Abstract
Background
To date, 39 SNPs have been associated with blood pressure (BP) or hypertension (HTN) in genome-wide association studies (GWAS) in Caucasians. Our hypothesis is that the loci/SNPs associated with BP/HTN are also associated with BP response to antihypertensive drugs.
Methods and Results
We assessed the association of these loci with BP response to atenolol or hydrochlorothiazide monotherapy in 768 hypertensive participants in the Pharmacogenomics Responses of Antihypertensive Responses (PEAR) study. Linear regression analysis was performed in Caucasians for each SNP in an additive model adjusting for baseline BP, age, gender and principal components for ancestry. Genetic scores were constructed to include SNPs with nominal associations and empirical p values were determined by permutation test. Genotypes of 37 loci were obtained from Illumina 50K cardiovascular or Omni1M GWAS chips. In Caucasians, no SNPs reached Bonferroni-corrected alpha of 0.0014, six reached nominal significance (p<0.05) and 3 were associated with atenolol BP response at p < 0.01. The genetic score of the atenolol BP lowering alleles was associated with response to atenolol (p =3.3*10−6 for SBP; p=1.6*10−6 for DBP). The genetic score of the HCTZ BP lowering alleles was associated with response to HCTZ (p = 0.0006 for SBP; p = 0.0003 for DBP). Both risk score p values were < 0.01 based on the empirical distribution from the permutation test.
Conclusions
These findings suggest selected signals from hypertension GWAS may predict BP response to atenolol and HCTZ when assessed through risk scoring.
Keywords: beta-blocker, diuretics, hypertension, pharmacogenetics, polymorphisms blood pressure
Introduction
Hypertension (HTN) places over 76 million Americans (and approximately one billion individuals worldwide) at substantially increased risk for stroke, coronary heart disease (including heart attack), renal failure, and heart failure, and is the most common chronic disease for which medications are prescribed.1 Five drug classes are considered appropriate first line therapy for HTN and are often defined by their effect on the renin angiotensin system (RAS; β-blockers, ACE inhibitors and angiotensin receptor blockers) or sodium and calcium (thiazide diuretics and calcium channel blockers). 2 Response rates to monotherapy with any given antihypertensive drug are only about 50%,3 and those who fail to respond to one category (e.g. those affecting RAS) class are more likely to respond to a drug with an alternate mechanism (~75% response rate).4, 5 Selection of the initial antihypertensive therapy is essentially by trial- and-error. The difficulty in determining the most appropriate antihypertensive drug for a specific patient likely contributes to the fact that less than half of the hypertensive patients in the U.S. (and worldwide) currently have their blood pressure (BP) controlled.6 Pharmacogenomics offers the clinical promise of individualization of therapy based on a person’s genetic make-up.
To date, 39 single nucleotide polymorphisms (SNPs) have been associated with blood pressure (BP) or hypertension in genome-wide association studies (GWAS) and meta-analyses in Caucasians.7-11 Our hypothesis is that the loci/SNPs associated with BP/HTN are also associated with BP response to antihypertensive drugs. To test this hypothesis, we assessed in the Pharmacogenomic Evaluation of Antihypertensive Responses (PEAR) study the association of these loci with BP response to two first-line antihypertensives with contrasting pharmacological mechanisms: atenolol (a cardioselective β-blocker) and hydrochlorothiazide (HCTZ, a thiazide diuretic).
Methods
Participants
The Pharmacogenomic Evaluation of Antihypertensive Responses (PEAR) study (clincaltrials.gov identifier NCT00246519) (n=768) was a randomized multi-center clinical trial examining the role of genetic variability on BP response to HCTZ and/or atenolol. 12 Males and females of any race between the ages of 17 and 65 with essential hypertension (clinic diastolic BP ≥ 90 mmHg, ≤ 110 mmHg) were recruited to participate at University of Florida (Gainesville, FL), Mayo Clinic (Rochester, MN), and Emory University (Atlanta, GA). The institutional review boards at each institution approved the protocol and each participant provided informed written consent prior to entry into the study. Those currently treated had their antihypertensive drug therapy tapered (as necessary) and discontinued. The mean washout period was 28 days. This washout period was deemed to sufficiently balance risks of non-treatment with a sufficient period to allow return to untreated BP because post-washout BP in previously treated participants were nearly identical to baseline BP of those who entered the trial untreated.
