Abstract
The study of angiogenesis is important to understanding a variety of human pathologies including cancer, cardiovascular and inflammatory diseases. In vivo angiogenesis assays can be costly and time-consuming, limiting their application in high-throughput studies. While traditional in vitro assays may overcome these limitations, they lack the ability to accurately recapitulate the main elements of the tissue microenvironment found in vivo, thereby limiting our ability to draw physiologically relevant biological conclusions. To bridge the gap between in vivo and in vitro angiogenesis assays, several microfluidic methods have been developed to generate in vitro assays that incorporate blood vessel models with physiologically relevant three-dimensional (3D) lumen structures. However, these models have not seen widespread adoption, which can be partially attributed to the difficulty in fabricating these structures. Here, we present a simple, accessible method that takes advantage of basic fluidic principles to create 3D lumens with circular cross-sectional geometries through ECM hydrogels that are lined with endothelial monolayers to mimic the structure of blood vessels in vitro. This technique can be used to pattern endothelial cell-lined lumens in different microchannel geometries, enabling increased flexibility for a variety of studies. We demonstrate the implementation and application of this technique to the study of angiogenesis in a physiologically relevant in vitro setting.
1. Introduction
Angiogenesis, the neovascularization of blood vessels from preexisting vasculature, is an important biological process involved in normal growth and development, as well as in various human pathologies including cancer, cardiovascular diseases, and inflammatory disorders. In cancer specifically, angiogenesis is necessary for tumors to grow beyond a critical size of a few millimeters. Without new vessel formation and proper blood supply, tumor cells too distant (> ~200 µm) from existing vessels would lack the supply of oxygen and nutrients essential for cell survival and proliferation [1]. Because of the importance of angiogenesis in tumor growth, metastasis, and overall cancer progression, therapeutic strategies have been developed around the concept of inhibiting angiogenesis with drugs and other angiostatic agents to restrict blood supply to the tumor. This is an area in drug discovery that continues to undergo intense study [2, 3]. The ability to study angiogenesis and investigate the effects of various factors on angiogenic responses is thus critical for furthering our understanding of the mechanisms of cancer development, as well as for the development of new and effective therapies.
Current angiogenesis assays span a wide range of methods that include in vivo preparations, organ cultures, and in vitro assays [4]. While in vivo methods such as the popular cranial window and dorsal skin chamber preparations have been instrumental in providing deep insights into the angiogenic process, these preparations are time-consuming, labor-intensive, expensive, and require significant skill in surgery, and thus are unsuitable as routine assays for widespread adoption or for high-throughput testing. Organ cultures such as the aortic ring and chick aortic arch assays are simpler preparations than in vivo methods, and maintain important elements of the complex tissue microenvironment, but tissue isolation, culture, and explant outgrowth of aortas can be time-consuming, challenging to do repeatedly and consistently, and difficult to scale up [5]. Thus, for high-throughput applications such as screening of large drug compound libraries and combinatorial testing of cellular and extracellular factors, a more suitable approach is to employ in vitro assays that rely on simple, accessible cell cultures and readily available substrates, and circumvent laborious lab procedures that involve the handling of animals and tissue explants. However, an often-cited major shortcoming of current in vitro assays is their inability to accurately recapitulate the main elements of the tissue microenvironment found in vivo, and this issue limits our ability to draw accurate biological conclusions. Therefore, an urgent need exists for the development of improved in vitro angiogenesis assays that can continue to offer high-throughput capacity and simple convenient operation while significantly enhancing the physiological relevance of the in vitro tissue microenvironment.
Recently, microfluidics technology has been applied to improve the spatiotemporal control of the cell or tissue microenvironment [6], thereby enabling the development of new and useful cell-based assays for biological research [7–10]. In particular, microfluidics has provided significant advances in new models of vascular function [11–15], with several notable designs specifically tailored to study the processes associated with angiogenesis [16–17]. While these methods have demonstrated the potential benefits of working at the microscale, and clearly offer more sophistication for studying angiogenesis, there remain significant challenges related to the fabrication and maintenance of blood vessel mimics that closely resemble three-dimensional vessel lumens in vivo. Furthermore, it is unclear whether these systems can be easily scalable for applications requiring increased throughput.
Here, we present a simple, accessible method that takes advantage of basic fluidic principles to create three-dimensional (3D) lumens with circular cross-sectional geometries lined with patent 3D endothelial monolayers applied to study angiogenesis in physiologically relevant in vitro microenvironments. The method requires only the use of a micropipette, and has the potential to be scaled into large arrays interfaced with automated liquid handlers as previously shown [18–20]. The method also allows incorporation of other cell types for organ-like cocultures that mimic the structure and organization of blood vessel lumens in vivo. We describe the development and application of 3D endothelial-lined lumens (ELL) with proper barrier function, biological response to VEGF gradients in the form of angiogenic sprouting, and further demonstrate the flexibility of the method for generating vessel networks on demand, without the need for re-designing molds from scratch.
