Abstract
How specific cell types can be directly converted into other distinct cell types is a matter of intense investigation with wide-ranging basic and biomedical implications. We show here that the removal of the histone 3 lysine 27 (H3K27) methyltransferase complex PRC2 (“Polycomb Repressor Complex 2”) permits ectopically expressed, neuron-type-specific transcription factors (“terminal selectors”) to convert C. elegans germ cells directly into specific neuron types. Terminal selector-induced germ cell-to-neuron conversion can not only be observed upon genome-wide loss of H3K27 methylation in PRC2(−) animals, but also upon genome-wide redistribution of H3K27 methylation patterns in animals which lack the H3K36 methyltransferase MES-4. Manipulation of the H3K27 methylation status not only permits conversion of germ cells into neurons, but also permits hlh-1/MyoD-dependent conversion of germ cells into muscle cells, indicating the PRC2 protects the germline from the aberrant execution of multiple distinct somatic differentiation programs. Taken together, our findings demonstrate that the normally multi-step process of development from a germ cell via a zygote to a terminally differentiated somatic cell type can be shortcut by providing an appropriate terminal selector transcription factor and manipulating histone methylation patterns.
INTRODUCTION
A plethora of transcription factors are known to be absolutely required for the induction of specific cellular differentiation programs. However, such transcription factors are often remarkably inefficient in imposing such a program on other cell types upon ectopic misexpression (Zhou and Melton, 2008). For example, ectopic misexpression of the CHE-1 zinc finger transcription factor, which is normally required to generate the ASE gustatory neuron type in C. elegans (Chang et al., 2003; Uchida et al., 2003), converts only a very small number of sensory neurons into ASE-like neurons; all other cell types are inert to the cell fate inducing ability of che-1 (Tursun et al., 2011).
To explore the context-dependency of che-1 activity, we considered the possibility that an inhibitory mechanism may exist to prevent che-1 from driving the ASE differentiation program in most other cell types. With this possibility in mind, we undertook a loss of function screen for genes whose knock-down enables che-1 to more broadly induce ASE-like fate in other cellular contexts. This RNAi-based screen identified a phylogenetically conserved histone chaperone, lin-53 (called Rbbp4 and Rbbp7 in vertebrates) whose removal permitted a direct, che-1-mediated conversion of mitotic germ cells into ASE-like neurons (Tursun et al., 2011).
In this paper, we explore the mechanistic basis of the conversion process by asking which other genes are involved in this process. We based our analysis on the well-documented observations that in vertebrates and invertebrates the histone chaperones LIN-53/Rbbp4,7 are components of many distinct multiprotein complexes with various functions in chromatin biology: the nucleosome remodeling and histone deacetylation (NURD) complex, the chromatin assembly factor (CAF) complex, the histone deacetylase corepressor complex Sin3, the histone acetyltransferase 1 (HAT1) complex, the nucleosome remodeling factor (NURF) complex, the Retinoblastoma-gene-containing repressor complex DRM (DP, Rb, and class B synMuv) and the Polycomb repressive complex 2 (PRC2)(Harrison et al., 2006; Loyola and Almouzni, 2004). The presence of LIN-53/Rbbp4,7 in these functionally very distinct complexes has been shown biochemically as well as through genetic analysis. We show here that the effect of lin-53 on germ cell-to-neuron conversion can be phenocopied by removal of the PRC2 complex and further characterize features of the cellular conversion process.
RESULTS AND DISCUSSION
Removal of PRC2 complex components allows for germ cell-to-neuron conversion
Our initial RNAi screen that uncovered lin-53 as a “brake” for converting germ cells to neurons (Tursun et al., 2011) did not reveal obvious, lin-53-like phenotypes with indigvidual members of the many complexes with which the LIN-53/Rbbp4,7 protein is known to associate. As a result, the mechanism by which LIN-53 operates to prevent a germ cell-to-neuron conversion remained an open question. However, negative results from this screen were difficult to interpret, mainly because RNAi of many of the various LIN-53 complex components resulted in infertility or early developmental defects, thereby precluding analysis of the germline.