Included participants were randomized to receive either HCTZ 12.5 mg daily or atenolol 50 mg daily for three weeks, followed by dose titration to 25 mg and 100 mg daily, respectively for SBP > 120 and DBP > 70. After nine weeks, BP response to monotherapy was assessed. The second study drug (alternate drug) was added in those patients with SBP > 120 mmHg or DBP > 70 mmHg (> 90% for both randomization arms), with dose titration after 3 weeks and response assessment after six more weeks, as in the first portion of the study. Participants were not given sodium restrictions, but were counseled to maintain consistent dietary intakes.
Blood pressure Phenotype
The primary BP phenotype for PEAR was home BP measured with an automated device (MicroLife 3AC1-PC, Minneapolis MN) provided to each participant. The device was set to measure BP in triplicate with each activation and to store the average systolic and diastolic BPs and the time of each set of measurements. Participants were required to take readings daily in the seated position in triplicate upon rising and before retiring at least five of seven days prior to the three visits: baseline (prior to randomization), assessment visit after monotherapy and assessment visit after add-on therapy. All BP readings since the previous study visit were downloaded to the computer by study coordinators. Home BP response to monotherapy was the primary response phenotype because it exhibits less variability and is superior to office BP in prediction of prognosis.13-15
Genotyping and QC steps
Thirty-nine SNPs/loci were identified from the BP GWAS studies and meta-analyses that have been published up to October 20117-11 as signals that reached genome-wide significance of p<5*10−8. In PEAR participants, genotypes of 37 SNPs (exact SNPs for 26 loci and proxies for 11 loci) were obtained from Illumina Human 50K cardiovascular Beadchip or Omni1M quad GWAS Beadchip. Two SNPs (rs4590817 and rs319690) were not covered on either platform (Table S1).
Illumina Human CVD 50K array
The HumanCVD50k SNP array 16 was a customized gene-centric array including ~2100 genes and ~50,000 SNPs genotyped using the Infinium II Assay (Illumina, San Diego, CA). Genotypes were called using GenomeStudio software version 2011.1 and the Genotyping Module version 1.9 calling algorithm (Illumina, San Diego, CA).
Participants were excluded if sample genotype call rates were below 95% and SNPs were excluded if genotype call rates were below 90%. The genotype data were not reclustered after QC filters, but the genotype and sample call rate was recalculated after QC. Sample contamination was detected by checking gender mismatches using X chromosome genotype data and cryptic relatedness was estimated by pairwise identity-by-descent (IBD) analysis implemented using PLINK (http://pngu.mgh.harvard.edu/purcell/plink/). After the QC procedures, the total SNP call rate in the remaining individuals was 99.799%. Hardy-Weinberg equilibrium was assessed with chi-square test if one degree of freedom.
Illumina Omni1M quad GWAS chip
Human Omni1M quad GWAS chip also used Infinium II assay and genotypes were called using BeadStudio software and GenTrain2 calling algorithm. The same QC procedures were carried out for the GWAS genotype data. After the QC procedures, the total SNP call rate in the remaining individuals was 99.519%. Principal component analysis was performed to assess the ancestral background. Participant’s self-identified race information was confirmed with principal component analysis for genetic ancestry and 464 participants were confirmed as European descendants and 304 as African descendants. Seven hundred and fifty nine participants were included in this analysis, including 461 Caucasians and 298 African Americans. Nine participants were excluded due to genotype QC issues or missing genotypes.