2. Materials and Methods
2.1 Device Fabrication
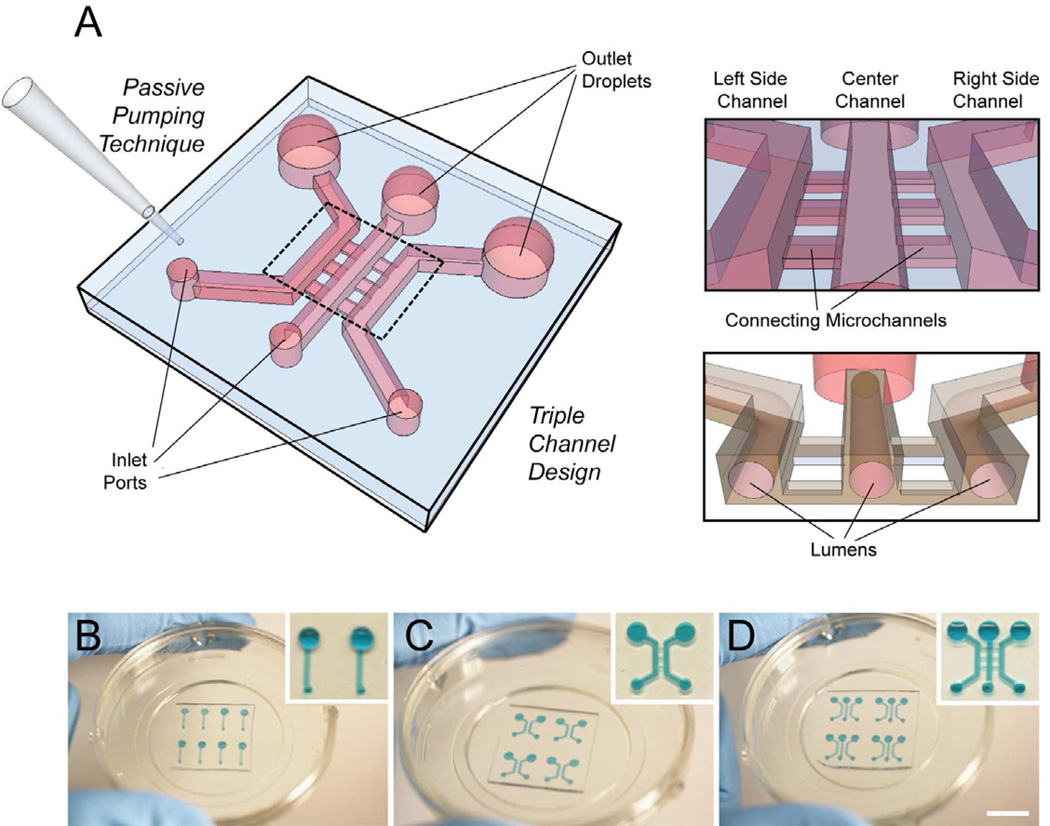
Polydimethylsiloxane (PDMS, Sylgard 184 Silicone Elastomer Kit, Dow Corning, Midland, MI) elastomer base and curing agent were mixed at a 10:1 ratio and degassed for 45 min under vacuum at room temperature. The degassed PDMS was then poured over SU-8 master molds that were generated using standard soft lithography methods [21]. PDMS was cured at 80°C for 4 h. Microchannel geometries used in these experiments varied depending on the application (Fig. 1).
Fig. 1.
Passive pumping-based microfluidic angiogenesis assay with 3D cylindrical lumens. (A) Illustration of a triple channel design with connecting microchannels. (B–D) Microchannel systems can be (B) single, (C) double, or (D) triple channel designs, and are arrayable. Devices are plasma-treated and bonded to glass-bottom Petri dishes. Scalebar ~ 10 mm.
2.2 Cell Culture
Human umbilical vein endothelial cells (HUVECs) were obtained from Lonza (Walkersville, MD, USA) and maintained with endothelial growth medium (EGM-2) with Bullet Kit (EGM-2MV; Lonza, Walkersville, MD, USA) on regular tissue culture flasks pre-coated with 2 µg/cm2 fibronectin (FN) (Sigma-Aldrich, St. Louis, MO, USA). 10T1/2 mouse smooth muscle cells were obtained from ATCC (Manassas, VA, USA) and maintained in Eagle’s Basal Medium (EBM) (Sigma, St. Louis, MO, USA) supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA) and 1% penicillin/streptomycin (Invitrogen, Carlsbad, CA, USA). The EBM-based media for 10T1/2 cells was changed to the EGM-2-based media for HUVECs 24 h prior to using cells in coculture experiments.
2.3 Hydrogel Preparation
A hydrogel consisting of extracellular matrix (ECM) proteins made of a final concentration of 6.0 mg/ml Type I collagen (rat tail, BD Biosciences, Bedfrod, MA, USA), and 25% Matrigel (BD Biosciences, Bedford, MA, USA) was used in all experiments. To make roughly 125µL of hydrogel solution, 1.56µL of a basic solution (5.0 N NaOH) and 20µL of 5X PBS was added to 78µL of the collagen I and incubated on ice for a period between 5–10 minutes to apply an additional nucleation phase prior to adding 25µL of Matrigel [22]. These volumes were scaled up to prepare the amount of hydrogel solution needed for a given experiment.