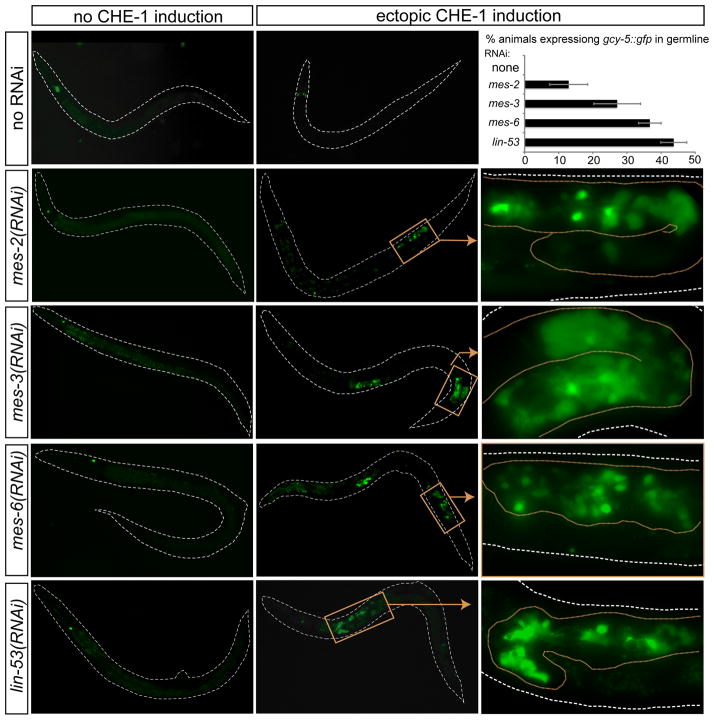
We focused our analysis on the well-characterized Polycomb Repressive Complex 2 (PRC2), which in vertebrates and Drosophila contains the LIN-53 orthologs Rbbp4,7 (CAF1 in Drosophila), the H3K27 methyltransferase Ezh2 (Enhancer of Zeste in Drosophila), the WD40 protein Eed (Extra sex combs in Drosophila) and other associated proteins (Kuzmichev et al., 2002; Margueron and Reinberg, 2011). Similarly, in C. elegans, the PRC2 complex has so far been shown to contain the H3K27 methyltransferase MES-2/Ezh2 and the two accessory proteins, MES-3 and the WD40 protein MES-6/Eed (Bender et al., 2004; Xu et al., 2001). Ectopic CHE-1 expression in mes-2 and mes-3 null mutants that lack both maternal and zygotic gene activity, did not induce neurons in the germline (data not shown), but this is due to the fact that the germline of such animals degenerates during larval stages (Capowski et al., 1991). In contrast, partial knockdown of mes-2, mes-3 and mes-6 by RNAi, in a genetic background that was not sensitized for RNAi improved fertility and viability of the dsRNA-treated animals, allowing production of more germ cells, and these germ cells appeared superficially normal (Suppl. Fig. 1). After feeding animals with dsRNA against mes-2, mes-3 and mes-6, we induced che-1 expression in the progeny of dsRNA-fed animals in all tissues through the heat-shock promoter, at about mid-larval stages. Feeding of control dsRNA or no dsRNA resulted in heat shock-induced che-1 being able to ectopically induce the ASE fate marker gcy-5 exclusively in a small number of head neurons. In contrast, RNAi of each member of the C. elegans PRC2 complex (mes-2/Ezh2; mes-3 and mes-6/Eed) resulted in che-1heat-shock -dependent gcy-5 expression in the germline (Fig. 1), providing the first hint that, as in lin-53(RNAi) animals, germ cells may have converted into ASE-like neurons. This effect is not the mere result of improved germline expression of che-1 as shown by antibody staining (Suppl. Fig. 2). Neuron-like conversion can not be observed in zygotic mes null mutant animals that still have maternal mes gene contribution (M+Z−), suggestion that partial, but not complete, elimination of maternal mes, achieved by RNAi, results in germ cell survival and susceptibility for conversion.
Fig. 1. Knockdown of members of the PRC2 complex allow che-1 mediated conversion of germ cells to neurons.
Larval progeny of RNAi-treated animals were analyzed for gcy-5::gfp (ASE fate marker; ntIs1 transgene) expression ~24 hours after heatshock induction of CHE-1 (otIs305 transgene). Right panels show blow-ups of boxed regions in middle panels, with germlines outlined by brown stippled lines. Top right panel shows penetrance of conversion phenotypes after che-1 induction at the ~L4 stage (at least 3 independent experiments, n=90–300 for each RNAi). The incomplete penetrance is most likely due to the incomplete effect of RNAi (as quantified in Suppl. Fig. 6). We had previously shown through antibody staining that the lin-53(RNAi) effect can not be explained by improved germline expression of che-1 from the heat-shock vector (Tursun et al., 2011). See also Experimental Procedures for more comments on transgene expression in the germline.
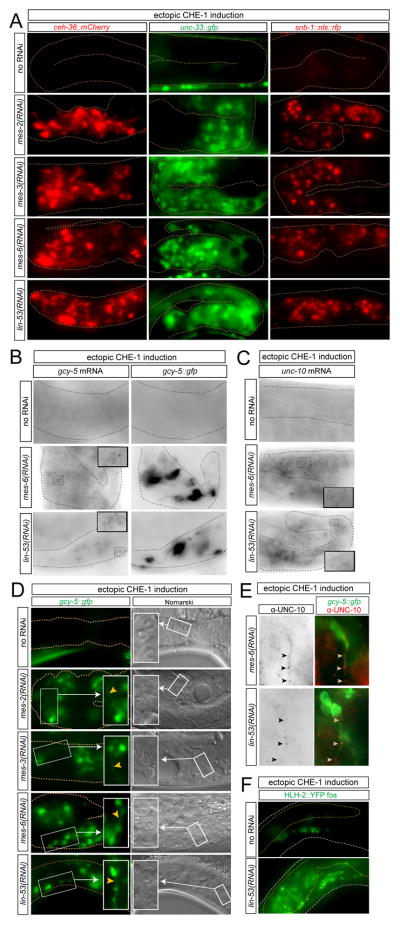
To study the cell fate conversion in more detail, we performed RNAi against mes-2, mes-3 and mes-6 and induced che-1 in a number of transgenic animals that express several reporter gene constructs. These included a second marker of ASE fate (ceh-36) and two panneuronal markers (unc-33 and snb-1). We found all these markers to be induced in the germline under these circumstances (Fig. 2A). Neuronal marker induction was not only observed on the level of reporter transgenes but was also confirmed by single molecule fluorescence in situ hybridization (smFISH)(Raj et al., 2008) which revealed induction of endogenous neuronal genes, normally expressed in ASE (gcy-5 and unc-10), in converted germ cells (Fig. 2B,C). Moreover, germ cell nuclei lost the characteristic fried-egg morphology and acquired a speckled nuclear morphology characteristic of neurons, and there was a concomitant loss of expression of the germ cell marker PGL-1(Fig. 2D; Suppl. Fig. 3). Most strikingly, marker gene-expressing cells extended cellular axo-dendritic-like projections, demonstrating that germ cells do not merely derepress marker genes but are also morphologically transformed into neurons (Fig. 2D). Theses extension show clusters of presynaptic proteins, as assessed by UNC-10/Rim antibody staining (Fig. 2E), corroborating the neuronal nature of these converted cells. All these che-1-dependent phenotypes in mes-2, mes-3 and mes-6 (RNAi) animals are highly similar to the phenotypes observed with lin-53(RNAi) (Fig. 1, Fig. 2)(Tursun et al., 2011).