Statistical Methods
The primary analysis was performed in Caucasians since the GWAS signals selected for this analysis arose from studies in Caucasians. Associations of these SNPs with SBP and DBP responses to atenolol or HCTZ monotherapy were evaluated using linear regression, adjusting for baseline BP, age, gender and the first 2 principal components for ancestry which correspond to European and African ancestry respectively. Previous analysis 17 of BP response in PEAR established that age, gender and baseline BP were significant demographic predictors of BP response, while common covariates such as BMI and smoking were not associated with BP response and therefore not included as covariates. P values of <0.0014 (0.05/37) were considered statistically significant. Additive mode of inheritance was assumed where the SNPs were coded as 0, 1 and 2 in the linear regression model. For the SNPs with p<0.05 in Caucasians, the association analysis of the index SNPs and the surrounding 100 kb region was also performed in African American participants to assess if an association with the same direction was present in participants with a different ancestral background. Particular attention was also given to SNPs with association in the opposite direction for HCTZ and atenolol, since the counter balancing mechanisms targeted by these agents lead to good responders to one of the drugs being poor responders to the other.18 For the SNPs that reached nominal significance, genetic scores were constructed by summing the alleles with the best BP response for inclusion in regression models (using the BP lowering allele as the coded allele). A weighted score method was also assessed based on the coefficient of BP response for each SNP. The participants with any missing genotypes would not get a score, therefore were not included in the score analysis. Since the BP response alleles are different for atenolol and HCTZ, separate genetic scores were constructed for atenolol and HCTZ monotherapy. To validate the association of the scores with the BP responses, we generated the distribution of the regression test statistics empirically by repetitively permutating the BP responses. The relationship between the BP responses and the scores was randomly shuffled 1000 times, and a regression model was fit for each permutation. The raw p values of the scores predicting BP response of the raw data was then compared to the empirical distribution of 1000 p values from the permutations. Analysis were performed using PLINK 19 assuming an additive mode of inheritance. The genetic score analysis and permutation test were performed in SAS version 9.3 (Cary, NC).
Results
The PEAR participants were Caucasian (60%) and African American (40%) hypertensive individuals, 53% were females. Participants had a mean age of 48.8 ± 9.2 years, and most were overweight or obese. The baseline characteristics of the 461 Caucasian and 298 African American participants assigned to atenolol and hydrochlorothiazide treatment are presented in Table 1.
Table 1.
Baseline characteristics of PEAR participants.
| Characteristics* | Caucasians (n=461) |
African Americans (n=298) | ||
|---|---|---|---|---|
| Atenolol (n = 233) |
HCTZ (n = 228) |
Atenolol (n = 150) |
HCTZ (n =148) |
|
| Age (mean ± SD) | 49.5 ± 9.5 | 50.0 ± 9.5 | 47.2 ± 8.5 | 47.4 ± 8.8 |
| Female gender (n, %) | 109 (46.8%) | 91 (40.0%) | 107 (71.3%) | 92 (62.2%) |
| BMI (kg/m2) | 30.3 ± 5.5 | 30.3 ± 4.9 | 31.6 ± 6.3 | 31.5 ± 5.4 |
| Duration of hypertension (years) |
7.0 ± 7.2 | 6.0 ± 7.1 | 6.4 ± 6.8 | 7.0 ± 7.7 |
| Taking at least 1 antihypertensive drug at entry |
180 (77.3%) | 164 (71.9%) | 105 (70.0%) | 110 (74.3%) |
| Smoking status | ||||
| Current smoker | 26 (11.2%) | 31 (13.6%) | 23 (15.3%) | 29 (19.6%) |
| Ex-smoker | 65 (27.9%) | 65 (28.5%) | 29 (19.3%) | 20 (13.5%) |
| Ever smoker | 91 (39.1%) | 96 (42.1%) | 52 (34.1%) | 49 (33.1%) |
| Baseline mean home-recorded BP | ||||
| Systolic (mmHg) | 145.0 ± 9.4 | 146.0 ± 10.0 | 145.1 ± 10.5 | 147.1 ± 11.5 |
| Diastolic (mmHg) | 92.8 ± 5.5 | 93.6 ± 5.5 | 93.9 ± 6.5 | 95.1 ± 6.6 |
Numeric characteristics were presented as mean± standard deviation and categorical variables were presented as number and percentages. Abbreviation: HCTZ: hydrochlorothiazide; BMI: body mass index. BP: blood pressure
After an average of 9 weeks of atenolol monotherapy, the mean blood pressure response was −11.0 ± 9.4/−9.9 ± 6.4 mmHg in Caucasians and −3.1± 10.6/−4.2 ± 6.7 mmHg in African American participants. Mean blood pressure response after HCTZ monotherapy was −7.7 ± 8.1/−4.3 ± 5.3 mmHg in Caucasians and −12.0 ± 9.7/7.1 ± 6.5 mmHg in African Americans. The distributions of BP response in Figure S1 demonstrate the large inter-individual variability in BP response to atenolol and HCTZ in both race groups.
Genotypes for 37 out of the 39 BP/HTN SNPs were available for PEAR participants from the two genotyping platforms. No SNPs deviated from Hardy-Weinberg equilibrium in Caucasians or African Americans.