2.4 Lumen Patterning
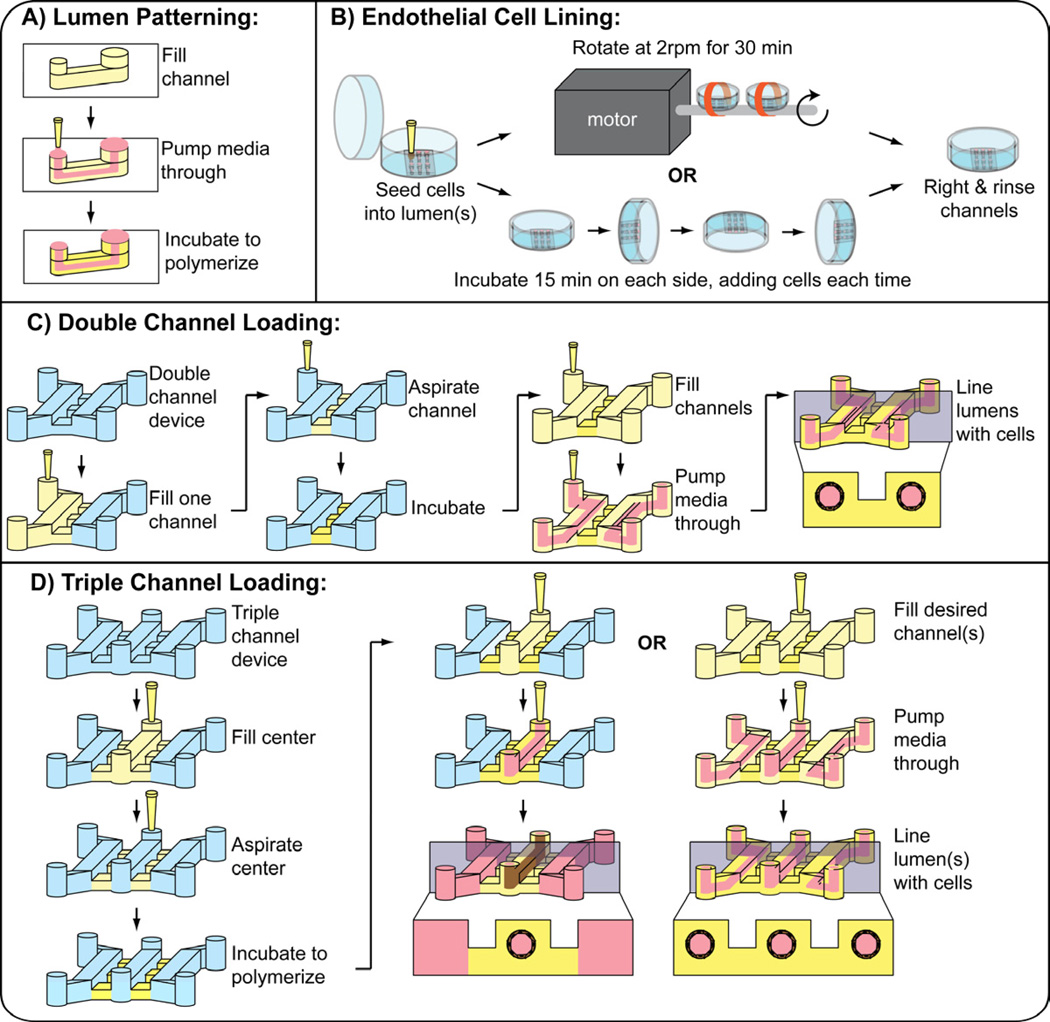
Prior to patterning lumens, devices were oxygen-plasma-treated to bond the PDMS channels to a glass surface (the inside of a glass-bottom Petri dish, MatTek, Ashland, ME, USA), and to render the inside of the chambers hydrophilic. The devices were coated with 100 µg/mL FN to facilitate adhesion of the hydrogel to the channel walls. After 20 min incubation at room temperature, the FN solution was aspirated from the channel. To pattern a lumen through a hydrogel in a single microchannel, viscous finger patterning was used as previously described [20]. Briefly, a pre-polymerized hydrogel solution was dispensed into a microchannel, and media was then dispensed through the hydrogel solution via surface tension-based passive pumping [23]. After incubating the hydrogel in the microchannels for 10 min at 37°C, polymerization of the hydrogel was completed, resulting in a patterned lumen through a hydrogel (Fig. 2A).
Fig. 2.
(A) Viscous finger patterning is used to pattern the initial lumen structure through an ECM hydrogel. A microchannel is filled with a pre-polymerized ECM hydrogel solution. Passive pumping is used to flow media through the ECM solution, pushing out the center. Following incubation for 10 minutes at 37 deg C to complete polymerization, a lumen is patterned through the hydrogel. (B) Methods used to line lumens with endothelial cells to generate ELLs. (C) Methods used to pattern two ELLs lumens in a double microchannel. (D) Methods used to pattern one or three ELL in a triple microchannel. Note: When patterning an interconnected network in double channels, the diffusion channels are the same height as the main channels. The entire channel is filled at once and viscous finger patterning is performed prior to incubation.
To pattern two or three lumens connected by a spacer hydrogel, a microfluidic design was used that consisted of two or three 500-µm tall single microchannels connected by three 100-µm tall perpendicular diffusion channels between each pair of microchannels. To pattern two lumens in a double microchannel (Fig. 2C), one of the channels was first filled with a hydrogel solution, which pinned at the outer edge of the diffusions channels due to surface tension. The hydrogel solution was then aspirated from the channel without any polymerization, leaving the solution behind only in the diffusion channels. The channels were incubated at 37°C for 10 min to polymerize the hydrogel. A hydrogel solution was then delivered into both of the channels and viscous finger patterning was performed, treating both of the channels as single microchannels, to pattern a lumen through the hydrogel solutions. The channels were incubated for 10 min at 37°C to complete polymerization. To pattern three lumens in a triple microchannel (Fig. 2D), the center channel was first filled with a hydrogel solution, which pinned at the outer edge of the diffusions channels on both sides due to surface tension. The hydrogel solution was then aspirated form the center channel without any polymerization, leaving the solution behind only in the diffusion channels. The channels were incubated at 37°C for 10 min to allow the solution to polymerize. A hydrogel solution was delivered into the three main channels and viscous finger patterning was performed, treating each of the connected microchannels as single microchannels, to pattern a lumen through the hydrogel solutions in each of the channels. The channels were incubated for 10 min at 37°C to complete polymerization. To pattern an interconnected network of lumens, two 500-µm tall single microchannels connected by three 500-µm tall perpendicular channels were used. The entire double microchannel was filled with a hydrogel solution. Viscous finger patterning was performed from both input ports. The hydrogel was patterned such that two main lumens were formed and interconnected by three perpendicular lumens.
For angiogenesis experiments, a single lumen in the center channel of a triple microchannel was patterned, leaving the two side channels open. Lumen patterning was achieved by the method described above, where culture media was added to the two side channels.
