Abstract
Skeletal muscle is a plastic organ that is maintained by multiple pathways regulating cell and protein turnover. During muscle atrophy, proteolytic systems are activated, and contractile proteins and organelles are removed, resulting in the shrinkage of muscle fibers. Excessive loss of muscle mass is associated with poor prognosis in several diseases, including myopathies and muscular dystrophies, as well as in systemic disorders such as cancer, diabetes, sepsis and heart failure. Muscle loss also occurs during aging. In this paper, we review the key mechanisms that regulate the turnover of contractile proteins and organelles in muscle tissue, and discuss how impairments in these mechanisms can contribute to muscle atrophy. We also discuss how protein synthesis and degradation are coordinately regulated by signaling pathways that are influenced by mechanical stress, physical activity, and the availability of nutrients and growth factors. Understanding how these pathways regulate muscle mass will provide new therapeutic targets for the prevention and treatment of muscle atrophy in metabolic and neuromuscular diseases.
Introduction
Muscles are the largest protein reservoir in the body. Muscles serve as a source of amino acids that can be used for energy production by various organs (including the heart, liver and brain) during catabolic periods, such as in cancer, sepsis, burn injury, heart failure and AIDS. However, excessive protein degradation in skeletal muscle, and the ensuing muscle loss (cachexia), is highly detrimental for the economy of the human body and can lead to death. Moreover, excessive loss of muscle mass is a poor prognostic indicator and can impair the efficacy of many different therapeutic treatments. Thus, cachexia ultimately aggravates diseases, and increases morbidity and mortality. The maintenance of healthy muscles is crucial for preventing metabolic disorders, maintaining healthy aging and providing energy to vital organs during stress conditions.
In adults, muscle mass and performance adapt to different pathophysiological conditions via activating pathways that regulate protein turnover. Muscle mass, like that of any other tissue, depends on protein turnover and cell turnover (Sartorelli and Fulco, 2004). In terms of muscle regeneration, the main players are satellite cells, a population of undifferentiated myogenic cells located between the basal lamina and the plasma membrane of muscle fibers. Although these cells play a major role in postnatal muscle growth, recent studies have challenged the idea that satellite cells are required for muscle mass maintenance and hypertrophy in adulthood (Amthor et al., 2009; Blaauw et al., 2009; McCarthy et al., 2011; Raffaello et al., 2010; Sartori et al., 2009).
Atrophy is defined as a decrease in the size of a tissue or organ due to cellular shrinkage; the decrease in cell size is caused by the loss of organelles, cytoplasm and proteins. This Review discusses the latest findings and emerging concepts related to pathways controlling muscle atrophy in physiological and pathological conditions. In particular, we focus on the ubiquitin-proteasome machinery and the autophagy-lysosome machinery, the two most important cell proteolytic systems that control protein turnover in muscle. The involvement of these systems in muscle physiopathology, as well as the signaling pathways controlling their activity, have been unraveled only in recent years, and evidence indicates that these two processes play a pivotal role in regulating overall muscle homeostasis.
The ubiquitin-proteasome system
In muscle, the ubiquitin-proteasome system is required to remove sarcomeric proteins upon changes in muscle activity. A decrease in muscle mass is associated with: (1) increased conjugation of ubiquitin to muscle proteins; (2) increased proteasomal ATP-dependent activity; (3) increased protein breakdown that can be efficiently blocked by proteasome inhibitors; and (4) upregulation of transcripts encoding ubiquitin, some ubiquitin-conjugating enzymes (E2), a few ubiquitin-protein ligases (E3) and several proteasome subunits (reviewed in Lecker et al., 2006) (Fig. 1). The human genome encodes more than 650 ubiquitin ligases (Lee and Goldberg, 2011), which are involved in the precise regulation of different cellular processes, and dramatic progress has been made recently in elucidating the roles of different E3s in regulating metabolism, transcription, cell cycle, oncogenesis and muscle size. Different E2–E3 pairs degrade different proteins; the specificity of E3s for certain groups of proteins provides exquisite selectivity to this degradation. The amount of different E2 and E3 proteins varies between tissues types and under different physiological conditions, but it is still unknown which specific E2s and E3s normally operate in muscle. Among the known E3s, only a few of them are muscle-specific and are upregulated during muscle loss (Sacheck et al., 2007).
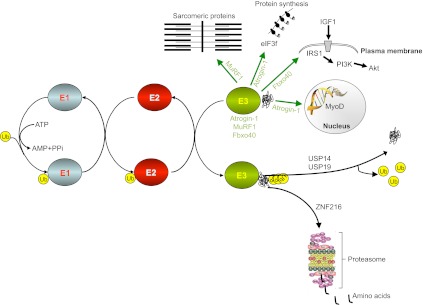
Fig. 1.
Ubiquitin-proteasome systems in muscle homeostasis. E1 enzymes activate ubiquitin proteins after the cleavage of ATP. The ubiquitin is then moved from E1 to members of the E2 enzyme class. The final ubiquitylation reaction is catalyzed by members of the E3 enzyme class. E3 binds to E2 and the protein substrate, inducing the transfer of ubiquitin from E2 to the substrate. Once the substrate is polyubiquitylated, it is docked to the proteasome for degradation. Note that polyubiquitin chains can be removed by de-ubiquitylating enzymes [ubiquitin-specific processing proteases (USPs)]. The components of this system that contribute to muscle wasting are depicted. ZNF216 is involved in the recognition and delivery to the proteasome of ubiquitylated proteins during muscle atrophy. Atrogin-1 regulates the half-life of the MyoD transcription factor and of eIF3f, which is crucial for protein synthesis. Fbxo40 regulates the half-life of IRS1, an essential factor for IGF1/insulin signaling, whereas MuRF1 regulates the half-life of several sarcomeric proteins. E3 ubiquitin ligases are depicted in green, with arrows pointing to their substrates. Note that ubiquitin ligases can have different cellular localizations and can shuttle into the nucleus. IRS1, insulin receptor substrate 1; Ub, ubiquitin.
A major contribution to the identification of which ubiquitin ligases are involved in muscle atrophy was provided by pioneering gene expression profiling performed independently by the groups of Alfred L. Goldberg and David J. Glass (Gomes et al., 2001; Bodine et al., 2001a). Comparing gene expression in different models of muscle atrophy led to the identification of a subset of genes that are commonly up- or downregulated in atrophying muscle. Because the diverse disease models used for these microarray experiments (including diabetes, cancer cachexia, chronic renal failure, fasting and denervation) have muscle atrophy in common, the commonly up- or downregulated genes are believed to regulate the loss of muscle components and are called atrophy-related genes or ‘atrogenes’ (Sacheck et al., 2007). Together, these findings revealed that muscle atrophy is an active process controlled by specific signaling pathways and transcriptional programs. Furthermore, the genes induced most strongly were found to encode two muscle-specific ubiquitin ligases, atrogin-1 (also known as MAFbx) and MuRF1 (Bodine et al., 2001a; Gomes et al., 2001) (Fig. 1). Notably, the transcription factor myogenin, an essential regulator of muscle development, is required for the maximal activation of these two genes, at least during denervation (Moresi et al., 2010).
Valuable information on the role of specific components of the ubiquitin-proteasome system in muscle was obtained by generating genetically modified animals (see Table 1). Mice lacking atrogin-1 and MuRF1 are resistant to muscle atrophy induced by denervation (Bodine et al., 2001a). Moreover, knockdown of atrogin-1 prevents muscle loss during fasting (Cong et al., 2011), whereas MuRF1 knockout mice (but not atrogin-1 knockout mice) are resistant to dexamethasone-induced muscle atrophy (Baehr et al., 2011). However, only a few muscle proteins have been identified as substrates for atrogin-1 thus far, and they all seem to be involved in growth-related processes or survival pathways. Atrogin-1 promotes degradation of MyoD, a key muscle transcription factor, and of eIF3f, an important activator of protein synthesis (Csibi et al., 2010; Tintignac et al., 2005) (Fig. 1). In the heart, atrogin-1 ubiquitylates and reduces the levels of calcineurin A, an important factor that triggers cardiac hypertrophy in response to pressure overload (Li et al., 2004). Conversely, MuRF1 was reported to interact and control the half-life of important muscle structural proteins, including troponin I (Kedar et al., 2004), myosin heavy chains (Clarke et al., 2007; Fielitz et al., 2007), myosin binding protein C and myosin light chain (Cohen et al., 2009) (Fig. 1).
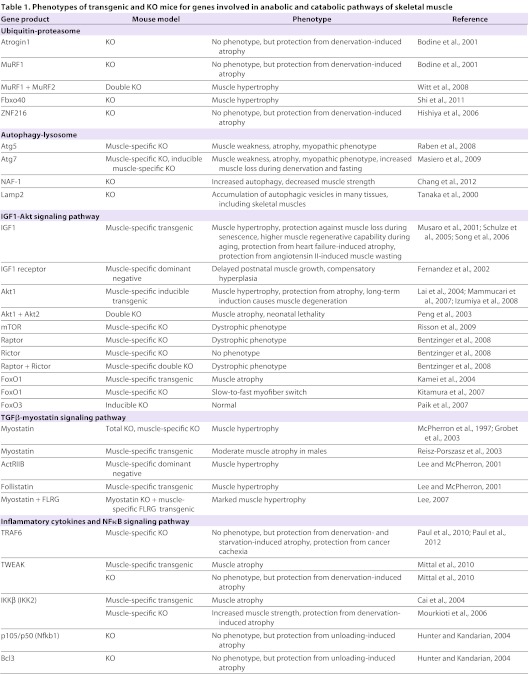
Table 1.
Phenotypes of transgenic and KO mice for genes involved in anabolic and catabolic pathways of skeletal muscle
Presumably, several other E3s are activated during atrophy that promote the clearance of soluble cell proteins and limit anabolic processes. A recent paper reported Trim32 as a crucial E3 ligase for the degradation of thin filaments (actin, tropomyosin and troponins), α-actinin and desmin (Cohen et al., 2012). However, Trim32 knockout mice are not protected from atrophy, but show an impairment in the recovery of muscle mass after atrophy (Kudryashova et al., 2012). Another ligase that seems to play an important role in atrophy is TRAF6 (Paul et al., 2010), an E3 ubiquitin ligase that mediates the conjugation of Lys63-linked polyubiquitin chains to its target proteins. Lys48-linked polyubiquitin chains are a signal for proteasome-dependent degradation, but they have other additional functions, such as regulating certain cell signaling pathways [e.g. modulating nuclear factor-κB (NFκB) activity] and being involved in autophagy-dependent cargo recognition via interacting with the scaffold protein p62 (also known as SQSTM1) (Kirkin et al., 2009; Komatsu et al., 2007; Pankiv et al., 2007). Notably, muscle-specific TRAF6 knockout mice have a decreased amount of polyubiquitylated proteins and almost no Lys63-polyubiquitylated proteins in starved muscles (Paul et al., 2012). TRAF6 knockout mice are resistant to muscle loss induced by denervation, cancer or starvation (Kumar et al., 2012; Paul et al., 2012; Paul et al., 2010). The mechanism of this protection involves both direct and indirect effects of TRAF6 on protein breakdown. In fact, TRAF6-mediated ubiquitylation is required for the optimal activation of JNK, AMPK, FoxO3 and NFκB (Paul et al., 2012). All of these factors are crucial regulators of atrogin-1 and MuRF1 expression (discussed in more detail below). Moreover, they also control the expression of several autophagy-related genes. Inhibition of TRAF6 reduces their maximal induction, thereby preserving muscle mass under catabolic conditions.
Another ligase that is involved in the ubiquitylation of filamin C, a muscle protein found in the Z-line, is CHIP (Arndt et al., 2010). However, CHIP-mediated ubiquitylation causes a lysosomal-dependent degradation of filamin C. Filamin is a protein that undergoes unfolding and refolding cycles during muscle contraction and is therefore prone to irreversible damage (Arndt et al., 2010). Alterations to filamin structure trigger the binding of the co-chaperone BAG3, which carries a complex made up of the chaperones Hsc70 and HspB8, as well as the ubiquitin ligase CHIP. CHIP ubiquitylates BAG3 and filamin, which are recognized and delivered to the autophagy system by p62 (Fig. 2) (Arndt et al., 2010). Interestingly, filamin B is controlled, at least during myogenesis, by another ubiquitin ligase, ASB2β, which is mainly expressed in muscle cells. In this case, the ubiquitylation of filamin B by ASB2β leads to proteasome-dependent degradation (Bello et al., 2009).