When the association of these 37 SNPs with SBP and DBP response to atenolol or HCTZ monotherapy was assessed, none of the SNPs reached a priori alpha of 0.0014, although six SNPs achieved nominal significance (p<0.05). The associations of these SNPs with monotherapy BP response in Caucasians are summarized in Table 2. The most interesting was an intergenic SNP rs1458038 near FGF5, which was associated with BP response to atenolol and HCTZ monotherapy, with genotype effects in the opposite direction (Figure 1). Participants with TT and TC genotypes of this common SNP responded better to atenolol than those with CC genotype (with SBP/DBP response of −12.9/−11.6 mmHg, −12.3/−10.6mmHg and −8.9/−8.5 mmHg for TT, TC and CC respectively, p =0.0067 for SBP, p = 0.018 for DBP), while the direction of association was the opposite for response to HCTZ, with −5.4/−2.6mmHg, −7.5/−4.3mmHg, −8.7/−4.8mmHg for TT, TC and CC respectively (p = 0.039 for SBP, p = 0.069 for DBP).
Table 2.
SNPs associated with BP response to atenolol or HCTZ monotherapy in Caucasians.
| SNP | Nearest Gene |
CHR | Position* | Alleles¶ | N | MAF | BP lowering allele§ |
Atenolol |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DBP |
SBP |
||||||||||||
| P | Beta | SE | P | Beta | SE | ||||||||
| rs1458038 | FGF5 | 4 | 81,383,747 | T/C | 233 | 0.37 | T | 0.02 | −1.42 | 0.60 | 0.0067 | −2.48 | 0.91 |
| rs871606 | CHIC2 | 4 | 54,494,002 | C/T | 233 | 0.10 | T | 0.0037 | −2.57 | 0.87 | 0.017 | −3.23 | 1.34 |
| rs2932538 | MOV10 | 1 | 113,018,066 | C/T | 233 | 0.24 | T | 0.005 | −1.84 | 0.66 | 0.06 | −1.89 | 1.01 |
| rs1799945 | HFE | 6 | 26,199,158 | G/C | 233 | 0.12 | G | 0.15 | −1.36 | 0.95 | 0.02 | −3.38 | 1.44 |
| SNP | Nearest Gene |
CHR | Position* | Alleles¶ | N | MAF | BP lowering allele§ |
HCTZ |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DBP |
SBP |
||||||||||||
| P | Beta | SE | P | Beta | SE | ||||||||
| rs1458038 | FGF5 | 4 | 81,383,747 | T/C | 228 | 0.37 | C | 0.07 | −0.95 | 0.52 | 0.039 | −1.55 | 0.75 |
| rs3184504 | SH2B3 | 12 | 110,368,991 | C/T | 228 | 0.49 | C | 0.013 | −1.28 | 0.51 | 0.02 | −1.73 | 0.74 |
| rs4551053 | |||||||||||||
| (proxy for rs11953630) |
EBF1 | 5 | 157,771,120 | A/G | 228 | 0.34 | G | 0.02 | −1.28 | 0.55 | 0.05 | −1.56 | 0.79 |
CHR: chromosome. MAF: minor allele frequency. HCTZ: hydrochlorothiazide. SBP: systolic blood pressure response in mmHg. DBP: diastolic blood pressure response in mmHg. P values were linear regression p adjusted for baseline BP, age, gender, principal components for ancestry. P values <0.05 were highlighted in bold. Beta indicates the blood pressure response (in mm Hg) for each BP lowering allele.
NCBI build 36 position.
Alleles were presented as major/minor alleles.
coded allele. Note: There are no missing genotypes for these six SNPs in the patients included in this table.
Figure 1.
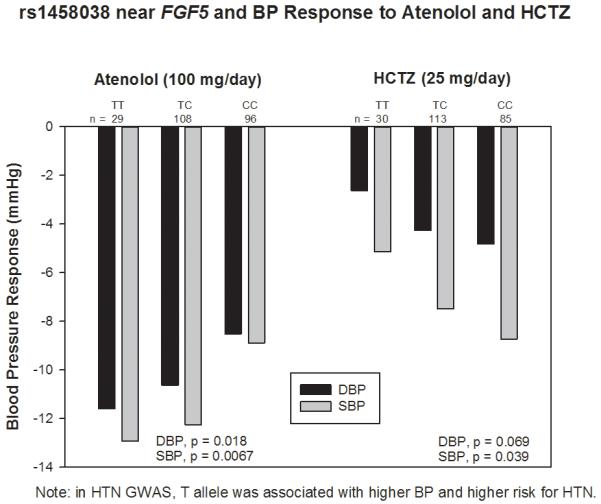
Hypertension SNP rs1458038 near FGF5 associated with BP response to atenolol or hydrochlorothiazide monotherapy among Caucasian hypertensive patients.