2.5 Endothelial Cell Seeding
To line the lumen with HUVECs and generate ELLs, two strategies were used to ensure uniform cell seeding (Fig. 2B). In the first strategy (motorized option), passive pumping was used to add 2 µL of a cell solution at 40,000 cells/µL to each lumen. With the lid of the Petri dish securely attached, the dish containing the microchannels was taped to the rod that is attached to a motor (BBQ Rotisserie Variable Speed Reversible Brushless Gear Motor, Wondermotor, CA, USA). This motor was placed inside an incubator at 37°C and tuned to rotate the dishes at 2RPM for 30 min to allow cell attachment. In the second strategy (manual option), a cell solution was prepared at 10,000 cells/µL and 2 µL were added to the lumens (Figure 1B). The channels were incubated for 15 min at 37°C to allow the cells to attach to the bottom of the lumen. 2 µL of the cell solution was added again to the channel via passive pumping, and the channels were rotated 90 degrees and incubated for 15 min at 37°C. This process was repeated with subsequent 90-deg rotations until all four sides of the lumen were coated [24]. The cell solution was kept on ice between cell seeding steps. After either procedure, channels were righted and incubated at 37°C for a minimum of 2 h before being perfused with fresh media via passive pumping to rinse unattached cells.
2.6 Endothelial Barrier Structure and Function
ELLs in single microchannels were cultured for 48 h with media replacements every 8–12 h. Permeability of the endothelial monolayers were determined by flowing FITC-labeled bovine serum albumin (BSA) (Molecular Probes, Eugene, OR, USA) into the lumens and calculating permeability coefficients using methods similar to those described by others [25–26]. Briefly, fluorescence images were taken at one minute intervals for 20 min. Samples were imaged with an Olympus IX81 (Center Valley, PA, USA) with a fluorescent attachment (BH2-RFL-T3, Center Valley, PA, USA), and images were acquired using SlideBook 5.0 (Intelligent Imagaing Innovations, Inc., Denver, CO, USA). The permeability coefficient P was calculated from the equation , where ΔI is the step increase in background intensity immediately after adding FITC-labeled BSA, is initial rate of increase in intensity (over the first 5 minutes) in the gel surrounding each lumen, and r is the radius of the lumen. The average permeability coefficient was determined by averaging coefficient values from 8 separate devices across 3 independent experiments.
To confirm that endothelial cells were forming cell junctions, ELLs were cultured for 48 h, fixed with 4% paraformaldehyde (PFA) (Fisher Scientific, Hampton, NH, USA), and stained for platelet endothelial cell adhesion molecule (PECAM-1; or CD31). ELLs were blocked with 3% BSA, immunolabeled with mouse anti-human CD31 antibodies (20 µg/mL, Santa Cruz Biotechnology, Inc, Santa Cruz, CA, USA), and fluorescently labeled with AlexaFlour 488 (1:200 dilution, Molecular Probes, Eugene, OR, USA). Hoechst 33342 (10 µg/mL, Molecular Probes) was used to stain the nuclei. Samples were imaged using an A1RSi confocal microscope (Nikon Instruments, Tokyo, Japan) and images were acquired using NIS Elements Advanced software (Nikon). Images files were converted using Fiji software, and volume-rendered images were generated using OsiriX.
2.7 Gradient Characterization
Characterization of a gradient in a triple microchannel was performed by patterning a lumen through and ECM hydrogel in the center chamber, and adding to one of the side chambers 50 ng/ml FITC-labeled dextran (40 kDa, Sigma-Aldrich, St. Louis, MO USA), a protein with a similar molecular weight to vascular endothelial growth factor (VEGF). Fluorescence intensity was monitored and measured every hour for 12 h. Samples were imaged with an Olympus IX81 with a fluorescent attachment (BH2-RFL-T3), and images were acquired using SlideBook 5.0.
2.8 Angiogenesis Assay: Procedure and Analysis
Experiments were performed by patterning an ELL in the center channel of triple microchannels and culturing under normal conditions for 24 h. 50 ng/ml VEGF (38.2 kDa, Sigma, St. Louis, MO, USA) was added to one of the side channels to generate a VEGF gradient that was maintained for a total of 48 h by replacing the media every 12 h. In coculture experiments with 10T1/2, 2µL of a 10T1/2 cell solution at 2×106 cells/mL were added to both side channels immediately after the channels were righted following cell lining. After 48 h, samples were imaged with an Olympus IX70 microscope (Center Valley, PA, USA) and images were acquired using MetaMorph 7.5 (Molecular Devices, LLC, Sunnyvale, CA, USA). To analyze angiogenic sprouting under different conditions, sprouts were counted and the average number of sprouts per channel was calculated across three independent experiments of three samples for each condition. Sprout length and area across three independent experiments were measured using Fiji software. A student T-test was used to determine significant differences between samples. P values less than 0.05 were considered to be significant.
To identify potential tip cell properties in the observed angiogenic sprouts, ELLs exposed to a VEGF gradient were fixed with 4% PFA and stained with a Jagged-1 antibody (1:50 dilution, Santa Cruz Biotechnology, Inc, Santa Cruz, CA, USA) and with Hoecsht stain (1:500). Secondary antibodies labeled with AlexaFlour-594 were used at a dilution of 1:200. Samples were imaged using a Nikon Eclipse Ti (Nikon) and images were acquired with NIS Elements D3.1 software (Nikon).
3. Results and Discussion
We developed a simple method for creating three-dimensional endothelial-lined lumens (ELLs) in ECM hydrogel-filled microchannels using basic fluidic principles, including passive pumping delivery and viscous fingering of fluids with different viscosities. The method uses a hydrogel solution which is comprised of 6.0 mg/ml Type I collagen and 25% Matrigel. As reported by others [25], lower hydrogel concentrations can often lead to cells invading into the hydrogel rather than forming an intact monolayer lining the hydrogel. In practice, patterning lumens through this hydrogel concentration consistently produced lumens of repeatable geometries and dimensions, with an average lumen diameter of 256 ± 21 µm. Using the described methods (Fig. 2), lumens could be independently patterned in a variety of channel geometries (single, double, or triple channels) as shown in Fig. 3. Each connected channel can be patterned independently enabling the flexibility to vary cell types, ECM composition, and treatments between connected channels, enabling a variety of studies. Additionally, interconnected ELL networks can be patterned, further increasing the flexibility of using these techniques for a variety of different studies (Fig. 4). Because all steps are performed with simple passive pumping regardless of channel geometry, this method can be interfaced with automated liquid handling systems, and thus has potential to be adopted for high-throughput screening applications.
