SUMMARY
The actin-bundling protein fascin is a key mediator of tumor invasion and metastasis and its activity drives filopodia formation, cell-shape changes and cell migration. Small-molecule inhibitors of fascin block tumor metastasis in animal models. Conversely, fascin deficiency might underlie the pathogenesis of some developmental brain disorders. To identify fascin-pathway modulators we devised a cell-based assay for fascin function and used it in a bidirectional drug screen. The screen utilized cultured fascin-deficient mutant Drosophila neurons, whose neurite arbors manifest the ‘filagree’ phenotype. Taking a repurposing approach, we screened a library of 1040 known compounds, many of them FDA-approved drugs, for filagree modifiers. Based on scaffold distribution, molecular-fingerprint similarities, and chemical-space distribution, this library has high structural diversity, supporting its utility as a screening tool. We identified 34 fascin-pathway blockers (with potential anti-metastasis activity) and 48 fascin-pathway enhancers (with potential cognitive-enhancer activity). The structural diversity of the active compounds suggests multiple molecular targets. Comparisons of active and inactive compounds provided preliminary structure-activity relationship information. The screen also revealed diverse neurotoxic effects of other drugs, notably the ‘beads-on-a-string’ defect, which is induced solely by statins. Statin-induced neurotoxicity is enhanced by fascin deficiency. In summary, we provide evidence that primary neuron culture using a genetic model organism can be valuable for early-stage drug discovery and developmental neurotoxicity testing. Furthermore, we propose that, given an appropriate assay for target-pathway function, bidirectional screening for brain-development disorders and invasive cancers represents an efficient, multipurpose strategy for drug discovery.
INTRODUCTION
A highly conserved actin-bundling protein, fascin has diverse roles in the developmental and physiological regulation of cellular morphology and function (Kureishy et al., 2002; Jayo and Parsons, 2010; Sedeh et al., 2010; Hashimoto et al., 2011). It is also implicated in human disease pathogenesis, under both loss-of-function and gain-of-function conditions, which motivated us to develop a fascin bioassay for drug discovery. Note that fascin is unrelated to either the fasciclins or neurofascin, which are members of the immunoglobulin cell-adhesion molecule superfamily. Fascin drives the formation of cell-membrane protrusions, including lamellipodia (Yamashiro et al., 1998), microspikes (Svitkina et al., 2003), filopodia (Vignjevic et al., 2006) and invadopodia (Li et al., 2010a), in part because F-actin bundles increase mechanical stiffness (Tseng et al., 2005; Vignjevic et al., 2006). In addition, fascin-mediated actin bundling and crosslinking, which are regulated by phosphorylation (Ono et al., 1997; Aratyn et al., 2007) and the extracellular matrix (ECM), enhance cell migration (Ono et al., 1997; Yamashiro et al., 1998; Anilkumar et al., 2003; Jawhari et al., 2003) and ECM degradation (Li et al., 2010a).
Mammals have three fascin-coding genes, of which Fascin-2 and Fascin-3 are expressed in narrow domains (Tubb et al., 2000; Tubb et al., 2002; Shin et al., 2010), whereas Fascin-1 is broadly and dynamically expressed. Fascin-1 is abundant early in development, especially in the central nervous system (CNS) and migrating cells, and is then downregulated as cells mature (De Arcangelis et al., 2004; Zhang et al., 2008; Zanet et al., 2009; Tang et al., 2010). In this paper, ‘fascin’ refers to the product of the Fascin-1 genes (FSCN1 in humans, MIM#602689; Fscn1 in mouse; and singed in Drosophila, FBgn0003447).
Fascin has a pivotal role in tumor invasion and metastasis (Machesky and Li, 2010), leading to the proposal that fascin-blocking drugs might prevent the spread of malignant cancers (Yoder et al., 2005; Hashimoto et al., 2011). Because most cancer-related deaths are due to metastases, there is an urgent need for development of anti-metastasis agents (Sporn, 1996; Sleeman and Steeg, 2010). For carcinomas from numerous organs, high fascin expression levels are associated with increased invasiveness and earlier patient death (Machesky and Li, 2010). Fascin is also involved in tissue infiltration by circulating tumor cells (Kim et al., 2009a). Similar associations have been reported for malignant glioblastomas (Peraud et al., 2003; Roma and Prayson, 2005; Gunal et al., 2008), which are prone to extensive dispersion within the CNS (Giese et al., 2003).