Fig. 2. Detailed characterization of germ cell-to-neuron conversion.
(A) The progeny of RNAi-treated animals were analyzed for the expression of several additional markers (ceh-36: otIs264; unc-33: otIs118; snb-1: otEx4445) after ~24 hours of heat-shock promoter-mediated che-1 induction at larval stages (otIs305 transgene). The penetrance of this phenotype ranged from 20% to 50% for the various markers (n = 30 to 60 for each marker, for each RNAi).
(B,C) Single molecule FISH shows induction of endogenous genes in converted germ cells. gcy-5 (B) and unc-10 (C) is induced in the germline of animals in which either lin-53 or the representative PRC2 complex component mes-6 was knocked down by RNAi and in which CHE-1 expression was ectopically induced. Individual mRNA molecules show up as individual black dots. In panel B, the gcy-5::gfp transgene (which only contains the gcy-5 promoter and will therefore not be picked up by the smFISH probes) allows to visualize individual, converted cells.
(D) Germ cells acquire a neuron-like morphology in terms of nuclear morphology (“fried egg” germ cell nuclei to “speckled” neuronal cell nuclei; right panels, including blow-up in white box) and axo/dendritic extensions (arrowheads; left panels, including blow-up in white box).
(E) Converted germ cells express the presynaptic protein UNC-10/Rim, which clusters along the length of a neuronal extension (arrowheads).
(F) Induction of the immature neuronal marker hlh-2, as assessed using a fosmid reporter transgene (otEx4720).
Focus of action of the PRC2 complex
mes-2/Ezh2 and mes-6/Eed are known to be broadly expressed in embryonic somatic cells as well as embryonic and adult germ cells (Holdeman et al., 1998; Korf et al., 1998). To analyze lin-53 expression, we generated a fosmid-based lin-53 reporter in which gfp was inserted into the lin-53 locus in the context of ~ 40 kb of genomic sequence, including the lin-53 locus and several genes up and downstream of the locus. Transgenic animals expressing this reporter show broad lin-53::gfp expression in all somatic tissue and the germline at all stages examined (Suppl. Fig. 4A,B). To test the most parsimonious model of PRC2 acting autonomously in the germ cells rather than in the surrounding somatic gonad to prevent che-1 induced germ cell-to-neuron conversion, we sought to eliminate PRC2 specifically in germ cells by using animals that lack the RNA-directed RNA polymerase rrf-1. rrf-1 is required for RNAi in many somatic cells (including the somatic gonad), but is not required for RNAi in the germline (Sijen et al., 2001). RNAi against mes-2,3,6 and lin-53 in a rrf-1(pk1417) mutant background will therefore eliminate gene function in germ cells but not in the somatic gonad. We found that in such animals, the che-1 induced conversion phenotype of mes(RNAi) and lin-53(RNAi) animals is still readily observable (Suppl. Fig. 4C,D).
Germ cell-to-neuron conversion occurs in the context of a global loss or global redistribution of H3K27 trimethylation
Previous studies have shown that genetic removal of mes-2, mes-3 and mes-6 results in a genome-wide loss of H3K27me3 in the germline which can be readily assessed by staining nuclei of mes-2, mes-3 or mes-6 mutant cells with H3K27me3 antibodies (Bender et al., 2004). We found that RNAi of not only mes-2, mes-3 or mes-6 but also lin-53(RNAi) causes a loss of H3K27me3 in germ cells (Suppl. Fig. 5). These results suggest that genome-wide H3K27me3 removal correlates with the susceptibility of germ cells to be converted into neurons, and they further underscore the expected phenotypic similarity of lin-53(RNAi) and mes-2/3/6(RNAi).
Recent work has shown that PRC2-mediated H3K27 methylation is antagonized by H3K36 methylation, suggesting that H3K36me is at least partially responsible for the precise genome-wide distribution of H3K27me3 (Gaydos et al., accompanying manuscript)(Yuan et al., 2011). In C. elegans, the histone methyltransferase MES-4 is responsible for all H3K36me2 and contributes to H3K36me3 (Rechtsteiner et al., 2010). As shown in the accompanying manuscript (and summarized in Suppl. Fig. 6), loss of mes-4 causes a genome-wide redistribution of H3K27me3, resulting in a net decrease of H3K27me3 on many somatic genes, including ASE-expressed and panneuronal genes. We found that in mes-4(RNAi) animals, germ cells also become susceptible to che-1-induced neuron conversion (Suppl. Fig. 6). Taken together, our data indicate that disruption of H3K27 methylation patterns, either through complete elimination or through genomic redistribution, renders the genome accessible to regulatory inputs that drive specific somatic cellular fates.