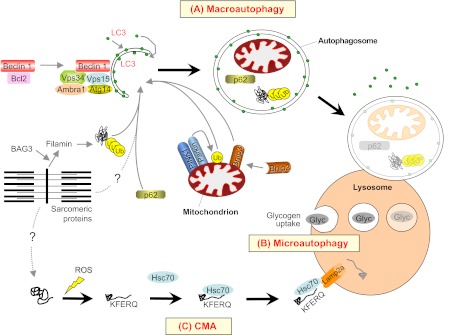
Fig. 2.
Macroautophagy, microautophagy and chaperone-mediated autophagy (CMA), and their contribution to protein degradation and organelle removal in skeletal muscle. (A) Macroautophagy is triggered by the activation of a regulatory complex (containing Vps34, Beclin 1, Vps15, Ambra1 and Atg14) that induces LC3 recruitment to the nascent autophagosome (isolation membrane). Selective removal of mitochondria (mitophagy; a specific form of macroautophagy) requires the PINK1-parkin complex and Bnip3 factors. Proteins that are committed for lysosomal degradation (including BAG3 and filamin, shown here) are labeled by polyubiquitin chains and delivered to the autophagosome by the p62 scaffold protein. (B) Microautophagy involves the direct engulfment of small portions of cytoplasm into lysosomes. Glycogen (Glyc) is reportedly taken up and broken down by microautophagy in skeletal muscle. (C) In CMA, proteins that are damaged by different agents, such as reactive oxygen species (ROS), expose a specific amino acid sequence (the KFERQ motif) that is recognized by the Hsc70 chaperone, which in turn delivers them to the lysosome via interaction with Lamp2a receptors. Dotted lines depict pathways whose molecular mechanisms and roles in adult skeletal muscle have not yet been fully defined.
In skeletal muscle, E3 ligases can also have important regulatory functions on signaling pathways. For example, it was recently found that the ubiquitin ligase Fbxo40 regulates anabolic signals (Shi et al., 2011). Fbxo40 ubiquitylates and affects the degradation of IRS1, a downstream effector of insulin-receptor-mediated signaling (Fig. 1). Inhibition of Fbxo40 by RNAi induces hypertrophy in myotubes, whereas Fbxo40 knockout mice display bigger muscle fibers (Shi et al., 2011).
Although some of the E3 ligases involved in muscle protein ubiquitylation and breakdown have now been identified, still little is known about how ubiquitylated proteins are recognized and delivered to the proteasome. A recent report identified ZNF216 as an important player in the recognition and delivery to the proteasome of ubiquitylated proteins during muscle atrophy (Fig. 1). Interestingly, ZNF216 is upregulated by FoxO transcription factors in atrophying muscles, and ZNF216-deficient mice are partially resistant to muscle loss during denervation. The absence of ZNF216 in muscle leads to the accumulation of polyubiquitylated proteins (Hishiya et al., 2006).
Much research in this field is focused on the ubiquitylation process, but little is known about the role of the de-ubiquitylating system and its contribution to muscle atrophy. The largest class of de-ubiquitylating enzymes are the ubiquitin-specific processing proteases (USPs) and, thus far, only two (USP14 and USP19) have been found to be upregulated in atrophying muscles (Fig. 1) (Combaret et al., 2005; Gomes et al., 2001). Knocking down USP19 expression in myotubes results in a decrease in protein degradation and reverts dexamethasone-induced loss of myosin heavy chain (Sundaram et al., 2009).
The autophagy-lysosome system
Autophagy plays a crucial role in the turnover of cell components both in constitutive conditions and in response to various stimuli, such as cellular stress, nutrient deprivation, amino acid starvation and cytokines (Mizushima et al., 2008). Three different mechanisms have been described in mammals for the delivery of the autophagic cargo to lysosomes: macroautophagy, chaperone-mediated autophagy (CMA) and microautophagy (Fig. 2). Thus far, most data on the role of the autophagic process in muscle are related to macroautophagy. Although it is still unknown whether microautophagy occurs in skeletal muscle, some findings indicate that microautophagy can participate in glycogen uptake into lysosomes when macroautophagy is blocked (Raben et al., 2008; Takikita et al., 2010). CMA has raised interest owing to its potential role in aging, neurodegenerative disorders and lysosomal storage diseases (Kon and Cuervo, 2010); however, whether it has roles in muscle homeostasis or atrophy is still largely unknown. In a study analyzing the contribution of CMA to protein breakdown in various organs of fasted rats, it was speculated that increased proteolysis in the liver and heart of fasted animals involves activation of CMA, but that this does not occur in skeletal muscles (Wing et al., 1991). The involvement of CMA in muscle homeostasis is an issue that needs to be addressed in detail in the coming years.
Autophagy was first described many years ago, but its involvement in muscle protein breakdown during atrophy was not recognized for a long time. Early evidence showed that lysosomal degradation contributes to protein breakdown in denervated muscle (Furuno et al., 1990; Schiaffino and Hanzlíková, 1972). Moreover, cathepsin L, a lysosomal protease, was shown to be upregulated during muscle atrophy (Deval et al., 2001). The development of molecular and imaging tools to follow autophagosome formation has greatly improved the characterization of autophagy in normal and atrophying muscles (Klionsky et al., 2008). In fact, analysis of different organs revealed that skeletal muscle is one of the tissues with the highest rates of vesicle formation during fasting. Interestingly, fast glycolytic muscles display a higher content of autophagosomes than slow β-oxidative muscles (Mizushima et al., 2004). It is now known that myofiber atrophy induced by in vivo overexpression of constitutively active FoxO3 requires autophagy, and siRNA-mediated knockdown of LC3 (a protein that contributes to autophagosome formation) partially prevents FoxO3-mediated muscle loss (Mammucari et al., 2007). Other genetic models have confirmed the role of autophagy during muscle atrophy. For example, oxidative stress induced by the muscle-specific expression of a mutant superoxide dismutase protein (SOD1G93A) causes muscle atrophy mainly by activating autophagy, and attenuation of autophagy by shRNA-mediated knockdown of LC3 preserves muscle mass in SOD1G93A transgenic mice (Dobrowolny et al., 2008). In addition, knockdown of the dihydropyridine receptor (DHPR), an L-type Ca2+ channel, in adult muscle leads to increased levels of cytosolic neuronal nitric oxide synthase (nNOS) and FoxO3 activation, which induce oxidative stress and autophagosome formation, respectively (Piétri-Rouxel et al., 2010).
Autophagy is primarily considered to be a non-selective degradation pathway, but the significance of more selective forms of autophagy is becoming increasingly evident. Indeed, autophagy can trigger the selective removal of specific organelles, such as mitochondria via mitophagy. In mammals, parkin, PINK1, Bnip3 and Bnip3L have been shown to regulate mitophagy, and inactivation of the genes encoding these proteins leads to abnormal mitochondria (Fig. 2) (Bothe et al., 2000; Hara et al., 2006). PINK1 recruits parkin to mitochondria, where parkin promotes mitophagy through ubiquitylation of outer mitochondrial membrane proteins that are recognized by p62, which brings autophagic vesicles to ubiquitylated mitochondrial proteins (Narendra and Youle, 2011; Youle and Narendra, 2011). Bnip3 and Bnip3L reportedly bind directly to LC3, and can therefore recruit the growing autophagosome to mitochondria (Hanna et al., 2012; Novak et al., 2010). In atrophying muscle, the mitochondrial network is dramatically remodeled following fasting or denervation, and autophagy via Bnip3 contributes to mitochondrial remodeling (Romanello et al., 2010; Romanello and Sandri, 2010). Expression of the fission machinery is sufficient to cause muscle wasting in mice, whereas inhibition of mitochondrial fission prevents muscle loss during denervation, indicating that disruption of the mitochondrial network is a crucial amplificatory loop of the muscle atrophy program (Romanello et al., 2010; Romanello and Sandri, 2010). Conversely, impairment of basal mitophagy is deleterious to muscle homeostasis, and leads to the accumulation of damaged and dysfunctional mitochondria (Grumati et al., 2010). Besides mitophagy, other forms of selective autophagy probably play important roles in the maintenance of skeletal muscle homeostasis. For example, a proper rate of nucleophagy seems essential for nuclear remodeling of muscle fibers, as indicated by nuclear envelopathies, a group of genetic disorders characterized by increased nuclear fragility and giant autophagosomes that contain nuclear components (Park et al., 2009).
Although available data are still limited to a few components of the autophagic machinery, the phenotypes of mice with muscle-specific inactivation of genes encoding autophagy-related proteins clearly demonstrate the essential role of autophagy in muscle homeostasis (see Fig. 2 and Table 1). Ablation of Atg7, the unique E1 enzyme of the autophagic machinery, causes disorganized sarcomeres and activation of the unfolded protein response, which in turn triggers myofiber degeneration; this phenotype is associated with complete inhibition of autophagosome formation, leading to abnormal mitochondria, oxidative stress and accumulation of polyubiquitylated proteins (Masiero et al., 2009). These Atg7-null mice are affected by muscle weakness and atrophy, and they display several signs of myopathy. Another study showed that suppression of autophagy exacerbates fasting- and denervation-induced atrophy in Atg7-null mice (Masiero and Sandri, 2010). A similar phenotype was observed in mice with muscle-specific ablation of Atg5, another crucial component of the autophagy machinery (Raben et al., 2008). Another recent mouse study revealed that nutrient-deprivation autophagy factor-1 (Naf-1), a Bcl-2-associated autophagy regulator, is required for the homeostatic maintenance of skeletal muscle. Naf1-null mice display muscle weakness and markedly decreased strength, accompanied by increased autophagy, dysregulation of calcium homeostasis and enlarged mitochondria (Chang et al., 2012).
Recent genetic studies by the group of Eric N. Olson revealed a key role for histone deacetylases (HDACs) in the control of skeletal muscle homeostasis and autophagic flux (Moresi et al., 2012; Moresi et al., 2010) (Table 1). Muscle-specific ablation of both HDAC1 and HDAC2 results in partial perinatal lethality, and HDAC1/2 double-knockout mice surviving postnatally develop a progressive myopathy characterized by impaired autophagy. HDAC1 and HDAC2 were found to regulate muscle autophagy by inducing the expression of autophagy genes. Notably, feeding HDAC1/2 knockout mice a high-fat diet releases the block in autophagy and prevents myopathy (Moresi et al., 2012). Another study by the same group also revealed roles for HDAC4 and HDAC5, as well as for the transcriptional muscle regulator myogenin, in modulating atrophy following denervation (Moresi et al., 2010). These studies demonstrated that myogenin activates the expression of the E3 ubiquitin ligases atrogin-1 and MuRF1, and that myogenin knockout mice fail to upregulate atrogin-1 and MuRF1 following denervation and are protected from atrophy. Similarly, mice lacking both HDAC4 and HDAC5 in skeletal muscle fail to upregulate myogenin and preserve muscle mass following denervation (Moresi et al., 2010).
The crucial role of the autophagy-lysosome system in skeletal muscles is confirmed by the fact that alterations to this process contribute to the pathogenesis of several genetic muscle diseases. Autophagy has a dual role in muscle homeostasis: it can be detrimental and contribute to muscle degeneration, but can also be a compensatory mechanism for cell survival. Congenital muscular dystrophies that are linked to the extracellular matrix (ECM) proteins collagen VI and laminin-2 illustrate the opposite effects that autophagy can have in skeletal muscles (Carmignac et al., 2011; Grumati et al., 2010). Our recent work indicated that autophagy plays a protective role against muscle weakness and wasting in Bethlem myopathy and Ullrich congenital muscular dystrophy, which are two inherited muscle disorders associated with collagen VI deficiency (Bernardi and Bonaldo, 2008; Grumati et al., 2010). A failure of the autophagic machinery is responsible for the inefficient removal and persistence of altered mitochondria in myofibers of Col6a1-null mice, and of Bethlem and Ullrich patients. Notably, reactivation of the autophagic flux in collagen-VI-deficient muscles (through diet or with pharmacological tools) can eliminate altered organelles and rescue the myopathic phenotype, showing promising therapeutic potential for counteracting muscle atrophy and weakness in these diseases (Grumati et al., 2010). Lack of collagen VI has a remarkable impact on molecules that are involved in the regulation of autophagy, decreasing beclin-1 and Bnip3 protein levels and causing persistent activation of the Akt [also known as protein kinase B (PKB)]-mTOR (mammalian target-of rapamycin) pathway, even during starvation (Grumati et al., 2010; Grumati et al., 2011a). However, the full details of the molecular axis that transduces collagen VI signals from the ECM to the autophagy machinery remain to be elucidated.