TRANSLATIONAL IMPACT.
Clinical issue
The findings reported here address two major clinical problems that do not initially seem related. First, invasive cancers, including most brain tumors, cause death due to invasion or migration, which are not inhibited by currently available cancer treatments. Second, developmental brain disorders are not treatable with drugs that enhance cognitive function. For both of these unmet medical needs, a major obstacle has been the lack of cellular bioassays for compound screening. The actin-bundling protein fascin links these two challenging clinical conditions: excess fascin promotes tumor invasion and metastasis, whereas insufficient fascin disrupts brain development. Thus, the fascin pathway represents a highly desirable drug target.
Results
The filagree phenotype of fascin-deficient mutant Drosophila neurons enabled the authors to develop a bidirectional in vitro cellular bioassay to screen for drugs that modify the fascin pathway. A library of 1040 known compounds (NINDS-II) was chosen on the basis of high molecular diversity, and was screened with the aim of identifying drugs that could be repurposed for new indications. Of these compounds, 81 were active as fascin-pathway modifiers. There was wide pharmacological and chemical-structure diversity in each set of active compounds (34 blockers and 48 enhancers), strongly suggesting that each set has multiple targets along the fascin pathway. Comparison of closely related compounds that differ in activity provided structure-activity relationship (SAR) hypotheses that can be tested in follow-up studies. Notably, all four of the statin compounds in the library caused a unique, reversible neurotoxic morphological effect characterized by intraneurite nodules containing aggregations of organelles [‘beads-on-a-string’ (BOS)]. Fascin deficiency enhances the sensitivity of neurons to BOS.
Implications and future directions
These findings introduce a conceptually simple cell-based fascin bioassay and apply it to identify many compounds and preliminary SAR information that can be pursued for drug development, either by repurposing or lead optimization. Fascin-pathway blockers could serve as anti-invasion and anti-metastasis agents for patients with malignant carcinomas or gliomas. Fascin-pathway enhancers could improve neurocognitive function and behavior in a subset of children with developmental brain disorders. The potential of fascin-pathway modifiers warrants testing in various mammalian models of fascin-sensitive human disease. In addition, bidirectional cell-based screening could be applied to other important biological pathways with dual functions in brain development and tumor invasion.
The authors propose that the statin-induced BOS they observed represents a cellular correlate of statin-associated cognitive side effects experienced by some patients on statins. Because cholesterol is an essential nutrient in Drosophila, this system provides a unique opportunity to study statin-mediated neurotoxicity separately from cholesterol biosynthesis. The authors’ demonstration that genetic background impacts the sensitivity of neurons to statin-induced BOS suggests experiments to identify potential human genetic risk factors for statin-associated cognitive deficits. More generally, this primary neuron culture system in Drosophila holds great promise as a neurotoxicity screening platform with the ability to identify biologically relevant gene-by-environment interactions.
The causal role of fascin in tumor phenotypes is supported by laboratory studies in which blocking of fascin expression reduced the invasive and/or metastatic properties of colon carcinoma (Hashimoto et al., 2007), glioblastoma multiforme (Hwang et al., 2008), gastric carcinoma (Fu et al., 2009; Kim et al., 2010) and breast carcinoma (Chen et al., 2010; Al-Alwan et al., 2011). Moreover, migrastatin-family compounds, which are potent inhibitors of tumor invasion and metastasis in the laboratory (Shan et al., 2005), bind to fascin and inhibit its actin-bundling activity (Chen et al., 2010). Thus, the experimental and clinical data strongly suggest that a fascin bioassay would be of great value for discovery of drugs with anti-invasion and/or anti-metastasis activity.