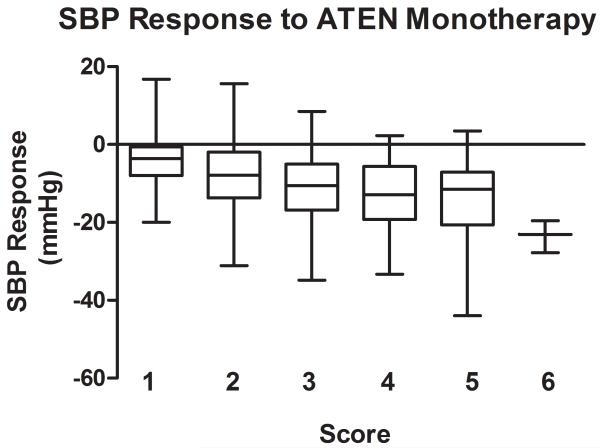
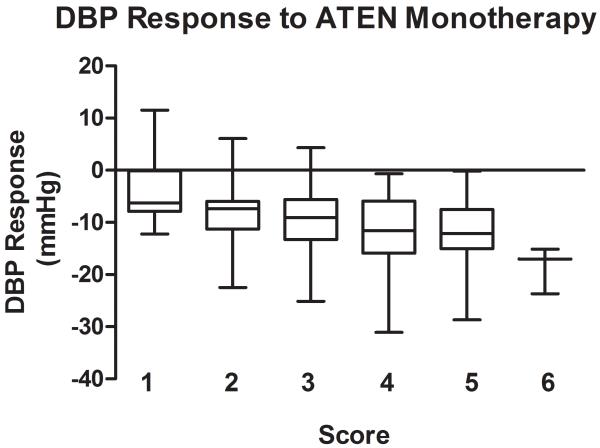
The genetic score comprised of the 4 BP lowering alleles in response to atenolol monotherapy showed stronger association, with p = 3.3*10−6 for SBP response and p=1.6*10−6 for DBP response. The raw p values are smaller than the minimum p values from the 1000 permutations (Table S2) and thus the significance was validated through the permutation tests. The BP responses to atenolol monotherapy by genetic score are shown in Figure 2 (A for SBP response, B for DBP response). In Caucasian hypertensive patients with only one of the 4 BP lowering alleles, the average BP response to atenolol monotherapy was −4.0/−3.3 mmHg, while the average BP response was −18.6/−23.5 mmHg in those with six BP lowering alleles. This genetic score explained 8.5% of SBP response and 8.2% of DBP response to atenolol monotherapy, respectively.
Figure 2.
Blood pressure response by genetic score of blood pressure lowering alleles in response to ATEN monotherapy. Blood pressure responses (mmHg) are presented in box plots where the whiskers indicating the minimum and maximum values. A. SBP; B. DBP
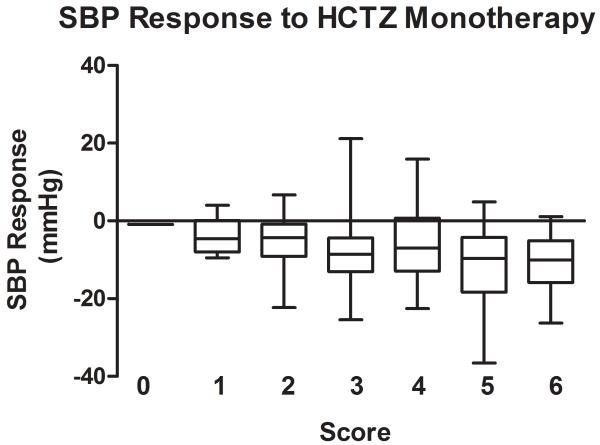
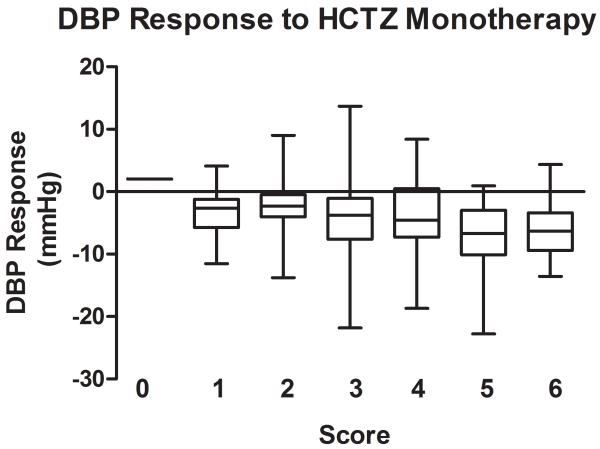
The association of the genetic score comprised of the 3 BP lowering alleles for HCTZ monotherapy was also stronger than those for the individual SNPs, with p of 0.0006 for SBP response and p of 0.0003 for DBP response to HCTZ monotherapy. The raw p values are < 0.01 based on the empirical distribution of the permutation test. The BP responses to HCTZ monotherapy by genetic score are shown in Figure 3 (A for SBP responses and B for DBP responses). This genetic score explained 4.3% for SBP response and 5.3% for DBP response to HCTZ monotherapy. The weighted scores yielded almost identical results and due to the simplicity of the unweighted scores, this was considered the preferred model.