Further studies revealed that abnormal regulation of autophagy is involved in the pathogenesis of other mouse models of muscular dystrophies. Unlike mice with collagen VI deficiency, dy3K/dy3K mice [which lack laminin-2 and represent a model for merosin-deficient congenital muscular dystrophy type 1A (MDC1A)] display a general upregulation of autophagy-related genes, and pharmacological inhibition of autophagy significantly improves their dystrophic phenotype (Carmignac et al., 2011). Therefore, inappropriate regulation of autophagy is pathogenic in the two most common forms of congenital muscular dystrophy, which are both linked to deficiency of ECM proteins (collagen IV or laminin-2). How the absence of either of these two ECM proteins results in such opposing effects on autophagy remains undefined, but it is clear that alterations to the main components of muscle endomysial ECM have a marked impact on the regulation of autophagy.
Accumulation of autophagosomes occurs in many myopathies and represents the major feature of a group of muscle disorders named autophagic vacuolar myopathies (Malicdan et al., 2008). These myopathies are characterized by mutation of genes encoding proteins involved in lysosomal function, and include Pompe disease (caused by a defect in lysosomal acid α-glucosidase), Danon disease (caused by mutations of the LAMP2 gene) and X-linked myopathy with excessive autophagy (XMEA; which is associated with mutations of the VMA21 gene) (Malicdan et al., 2008; Ramachandran et al., 2009). It remains to be established whether accumulation of autophagosomes in these myopathies contributes to muscle damage or, conversely, whether it is a compensatory effect. Indeed, for many years the pathogenic defects of Pompe disease were attributed to impairment and rupture of lysosomes. However, this view was challenged by the newly proposed idea that the massive accumulation of autophagosomes (resulting from defective autophagy) is the main event causing myofibrillar disorganization and altered endocytic trafficking (Fukuda et al., 2006a; Fukuda et al., 2006b). Recently, additional inherited muscle disorders were shown to be related to increased autophagy. For example, mutations that inactivate MTMR14 (also known as Jumpy), a phosphatase counteracting the action of Vps34 in autophagosome formation and reducing the autophagy flux, have been associated with centronuclear myopathy (Vergne et al., 2009).
Together, the above findings in normal and diseased muscle clearly indicate that a proper balance of the autophagic flux is essential for maintaining healthy skeletal muscle, and that unbalanced autophagy is a main pathogenic mechanism in many muscle diseases. Thus, too much autophagy impairs myofiber homeostasis, causing excessive removal of cellular components that are needed for normal activities and leading to muscle atrophy when excessive catabolic activity is sustained for long periods. Insufficient autophagy also impairs myofiber homeostasis, leading to accumulation of damaged or dysfunctional cell components, with structural and functional impairment causing muscle weakness.
Besides its obvious pathogenic role in several muscle diseases, it is also conceivable that a slight and chronic unbalance of the autophagic process might significantly contribute to sarcopenia, the excessive loss of muscle mass that occurs in the elderly. An abnormal regulation of autophagy in aged individuals might interfere with the contractile properties of myofibers and render them less stable and more susceptible to contraction-induced damage, eventually leading to muscle atrophy (Cuervo, 2008). Although few studies have investigated autophagy in skeletal muscle of aged individuals, recent data confirmed that unbalanced autophagy might contribute to sarcopenia. Findings in rat muscles showed an age-related decline in autophagic degradation and a concomitant age-related increase in oxidative damage and apoptosis, which were both negatively correlated with autophagy. Interestingly, a constant autophagic stimulus, such as caloric restriction, was found to ameliorate the physiological state of muscles during aging (Wohlgemuth et al., 2009). During aging, there is also a progressive deterioration of mitochondrial function and activation of autophagy. Forced expression of proliferator-activated receptor gamma coactivator-1α (PGC1α; a master gene of mitochondrial biogenesis) ameliorates loss of muscle mass and prevents the age-related increase in autophagy (Wenz et al., 2009). An interesting link between autophagy and aging was recently described in Drosophila, where the maintenance of a normal autophagic flux in aged muscles was found to induce a beneficial extension of lifespan. Selective activation of autophagy in skeletal muscle via FoxO prevents accumulation of protein aggregates and, consequently, muscle weakness, thus providing the first genetic evidence correlating FoxO, autophagy and healthy muscles to lifespan extension (Demontis and Perrimon, 2010).
Other recent work has demonstrated a link between physical exercise and autophagy in muscle. We have shown that physical exercise is very effective in stimulating autophagy in skeletal muscles, and that the accompanying clearance of damaged cell components and dysfunctional mitochondria is crucial for muscle homeostasis (Grumati et al., 2011b). Further work extended these observations and demonstrated that exercise-induced autophagy plays an important and previously unrecognized role in muscle metabolism (He et al., 2012). These findings provide a mechanism for explaining the well-known beneficial effects of physical activity in healthy individuals.
Signaling pathways regulating muscle atrophy
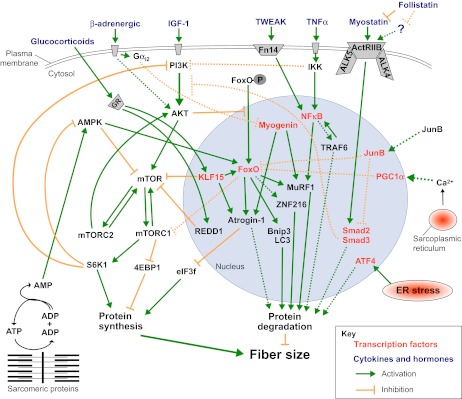
Many recent findings have highlighted a complex scenario whereby an intricate network of signaling pathways regulates the size of myofibers and the contractile performance of muscle. Intriguingly, these different pathways crosstalk and modulate one another at different levels, coordinating protein synthesis and degradation simultaneously (Fig. 3). Below, we discuss the involvement of four major pathways (IGF1-Akt-FoxO, myostatin, NFκB and glucocorticoids) in muscle atrophy.
Fig. 3.
Major pathways that control muscle fiber size. Protein synthesis and degradation are regulated by several different stimuli, which activate multiple signaling pathways, many of which converge at common intermediates and/or crosstalk with one another. Many of the components shown here could be promising therapeutic targets. See the main text for further details. Dotted lines depict pathways whose molecular mechanisms and role in adult skeletal muscle have yet to be completely defined. GR, glucocorticoid receptor.
IGF1-Akt-FoxO signaling
Insulin-like growth factor 1 (IGF1), a circulating growth factor, is also produced locally by many tissues, including skeletal muscle (reviewed in Schiaffino and Mammucari, 2011). Muscle-specific overexpression of a locally acting IGF1 isoform in mice showed that localized IGF1 expression sustains muscle growth and regeneration (Musarò et al., 2001) (Table 1). Indeed, these mice have muscle hypertrophy and display higher regenerative capacity and maintenance of muscle mass during aging (Musarò et al., 2001). Other studies have shown that IGF1 and/or insulin signaling can suppress protein breakdown while promoting muscle growth (Monier et al., 1983; Rommel et al., 2001; Sacheck et al., 2004). Furthermore, IGF1 transgenic mice are resistant to muscle atrophy induced by angiotensin treatment, and to cardiac cachexia (Schulze et al., 2005; Song et al., 2005). Furthermore, local IGF1 injection is sufficient to block disuse atrophy (Stitt et al., 2004). In these models of muscle loss, IGF1 completely suppressed the induction of the two crucial ubiquitin ligases.
Additional data supporting the role of the IGF1 pathway in regulating muscle atrophy derive from studies of Akt. Electroporation of constitutively active Akt in adult myofibers completely blocks muscle atrophy induced by denervation (Bodine et al., 2001b). Generation of transgenic mice overexpressing Akt in skeletal muscle further demonstrated the crucial role of Akt in regulating muscle size (Table 1). These transgenic mice display muscle hypertrophy and protection from denervation-induced atrophy (Izumiya et al., 2008; Lai et al., 2004; Mammucari et al., 2007), proving that the Akt pathway can promote muscle growth and simultaneously block protein degradation (Lai et al., 2004; Sartori et al., 2009). A more recent study indicated that the IGF1-Akt axis can control myofibril growth and maintenance by a mechanism acting through nebulin and N-WASP (Takano et al., 2010).
Akt controls both protein synthesis, via mTOR, and protein degradation, via transcription factors of the FoxO family (Fig. 3). mTOR is a crucial kinase downstream of insulin and nutrient-sensitive pathways required for cell growth. mTOR is part of two different complexes, the rapamycin-sensitive TORC1 complex, which contains Raptor, and the rapamycin-insensitive TORC2 complex, which contains Rictor (for a review, see Laplante and Sabatini, 2012). The phenotypes of mice with muscle-specific ablation of mTOR, Raptor and Rictor provide valuable information (Table 1): Rictor knockout muscles do not show any overt phenotype, but Raptor- and mTOR-deficient mice display a progressive muscular dystrophy phenotype (Bentzinger et al., 2008; Raben et al., 2008; Risson et al., 2009).
The upregulation of atrogin-1, MuRF1 and several autophagy-related genes is normally blocked by Akt through negative regulation of FoxO transcription factors (Lee et al., 2004; Sandri et al., 2004; Stitt et al., 2004). The FoxO family in skeletal muscle is comprised of three isoforms: FoxO1, FoxO3 and FoxO4. Akt phosphorylates FoxO proteins, promoting their export from the nucleus to the cytoplasm. As predicted, the reduced activity of the Akt pathway observed in different models of muscle atrophy results in decreased levels of phosphorylated FoxO in the cytoplasm and a marked increase of nuclear FoxO (reviewed in Calnan and Brunet, 2008) (Fig. 3). The translocation and activity of FoxO members is required for the upregulation of atrogin-1 and MuRF1, and FoxO3 is sufficient to promote atrogin-1 expression and muscle atrophy when transfected into skeletal muscles in vivo (Sandri et al., 2004). FoxO1 transgenic mice show markedly reduced muscle mass and fiber atrophy, further supporting the notion that FoxO proteins are sufficient to promote muscle loss (Kamei et al., 2004; Southgate et al., 2007). In contrast, FoxO knockdown by RNAi can block the upregulation of atrogin-1 expression during atrophy and muscle loss (Liu et al., 2007; Sandri et al., 2004).
Crosstalk between protein breakdown and protein synthesis is not limited to Akt, but also involves FoxO. Activation of FoxO in Drosophila muscle upregulates 4E-BP1 (Demontis and Perrimon, 2010) and represses mTOR via sestrin (Lee et al., 2010). Consistently, in mammals, FoxO3 reduces total protein synthesis in adult muscle (Reed et al., 2012). Thus, when Akt is active, protein breakdown is suppressed, and when FoxO is induced, protein synthesis is further suppressed. This is not trivial, because FoxO activity is regulated by several different post-translational modifications, including phosphorylation, acetylation, and mono-and polyubiquitylation (Huang and Tindall, 2007). Indeed, recent evidence suggests that acetylation negatively regulates FoxO3 activity, whereas it has no effect on FoxO1 (Senf et al., 2011). Mutants of FoxO3 that mimic acetylation show a cytosolic localization and a lower ability to induce transcription of the gene encoding atrogin-1, and to induce muscle atrophy (Bertaggia et al., 2012). Most of these regulatory mechanisms are Akt-independent and might play a role in muscle atrophy induced by oxidative or energy stress (see below).