Mitotic cycling is not required for CHE-1-driven germ cell-to-neuron conversion
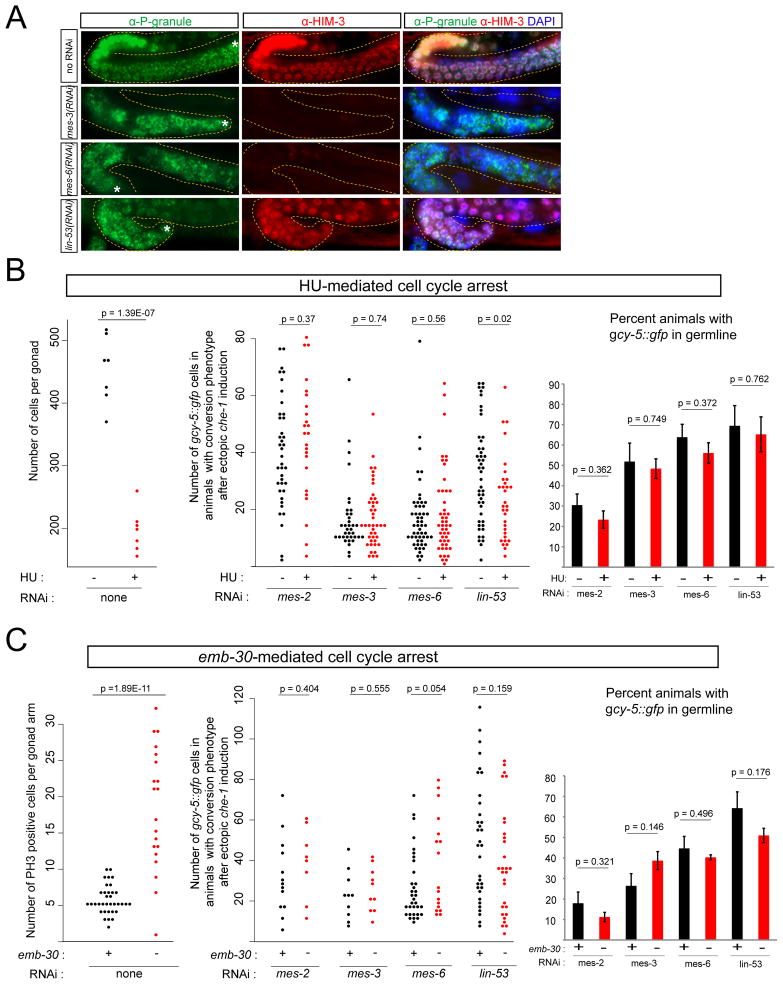
Having established the importance of PRC2 in the germ cell-to-neuron conversion process, we next asked whether PRC2(-) germ cells need to be in a specific cellular state to be converted into neuron types. In wild-type animals, germ cells are in various states of mitotic and ensuing meiotic maturation. We could rule out that being in a meiotic state is required for che-1-induced neuron conversion, since we found that RNAi against PRC2 components prevents meiotic entry of germ cells, as deduced by a lack of staining of the meiotic marker HIM-3 (Fig. 3A).
Fig. 3. Mitotic germ cells are converted but mitosis is not required.
(A) RNAi knockdown of MES proteins inhibits entry into meiosis, as assessed by staining with the meiotic marker HIM-3. Knockdown of lin-53 does not completely abolish entry into meiosis. Germ cells were still present in all cases, as assessed by staining with a P-granule specific antibody (OIC1D4). In the case of mes-3(RNAi) and mes-6(RNAi), 80-90% of animals that stained positive for OIC1D4 did not contain HIM-3 positive cells. The remaining 10–20% animals that expressed HIM-3 had healthier-appearing gonads, indicating that RNAi knockdown was inefficient in these worms. Over 90% of lin-53(RNAi) animals that stained positive for OIC1D4, also expressed HIM-3. Asterisks point out the distal most part of the gonad in the image.
(B) Cell cycle arrest by hydroxyurea (HU) treatment. Left panel: reduced number of germ cells in gonads of HU treated animals. Middle panel: expressivity of germline conversion remained unchanged after HU-mediated cell cycle arrest. HU “−” or “+” indicates “no HU treatment” or “5 hours HU treatment” (see Experimental Procedures). Each dot represents an individual animal. The effectiveness of HU treatment was also assessed by the lack of EdU staining (Suppl. Fig. 7). Right panel: The penetrance (i.e. number of animals displaying phenotype) of conversion is also not significantly altered.
(C) Cell cycle arrest by shifting emb-30(tn377ts) mutants to the non-permissive temperature. Left panel: on average, there were about 3 times more germ cells in metaphase after an 8 hour temperature shift. Middle panel: expressivity of germline conversion upon ectopic che-1 induction remained unchanged after emb-30-mediated cell cycle arrest. Each dot represents an individual animal (scored ~24 hours after che-1 induction at the L4 stage). Right panel: The penetrance (i.e. number of animals displaying phenotype) of conversion is also not significantly altered.