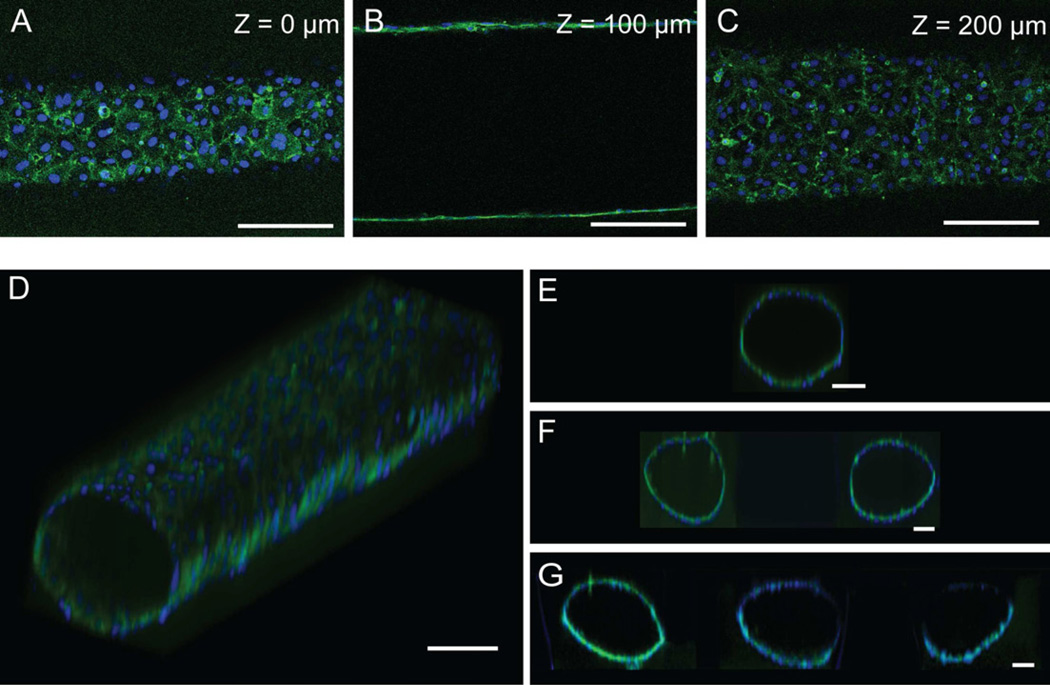
Fig. 3.
ELLs were cultured for 48hours and stained for CD31 (green) and nuclei (blue). (A)–(C) Confocal slices of an ELL at the specified height. (D) Volume-rendered image of an ELL in a single microchannel. (E) Volume-rendered cross section of an ELL in a single microchannel. (F) Volume-rendered cross section of two ELLs in a double microchannel. (G) Volume-rendered cross section of three ELLs in a triple microchannel. Scale bar represents 100µm.
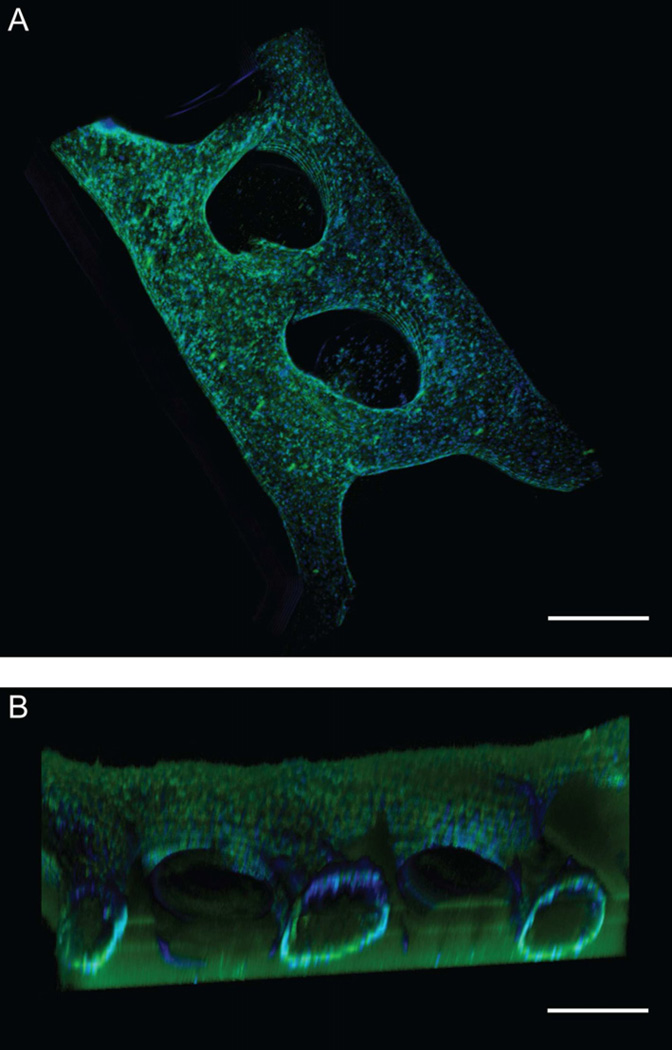
Fig. 4.
(A) Two interconnected ELLs were patterned in a double microchannel. (B) Volume-rendered cross-sectional view. Scale bar represents 500µm.