Other findings have revealed a connection between AMPK and FoxO3 (Fig. 3). AMPK phosphorylates several Akt-independent sites on FoxO3, thereby stimulating its transcriptional activity (Greer et al., 2007a; Greer et al., 2007b). Indeed, treatment of muscle cultures with AICAR, an activator of AMPK, increases protein breakdown and atrogin-1 expression via the FoxO family (Nakashima and Yakabe, 2007). We recently found that FoxO3 is activated via AMPK in myofibers to induce the expression of atrogin-1 and MuRF1 in conditions of energy stress (Romanello et al., 2010). Activation of AMPK also leads to induction of some autophagy-related genes, such as those coding for LC3 and Bnip3.
Increased oxidative stress occurs during denervation and hindlimb suspension. During these disuse conditions, nNOS moves from the sarcolemma, where it is bound to the dystrophin-glycoprotein complex, to the cytosol. Free cytosolic nNOS induces oxidative stress and enhances FoxO3-mediated transcription of atrogin-1 and MuRF1, thereby causing muscle loss (Suzuki et al., 2007). Interestingly, the NFκB pathway is not involved in nNOS-mediated muscle atrophy (Suzuki et al., 2007). Similarly, when DHPR is reduced in adult muscle by RNA interference (RNAi), it also triggers atrophy via nNOS relocalization and FoxO3 activation (Piétri-Rouxel et al., 2010). However, in this latter setting, the genes upregulated by FoxO3 are those coding for the autophagy regulators LC3, Vps34 and Bnip3, and the gene encoding the lysosomal enzyme cathepsin L. In humans, the diaphragm of patients that are mechanically ventilated undergoes rapid atrophy by activating different proteolytic systems, including autophagy (Levine et al., 2008). Prolonged mechanical ventilation induces diaphragm disuse that, in turn, leads to activation of autophagy through Akt inhibition and FoxO1 induction. Interestingly, oxidative stress is increased and therefore contributes to FoxO activation in this setting of disuse-mediated atrophy.
FoxO activity is also modulated by direct or indirect actions of cofactors and by interacting with other transcription factors. FoxO has been found to interact with PGC1α, a critical cofactor for mitochondrial biogenesis (Puigserver et al., 2003; Wu et al., 1999). Maintaining high levels of PGC1α during catabolic conditions (either in transgenic mice or by transfecting adult myofibers) spares muscle mass during denervation, fasting, heart failure, aging and sarcopenia – similar to the effect observed with expression of constitutively active FoxO3 (Geng et al., 2011; Sandri et al., 2006; Wenz et al., 2009). Similar beneficial effects were recently obtained by overexpression of PGC1β, a homolog of PGC1α (Brault et al., 2010). The positive action on muscle mass of these cofactors is due to the inhibition of autophagy-lysosome and ubiquitin-proteasome degradation systems. PGC1α and PGC1β reduce protein breakdown by inhibiting the transcriptional activity of FoxO3 and NFκB, but they do not affect protein synthesis. Thus, these cofactors prevent the excessive activation of proteolytic systems by inhibiting the action of the pro-atrophy transcription factors without perturbing the translational machinery.
We recently reported that the transcription factor JunB blocks atrophy and promotes hypertrophy in adult mouse muscles (Raffaello et al., 2010). Indeed, JunB can block myofiber atrophy of denervated tibialis anterior muscles and cultured myotubes induced by FoxO3 overexpression, dexamethasone treatment or starvation. In these conditions, JunB prevents the activation of atrogin-1 and partially that of MuRF1, thereby reducing the increase in overall protein degradation induced by activated FoxO3. Further analysis revealed that JunB does not inhibit FoxO3-mediated activation of the autophagy-lysosome system, but only the ubiquitin-proteasome degradation, through inhibiting atrogin-1 and MuRF1 induction during catabolic conditions. In fact, JunB directly binds FoxO3, thereby preventing its recruitment to the promoters of key atrogenes. Moreover, JunB overexpression is sufficient to induce dramatic hypertrophy of myotubes and of adult muscle. These hypertrophic changes depend on increased protein synthesis, without affecting the basal rate of protein degradation. The growth-promoting effects mediated by JunB in muscle resemble the effects of inhibiting the transforming grown factor-β (TGFβ) pathway (Sartori et al., 2009; Trendelenburg et al., 2009). Indeed, JunB overexpression markedly suppresses myostatin expression in transfected myotubes and decreases the phosphorylation of Smad3, the transcription factor downstream of the myostatin-TGFβ signaling pathway (Raffaello et al., 2010).
Inflammatory cytokines and NFκB signaling
NFκB transcription factors, which play major roles as mediators of immunity and inflammation, are also expressed in skeletal muscle, where they mediate the effect of inflammatory cytokines, particularly tumor necrosis factor-α (TNFα), on muscle wasting and cachexia (reviewed in Peterson et al., 2011) (Fig. 3). In the inactive state, NFκB is sequestered in the cytoplasm by a family of inhibitory proteins called IκB. In response to TNFα, the IκB kinase (IKKβ) complex phosphorylates IκB, resulting in its ubiquitylation and proteasomal degradation. This leads to nuclear translocation of NFκB and activation of NFκB-mediated gene transcription (Peterson et al., 2011).
Muscle-specific overexpression of IKKβ in transgenic mice leads to severe muscle wasting mediated, at least in part, by the ubiquitinligase MuRF1, but not by atrogin-1 (Cai et al., 2004) (Table 1). In contrast, muscle-specific inhibition of NFκB by transgenic expression of a constitutively active IκB mutant does not induce an overt phenotype, but denervation atrophy is substantially reduced (Judge et al., 2007). Muscle atrophy induced by hindlimb unloading is likewise reduced in mice deficient for the p105/p50 subunit of NFκB (Hunter and Kandarian, 2004). However, TNFα and pro-inflammatory cytokines also cause insulin resistance and suppression of the IGF1-Akt pathway (de Alvaro et al., 2004; Dogra et al., 2007; Hirosumi et al., 2002). Therefore, Akt phosphorylation should always be explored when the NFκB pathway is perturbed, because Akt inhibition can substantially contribute to muscle atrophy. This concept is supported by the phenotype of IKKβ conditional knockout mice, which are resistant to muscle atrophy but show hyperphosphorylation of Akt (Mourkioti et al., 2006). The significance of decreased muscle atrophy following IKKβ ablation and the degree to which this effect is Akt dependent remain unclear. Nevertheless, these findings highlight the relevance of the crosstalk between the two pathways, and future studies are needed to elucidate the respective contributions of the IKKβ-NFκB and Akt-FoxO pathways to muscle atrophy.
A recent study revealed an unexpected connection between TNFα signaling and myogenin on MuRF1 and atrogin-1 expression: TNFα treatment causes upregulation of myogenin, MuRF1 and atrogin-1. Interestingly, a G-protein-coupled receptor blocks the TNFα-mediated myogenin upregulation by activating Gαi2 (Minetti et al., 2011) and the expression of muscle-specific ubiquitin ligases. However, the precise mechanisms of TNFα-mediated myogenin regulation, the interplay with Gαi2 and the implications for muscle wasting are still far from fully understood.
TNF-like weak inducer of apoptosis (TWEAK) is a member of the TNF superfamily and was recently found to induce muscle atrophy (Dogra et al., 2007; Mittal et al., 2010). TWEAK acts on responsive cells by binding to fibroblast growth factor-inducible 14 (Fn14), a small cell-surface receptor (Fig. 3). Fn14 is upregulated in denervated muscle, allowing NFκB activation and consequently MuRF1 (but not atrogin-1) expression (Mittal et al., 2010). TWEAK knockout mice display less atrophy than wild-type mice during denervation, as well as reduced NFκB activation and MuRF1 expression (Table 1). However, Fn14 levels do not increase in all conditions of muscle atrophy; for instance, Fn14 is not induced by dexamethasone treatment. Another important player in NFκB signaling is the ubiquitin ligase TRAF6, which is required for Fn14 upregulation during fasting (Paul et al., 2012). As noted earlier, TRAF6 is also required for the activation of FoxO3 and AMPK in starved muscles, and for the induction of ubiquitin-proteasome and autophagy-lysosome systems (Paul et al., 2012).
Myostatin and crosstalk with Akt
Myostatin, a member of the TGFβ family, is expressed and secreted predominantly by skeletal muscle, and functions as a negative regulator of muscle growth. Mutations of the myostatin gene lead to a hypertrophic phenotype in mice, sheep and cattle, and a loss-of-function mutation in the human myostatin gene was also found to induce increased muscle mass (Clop et al., 2006; Lee and McPherron, 2001; McPherron and Lee, 1997; Schuelke et al., 2004). The increase in muscle mass caused by loss of myostatin is a consequence of both hyperplasia (i.e. increased cell number) and hypertrophy (i.e. increased cell size) (Lee, 2004; Lee and McPherron, 1999; McPherron et al., 1997).
In contrast, the capacity of myostatin activation to trigger muscle atrophy is not obvious. Although an early report described severe (even fatal) atrophy when Chinese hamster ovary (CHO) cells engineered to express myostatin were injected into mouse skeletal muscle (Zimmers et al., 2002), these findings were not confirmed in transgenic mice. Muscle-specific overexpression of myostatin leads to only 20% atrophy (i.e. a 20% decrease in muscle mass) in males and no phenotype in females (Reisz-Porszasz et al., 2003) (Table 1). Electroporation experiments showed that myostatin expression in adult muscle induces a degree of atrophy comparable to that observed in transgenic mice (Durieux et al., 2007). These findings suggest that the CHO cells used by Zimmers et al. contributed to muscle atrophy by secreting cachectic factors, and that the myostatin pathway synergizes with other pathways. However, in muscle cell cultures, myostatin was reported to upregulate essential atrophy-related ubiquitin ligases; notably, this regulation was found to be FoxO dependent (McFarlane et al., 2006). In fact, myostatin treatment blocks the IGF1-PI3K-Akt pathway and activates FoxO1, allowing increased expression of atrogin-1. This crosstalk between the two pathways does not require NFκB, whose inhibition does not prevent upregulation of atrogin-1 (McFarlane et al., 2006). Similarly, in cardiac cells, myostatin abrogates phenylephrine hypertrophic effects through Akt inhibition (Morissette et al., 2006). Furthermore, myostatin expression is controlled by FoxO1, supporting the concept that the myostatin pathway synergizes with Akt-FoxO signaling (Allen and Unterman, 2007).
Recent reports dissected the myostatin pathway both in vitro and in vivo, and showed that Smad2 and Smad3 are the transcription factors mediating myostatin effects on muscle mass (Lokireddy et al., 2011; Sartori et al., 2009; Trendelenburg et al., 2009) (Fig. 3). The downstream targets of Smad2 and Smad3 are not yet known, so the mechanisms of Smad-dependent atrophy remain to be established. Importantly, Smads can recognize the DNA sequence CAGAC, but their affinity is not sufficient to support unassisted binding to DNA. In addition, if their binding affinity for this simple sequence were higher, Smad proteins would decorate entire chromosomes. Therefore, activated Smads must associate with different DNA-binding cofactors for the recognition and regulation of specific target genes (Massagué et al., 2005). Interestingly, members of the FoxO family were found to play such a role (Gomis et al., 2006). Recent reports have identified an inhibitory effect of myostatin-Smad2/3 signaling on the IGF1-Akt-mTOR pathway (Amirouche et al., 2009; Sartori et al., 2009; Trendelenburg et al., 2009), thereby supporting the idea that there is synergy between FoxOs and Smads to activate an atrophic program. However, the growth-promoting effects of myostatin inhibition seem to be independent of the Akt pathway (Goncalves et al., 2010). The mechanism through which myostatin inhibition leads to hypertrophy is a key issue that needs to be addressed to better understand the therapeutic potential of anti-myostatin drugs. A recent study showed that inhibition of myostatin by soluble ActRIIB prevents and fully reverses skeletal muscle loss and atrophy of the heart in tumor-bearing animals (Zhou et al., 2010). Such treatment dramatically prolongs the survival of these animals, suggesting potential therapeutic efficacy in patients with cancer cachexia.