Cell division has been proposed to be an important mediator of transitions between different states of gene expression and transcription factor-induced cellular reprogramming is indeed aided by cells being mitotically active (Egli et al., 2008; Hanna et al., 2009). We therefore asked whether the susceptibility of PRC2(RNAi) mitotic germ cells to conversion requires the mitosis process per se. To test this notion, we treated worms with dsRNA, then arrested the cell cycle before inducing che-1heat-shock, and asked whether arrested cells are still convertible. Cell cycle arrest was achieved in two independent ways. First, we blocked the cell cycle chemically through hydroxyurea treatment. Hydroxyurea arrests the cell cycle in S-phase, as previously documented in many defined settings, including the C. elegans germline (Fox et al., 2011). We confirmed the effect of hydroxyurea by counting the reduction of germ cell number and by observing the loss of EdU incorporation (Fig. 3B). We found that PRC2(RNAi) cells treated with hydroxyurea can still be converted into ASE-like neurons through heat-shock induction of che-1 (Fig. 3B).
As an independent approach to investigate the role of the cell cycle, we blocked the cell cycle genetically through the use of a temperature-sensitive allele, tn377, of the cell cycle regulator emb-30, an anaphase-promoting complex/cyclosome component (Furuta et al., 2000). We grew PRC2(RNAi)-treated emb-30(tn377ts) animals at 15° C and inactivated emb-30 by shifting worms to 25° C eight hours before che-1 heat shock induction. We confirmed that within these eight hours an increased number of germ cells indeed become mitotically arrested through staining with PH3, a marker of metaphase (Fig. 3C). We found that che-1 could still convert germ cells into neurons if PRC2 had been knocked down (Fig. 3C). We conclude from these results that PRC2(−) cells can be directly reprogrammed into neurons without the need to pass through the cell cycle.
The germ cell-to-neuron conversion process passes through an immature neuronal stage
Our previous phenotypic characterization of converted neurons in the germline of PRC2(RNAi) animals after che-1 expression focused on examination of terminal markers of ASE neurons, but did not examine possible intermediary stages of the conversion process. Unlike in vertebrates where many markers of immature neurons are known, the only broadly expressed, early neuronal marker in C. elegans that we are aware of is the bHLH cofactor hlh-2/Daughterless. hlh-2 is expressed broadly in the developing nervous system during mid-embryogenesis but its expression fades in postmitotic neurons (Krause et al., 1997). This is consistent with the activity of lineage specific, proneural bHLH partners of hlh-2 which, in most organisms studied, including C. elegans, operate transiently during development to ensure the induction of neuronal fate (e.g. Poole et al., 2011). hlh-2 is not expressed in the germline of wild-type animals (with or without che-1 induction), nor in PRC2(RNAi) animals, but it is transiently induced in the converted neurons of PRC2(RNAi); che-1(hs) animals (Fig. 2F).
Specificity of somatic fate conversion
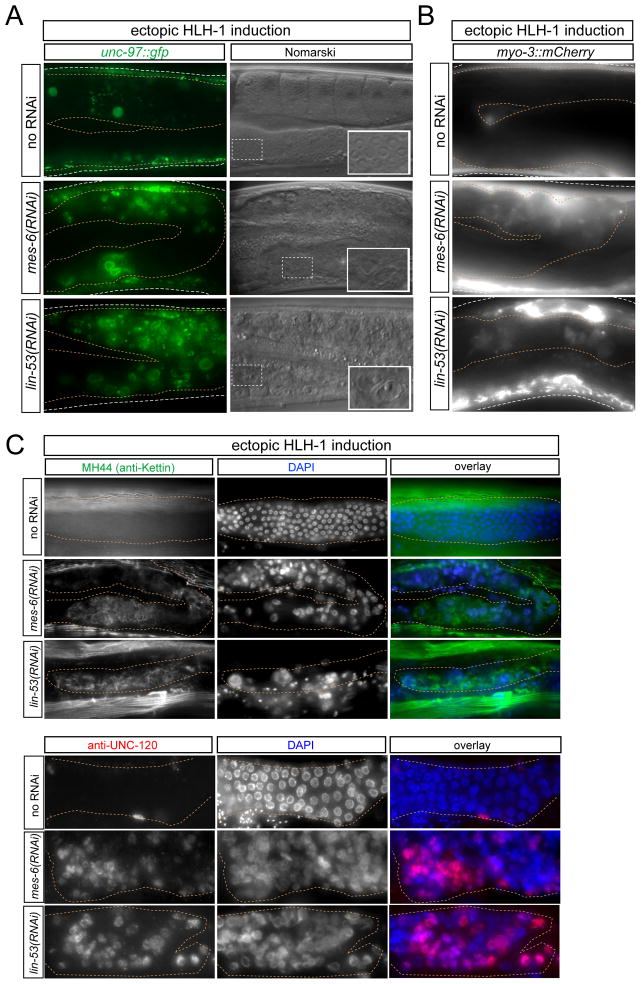
We next asked whether removal of PRC2 components makes mitotic germ cells only susceptible to be driven to a neuronal fate or whether non-neuronal, somatic fates can also be induced. To this end, we turned to the C. elegans MyoD homolog hlh-1, a factor that acts, in analogy to neuronal terminal selectors, as a direct regulator of terminal muscle features (Lei et al., 2010). Previously, we found that in lin-53(RNAi) animals, ectopic induction of hlh-1 was not able to convert mitotic germ cells to muscle (Tursun et al., 2011). To probe this issue further, we generated new hlh-1 transgenic lines that are less repetitive in nature than the ones previously used and therefore less prone to partial or complete silencing in the germline. We found that upon removal of the PRC2 component mes-6 or lin-53, hlh-1 is indeed able to convert germ cells directly into muscle-like cells, as assessed by the induction of transgenic markers for two distinct muscle proteins, UNC-97/PINCH (muscle dense body component which also localizes to muscle nuclei)(Hobert et al., 1999), and muscle myosin (myo-3)(Fire and Waterston, 1989). Moreover, we observed induction of two additional muscle proteins by antibody staining (Fig. 4), namely, Kettin, a normal components of myofibrils (Ono et al., 2006) and the transcription factor UNC-120/SRF, which is one of the three components (with HLH-1 and HND-1) of a the muscle-specific regulatory signature (Fukushige et al., 2006). A morphological transformation was also observed in the form of converted cells displaying a distinctive muscle nuclear morphology, based on size and perinuclear localization of UNC-97 (Fig. 4).