Fascin is also required for normal brain development (Yamakita et al., 2009) (R.K. and L.L.R., unpublished data), plausibly by regulating neuronal differentiation (Deinhardt et al., 2011; Marín-Vicente et al., 2011). Fascin insufficiency or dysregulation might underlie disorders of brain development and plasticity, resulting in intellectual disability (Kraft et al., 2006). Fascin regulation is likely to be faulty in the brain-development disorders Rubinstein-Taybi syndrome (Roelfsema and Peters, 2007) and tuberous sclerosis (Ess, 2006), which are caused by mutations in CREBBP (MIM#600140) and TSC1 (MIM#605284) or TSC2 (MIM#191092), respectively. FSCN1 is an upregulated transcriptional target of CREB binding protein (CREBBP) (Megiorni et al., 2005), whereas fascin protein is a target of the TSC1-TSC2 complex (Gan et al., 2008).
The connection between fascin and brain plasticity has also been revealed by unbiased proteomics screens. Reduced fascin levels were found in two mouse models of absence epilepsy (Ryu et al., 2007; Ryu et al., 2008) and after long-term memory induction (Li et al., 2010b). These data suggest that fascin levels are downregulated by neural activity, perhaps to permit synapse-structure changes. By contrast, fascin levels were elevated in a polytransgenic mouse model of Down syndrome (Shin et al., 2007). This could be a molecular feature of the brain-development disorder or early-onset neurodegeneration, or both. In a neuron-culture model of neuroprotection, rapid induction of ischemic tolerance was associated with ubiquitylation of fascin, which was subsequently degraded, as well as with transient retraction of dendritic spines. This is consistent with the idea that fascin removal allows dissolution of actin bundles, thereby accelerating synapse remodeling (Meller, 2009). Finally, in a canine model of aging, slowing of cognitive decline by environment and diet was associated with reduced levels of fascin carbonyl, a marker of oxidation (Opii et al., 2008). This suggests that preventing fascin oxidation contributes to better cognitive performance. These reports point to a role for fascin in regulating neuronal differentiation and synaptic plasticity, which are disrupted in brain-development disorders (Johnston, 2004) and vulnerable during aging (Burke and Barnes, 2006). Hence, pharmacological enhancers of fascin expression or function could be beneficial for diverse neurodevelopmental and cognitive or behavioral conditions.
Loss-of-function Fascin-1 mutations are available in Drosophila melanogaster and Mus musculus. The targeted Fscn1 disruption in the mouse causes structural brain abnormalities and high rates of neonatal death (Yamakita et al., 2009). Drosophila has a single fascin-coding gene (Bryan et al., 1993; Kureishy et al., 2002), named singed for the gnarled bristles of mutant flies (Bender, 1960; Cant et al., 1994; Tilney et al., 1995; Wulfkühle et al., 1998). Wild-type singed function is also essential for oogenesis (Cant et al., 1994), blood cell migration (Zanet et al., 2009) and some aspects of brain development (R.K. and L.L.R., unpublished data), but not for viability (Bender, 1960). We discovered that singed mutations cause a neuronal morphogenesis defect, ‘filagree’, when developing mutant CNS neurons are cultured in vitro (Kraft et al., 2006). Filagree neurite arbors have exaggerated clockwise curvature and erratic variation in neurite caliber, but no reduction in length, branching or axon-dendrite ratio (Kraft et al., 2006). Filagree neurons have striking disruptions of their actin cytoskeleton which, like other singed phenotypes, can be explained by failure of actin bundling (Cant et al., 1994; Guild et al., 2003; Kraft et al., 2006; Zanet et al., 2009). Because decreasing fascin function causes increasing severity of the filagree phenotype, we reasoned that cultured neurons could provide a cellular bioassay for fascin function.
The use of a Drosophila cell-based fascin bioassay for drug discovery is justified by the phylogenetic conservation of fascin (Hashimoto et al., 2011) and of the pathways underlying cognition (Greenspan and Dierick, 2004; Inlow and Restifo, 2004; Bolduc and Tully, 2009), tumor invasion and metastasis (Miles et al., 2011). Here we present the results of a bidirectional screen for pharmacological modifiers of the filagree neuronal morphogenesis defect.