Glucocorticoid-induced muscle atrophy and the control of protein homeostasis
Glucocorticoid levels are increased in many pathological conditions associated with muscle loss. Consistently, glucocorticoid treatment induces atrogin-1 and MuRF1 expression and muscle wasting in cell culture and in vivo (Bodine et al., 2001b; Clarke et al., 2007; Sacheck et al., 2004; Sandri et al., 2004; Schakman et al., 2008). In contrast, adrenalectomy or treatment with a glucocorticoid receptor antagonist attenuates muscle loss in some diseases (Menconi et al., 2007; Schakman et al., 2008). The mechanisms of glucocorticoid-mediated muscle atrophy were recently unraveled (Fig. 3). Once in the nucleus, the glucocorticoid receptor activates the expression of two target genes, encoding REDD1 and KLF15 (Shimizu et al., 2011). REDD1 inhibits mTOR activity by sequestering 14-3-3 and increasing TSC1/2 activity. Inhibition of mTOR is permissive for the activation of an atrophy program via KLF15. Indeed, mTOR activation attenuates glucocorticoid-induced muscle atrophy. KLF15 is a transcription factor that is involved in several metabolic processes in skeletal muscle, for instance in the upregulation of the branched-chain aminotransferase 2 (BCAT2). KLF15 participates in muscle catabolism via transcriptional regulation of FoxO1, atrogin-1 and MuRF1. Moreover, KLF15 negatively affects mTOR through upregulation of BCAT2, which in turn induces branched-chain amino acid degradation. Interestingly, FoxO1 and glucocorticoid receptor cooperate to upregulate MuRF1 expression (Waddell et al., 2008).
Therapeutic perspectives for counteracting muscle atrophy
The mechanisms controlling muscle mass have attracted increasing interest in the scientific community because increased understanding of these can potentially help to tackle various clinical problems, including aging, the prognosis of many diseases, quality of life and sports medicine. The results of recent research offer new and exciting perspectives to the field, and have introduced several stimulating questions to the community, setting the foundation for future studies that will hopefully identify new therapeutic targets and drugs.
Several potentially interesting therapeutic targets have already been identified, although an effective drug that can counteract muscle wasting is not yet clinically available. The most interesting targets belong to the anabolic pathways and the ubiquitin-proteasome system. As we have discussed, Akt and its downstream targets seem to be at the intersection of several different pathways (see below), including β-adrenergic signaling, myostatin and JNK. Moreover, the IGF1-Akt axis is unique in that it controls both protein synthesis and protein degradation. Therefore, IGF1 analogs might be extremely useful for counteracting muscle loss and weakness. However, the same pathway plays major roles in other biological processes, including cell survival, and in other contexts it can promote tumorigenesis. Thus, the development of a new generation of IGF1 mimetics that specifically activate the Akt pathway in skeletal muscle is a goal for the field. Even if these are successfully developed, however, it should be considered that prolonged inhibition of protein degradation can have a major impact on protein quality control. Such agents could have major drawbacks, such as promoting the accumulation of misfolded or aggregate-prone proteins (Grumati et al., 2010; Masiero et al., 2009).
β-adrenergic agonists such as clenbuterol are considered pro-growth and anti-atrophic drugs. Most effects of clenbuterol are mediated by activating Akt-mTOR signaling (Kline et al., 2007), so the concerns associated with IGF1 stimulation can also be applied to β-adrenergic agonists. However, as noted earlier, a recent report revealed that the β-adrenergic signal might act via a different mechanism (involving hypertrophy induced by a G-protein-coupled receptor and Gαi2 through inhibition of GSK-3β and activation of p70S6K1) that is independent of the PI3K-Akt axis (Minetti et al., 2011). Notably, the contribution of p70S6K1 and its downstream target S6 to protein synthesis in muscle is uncertain, because S6K1 knockout mice show no impairment of polysome formation, protein synthesis or protein degradation (Mieulet et al., 2007). Therefore, S6 phosphorylation should not be considered a marker of protein synthesis, and we need a better understanding of which mTOR downstream targets are crucial regulators of protein synthesis in order to evaluate the role of this pathway in muscle growth and its potential in therapeutic approaches.
Another important factor that is currently being explored as a means to counteract muscle loss is myostatin. A recent report that inhibition of myostatin by using soluble ActRIIB prevents and fully reverses skeletal muscle loss, and prolongs the survival of tumor-bearing animals (Zhou et al., 2010), has opened the field to myostatin inhibition as a promising therapeutic approach. Despite the well-established growth-promoting action of myostatin inhibition, the anti-atrophic effect of myostatin blockade remains uncertain. In fact, expression of dominant-negative ActRIIB, or knockdown of Smad2 and Smad3, cannot prevent muscle loss during denervation (Sartori et al., 2009). Therefore, targeting the myostatin pathway might be better suited for muscle rehabilitation after injury than for prevention of muscle loss.
The last category of drug targets is represented by the proteasome system. Proteasome inhibitors have been successfully used to block atrophy in different animal models (Caron et al., 2011; Jamart et al., 2011; Supinski et al., 2009). However, the ubiquitin-proteasome system regulates many relevant biological processes and its prolonged inhibition might be detrimental for muscle cells. Indeed, patients chronically treated with bortezomib, a proteasome inhibitor that was approved by the FDA to treat multiple myeloma, display cardiac complications (Orciuolo et al., 2007). Therefore, more specific approaches that can target the ubiquitin ligases involved in ubiquitylation and degradation of sarcomeric proteins should be pursued. Among the different ubiquitin ligases, MuRF1 and TRAF6 seem interesting candidates for developing specific inhibitors. However, ablation of MuRF1 or TRAF6 only partially protects from muscle loss during denervation in mice (Bodine et al., 2001a; Paul et al., 2010), indicating that other ubiquitin ligases are also involved in protein degradation. Moreover, it is still unknown whether different muscle atrophic conditions recruit different sets of ubiquitin ligases. Nonetheless, the findings of these last few years have greatly enhanced our knowledge of protein synthesis and degradation in skeletal muscle, and there is increasing hope that it will be possible to develop efficient therapeutic approaches for counteracting muscle wasting in the near future.
Acknowledgments
We apologize to colleagues whose studies were not cited owing to space limitations.
Footnotes
FUNDING
Our work is supported by grants from Telethon-Italy (GGP10225 and GGP11082 to P.B.; TCP04009 to M.S.), from the European Union (MYOAGE, contract: 223576 of FP7 to M.S.; BIO-NMD FP7-HEALTH-241665 to P.B.), ERC (MyoPHAGY to M.S.), the Italian Ministry of Education, University and Research (to P.B. and M.S.) and CARIPARO (to M.S. and P.B.).
REFERENCES
- Allen D. L., Unterman T. G. (2007). Regulation of myostatin expression and myoblast differentiation by FoxO and SMAD transcription factors. Am. J. Physiol. Cell. Physiol. 292, C188–C199 [DOI] [PubMed] [Google Scholar]
- Amirouche A., Durieux A. C., Banzet S., Koulmann N., Bonnefoy R., Mouret C., Bigard X., Peinnequin A., Freyssenet D. (2009). Down-regulation of Akt/mammalian target of rapamycin signaling pathway in response to myostatin overexpression in skeletal muscle. Endocrinology 150, 286–294 [DOI] [PubMed] [Google Scholar]
- Amthor H., Otto A., Vulin A., Rochat A., Dumonceaux J., Garcia L., Mouisel E., Hourdé C., Macharia R., Friedrichs M., et al. (2009). Muscle hypertrophy driven by myostatin blockade does not require stem/precursor-cell activity. Proc. Natl. Acad. Sci. USA 106, 7479–7484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt V., Dick N., Tawo R., Dreiseidler M., Wenzel D., Hesse M., Fürst D. O., Saftig P., Saint R., Fleischmann B. K., et al. (2010). Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr. Biol. 20, 143–148 [DOI] [PubMed] [Google Scholar]
- Baehr L. M., Furlow J. D., Bodine S. C. (2011). Muscle sparing in muscle RING finger 1 null mice: response to synthetic glucocorticoids. J. Physiol. 589, 4759–4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello N. F., Lamsoul I., Heuzé M. L., Métais A., Moreaux G., Calderwood D. A., Duprez D., Moog-Lutz C., Lutz P. G. (2009). The E3 ubiquitin ligase specificity subunit ASB2beta is a novel regulator of muscle differentiation that targets filamin B to proteasomal degradation. Cell Death Differ. 16, 921–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentzinger C. F., Romanino K., Cloëtta D., Lin S., Mascarenhas J. B., Oliveri F., Xia J., Casanova E., Costa C. F., Brink M., et al. (2008). Skeletal muscle-specific ablation of raptor, but not of rictor, causes metabolic changes and results in muscle dystrophy. Cell Metab. 8, 411–424 [DOI] [PubMed] [Google Scholar]
- Bernardi P., Bonaldo P. (2008). Dysfunction of mitochondria and sarcoplasmic reticulum in the pathogenesis of collagen VI muscular dystrophies. Ann. N. Y. Acad. Sci. 1147, 303–311 [DOI] [PubMed] [Google Scholar]
- Bertaggia E., Coletto L., Sandri M. (2012). Posttranslational modifications control FoxO3 activity during denervation. Am. J. Physiol. Cell. Physiol. 302, C587–C596 [DOI] [PubMed] [Google Scholar]
- Blaauw B., Canato M., Agatea L., Toniolo L., Mammucari C., Masiero E., Abraham R., Sandri M., Schiaffino S., Reggiani C. (2009). Inducible activation of Akt increases skeletal muscle mass and force without satellite cell activation. FASEB J. 23, 3896–3905 [DOI] [PubMed] [Google Scholar]
- Bodine S. C., Latres E., Baumhueter S., Lai V. K., Nunez L., Clarke B. A., Poueymirou W. T., Panaro F. J., Na E., Dharmarajan K., et al. (2001a). Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294, 1704–1708 [DOI] [PubMed] [Google Scholar]
- Bodine S. C., Stitt T. N., Gonzalez M., Kline W. O., Stover G. L., Bauerlein R., Zlotchenko E., Scrimgeour A., Lawrence J. C., Glass D. J., et al. (2001b). Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat. Cell Biol. 3, 1014–1019 [DOI] [PubMed] [Google Scholar]
- Bothe G. W., Haspel J. A., Smith C. L., Wiener H. H., Burden S. J. (2000). Selective expression of Cre recombinase in skeletal muscle fibers. Genesis 26, 165–166 [PubMed] [Google Scholar]
- Brault J. J., Jespersen J. G., Goldberg A. L. (2010). Peroxisome proliferator-activated receptor gamma coactivator 1alpha or 1beta overexpression inhibits muscle protein degradation, induction of ubiquitin ligases, and disuse atrophy. J. Biol. Chem. 285, 19460–19471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D., Frantz J. D., Tawa N. E., Jr, Melendez P. A., Oh B. C., Lidov H. G., Hasselgren P. O., Frontera W. R., Lee J., Glass D. J., et al. (2004). IKKbeta/NF-kappaB activation causes severe muscle wasting in mice. Cell 119, 285–298 [DOI] [PubMed] [Google Scholar]
- Calnan D. R., Brunet A. (2008). The FoxO code. Oncogene 27, 2276–2288 [DOI] [PubMed] [Google Scholar]