Fig. 4. Conversion of germ cells into muscle cells.
The progeny of RNAi-treated animals were analyzed for muscle marker expression ~24 hours after heat-shock promoter-mediated hlh-1 induction at larval stages (otEx4945 transgene).
(A) Induction of the LIM domain protein UNC-97, as observed with an unc-97::gfp translational fusion transgene mgIs25. UNC-97 protein is known to localize in muscle cells to both dense bodies (cellular attachment structures) as well as the nuclear periphery (Hobert et al., 1999). Some converted nuclei also showed a muscle-like morphology based on size and localization of UNC-97 at the nuclear periphery.
(B) Induction of the myosin gene, as assessed with the myo-3 transgene otIs377.
(C) Induction of the myofibrillar, actin-binding Kettin protein as assessd with antibody MH44 and of the myogenic transcription factor UNC-120, as assessed by anti-UNC120 antibody staining.
Conclusions
Based on the phenotypic similarities of removal of lin-53 and the mes-2, mes-3 and mes-6 genes, as well as the physical association of fly and vertebrate orthologs of their protein products (Kuzmichev et al., 2002; Margueron and Reinberg, 2011), we conclude that the lin-53 phenotype that we reported previously is likely the result of the functional disruption of the PRC2 complex. The conserved enzymatic role of PRC2 is the deposition of H3K27 di- and tri-methyl marks, which are associated with developmentally regulated gene repression. PRC2 has numerous intricate biological roles that vary depending on cellular and temporal developmental contexts (Margueron and Reinberg, 2011; Zhang et al., 2011). For example, in C. elegans, PRC2 has been shown to play a role in restricting the plasticity of somatic cells in the developing embryo (Yuzyuk et al., 2009). Our findings suggest that PRC2 defines a chromatin state in germ cells that protects the genome from aberrant regulatory inputs. Disruption of this chromatin state renders germ cells susceptible to direct, cell cycle-independent conversion into differentiated somatic cell types. These findings provide a conceptual framework for understanding the cellular context-dependency of transcription factors which may be dictated by “protective” chromatin states. Protective chromatin states may be distinct in different cell types since the loss of PRC2 only makes germ cells but not other somatic cell types susceptible to cellular conversion. Recent work in Drosophila illustrates that repressed chromatin has distinct molecular signatures (van Steensel, 2011) and these may be utilized in a cell-type specific manner.
Our findings can also be viewed from the perspective of the multi-step process of development from a germ cell via a zygote to a differentiated somatic cell type. We show that this process can be dramatically shortcut through the manipulation of chromatin modification patterns and provision of terminal selector transcription factors. The deposition of chromatin marks to specific genomic regions and the choice of a specific terminal selector transcription factor can be viewed as the ultimate goal for cells to achieve during development to adopt their final identity.
EXPERIMENTAL PROCEDURES
Strains and transgenes
The following strains and transgenes were used:
OH9846: otIs305 [hsp16-2prom::che-1::2xFLAG; rol-6(d)]; ntIs1 [gcy-5::gfp; lin-15(+)]
SS186: mes-2(bn11) unc-4(e120)/mnC1 dpy-10(e128) unc-52(e444)II
SS262: mes-3(bn35) dpy-5(e61) I; sDp2(l;f)
OH9209: otIs264 [ceh-36::tagrfp]
OH10596: otEx4720 [hlh-2fosmid::yfp; rol-6(d)]
OH439: otIs118 [unc-33::gfp]
OH10003: pha-1; otEx4445[snb-1::NLSrfp; pBX]
DG627: emb-30(tn377)III
NL2096: rrf-1(pk1417)I
OH10993: otEx4944 [lin-53fosmid::gfp; rol-6(d)]
OH10995: otIs377 [myo-3::mCherry]
OH10994: otEx4945 [hsp16-2prom::hlh-1::2xFLAG; rol-6(d)]; mgIs25[unc-97::gfp]
Like the che-1 heat shock array otIs305 (and other previously described che-1 arrays; Tursun et al., 2011), the hlh-1 heat shock array otEx4945 is a complex array, generated by co-injection of PvuII-digested, bacterial genomic DNA (~150 ng/μl), the hlh-1 expression construct (0.5 ng/μl) and pRF4 (2 ng/μl). In contrast to simple arrays, such complex arrays are not normally silenced in the germline (Tursun et al., 2009, L. Cochella, B.T. and O.H., unpublished data). The previously used hlh-1 transgene (Tursun et al., 2011) is a simple array (Fukushige and Krause, 2005). The lin-53 fosmid reporter was generated with 10 ng/μl lin-53::gfp fosmid, 2 ng/μl pRF4, 135 ng/μl PvuII-digested, bacterial genomic DNA. The hlh-2 fosmid reporter was a kind gift from the Greenwald lab.