RESULTS
Drug screen design using the fascin bioassay
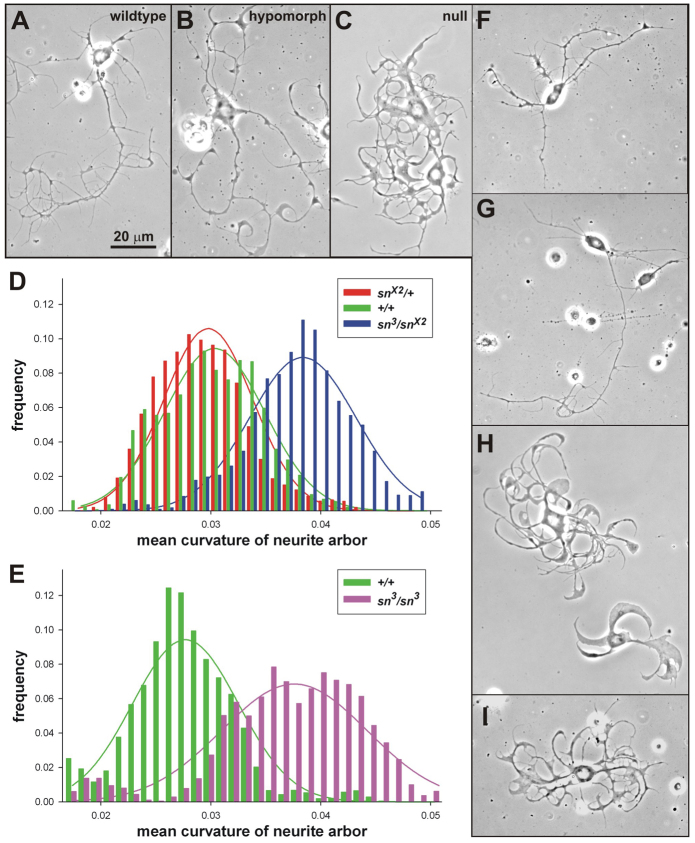
Filagree is a highly penetrant, quantifiable, pan-neuronal phenotype of fascin-deficient singed-mutant neurons cultured from the developing CNS of Drosophila mature larvae or young pupae (Fig. 1A–C) (Kraft et al., 2006). This developmental interval corresponds to high levels of singed transcript accumulation in wild-type CNS (Kaitlin L. Bergfield, R.K. and L.L.R., unpublished). The filagree phenotype is so distinctive and so consistent that trained observers can easily distinguish singed-mutant versus wild-type population cultures, typically containing 1500–2000 neurons each. In fact, both humans and a computational method for neurite-curvature quantification can classify individual photomicrographs of mutant or wild-type neurons with =90% accuracy (Kraft et al., 2006). To anticipate how much restoration of function would be required to rescue the fascin-deficient filagree phenotype, we asked whether filagree is recessive, like other singed phenotypes. We quantified the neurite-curvature distributions of wild-type and snX2/+ cultured larval CNS neurons, which have 100% and 50% of normal fascin function, respectively (Fig. 1D). The two distributions were statistically indistinguishable, whereas neurons from sn3/snX2 (a near-null genotype) and snX2/+ were very different (Welch’s t-test, P<1×10−18; Fig. 1D). Therefore, the results show that the singed neurite-curvature phenotype is recessive and that rescue of singed-mutant neurite-arbor morphology requires no more than 50% of wild-type fascin function.
Fig. 1.
Genetic and pharmacological modification of neurite curvature. (A–C,F–I) Phase-contrast images (60×) of neurons cultured for 3 d.i.v. from the CNS of wandering third instar larvae. Magnification is the same throughout. (A–C) Loss-of-function singed mutations increase neurite curvature and disrupt proximal-to-distal tapering. (A) Wild-type (sn+/sn+), OreR-C laboratory strain. (B) A partial loss-of-function (hypomorphic) mutation (sn3/sn3) causes a moderate filagree phenotype. (C) A null mutation (snX2/Y) causes a severe filagree phenotype. (D,E) Mean curvature distributions of neurite arbors of cultured neurons with differing genotypes, plotted with soft binning. Normal-distribution curves were fit to each population and scaled (y-axis) to the corresponding histograms. (D) Genetically marked γ-MB neurons. The increased curvature of sn3/snX2 (blue) neurons is easily seen. The similarity of snX2/+(red) and sn+/sn+ (green) curvature distributions demonstrates that filagree is recessive. (E) Random CNS neurons. The mean neurite curvature distribution of sn3/sn3 mutant neurons (magenta) is significantly increased over that of wild-type (sn+/sn+, OreR-C) neurons (green). (F–I) Exposure to drugs in vitro can modify the filagree phenotype of fascin-deficient sn3/sn3 neurons. (F,G) Two examples of filagree decreasers (fascin-pathway enhancers): estradiol propionate, 50 μM (E) and Anisindione, 50 μM (G). Neurite curvature, especially of terminal neurites, is reduced and the smooth proximal-to-distal tapering is restored. (H,I) Two examples of filagree increasers (fascin-pathway blockers): griseofulvin, 50 μM (H) and acetyltryptophan, 50 μM (I). Note the exaggerated neurite curvature and frequent expansions of neurite width.