- Carmignac V., Svensson M., Körner Z., Elowsson L., Matsumura C., Gawlik K. I., Allamand V., Durbeej M. (2011). Autophagy is increased in laminin α2 chain-deficient muscle and its inhibition improves muscle morphology in a mouse model of MDC1A. Hum. Mol. Genet. 20, 4891–4902 [DOI] [PubMed] [Google Scholar]
- Caron A. Z., Haroun S., Leblanc E., Trensz F., Guindi C., Amrani A., Grenier G. (2011). The proteasome inhibitor MG132 reduces immobilization-induced skeletal muscle atrophy in mice. BMC Musculoskelet. Disord. 12, 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang N. C., Nguyen M., Bourdon J., Risse P. A., Martin J., Danialou G., Rizzuto R., Petrof B. J., Shore G. C. (2012). Bcl-2-associated autophagy regulator Naf-1 required for maintenance of skeletal muscle. Hum. Mol. Genet. 21, 2277–2287 [DOI] [PubMed] [Google Scholar]
- Clarke B. A., Drujan D., Willis M. S., Murphy L. O., Corpina R. A., Burova E., Rakhilin S. V., Stitt T. N., Patterson C., Latres E., et al. (2007). The E3 Ligase MuRF1 degrades myosin heavy chain protein in dexamethasone-treated skeletal muscle. Cell Metab. 6, 376–385 [DOI] [PubMed] [Google Scholar]
- Clop A., Marcq F., Takeda H., Pirottin D., Tordoir X., Bibé B., Bouix J., Caiment F., Elsen J. M., Eychenne F., et al. (2006). A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat. Genet. 38, 813–818 [DOI] [PubMed] [Google Scholar]
- Cohen S., Brault J. J., Gygi S. P., Glass D. J., Valenzuela D. M., Gartner C., Latres E., Goldberg A. L. (2009). During muscle atrophy, thick, but not thin, filament components are degraded by MuRF1-dependent ubiquitylation. J. Cell Biol. 185, 1083–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S., Zhai B., Gygi S. P., Goldberg A. L. (2012). Ubiquitylation by Trim32 causes coupled loss of desmin, Z-bands, and thin filaments in muscle atrophy. J. Cell Biol. 198, 575–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combaret L., Adegoke O. A., Bedard N., Baracos V., Attaix D., Wing S. S. (2005). USP19 is a ubiquitin-specific protease regulated in rat skeletal muscle during catabolic states. Am. J. Physiol. Endocrinol. Metab. 288, E693–E700 [DOI] [PubMed] [Google Scholar]
- Cong H., Sun L., Liu C., Tien P. (2011). Inhibition of atrogin-1/MAFbx expression by adenovirus-delivered small hairpin RNAs attenuates muscle atrophy in fasting mice. Hum. Gene Ther. 22, 313–324 [DOI] [PubMed] [Google Scholar]
- Csibi A., Cornille K., Leibovitch M. P., Poupon A., Tintignac L. A., Sanchez A. M., Leibovitch S. A. (2010). The translation regulatory subunit eIF3f controls the kinase-dependent mTOR signaling required for muscle differentiation and hypertrophy in mouse. PLoS ONE 5, e8994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo A. M. (2008). Autophagy and aging: keeping that old broom working. Trends Genet. 24, 604–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Alvaro C., Teruel T., Hernandez R., Lorenzo M. (2004). Tumor necrosis factor alpha produces insulin resistance in skeletal muscle by activation of inhibitor kappaB kinase in a p38 MAPK-dependent manner. J. Biol. Chem. 279, 17070–17078 [DOI] [PubMed] [Google Scholar]
- Demontis F., Perrimon N. (2010). FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell 143, 813–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deval C., Mordier S., Obled C., Bechet D., Combaret L., Attaix D., Ferrara M. (2001). Identification of cathepsin L as a differentially expressed message associated with skeletal muscle wasting. Biochem. J. 360, 143–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrowolny G., Aucello M., Rizzuto E., Beccafico S., Mammucari C., Boncompagni S., Belia S., Wannenes F., Nicoletti C., Del Prete Z., et al. (2008). Skeletal muscle is a primary target of SOD1G93A-mediated toxicity. Cell Metab. 8, 425–436 [DOI] [PubMed] [Google Scholar]
- Dogra C., Changotra H., Wedhas N., Qin X., Wergedal J. E., Kumar A. (2007). TNF-related weak inducer of apoptosis (TWEAK) is a potent skeletal muscle-wasting cytokine. FASEB J. 21, 1857–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durieux A. C., Amirouche A., Banzet S., Koulmann N., Bonnefoy R., Pasdeloup M., Mouret C., Bigard X., Peinnequin A., Freyssenet D. (2007). Ectopic expression of myostatin induces atrophy of adult skeletal muscle by decreasing muscle gene expression. Endocrinology 148, 3140–3147 [DOI] [PubMed] [Google Scholar]
- Fernández A. M., Dupont J., Farrar R. P., Lee S., Stannard B., Le Roith D. (2002). Muscle-specific inactivation of the IGF-I receptor induces compensatory hyperplasia in skeletal muscle. J. Clin. Invest. 109, 347–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielitz J., Kim M. S., Shelton J. M., Latif S., Spencer J. A., Glass D. J., Richardson J. A., Bassel-Duby R., Olson E. N. (2007). Myosin accumulation and striated muscle myopathy result from the loss of muscle RING finger 1 and 3. J. Clin. Invest. 117, 2486–2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T., Ewan L., Bauer M., Mattaliano R. J., Zaal K., Ralston E., Plotz P. H., Raben N. (2006a). Dysfunction of endocytic and autophagic pathways in a lysosomal storage disease. Ann. Neurol. 59, 700–708 [DOI] [PubMed] [Google Scholar]
- Fukuda T., Roberts A., Ahearn M., Zaal K., Ralston E., Plotz P. H., Raben N. (2006b). Autophagy and lysosomes in Pompe disease. Autophagy 2, 318–320 [DOI] [PubMed] [Google Scholar]
- Furuno K., Goodman M. N., Goldberg A. L. (1990). Role of different proteolytic systems in the degradation of muscle proteins during denervation atrophy. J. Biol. Chem. 265, 8550–8557 [PubMed] [Google Scholar]
- Geng T., Li P., Yin X., Yan Z. (2011). PGC-1α promotes nitric oxide antioxidant defenses and inhibits FOXO signaling against cardiac cachexia in mice. Am. J. Pathol. 178, 1738–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes M. D., Lecker S. H., Jagoe R. T., Navon A., Goldberg A. L. (2001). Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc. Natl. Acad. Sci. USA 98, 14440–14445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomis R. R., Alarcón C., He W., Wang Q., Seoane J., Lash A., Massagué J. (2006). A FoxO-Smad synexpression group in human keratinocytes. Proc. Natl. Acad. Sci. USA 103, 12747–12752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves M. D., Pistilli E. E., Balduzzi A., Birnbaum M. J., Lachey J., Khurana T. S., Ahima R. S. (2010). Akt deficiency attenuates muscle size and function but not the response to ActRIIB inhibition. PLoS ONE 5, e12707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer E. L., Dowlatshahi D., Banko M. R., Villen J., Hoang K., Blanchard D., Gygi S. P., Brunet A. (2007a). An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr. Biol. 17, 1646–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer E. L., Oskoui P. R., Banko M. R., Maniar J. M., Gygi M. P., Gygi S. P., Brunet A. (2007b). The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J. Biol. Chem. 282, 30107–30119 [DOI] [PubMed] [Google Scholar]
- Grobet L., Pirottin D., Farnir F., Poncelet D., Royo L. J., Brouwers B., Christians E., Desmecht D., Coignoul F., Kahn R., et al. (2003). Modulating skeletal muscle mass by postnatal, muscle-specific inactivation of the myostatin gene. Genesis 35, 227–238 [DOI] [PubMed] [Google Scholar]
- Grumati P., Coletto L., Sabatelli P., Cescon M., Angelin A., Bertaggia E., Blaauw B., Urciuolo A., Tiepolo T., Merlini L., et al. (2010). Autophagy is defective in collagen VI muscular dystrophies, and its reactivation rescues myofiber degeneration. Nat. Med. 16, 1313–1320 [DOI] [PubMed] [Google Scholar]
- Grumati P., Coletto L., Sandri M., Bonaldo P. (2011a). Autophagy induction rescues muscular dystrophy. Autophagy 7, 426–428 [DOI] [PubMed] [Google Scholar]
- Grumati P., Coletto L., Schiavinato A., Castagnaro S., Bertaggia E., Sandri M., Bonaldo P. (2011b). Physical exercise stimulates autophagy in normal skeletal muscles but is detrimental for collagen VI-deficient muscles. Autophagy 7, 1415–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna R. A., Quinsay M. N., Orogo A. M., Giang K., Rikka S., Gustafsson A. B. (2012). Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. J. Biol. Chem. 287, 19094–19104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., et al. (2006). Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441, 885–889 [DOI] [PubMed] [Google Scholar]
- He C., Bassik M. C., Moresi V., Sun K., Wei Y., Zou Z., An Z., Loh J., Fisher J., Sun Q., et al. (2012). Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature 481, 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirosumi J., Tuncman G., Chang L., Görgün C. Z., Uysal K. T., Maeda K., Karin M., Hotamisligil G. S. (2002). A central role for JNK in obesity and insulin resistance. Nature 420, 333–336 [DOI] [PubMed] [Google Scholar]
- Hishiya A., Iemura S., Natsume T., Takayama S., Ikeda K., Watanabe K. (2006). A novel ubiquitin-binding protein ZNF216 functioning in muscle atrophy. EMBO J. 25, 554–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Tindall D. J. (2007). Dynamic FoxO transcription factors. J. Cell Sci. 120, 2479–2487 [DOI] [PubMed] [Google Scholar]
- Hunter R. B., Kandarian S. C. (2004). Disruption of either the Nfkb1 or the Bcl3 gene inhibits skeletal muscle atrophy. J. Clin. Invest. 114, 1504–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumiya Y., Hopkins T., Morris C., Sato K., Zeng L., Viereck J., Hamilton J. A., Ouchi N., LeBrasseur N. K., Walsh K. (2008). Fast/Glycolytic muscle fiber growth reduces fat mass and improves metabolic parameters in obese mice. Cell Metab. 7, 159–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamart C., Raymackers J. M., Li An G., Deldicque L., Francaux M. (2011). Prevention of muscle disuse atrophy by MG132 proteasome inhibitor. Muscle Nerve 43, 708–715 [DOI] [PubMed] [Google Scholar]
- Judge A. R., Koncarevic A., Hunter R. B., Liou H. C., Jackman R. W., Kandarian S. C. (2007). Role for IkappaBalpha, but not c-Rel, in skeletal muscle atrophy. Am. J. Physiol. Cell. Physiol. 292, C372–C382 [DOI] [PubMed] [Google Scholar]
- Kamei Y., Miura S., Suzuki M., Kai Y., Mizukami J., Taniguchi T., Mochida K., Hata T., Matsuda J., Aburatani H., et al. (2004). Skeletal muscle FOXO1 (FKHR) transgenic mice have less skeletal muscle mass, down-regulated Type I (slow twitch/red muscle) fiber genes, and impaired glycemic control. J. Biol. Chem. 279, 41114–41123 [DOI] [PubMed] [Google Scholar]
- Kedar V., McDonough H., Arya R., Li H. H., Rockman H. A., Patterson C. (2004). Muscle-specific RING finger 1 is a bona fide ubiquitin ligase that degrades cardiac troponin I. Proc. Natl. Acad. Sci. USA 101, 18135–18140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V., Lamark T., Sou Y. S., Bjørkøy G., Nunn J. L., Bruun J. A., Shvets E., McEwan D. G., Clausen T. H., Wild P., et al. (2009). A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 33, 505–516 [DOI] [PubMed] [Google Scholar]
- Kitamura T., Kitamura Y. I., Funahashi Y., Shawber C. J., Castrillon D. H., Kollipara R., DePinho R. A., Kitajewski J., Accili D. (2007). A Foxo/Notch pathway controls myogenic differentiation and fiber type specification. J. Clin. Invest. 117, 2477–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline W. O., Panaro F. J., Yang H., Bodine S. C. (2007). Rapamycin inhibits the growth and muscle-sparing effects of clenbuterol. J. Appl. Physiol. 102, 740–747 [DOI] [PubMed] [Google Scholar]
- Klionsky D. J., Abeliovich H., Agostinis P., Agrawal D. K., Aliev G., Askew D. S., Baba M., Baehrecke E. H., Bahr B. A., Ballabio A., et al. (2008). Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4, 151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Koike M., Sou Y. S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S., et al. (2007). Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131, 1149–1163 [DOI] [PubMed] [Google Scholar]
