Q, Stamenkovic I. Transforming growth factor-beta facilitates breast carcinoma metastasis by promoting tumor cell survival. Clin Exp Metastasis. 2004;21:235–242. doi: 10.1023/b:clin.0000037705.25256.d3. [DOI] [PubMed] [Google Scholar]