A bidirectional drug-screen design requires that the baseline neuronal morphology be intermediate and distinct from both wild type (Fig. 1A) and singed-null (snX2/Y; Fig. 1C), allowing detection of both drug-induced worsening and drug-induced rescue of the filagree defect (Fig. 1B). The neurite-curvature distributions of the severe hypomorph,sn3/sn3 (Fig. 1B), and the wild-type (Fig. 1A) were significantly different (Welch’s t-test, P<1×10−13; Fig. 1E), with greater overlap of distributions than between wild type and sn3/snX2 (Kraft et al., 2006). This is consistent with the relative function among the genotypes. Moreover, the sn3 mutation leaves the fascin open reading frame intact, allowing small amounts of normal protein to be produced (Paterson and O’Hare, 1991; Cant et al., 1994). Thus, sn3/sn3 neurons have a desirable neuronal phenotype, and contain some wild-type fascin protein that can serve as a drug target.
The striking nature of the filagree defect makes it suitable for use in a modifier screen based on direct phenotype observation. Using an approach that parallels genetic-modifier screens commonly performed with model organisms, we conducted a chemical screen to identify small molecules that induce obvious changes in neuronal phenotype. Compounds were tested at 10 μM and 50 μM by adding them to the cultures at the time of plating the dissociated neurons, and evaluating their effects on the filagree phenotype after 3 days in vitro (d.i.v.). Thus, the neurons were exposed to the drug during the entire period of neurite-arbor morphogenesis. To facilitate the drug-screening process, we replaced quantitative analysis of immunostained images of randomly sampled neurons with holistic scoring by phase-contrast microscopy of populations of living cells, typically 1500–2000 neurons per ∼50-mm2 culture well.
Selection and diversity analysis of the NINDS-II compound collection
Inspired by striking examples of new uses for existing drugs, we took a ‘repurposing’ approach, also called ‘repositioning’ or ‘indication switch’ (Ashburn and Thor, 2004; Dueñas-González et al., 2008). For example, minoxidil was developed as an oral anti-hypertensive drug, but is now most popular as a topical treatment (e.g. Rogaine®) for hair loss (Zins, 1988). More dramatic is the repurposing of thalidomide, much-maligned for its teratogenic effects when taken during pregnancy (Ito and Handa, 2012), but now used to treat multiple myeloma, a bone marrow malignancy (Palumbo et al., 2008). Repurposing screens have the advantage of testing compounds whose pharmacology and side-effect profiles are partially understood.
Because fascin acts in diverse pathways (Jayo and Parsons, 2010; Hashimoto et al., 2011), modification of the filagree defect could occur through several potential drug targets, including but not limited to fascin itself. This called for a chemically diverse screening library. The new indications, including brain tumors and neurodevelopmental disorders, made good representation of neuroactive compounds highly desirable. A collaboration between the National Institute of Neurological Disorders and Stroke (NINDS) and the private sector (Heemskerk, 2005) produced a screening library of 1040 compounds (NINDS-II; supplementary material Table S1), comprised primarily of FDA-approved drugs for diverse indications, as well as natural products and laboratory reagents; many of these compounds are neuroactive. Screens of the NINDS-II library with various assays and protocols identified drugs with potential utility for Huntington’s disease, spinomuscular atrophy, amyotrophic lateral sclerosis (ALS), stroke and familial dysautonomia (Aiken et al., 2004; Piccioni et al., 2004; Slaugenhaupt et al., 2004; Stavrovskaya et al., 2004; Rothstein et al., 2005; Vincent et al., 2005; Wang et al., 2005a; Wang et al., 2005b; Desai et al., 2006). The antibiotic ceftriaxone was protective in diverse neurodegeneration assays, leading to Phase III clinical trials for ALS (Traynor et al., 2006) (ClinicalTrials.gov identifier NCT00349622). This library does not contain any migrastatin-family compounds.