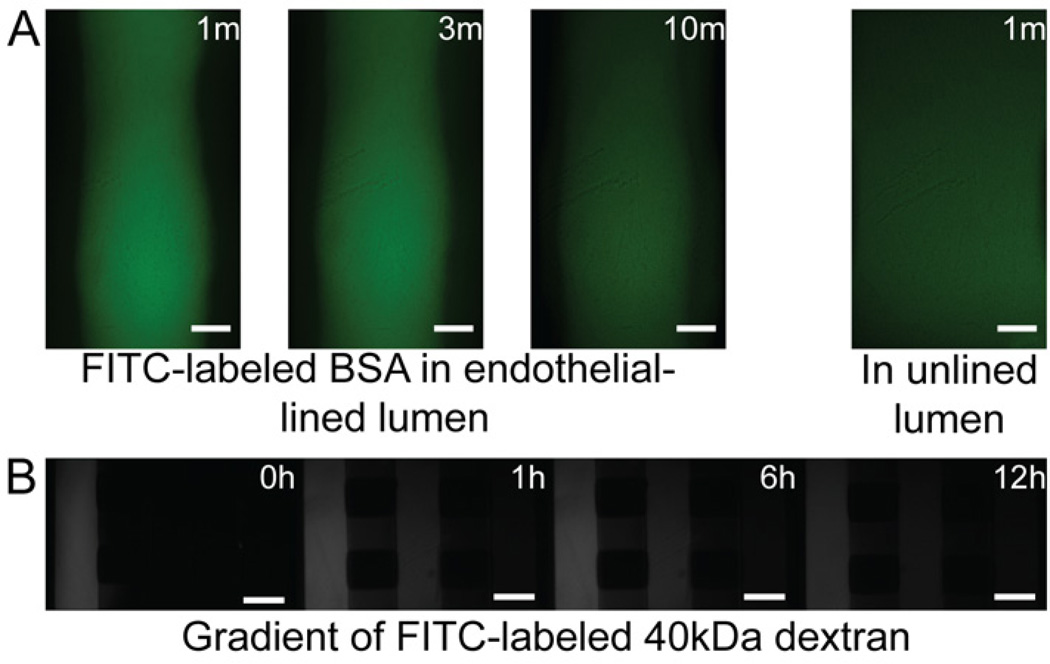
Endothelial structural integrity of the 3D ELLs were confirmed by the presence of CD31 (or PECAM-1) at cell-cell junctions throughout the monolayer (Fig. 3, 4). To confirm proper endothelial barrier function, permeability of the microvessels was measured by flowing FITC-labeled BSA through the lumens of the ELLs (Fig. 5A), and was found to be 4.73 (±0.75) × 10−6 cm/s, comparable to permeability coefficients found for other in vitro endothelial monolayers [25],[27–28]
Fig. 5.
(A) Images of an ELL and an unlined lumen at the specified time point after the addition of FITC-labeled BSA. Scale bar represents 100µm. (B) Fluorescent images taken at the specified time point following the addition of 50ng/ml of 40kDa FITC-labeled Dextran to the left channel in a triple microchannel. Scale bar represents 500µm.
Vascular endothelial growth factor (VEGF) plays a role in vessel formation, endothelial barrier function, and is also known to induce angiogenic sprouting by stimulating tip cells that respond by migrating up a VEGF concentration gradient [29]. To assess the response of the ELLs to a VEGF gradient, we performed a set of experiments in triple microchannels with an ELL patterned through an ECM hydrogel in the center channel. We first characterized gradient formation and stability using a 40-kDa FITC-labeled dextran as a model for VEGF. Dextran added to one of the side channels diffused from the source channel toward the sink channel, and developed a stable gradient after ~1 h that was maintained for at least 12 h (Fig 5B). In practice, the gradient can be maintained for extended periods of time by replacing VEGF and media roughly every 12 h [30].
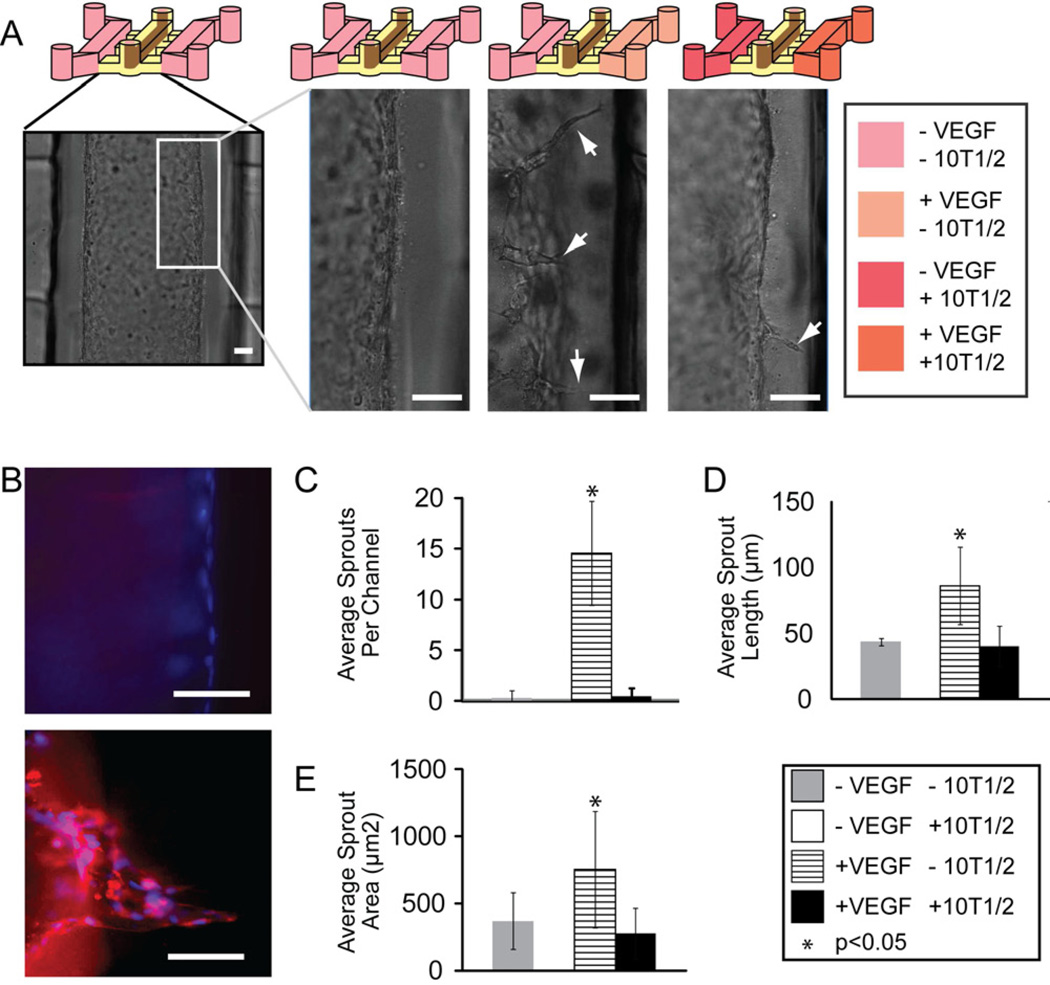
We next exposed patterned ELLs to a VEGF gradient for 24 h, and observed sprouting toward the direction of the VEGF source (Fig. 6A). Endothelial sprouts were found to express upregulation of jagged-1, a known endothelial tip cell marker [31], compared to control microvessels that were not cultured in the presence of a VEGF gradient (Fig. 6B). When the gradient was maintained for 48 h, there was a significant increase in the number of sprouts compared to control ELLs where no VEGF gradient was applied (Fig. 6C). Sprout length and sprout area were also significantly larger in ELLs exposed to a VEGF gradient, compared to controls (Fig. 6D–E).