Figure 3.
Blood pressure response by genetic score of blood pressure lowering alleles in response to HCTZ monotherapy. Blood pressure responses (mmHg) are presented in box plots where the whiskers indicating the minimum and maximum values. A. SBP; B. DBP
Associations of these six SNPs and the SNPs in the same region with the BP response in African Americans were also evaluated. None of the Caucasian index SNPs achieved nominal significance in African Americans with a directional effect the same as in Caucasians. rs871606 near CHIC2 was marginally associated with DBP response to atenolol monotherapy in African Americans (DBP, p = 0.06, beta=1.89), with the same direction of effect as in Caucasians. One SNP had a directionally opposite effect association in African Americans as in Caucasians: the C allele of rs3184504 in SH2B3 (nonsynonymous, T>C, Trp > Arg) was associated with better BP reduction in Caucasians in response to HCTZ (show in Table 2). However in African Americans treated with HCTZ, the C allele of this SNP is associated with a slight increase in blood pressure (DBP, p = 0.047, beta=0.21).
When the 100kb regions flanking the index SNPs were evaluated in African Americans, two different SNPs (rs12069113, rs12076902) near MOV10 were found to be nominally associated with BP response to atenolol (DBP, p=0.042 and 0.02, respectively. Figure S2). These two SNP were in high LD with each other (r2 = 0.78 and 1 in Caucasians and African Americans, respectively) but in very low LD with the Caucasian index SNP, rs2932538, in both populations (r2 = 0.01 and 0.02 in Caucasians and African Americans, respectively). No stronger signals were found in the African Americans in the regions flanking the Caucasian index SNPs. There was also no evidence that the genetic scores for either atenolol or HCTZ BP lowering alleles in Caucasians are associated with BP response in African Americans, p = 0.49 for SBP and p = 0.49 for DBP in response to HCTZ and p = 0.59 for SBP and p = 0.42 for DBP in response to atenolol monotherapy, respectively.
Discussion
Among 37 SNPs previously documented to influence BP, none achieved a priori alpha values for association with BP response to atenolol or HCTZ. However six SNPs were nominally associated with BP response in Caucasians. Two of these SNPs were also marginally associated with response in African Americans for the same drug, one with the same direction of effect and the other with the opposite direction of effect. Lack of association of these index SNPs in African Americans is not surprising due to different LD structures between these two populations.
Of interest is that gene score analyses for both atenolol and HCTZ BP lowering alleles revealed that the scores were more strongly associated with the BP response than the individual SNPs. This is not surprising for a phenotype with multiple genetic contributors, as is expected for the antihypertensive response. The percentage of variance of BP response explained by these genetic scores ranged from 4.3 ~ 8.5%, which are larger effect sizes than the genetic risk scores constructed to predict the untreated BP in the HTN GWAS studies (around 116 independent loci collectively explain ~ 2.2% of the phenotypic variance for SBP and DBP).7 This is consistent with numerous studies indicating that pharmacogenetic effect sizes are larger than disease genetic effect sizes. These data also provide insight into how genetic risk scores might be built to guide selection of an antihypertensive drug based on genetic information. In order to make sure the associations of these SNPs were not due to multicollinearity with the baseline BP, we also tested the association without adjusting for baseline BP and found that the results were essentially identical. In addition, we also tested the association of these SNPs with baseline BP and not surprisingly, given our small sample size, and limited BP range (DBP 90-110 mmHg) there was not a significant association with baseline BP. This indicates that the associations presented are not likely to be due to collinearity with the baseline BP.