The lin-53 fosmid reporter was generated by fosmid recombineering (Tursun et al., 2009), using fosmid WRM0634aA12. This transgene does not rescue the putative null (n3368). Primer sequences are available upon request. The resulting transgene, again generated as a complex array, is called otEx4944.
Antibody staining, smFISH and microscopy
For antibody staining, a freeze crack protocol was used (Duerr, 2006). A detailed protocol, including the antibodies used, can be found in the Supplementary Material. smFISH was done using Custom Stellaris™ FISH probes, purchased from Biosearch Technologies and staining was done according to the manufacturers protocol. μManager was used for image acquisition and processing (Edelstein et al., 2010).
RNA interference
RNAi was done as previously described (Tursun et al., 2011). In brief, transgenic worms expressing heat-shock inducible che-1 or hlh-1 in either the N2 wildtype background or mutant backgrounds (emb-30 or rrf-1) were transferred to plates that were seeded with bacteria containing specific RNAi clones against either lin-53, mes-2, mes-3 or mes-6 at the L4 stage. In contrast to mes null mutant animals whose germline degenerates, F1 progeny of RNAi-treated animals contained more germ cells and these germ cells appeared superficially normal, as assessed by staining with germ cell markers (Suppl. Fig. 6 and shown as controls Fig. 2C and Fig. 3A). These worms were heat-shocked at the L3 to young adult stage by incubating them at 37°C for 30 min. Heat-shocked worms were kept at 25°C overnight and scored the following day. emb-30 mutants were grown at 15°C, shifted to 25°C for 8hr when F1 progeny on RNAi plates were at the L3-young adult stage, heat shocked at 37°C, then kept overnight at 25°C and scored the next day.
Cell cycle arrest by hydroxyurea treatment
HU treatment was performed as described previously (Fox et al., 2011). In brief, plates were seeded with MG1693 bacteria that incorporated 5-ethynyl-2′-deoxyuridine. To access cell cycle arrest, hydroxyurea, at a final concentration of 250 μM, was added to some of the plates. L4 animals, grown on OP50, were moved to the HU treated and untreated EdU-labeled bacteria plates. After 5 hours, these animals were washed off, freeze cracked on poly-L-lysine coated slides, fixed with 3% paraformaldehyde, and DAPI stained. Following this, the EdU detection reaction, which labels EdU with an alexa-fluor dye, was performed using an EdU labeling kit (Invitrogen).
Supplementary Material
Highlights.
Selector-type transcription factors cannot convert germ cells into neurons or muscle
Loss of H3K27me3 allows direct, selector-driven germ cell conversion to neurons or muscle
H3K27me3 redistribution in H3K36me(−) animals also permits germ cell conversion
Mitotic cycling is not required for induced germ cell conversion
Acknowledgments
We thank Tim Schedl, Susan Strome and Iva Greenwald for comments on the manuscript, Tim Schedl and his laboratory for technical advice, Monique Zetka, Susan Strome, Pamela Hoppe, Iva Greenwald and Michael Krause for reagents and Qi Chen for expert assistance in strain generation. This study was supported by the NIH (1R21NS076191-01; R01NS039996-05; R01NS050266-03) and by the Leona M. and Harry B. Helmsley Charitable Trust. T.P. was funded by an NSF predoctoral fellowship and B. T. was funded by a Francis Goelet Postdoctoral Fellowship. O.H. is an Investigator of the HHMI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bender LB, Cao R, Zhang Y, Strome S. The MES-2/MES-3/MES-6 complex and regulation of histone H3 methylation in C. elegans. Curr Biol. 2004;14:1639–1643. doi: 10.1016/j.cub.2004.08.062. [DOI] [PubMed] [Google Scholar]
- Capowski EE, Martin P, Garvin C, Strome S. Identification of grandchildless loci whose products are required for normal germ-line development in the nematode Caenorhabditis elegans. Genetics. 1991;129:1061–1072. doi: 10.1093/genetics/129.4.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Johnston RJ, Jr, Hobert O. A transcriptional regulatory cascade that controls left/right asymmetry in chemosensory neurons of C. elegans. Genes Dev. 2003;17:2123–2137. doi: 10.1101/gad.1117903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerr JS. Immunohistochemistry. WormBook. 2006:1–61. doi: 10.1895/wormbook.1.105.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer control of microscopes using microManager. Curr Protoc Mol Biol. 2010;Chapter 14(Unit14):20. doi: 10.1002/0471142727.mb1420s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egli D, Birkhoff G, Eggan K. Mediators of reprogramming: transcription factors and transitions through mitosis. Nat Rev Mol Cell Biol. 2008;9:505–516. doi: 10.1038/nrm2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Waterston RH. Proper expression of myosin genes in transgenic nematodes. Embo J. 1989;8:3419–3428. doi: 10.1002/j.1460-2075.1989.tb08506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox PM, Vought VE, Hanazawa M, Lee MH, Maine EM, Schedl T. Cyclin E and CDK-2 regulate proliferative cell fate and cell cycle progression in the C. elegans germline. Development. 2011;138:2223–2234. doi: 10.1242/dev.059535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushige T, Brodigan TM, Schriefer LA, Waterston RH, Krause M. Defining the transcriptional redundancy of early bodywall muscle development in C. elegans: evidence for a unified theory of animal muscle development. Genes Dev. 2006;20:3395–3406. doi: 10.1101/gad.1481706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushige T, Krause M. The myogenic potency of HLH-1 reveals widespread developmental plasticity in early C. elegans embryos. Development. 2005;132:1795–1805. doi: 10.1242/dev.01774. [DOI] [PubMed] [Google Scholar]