- Kon M., Cuervo A. M. (2010). Chaperone-mediated autophagy in health and disease. FEBS Lett. 584, 1399–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudryashova E., Kramerova I., Spencer M. J. (2012). Satellite cell senescence underlies myopathy in a mouse model of limb-girdle muscular dystrophy 2H. J. Clin. Invest. 122, 1764–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Bhatnagar S., Paul P. K. (2012). TWEAK and TRAF6 regulate skeletal muscle atrophy. Curr. Opin. Clin. Nutr. Metab. Care 15, 233–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai K. M., Gonzalez M., Poueymirou W. T., Kline W. O., Na E., Zlotchenko E., Stitt T. N., Economides A. N., Yancopoulos G. D., Glass D. J. (2004). Conditional activation of akt in adult skeletal muscle induces rapid hypertrophy. Mol. Cell. Biol. 24, 9295–9304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M., Sabatini D. M. (2012). mTOR signaling in growth control and disease. Cell 149, 274–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecker S. H., Goldberg A. L., Mitch W. E. (2006). Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J. Am. Soc. Nephrol. 17, 1807–1819 [DOI] [PubMed] [Google Scholar]
- Lee S. J. (2004). Regulation of muscle mass by myostatin. Annu. Rev. Cell Dev. Biol. 20, 61–86 [DOI] [PubMed] [Google Scholar]
- Lee S. J. (2007). Quadrupling muscle mass in mice by targeting TGF-beta signaling pathways. PLoS ONE 2, e789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D., Goldberg A. (2011). Atrogin1/MAFbx: what atrophy, hypertrophy, and cardiac failure have in common. Circ. Res. 109, 123–126 [DOI] [PubMed] [Google Scholar]
- Lee S. J., McPherron A. C. (1999). Myostatin and the control of skeletal muscle mass. Curr. Opin. Genet. Dev. 9, 604–607 [DOI] [PubMed] [Google Scholar]
- Lee S. J., McPherron A. C. (2001). Regulation of myostatin activity and muscle growth. Proc. Natl. Acad. Sci. USA 98, 9306–9311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. W., Dai G., Hu Z., Wang X., Du J., Mitch W. E. (2004). Regulation of muscle protein degradation: coordinated control of apoptotic and ubiquitin-proteasome systems by phosphatidylinositol 3 kinase. J. Am. Soc. Nephrol. 15, 1537–1545 [DOI] [PubMed] [Google Scholar]
- Lee J. H., Budanov A. V., Park E. J., Birse R., Kim T. E., Perkins G. A., Ocorr K., Ellisman M. H., Bodmer R., Bier E., et al. (2010). Sestrin as a feedback inhibitor of TOR that prevents age-related pathologies. Science 327, 1223–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine S., Nguyen T., Taylor N., Friscia M. E., Budak M. T., Rothenberg P., Zhu J., Sachdeva R., Sonnad S., Kaiser L. R., et al. (2008). Rapid disuse atrophy of diaphragm fibers in mechanically ventilated humans. N. Engl. J. Med. 358, 1327–1335 [DOI] [PubMed] [Google Scholar]
- Li H. H., Kedar V., Zhang C., McDonough H., Arya R., Wang D. Z., Patterson C. (2004). Atrogin-1/muscle atrophy F-box inhibits calcineurin-dependent cardiac hypertrophy by participating in an SCF ubiquitin ligase complex. J. Clin. Invest. 114, 1058–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. M., Yang Z., Liu C. W., Wang R., Tien P., Dale R., Sun L. Q. (2007). Effect of RNA oligonucleotide targeting Foxo-1 on muscle growth in normal and cancer cachexia mice. Cancer Gene Ther. 14, 945–952 [DOI] [PubMed] [Google Scholar]
- Lokireddy S., McFarlane C., Ge X., Zhang H., Sze S. K., Sharma M., Kambadur R. (2011). Myostatin induces degradation of sarcomeric proteins through a Smad3 signaling mechanism during skeletal muscle wasting. Mol. Endocrinol. 25, 1936–1949 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Malicdan M. C., Noguchi S., Nonaka I., Saftig P., Nishino I. (2008). Lysosomal myopathies: an excessive build-up in autophagosomes is too much to handle. Neuromuscul. Disord. 18, 521–529 [DOI] [PubMed] [Google Scholar]
- Mammucari C., Milan G., Romanello V., Masiero E., Rudolf R., Del Piccolo P., Burden S. J., Di Lisi R., Sandri C., Zhao J., et al. (2007). FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 6, 458–471 [DOI] [PubMed] [Google Scholar]
- Masiero E., Sandri M. (2010). Autophagy inhibition induces atrophy and myopathy in adult skeletal muscles. Autophagy 6, 307–309 [DOI] [PubMed] [Google Scholar]
- Masiero E., Agatea L., Mammucari C., Blaauw B., Loro E., Komatsu M., Metzger D., Reggiani C., Schiaffino S., Sandri M. (2009). Autophagy is required to maintain muscle mass. Cell Metab. 10, 507–515 [DOI] [PubMed] [Google Scholar]
- Massagué J., Seoane J., Wotton D. (2005). Smad transcription factors. Genes Dev. 19, 2783–2810 [DOI] [PubMed] [Google Scholar]
- McCarthy J. J., Mula J., Miyazaki M., Erfani R., Garrison K., Farooqui A. B., Srikuea R., Lawson B. A., Grimes B., Keller C., et al. (2011). Effective fiber hypertrophy in satellite cell-depleted skeletal muscle. Development 138, 3657–3666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane C., Plummer E., Thomas M., Hennebry A., Ashby M., Ling N., Smith H., Sharma M., Kambadur R. (2006). Myostatin induces cachexia by activating the ubiquitin proteolytic system through an NF-kappaB-independent, FoxO1-dependent mechanism. J. Cell. Physiol. 209, 501–514 [DOI] [PubMed] [Google Scholar]
- McPherron A. C., Lee S. J. (1997). Double muscling in cattle due to mutations in the myostatin gene. Proc. Natl. Acad. Sci. USA 94, 12457–12461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherron A. C., Lawler A. M., Lee S. J. (1997). Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature 387, 83–90 [DOI] [PubMed] [Google Scholar]
- Menconi M., Fareed M., O’Neal P., Poylin V., Wei W., Hasselgren P. O. (2007). Role of glucocorticoids in the molecular regulation of muscle wasting. Crit. Care Med. 35, S602–S608 [DOI] [PubMed] [Google Scholar]
- Mieulet V., Roceri M., Espeillac C., Sotiropoulos A., Ohanna M., Oorschot V., Klumperman J., Sandri M., Pende M. (2007). S6 kinase inactivation impairs growth and translational target phosphorylation in muscle cells maintaining proper regulation of protein turnover. Am. J. Physiol. Cell. Physiol. 293, C712–C722 [DOI] [PubMed] [Google Scholar]
- Minetti G. C., Feige J. N., Rosenstiel A., Bombard F., Meier V., Werner A., Bassilana F., Sailer A. W., Kahle P., Lambert C., et al. (2011). Gαi2 signaling promotes skeletal muscle hypertrophy, myoblast differentiation, and muscle regeneration. Sci. Signal. 4, ra80. [DOI] [PubMed] [Google Scholar]
- Mittal A., Bhatnagar S., Kumar A., Lach-Trifilieff E., Wauters S., Li H., Makonchuk D. Y., Glass D. J., Kumar A. (2010). The TWEAK-Fn14 system is a critical regulator of denervation-induced skeletal muscle atrophy in mice. J. Cell Biol. 188, 833–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Yamamoto A., Matsui M., Yoshimori T., Ohsumi Y. (2004). In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell 15, 1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Levine B., Cuervo A. M., Klionsky D. J. (2008). Autophagy fights disease through cellular self-digestion. Nature 451, 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier S., Le Cam A., Le Marchand-Brustel Y. (1983). Insulin and insulin-like growth factor I. Effects on protein synthesis in isolated muscles from lean and goldthioglucose-obese mice. Diabetes 32, 392–397 [DOI] [PubMed] [Google Scholar]
- Moresi V., Williams A. H., Meadows E., Flynn J. M., Potthoff M. J., McAnally J., Shelton J. M., Backs J., Klein W. H., Richardson J. A., et al. (2010). Myogenin and class II HDACs control neurogenic muscle atrophy by inducing E3 ubiquitin ligases. Cell 143, 35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moresi V., Carrer M., Grueter C. E., Rifki O. F., Shelton J. M., Richardson J. A., Bassel-Duby R., Olson E. N. (2012). Histone deacetylases 1 and 2 regulate autophagy flux and skeletal muscle homeostasis in mice. Proc. Natl. Acad. Sci. USA 109, 1649–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morissette M. R., Cook S. A., Foo S., McKoy G., Ashida N., Novikov M., Scherrer-Crosbie M., Li L., Matsui T., Brooks G., et al. (2006). Myostatin regulates cardiomyocyte growth through modulation of Akt signaling. Circ. Res. 99, 15–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourkioti F., Kratsios P., Luedde T., Song Y. H., Delafontaine P., Adami R., Parente V., Bottinelli R., Pasparakis M., Rosenthal N. (2006). Targeted ablation of IKK2 improves skeletal muscle strength, maintains mass, and promotes regeneration. J. Clin. Invest. 116, 2945–2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musarò A., McCullagh K., Paul A., Houghton L., Dobrowolny G., Molinaro M., Barton E. R., Sweeney H. L., Rosenthal N. (2001). Localized Igf-1 transgene expression sustains hypertrophy and regeneration in senescent skeletal muscle. Nat. Genet. 27, 195–200 [DOI] [PubMed] [Google Scholar]
- Nakashima K., Yakabe Y. (2007). AMPK activation stimulates myofibrillar protein degradation and expression of atrophy-related ubiquitin ligases by increasing FOXO transcription factors in C2C12 myotubes. Biosci. Biotechnol. Biochem. 71, 1650–1656 [DOI] [PubMed] [Google Scholar]
- Narendra D. P., Youle R. J. (2011). Targeting mitochondrial dysfunction: role for PINK1 and Parkin in mitochondrial quality control. Antioxid. Redox Signal. 14, 1929–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak I., Kirkin V., McEwan D. G., Zhang J., Wild P., Rozenknop A., Rogov V., Löhr F., Popovic D., Occhipinti A., et al. (2010). Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 11, 45–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orciuolo E., Buda G., Cecconi N., Galimberti S., Versari D., Cervetti G., Salvetti A., Petrini M. (2007). Unexpected cardiotoxicity in haematological bortezomib treated patients. Br. J. Haematol. 138, 396–397 [DOI] [PubMed] [Google Scholar]
- Paik J. H., Kollipara R., Chu G., Ji H., Xiao Y., Ding Z., Miao L., Tothova Z., Horner J. W., Carrasco D. R., et al. (2007). FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell 128, 309–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S., Clausen T. H., Lamark T., Brech A., Bruun J. A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. (2007). p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–24145 [DOI] [PubMed] [Google Scholar]
- Park Y. E., Hayashi Y. K., Bonne G., Arimura T., Noguchi S., Nonaka I., Nishino I. (2009). Autophagic degradation of nuclear components in mammalian cells. Autophagy 5, 795–804 [DOI] [PubMed] [Google Scholar]
- Paul P. K., Gupta S. K., Bhatnagar S., Panguluri S. K., Darnay B. G., Choi Y., Kumar A. (2010). Targeted ablation of TRAF6 inhibits skeletal muscle wasting in mice. J. Cell Biol. 191, 1395–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul P. K., Bhatnagar S., Mishra V., Srivastava S., Darnay B. G., Choi Y., Kumar A. (2012). The E3 ubiquitin ligase TRAF6 intercedes in starvation-induced skeletal muscle atrophy through multiple mechanisms. Mol. Cell. Biol. 32, 1248–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X. D., Xu P. Z., Chen M. L., Hahn-Windgassen A., Skeen J., Jacobs J., Sundararajan D., Chen W. S., Crawford S. E., Coleman K. G., et al. (2003). Dwarfism, impaired skin development, skeletal muscle atrophy, delayed bone development, and impeded adipogenesis in mice lacking Akt1 and Akt2. Genes Dev. 17, 1352–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson J. M., Bakkar N., Guttridge D. C. (2011). NF-κB signaling in skeletal muscle health and disease. Curr. Top. Dev. Biol. 96, 85–119 [DOI] [PubMed] [Google Scholar]