The finding of SNP rs1458038 near FGF5 is perhaps most intriguing single SNP finding since it was associated with BP responses to both ATEN and HCTZ, but with the effects in opposite directions. This represents a form of replication since the HCTZ-treated participants were an independent group, and the opposite direction of effect is consistent with our expectations on genetic associations for the two drugs. The effect sizes of BP response for this SNP ranged from 1.0 to 2.5 mmHg per allele (double that for variant homozygotes). Differences of 2 to 3 mmHg SBP reduction have been shown to reduce stroke risk by 20 ~ 30% in a meta-analysis of nine randomized controlled trials of antihypertensive drugs,20 suggesting the associations noted here would be considered clinically significant.
In a HTN GWAS study,7 the T allele of rs1458038 near FGF5 was associated with higher BP and higher risk for HTN in Caucasian individuals. This SNP was also strongly associated with BP in East Asians and South Asians and only marginally in African Americans. Our study indicated that Caucasian hypertensive individuals with the risk allele for HTN (T) might respond better to atenolol, than to HCTZ. FGF5 encodes fibroblast growth factor 5, which is involved in a variety of biological processes, including embryonic development, cell growth, morphogenesis, tissue repair, tumor growth and invasion. It is not immediately clear how this gene is involved in BP regulation, but the association with atenolol responsiveness suggests this gene might be a marker for high sympathetic nervous system and renin-angiotensin activity and unraveling the function of this gene may provide additional insights.
Conclusions
We tested 37 SNPs/loci previously associated with BP in GWAS for association with antihypertensive response. Our results showed that the genetic loci associated with BP and/or HTN are not strongly associated with the BP response to commonly prescribed antihypertensives individually, with the most compelling data for a single SNP being one near FGF5. However, collectively a selection of these loci may contribute moderately to the BP response to the antihypertensives. This highlights the potential for building response scores for clinical prediction of response.
Supplementary Material
Acknowledgments
We acknowledge and thank the valuable contributions of the PEAR study participants, support staff, and study physicians: Drs. R. Whit Curry, Karen Hall, Frederic Rabari-Oskoui, Dan Rubin, and Siegfried Schmidt.
Funding Sources: PEAR was supported by the National Institute of Health Pharmacogenetics Research Network grant U01-GM074492 and the National Center for Advancing Translational Sciences under the award number UL1 TR000064 (University of Florida); UL1 TR000454 (Emory University) and UL1 TR000135 (Mayo Clinic) and funds from the Mayo Foundation. This research was also supported by K23 HL091120 (A.L.B.) and K23 HL086558 (R.M.C-D).
Footnotes
Conflict of Interest Disclosures: YG, CWM, ZY, WH, TYL, KRB declares no conflict of interest. RMC, ALB, ABC, JGG, EB, STT and JAJ received funding from NIH. RMC also received funding from Women’s Health Initiative. JGG received funding from Janssen Pharmaceuticals, Inc., has Speaker’s Bureau appointment from Boehringer-Ingelheim and is a consultant for Forest Pharmaceuticals and Boehringer-Ingelheim. EB received honoraria from the Foundation of Rome.