Fig. 6.
(A) Sprouting was induced on one side of an ELL patterned in the center channel of a triple microchannel when 50ng/ml of VEGF was added to the right side channel to generate a VEGF gradient. VEGF gradient was started 24 hours after HUVEC seeding and maintained for 48 hours. Scale bar represents 50µm. (B) Fluorescent images of the wall of an ELL (top) and an ELL exposed to a VEGF gradient (bottom) stained for Jagged-1 (red) and nuclei (blue). Scale bar represents 50µm. (C) Quantitative analysis of endothelial sprout number, (D) sprout length, and (E) sprout area when ELLs were exposed to a variety of conditions. Error bars represent standard deviation.
In addition to using the side channels to observe the effects of applied VEGF or other treatments to microvessels, the influence of cocultured cells on the ELLs was observed. When 10T1/2 cells were cocultured in connecting side channels of a triple microchannel with an ELL patterned in the center channel, and simultaneously in the presence of a VEGF gradient, the number, length and area of endothelial sprouts observed was markedly reduced compared to ELLs in the presence of a VEGF gradient alone (Figure 6C–E). This is in good agreement with results presented by Chung and co-workers, who showed that coculture of endothelial cells with smooth muscle cells (10T1/2) reduced endothelial migration into a scaffold in the presence of a VEGF gradient [16].
4. Conclusions
We have presented a simple method to pattern 3D lumen structures through ECM hydrogels that can be lined with endothelial cells to mimic vessel structures in a variety of microchannel geometries. We have shown that these methods offer the flexibility to pattern ELLs in a variety of geometries on demand without the need to redesign molds from scratch, an attractive feature not available with other recently developed methods used to pattern cylindrical ELLs. Additionally, we have demonstrated the ability to add reagents or different cell types in coculture with 3D ELLs. The ELLs in this system display normal barrier structure and function, as well as expected response to VEGF treatment and coculture with smooth muscle cells. The techniques described here offer a unique balance of accessibility, ease of use, and anatomical accuracy that is an improvement over current quantitative angiogenesis assays.
Acknowledgements
The authors thank Jacob Zimmerman and Neil Zimmerman for valuable input in developing the motorized cell seeding option and Erin Gemperline for assistance with image analysis. This work was supported in part by the NIH grants R33CA137673 and T32HL007889, and the Korea Research Foundation Grant KRF-2008-220-D001331-019. David J. Beebe has an ownership interest in Bellbrook Labs LLC, which has licensed technology reported in this publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Carmeliet P, Jain R. Angiogenesis in cancer and other diseases. Nature. 2000;407:249–257. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- 2.Gerstner ER, Duda DG, di Tomaso E, Ryg PA, Loeffler JS, Sorensen AG, et al. VEGF inhibitors in the treatment of cerebral edema in patients with brain cancer. Nat Rev Clin Oncol. 2009;6:229–236. doi: 10.1038/nrclinonc.2009.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chi AS, Sorensen AG, Jain RK, Batchelor TT. Angiogenesis as a therapeutic target in malignant gliomas. Oncologist. 2009;14:621–636. doi: 10.1634/theoncologist.2008-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jain R, Schlenger K, Hockel M, Yuan F. Quantitative angiogenesis assays: progress and problems. Nat Med. 1997;3:1203–1208. doi: 10.1038/nm1197-1203. [DOI] [PubMed] [Google Scholar]
- 5.Auerbach R, Lewis R, Shinners B, Kubai L, Akhtar N. Angiogenesis assays: A critical overview. Clin Chem. 2003;49:32–40. doi: 10.1373/49.1.32. [DOI] [PubMed] [Google Scholar]
- 6.Young EWK, Beebe DJ. Fundamentals of microfluidic cell culture in controlled microenvironments. Chem Soc Rev. 2010;39:1036–1048. doi: 10.1039/b909900j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbulovic-Nad I, Au SH, Wheeler AR. A microfluidic platform for complete mammalian cell culture. Lab Chip. 2010;10:1536–1542. doi: 10.1039/c002147d. [DOI] [PubMed] [Google Scholar]
- 8.Su X, Young EWK, Underkofler HAS, Kamp TJ, January CT, Beebe DJ. Microfluidic cell culture and its application in high-throughput drug screening: cardiotoxicity assay for hERG channels. J Biomol Screen. 2011;16:101–111. doi: 10.1177/1087057110386218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lecault V, VanInsberghe M, Sekulovic S, Knapp DJHF, Wohrer S, Bowden W, et al. High-throughput analysis of single hematopoietic stem cell proliferation in microfluidic cell culture arrays. Nat Methods. 2011;8:581–586. doi: 10.1038/nmeth.1614. [DOI] [PubMed] [Google Scholar]