- Furuta T, Tuck S, Kirchner J, Koch B, Auty R, Kitagawa R, Rose AM, Greenstein D. EMB-30: an APC4 homologue required for metaphase-to-anaphase transitions during meiosis and mitosis in Caenorhabditis elegans. Mol Biol Cell. 2000;11:1401–1419. doi: 10.1091/mbc.11.4.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, van Oudenaarden A, Jaenisch R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MM, Ceol CJ, Lu X, Horvitz HR. Some C. elegans class B synthetic multivulva proteins encode a conserved LIN-35 Rb-containing complex distinct from a NuRD-like complex. Proc Natl Acad Sci U S A. 2006;103:16782–16787. doi: 10.1073/pnas.0608461103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O, Moerman DG, Clark KA, Beckerle MC, Ruvkun G. A conserved LIM protein that affects muscular adherens junction integrity and mechanosensory function in Caenorhabditis elegans. J Cell Biol. 1999;144:45–57. doi: 10.1083/jcb.144.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdeman R, Nehrt S, Strome S. MES-2, a maternal protein essential for viability of the germline in Caenorhabditis elegans, is homologous to a Drosophila Polycomb group protein. Development. 1998;125:2457–2467. doi: 10.1242/dev.125.13.2457. [DOI] [PubMed] [Google Scholar]
- Korf I, Fan Y, Strome S. The Polycomb group in Caenorhabditis elegans and maternal control of germline development. Development. 1998;125:2469–2478. doi: 10.1242/dev.125.13.2469. [DOI] [PubMed] [Google Scholar]
- Krause M, Park M, Zhang JM, Yuan J, Harfe B, Xu SQ, Greenwald I, Cole M, Paterson B, Fire A. A C. elegans E/Daughterless bHLH protein marks neuronal but not striated muscle development. Development. 1997;124:2179–2189. doi: 10.1242/dev.124.11.2179. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei H, Fukushige T, Niu W, Sarov M, Reinke V, Krause M. A widespread distribution of genomic CeMyoD binding sites revealed and cross validated by ChIP-Chip and ChIP-Seq techniques. PLoS ONE. 2010;5:e15898. doi: 10.1371/journal.pone.0015898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyola A, Almouzni G. Histone chaperones, a supporting role in the limelight. Biochim Biophys Acta. 2004;1677:3–11. doi: 10.1016/j.bbaexp.2003.09.012. [DOI] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K, Yu R, Mohri K, Ono S. Caenorhabditis elegans kettin, a large immunoglobulin-like repeat protein, binds to filamentous actin and provides mechanical stability to the contractile apparatuses in body wall muscle. Mol Biol Cell. 2006;17:2722–2734. doi: 10.1091/mbc.E06-02-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole RJ, Bashllari E, Cochella L, Flowers EB, Hobert O. A Genome-Wide RNAi Screen for Factors Involved in Neuronal Specification in Caenorhabditis elegans. PLoS Genet. 2011;7:e1002109. doi: 10.1371/journal.pgen.1002109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechtsteiner A, Ercan S, Takasaki T, Phippen TM, Egelhofer TA, Wang W, Kimura H, Lieb JD, Strome S. The histone H3K36 methyltransferase MES-4 acts epigenetically to transmit the memory of germline gene expression to progeny. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- Tursun B, Cochella L, Carrera I, Hobert O. A toolkit and robust pipeline for the generation of fosmid-based reporter genes in C. elegans. PLoS ONE. 2009;4:e4625. doi: 10.1371/journal.pone.0004625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tursun B, Patel T, Kratsios P, Hobert O. Direct conversion of C. elegans germ cells into specific neuron types. Science. 2011;331:304–308. doi: 10.1126/science.1199082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida O, Nakano H, Koga M, Ohshima Y. The C. elegans che-1 gene encodes a zinc finger transcription factor required for specification of the ASE chemosensory neurons. Development. 2003;130:1215–1224. doi: 10.1242/dev.00341. [DOI] [PubMed] [Google Scholar]
- van Steensel B. Chromatin: constructing the big picture. Embo J. 2011;30:1885–1895. doi: 10.1038/emboj.2011.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Fong Y, Strome S. The Caenorhabditis elegans maternal-effect sterile proteins, MES-2, MES-3, and MES-6, are associated in a complex in embryos. Proc Natl Acad Sci U S A. 2001;98:5061–5066. doi: 10.1073/pnas.081016198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem. 2011;286:7983–7989. doi: 10.1074/jbc.M110.194027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzyuk T, Fakhouri TH, Kiefer J, Mango SE. The polycomb complex protein mes-2/E(z) promotes the transition from developmental plasticity to differentiation in C. elegans embryos. Dev Cell. 2009;16:699–710. doi: 10.1016/j.devcel.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Jones A, Sun CW, Li C, Chang CW, Joo HY, Dai Q, Mysliwiec MR, Wu LC, Guo Y, et al. PRC2 complexes with JARID2, MTF2, and esPRC2p48 in ES cells to modulate ES cell pluripotency and somatic cell reprogramming. Stem Cells. 2011;29:229–240. doi: 10.1002/stem.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Melton DA. Extreme makeover: converting one cell into another. Cell Stem Cell. 2008;3:382–388. doi: 10.1016/j.stem.2008.09.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.