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Roger VL, Go AS, Lloyd-Jones DM, Adams RJ, Berry JD, Brown TM, et al. Heart disease and stroke statistics--2011 update: A report from the american heart association. Circulation. 2011;123:e18–e209. doi: 10.1161/CIR.0b013e3182009701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chobanian AV, Bakris GL, Black HR, Cushman WC, Green LA, Izzo JL, Jr., et al. The seventh report of the joint national committee on prevention, detection, evaluation, and treatment of high blood pressure: The jnc 7 report. JAMA. 2003;289:2560–2572. doi: 10.1001/jama.289.19.2560. [DOI] [PubMed] [Google Scholar]
- 3.Materson BJ, Reda DJ, Cushman WC, Massie BM, Freis ED, Kochar MS, et al. Single-drug therapy for hypertension in men. A comparison of six antihypertensive agents with placebo. The department of veterans affairs cooperative study group on antihypertensive agents. N Engl J Med. 1993;328:914–921. doi: 10.1056/NEJM199304013281303. [DOI] [PubMed] [Google Scholar]
- 4.Materson BJ, Reda DJ, Preston RA, Cushman WC, Massie BM, Freis ED, et al. Response to a second single antihypertensive agent used as monotherapy for hypertension after failure of the initial drug. Department of veterans affairs cooperative study group on antihypertensive agents. Arch Intern Med. 1995;155:1757–1762. [PubMed] [Google Scholar]
- 5.Dickerson JE, Hingorani AD, Ashby MJ, Palmer CR, Brown MJ. Optimisation of antihypertensive treatment by crossover rotation of four major classes. Lancet. 1999;353:2008–2013. doi: 10.1016/s0140-6736(98)07614-4. [DOI] [PubMed] [Google Scholar]
- 6.Roger VL, Go AS, Lloyd-Jones DM, Benjamin EJ, Berry JD, Borden WB, et al. Heart disease and stroke statistics--2012 update: A report from the american heart association. Circulation. 2012;125:e2–e220. doi: 10.1161/CIR.0b013e31823ac046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson AD, Newton-Cheh C, Chasman DI, Ehret GB, Johnson T, Rose L, et al. Association of hypertension drug target genes with blood pressure and hypertension in 86,588 individuals. Hypertension. 2011;57:903–910. doi: 10.1161/HYPERTENSIONAHA.110.158667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wain LV, Verwoert GC, O’Reilly PF, Shi G, Johnson T, Johnson AD, et al. Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nat Genet. 2011;43:1005–1011. doi: 10.1038/ng.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson JA, Boerwinkle E, Zineh I, Chapman AB, Bailey K, Cooper-DeHoff RM, et al. Pharmacogenomics of antihypertensive drugs: Rationale and design of the pharmacogenomic evaluation of antihypertensive responses (pear) study. Am Heart J. 2009;157:442–449. doi: 10.1016/j.ahj.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ragot S, Genès N, Vaur L, Herpin D. Comparison of three blood pressure measurement methods for the evaluation of two antihypertensive drugs: Feasibility, agreement, and reproducibility of blood pressure response. Am J Hypertens. 2000;13:632–639. doi: 10.1016/s0895-7061(99)00258-7. [DOI] [PubMed] [Google Scholar]
- 14.Turner ST, Schwartz GL, Chapman AB, Beitelshees AL, Gums JG, Cooper-Dehoff RM, et al. Power to identify a genetic predictor of antihypertensive drug response using different methods to measure blood pressure response. J Transl Med. 2012;10:47. doi: 10.1186/1479-5876-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stergiou GS, Baibas NM, Gantzarou AP, Skeva II, Kalkana CB, Roussias LG, et al. Reproducibility of home, ambulatory, and clinic blood pressure: Implications for the design of trials for the assessment of antihypertensive drug efficacy. Am J Hypertens. 2002;15:101–104. doi: 10.1016/s0895-7061(01)02324-x. [DOI] [PubMed] [Google Scholar]
- 16.Keating BJ, Tischfield S, Murray SS, Bhangale T, Price TS, Glessner JT, et al. Concept, design and implementation of a cardiovascular gene-centric 50 k snp array for large-scale genomic association studies. PLoS One. 2008;3:e3583. doi: 10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turner ST, Schwartz GL, Chapman AB, Beitelshees AL, Gums JG, Cooper-DeHoff RM, et al. Plasma renin activity predicts blood pressure responses to beta-blocker and thiazide diuretic as monotherapy and add-on therapy for hypertension. Am J Hypertens. 2010;23:1014–1022. doi: 10.1038/ajh.2010.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laragh JH, Lamport B, Sealey J, Alderman MH. Diagnosis ex juvantibus. Individual response patterns to drugs reveal hypertension mechanisms and simplify treatment. Hypertension. 1988;12:223–226. doi: 10.1161/01.hyp.12.3.223. [DOI] [PubMed] [Google Scholar]
- 19.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. Plink: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Staessen JA, Wang JG, Thijs L. Cardiovascular protection and blood pressure reduction: A meta-analysis. Lancet. 2001;358:1305–1315. doi: 10.1016/S0140-6736(01)06411-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.