- 10.Young EWK, Pak C, Kahl BS, Yang DT, Callander NS, Miyamoto S, et al. Microscale functional cytomics for studying hematologic cancers. Blood. 2012;119:E76–E85. doi: 10.1182/blood-2011-10-384347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shao J, Wu L, Wu J, Zheng Y, Zhao H, Jin Q, et al. Integrated microfluidic chip for endothelial cells culture and analysis exposed to a pulsatile and oscillatory shear stress. Lab Chip. 2009;9:3118–3125. doi: 10.1039/b909312e. [DOI] [PubMed] [Google Scholar]
- 12.Young EWK, Simmons CA. Macro- and microscale fluid flow systems for endothelial cell biology. Lab Chip. 2010;10:143–160. doi: 10.1039/b913390a. [DOI] [PubMed] [Google Scholar]
- 13.Young EWK, Watson MWL, Srigunapalan S, Wheeler AR, Simmons CA. Technique for real-time measurements of endothelial permeability in a microfluidic membrane chip using laser-induced fluorescence detection. Anal Chem. 2010;82:808–816. doi: 10.1021/ac901560w. [DOI] [PubMed] [Google Scholar]
- 14.Srigunapalan S, Lam C, Wheeler AR, Simmons CA. A microfluidic membrane device to mimic critical components of the vascular microenvironment. Biomicrofluidics. 2011;5 doi: 10.1063/1.3530598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong KHK, Chan JM, Kamm RD, Tien J. Microfluidic models of vascular functions. Annu Rev of Biomed Eng. 2012;14:205–230. doi: 10.1146/annurev-bioeng-071811-150052. [DOI] [PubMed] [Google Scholar]
- 16.Chung S, Sudo R, Mack P, Wan C, Vickerman V, Kamm R. Cell migration into scaffolds under co-culture conditions in a microfluidic platform. Lab Chip. 2009;9:269–275. doi: 10.1039/b807585a. [DOI] [PubMed] [Google Scholar]
- 17.Song JW, Munn LL. Fluid forces control endothelial sprouting. PNAS. 2011;108:15342–15347. doi: 10.1073/pnas.1105316108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyvantsson I, Warrick J, Hayes S, Skoien A, Beebe DJ. Automated cell culture in high density tubeless microfluidic device arrays. Lab Chip. 2008;8:717–724. doi: 10.1039/b715375a. [DOI] [PubMed] [Google Scholar]
- 19.Puccinelli J, Su X, Beebe DJ. Automated high-throughput microchannel assays for cell biology: operational optimization and characterization. JALA. 2010;15:25–32. doi: 10.1016/j.jala.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bischel L, Lee S, Beebe DJ. A practical method for patterning lumens through ECM hydrogels via viscous fingering patterning. JALA. 2011;17(2):96–103. doi: 10.1177/2211068211426694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jo B, Lerberghe LV, Motsegood K, Beebe D. Three-dimensional micro-channel fabrication in polydimethylsiloxane (PDMS) elastomer. J Microelectromech S. 2000;9:76–81. [Google Scholar]
- 22.Sung K, Su G, Pehlke C, Trier S, Eliceiri K, Keely P, et al. Control of 3-dimensional collagen matrix polymerization for reproducible human mammary fibroblast cell culture in microfluidic devices. Biomaterials. 2009;30:4833–4841. doi: 10.1016/j.biomaterials.2009.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walker G, Beebe DJ. A passive pumping method for microfluidic devices. Lab Chip. 2002;2:131–134. doi: 10.1039/b204381e. [DOI] [PubMed] [Google Scholar]
- 24.Fiddes LK, Raz N, Srigunapalan S, Tumarkan E, Simmons CA, Wheeler AR, et al. A circular cross-section PDMS microfluidics system for replication of cardiovascular flow conditions. Biomaterials. 2010;31:3459–3464. doi: 10.1016/j.biomaterials.2010.01.082. [DOI] [PubMed] [Google Scholar]
- 25.Chrobak K, Potter D, Tien J. Formation of perfused, functional microvascular tubes in vitro. Microvasc Res. 2006;71:185–196. doi: 10.1016/j.mvr.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 26.Huxley V, Curry F, Adamson R. Quantitative fluorescence microscopy on single capillaries - alpha-lactalbumin transport. Am J Physiol. 1987;252:H188–H197. doi: 10.1152/ajpheart.1987.252.1.H188. [DOI] [PubMed] [Google Scholar]
- 27.Mehta D, Malik A. Signaling mechanisms regulating endothelial permeability. Physiol Rev. 2006;86:279–367. doi: 10.1152/physrev.00012.2005. [DOI] [PubMed] [Google Scholar]
- 28.Zheng Y, Chen J, Craven M, Choi NW, Totorica S, Diaz-Santana A, et al. In vitro microvessels for the study of angiogenesis and thrombosis. PNAS. 2012;109:9342–9347. doi: 10.1073/pnas.1201240109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerhardt H, Golding M, Fruttiger M, Ruhrberg C, Lundkvist A, Abramsson A, et al. VEGF guides angiogenic sprouting utilizing endothelial tip cell filopodia. J Cell Biol. 2003;161:1163–1177. doi: 10.1083/jcb.200302047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abhyankar V, Lokuta M, Huttenlocher A, Beebe DJ. Characterization of a membrane-based gradient generator for use in cell-signaling studies. Lab Chip. 2006;6:389–393. doi: 10.1039/b514133h. [DOI] [PubMed] [Google Scholar]
- 31.Sainson RCA, Johnston DA, Chu HC, Holderfield MT, Nakatsu MN, Crampton SP, et al. TNF primes endothelial cells for angiogenic sprouting by inducing a tip cell phenotype. Blood. 2008;111:4997–5007. doi: 10.1182/blood-2007-08-108597. [DOI] [PMC free article] [PubMed] [Google Scholar]