- Piétri-Rouxel F., Gentil C., Vassilopoulos S., Baas D., Mouisel E., Ferry A., Vignaud A., Hourdé C., Marty I., Schaeffer L., et al. (2010). DHPR alpha1S subunit controls skeletal muscle mass and morphogenesis. EMBO J. 29, 643–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P., Rhee J., Donovan J., Walkey C. J., Yoon J. C., Oriente F., Kitamura Y., Altomonte J., Dong H., Accili D., et al. (2003). Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature 423, 550–555 [DOI] [PubMed] [Google Scholar]
- Raben N., Hill V., Shea L., Takikita S., Baum R., Mizushima N., Ralston E., Plotz P. (2008). Suppression of autophagy in skeletal muscle uncovers the accumulation of ubiquitinated proteins and their potential role in muscle damage in Pompe disease. Hum. Mol. Genet. 17, 3897–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaello A., Milan G., Masiero E., Carnio S., Lee D., Lanfranchi G., Goldberg A. L., Sandri M. (2010). JunB transcription factor maintains skeletal muscle mass and promotes hypertrophy. J. Cell Biol. 191, 101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran N., Munteanu I., Wang P., Aubourg P., Rilstone J. J., Israelian N., Naranian T., Paroutis P., Guo R., Ren Z. P., et al. (2009). VMA21 deficiency causes an autophagic myopathy by compromising V-ATPase activity and lysosomal acidification. Cell 137, 235–246 [DOI] [PubMed] [Google Scholar]
- Reed S. A., Sandesara P. B., Senf S. M., Judge A. R. (2012). Inhibition of FoxO transcriptional activity prevents muscle fiber atrophy during cachexia and induces hypertrophy. FASEB J. 26, 987–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisz-Porszasz S., Bhasin S., Artaza J. N., Shen R., Sinha-Hikim I., Hogue A., Fielder T. J., Gonzalez-Cadavid N. F. (2003). Lower skeletal muscle mass in male transgenic mice with muscle-specific overexpression of myostatin. Am. J. Physiol. Endocrinol. Metab. 285, E876–E888 [DOI] [PubMed] [Google Scholar]
- Risson V., Mazelin L., Roceri M., Sanchez H., Moncollin V., Corneloup C., Richard-Bulteau H., Vignaud A., Baas D., Defour A., et al. (2009). Muscle inactivation of mTOR causes metabolic and dystrophin defects leading to severe myopathy. J. Cell Biol. 187, 859–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanello V., Sandri M. (2010). Mitochondrial biogenesis and fragmentation as regulators of muscle protein degradation. Curr. Hypertens. Rep. 12, 433–439 [DOI] [PubMed] [Google Scholar]
- Romanello V., Guadagnin E., Gomes L., Roder I., Sandri C., Petersen Y., Milan G., Masiero E., Del Piccolo P., Foretz M., et al. (2010). Mitochondrial fission and remodelling contributes to muscle atrophy. EMBO J. 29, 1774–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rommel C., Bodine S. C., Clarke B. A., Rossman R., Nunez L., Stitt T. N., Yancopoulos G. D., Glass D. J. (2001). Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat. Cell Biol. 3, 1009–1013 [DOI] [PubMed] [Google Scholar]
- Sacheck J. M., Ohtsuka A., McLary S. C., Goldberg A. L. (2004). IGF-I stimulates muscle growth by suppressing protein breakdown and expression of atrophy-related ubiquitin ligases, atrogin-1 and MuRF1. Am. J. Physiol. Endocrinol. Metab. 287, E591–E601 [DOI] [PubMed] [Google Scholar]
- Sacheck J. M., Hyatt J. P., Raffaello A., Jagoe R. T., Roy R. R., Edgerton V. R., Lecker S. H., Goldberg A. L. (2007). Rapid disuse and denervation atrophy involve transcriptional changes similar to those of muscle wasting during systemic diseases. FASEB J. 21, 140–155 [DOI] [PubMed] [Google Scholar]
- Sandri M., Sandri C., Gilbert A., Skurk C., Calabria E., Picard A., Walsh K., Schiaffino S., Lecker S. H., Goldberg A. L. (2004). Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 117, 399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri M., Lin J., Handschin C., Yang W., Arany Z. P., Lecker S. H., Goldberg A. L., Spiegelman B. M. (2006). PGC-1alpha protects skeletal muscle from atrophy by suppressing FoxO3 action and atrophy-specific gene transcription. Proc. Natl. Acad. Sci. USA 103, 16260–16265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartorelli V., Fulco M. (2004). Molecular and cellular determinants of skeletal muscle atrophy and hypertrophy. Sci. STKE 2004, re11. [DOI] [PubMed] [Google Scholar]
- Sartori R., Milan G., Patron M., Mammucari C., Blaauw B., Abraham R., Sandri M. (2009). Smad2 and 3 transcription factors control muscle mass in adulthood. Am. J. Physiol. Cell Physiol. 296, C1248–C1257 [DOI] [PubMed] [Google Scholar]
- Schakman O., Gilson H., Thissen J. P. (2008). Mechanisms of glucocorticoid-induced myopathy. J. Endocrinol. 197, 1–10 [DOI] [PubMed] [Google Scholar]
- Schiaffino S., Hanzlíková V. (1972). Studies on the effect of denervation in developing muscle. II. The lysosomal system. J. Ultrastruct. Res. 39, 1–14 [DOI] [PubMed] [Google Scholar]
- Schiaffino S., Mammucari C. (2011). Regulation of skeletal muscle growth by the IGF1-Akt/PKB pathway: insights from genetic models. Skelet. Muscle 1, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuelke M., Wagner K. R., Stolz L. E., Hübner C., Riebel T., Kömen W., Braun T., Tobin J. F., Lee S. J. (2004). Myostatin mutation associated with gross muscle hypertrophy in a child. N. Engl. J. Med. 350, 2682–2688 [DOI] [PubMed] [Google Scholar]
- Schulze P. C., Fang J., Kassik K. A., Gannon J., Cupesi M., MacGillivray C., Lee R. T., Rosenthal N. (2005). Transgenic overexpression of locally acting insulin-like growth factor-1 inhibits ubiquitin-mediated muscle atrophy in chronic left-ventricular dysfunction. Circ. Res. 97, 418–426 [DOI] [PubMed] [Google Scholar]
- Senf S. M., Sandesara P. B., Reed S. A., Judge A. R. (2011). p300 Acetyltransferase activity differentially regulates the localization and activity of the FOXO homologues in skeletal muscle. Am. J. Physiol. Cell Physiol. 300, C1490–C1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Luo L., Eash J., Ibebunjo C., Glass D. J. (2011). The SCF-Fbxo40 complex induces IRS1 ubiquitination in skeletal muscle, limiting IGF1 signaling. Dev. Cell 21, 835–847 [DOI] [PubMed] [Google Scholar]
- Shimizu N., Yoshikawa N., Ito N., Maruyama T., Suzuki Y., Takeda S., Nakae J., Tagata Y., Nishitani S., Takehana K., et al. (2011). Crosstalk between glucocorticoid receptor and nutritional sensor mTOR in skeletal muscle. Cell Metab. 13, 170–182 [DOI] [PubMed] [Google Scholar]
- Song Y. H., Li Y., Du J., Mitch W. E., Rosenthal N., Delafontaine P. (2005). Muscle-specific expression of IGF-1 blocks angiotensin II-induced skeletal muscle wasting. J. Clin. Invest. 115, 451–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southgate R. J., Neill B., Prelovsek O., El-Osta A., Kamei Y., Miura S., Ezaki O., McLoughlin T. J., Zhang W., Unterman T. G., et al. (2007). FOXO1 regulates the expression of 4E-BP1 and inhibits mTOR signaling in mammalian skeletal muscle. J. Biol. Chem. 282, 21176–21186 [DOI] [PubMed] [Google Scholar]
- Stitt T. N., Drujan D., Clarke B. A., Panaro F., Timofeyva Y., Kline W. O., Gonzalez M., Yancopoulos G. D., Glass D. J. (2004). The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol. Cell 14, 395–403 [DOI] [PubMed] [Google Scholar]
- Sundaram P., Pang Z., Miao M., Yu L., Wing S. S. (2009). USP19-deubiquitinating enzyme regulates levels of major myofibrillar proteins in L6 muscle cells. Am. J. Physiol. Endocrinol. Metab. 297, E1283–E1290 [DOI] [PubMed] [Google Scholar]
- Supinski G. S., Vanags J., Callahan L. A. (2009). Effect of proteasome inhibitors on endotoxin-induced diaphragm dysfunction. Am. J. Physiol. Lung Cell Mol. Physiol. 296, L994–L1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N., Motohashi N., Uezumi A., Fukada S., Yoshimura T., Itoyama Y., Aoki M., Miyagoe-Suzuki Y., Takeda S. (2007). NO production results in suspension-induced muscle atrophy through dislocation of neuronal NOS. J. Clin. Invest. 117, 2468–2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano K., Watanabe-Takano H., Suetsugu S., Kurita S., Tsujita K., Kimura S., Karatsu T., Takenawa T., Endo T. (2010). Nebulin and N-WASP cooperate to cause IGF-1-induced sarcomeric actin filament formation. Science 330, 1536–1540 [DOI] [PubMed] [Google Scholar]
- Takikita S., Schreiner C., Baum R., Xie T., Ralston E., Plotz P. H., Raben N. (2010). Fiber type conversion by PGC-1α activates lysosomal and autophagosomal biogenesis in both unaffected and Pompe skeletal muscle. PLoS ONE 5, e15239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Guhde G., Suter A., Eskelinen E. L., Hartmann D., Lüllmann-Rauch R., Janssen P. M., Blanz J., von Figura K., Saftig P. (2000). Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature 406, 902–906 [DOI] [PubMed] [Google Scholar]
- Tintignac L. A., Lagirand J., Batonnet S., Sirri V., Leibovitch M. P., Leibovitch S. A. (2005). Degradation of MyoD mediated by the SCF (MAFbx) ubiquitin ligase. J. Biol. Chem. 280, 2847–2856 [DOI] [PubMed] [Google Scholar]
- Trendelenburg A. U., Meyer A., Rohner D., Boyle J., Hatakeyama S., Glass D. J. (2009). Myostatin reduces Akt/TORC1/p70S6K signaling, inhibiting myoblast differentiation and myotube size. Am. J. Physiol. Cell Physiol. 296, C1258–C1270 [DOI] [PubMed] [Google Scholar]
- Vergne I., Roberts E., Elmaoued R. A., Tosch V., Delgado M. A., Proikas-Cezanne T., Laporte J., Deretic V. (2009). Control of autophagy initiation by phosphoinositide 3-phosphatase Jumpy. EMBO J. 28, 2244–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell D. S., Baehr L. M., van den Brandt J., Johnsen S. A., Reichardt H. M., Furlow J. D., Bodine S. C. (2008). The glucocorticoid receptor and FOXO1 synergistically activate the skeletal muscle atrophy-associated MuRF1 gene. Am. J. Physiol. Endocrinol. Metab. 295, E785–E797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenz T., Rossi S. G., Rotundo R. L., Spiegelman B. M., Moraes C. T. (2009). Increased muscle PGC-1alpha expression protects from sarcopenia and metabolic disease during aging. Proc. Natl. Acad. Sci. USA 106, 20405–20410 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wing S. S., Chiang H. L., Goldberg A. L., Dice J. F. (1991). Proteins containing peptide sequences related to Lys-Phe-Glu-Arg-Gln are selectively depleted in liver and heart, but not skeletal muscle, of fasted rats. Biochem. J. 275, 165–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt C. C., Witt S. H., Lerche S., Labeit D., Back W., Labeit S. (2008). Cooperative control of striated muscle mass and metabolism by MuRF1 and MuRF2. EMBO J. 27, 350–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlgemuth S. E., Seo A. Y., Marzetti E., Lees H. A., Leeuwenburgh C. (2010). Skeletal muscle autophagy and apoptosis during aging: effects of calorie restriction and life-long exercise. Exp. Gerontol. 45, 138–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R. C., et al. (1999). Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC- 1. Cell 98, 115–124 [DOI] [PubMed] [Google Scholar]
- Youle R. J., Narendra D. P. (2011). Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 12, 9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Wang J. L., Lu J., Song Y., Kwak K. S., Jiao Q., Rosenfeld R., Chen Q., Boone T., Simonet W. S., et al. (2010). Reversal of cancer cachexia and muscle wasting by ActRIIB antagonism leads to prolonged survival. Cell 142, 531–543 [DOI] [PubMed] [Google Scholar]
- Zimmers T. A., Davies M. V., Koniaris L. G., Haynes P., Esquela A. F., Tomkinson K. N., McPherron A. C., Wolfman N. M., Lee S. J. (2002). Induction of cachexia in mice by systemically administered myostatin. Science 296, 1486–1488 [DOI] [PubMed] [Google Scholar]