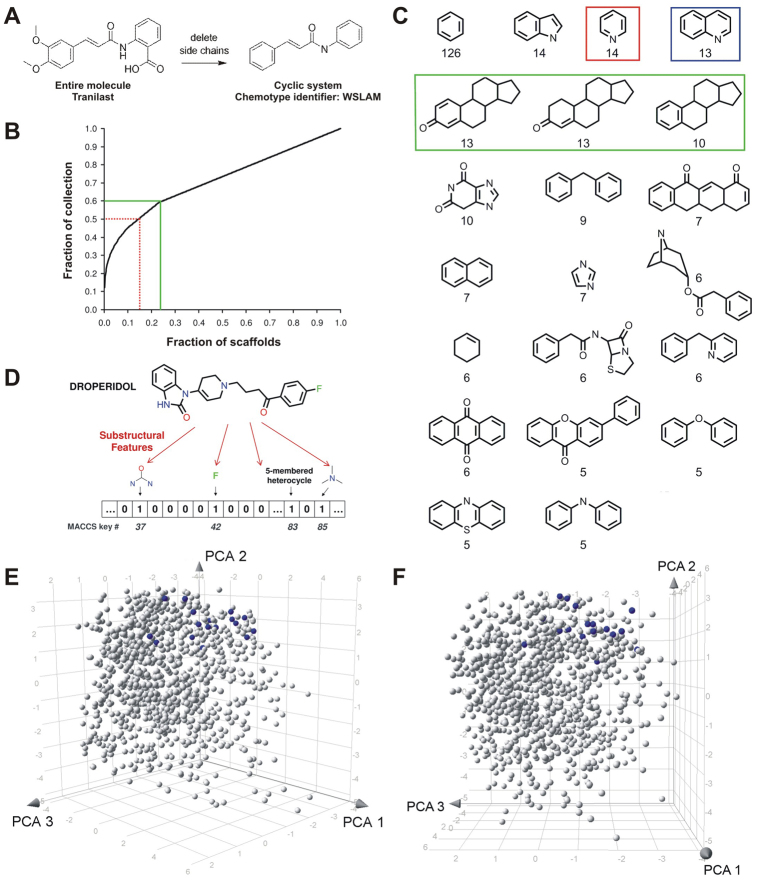
We evaluated the chemical diversity of the NINDS-II compounds (Fig. 2) based on: (i) the types and distribution of molecular scaffolds (Bemis and Murcko, 1996; Singh et al., 2009), (ii) measures of molecular similarity based on shared substructural features (Willett et al., 1998) and (iii) the chemical-space distribution of the compounds (Maggiora and Shanmugasundaram, 2011). The 1040 compounds of the collection are distributed over 617 scaffolds (Fig. 2B), of which 551 are cyclic systems. The highly populated scaffolds (Fig. 2C) are indoles, pyridines, quinolines and sterols, common among drug-like small molecules. Indicative of high diversity, 77% (424) of the cyclic scaffolds are singletons, i.e. populated by a single compound. From the perspective of the compounds, ∼60% of them cover ∼23% of the cyclic scaffolds. In other words, this relatively small collection allows sampling of a relatively large number of scaffolds. The molecular fingerprint, based on 166 structural features (Fig. 2D), of each compound was compared pair-wise with all others, and the Tanimoto coefficients were computed as measures of similarity (Maggiora and Shanmugasundaram, 2011). Overall molecular diversity is inversely proportional to the average Tanimoto similarity for all pair-wise comparisons. For the NINDS-II library, low similarity values (mean 0.295; median 0.284) reflect high diversity and compare favorably to those of the ∼1500-compound DrugBank database (Wishart et al., 2008; Singh et al., 2009). The distribution of compounds in ‘chemical space,’ where the similarity of any pair is inversely related to the distance between them (Maggiora and Shanmugasundaram, 2011), was represented in a three-dimensional (3D) plot (Fig. 2E,F). The compounds are spread throughout the space, reflecting the high diversity of the collection. In summary, the high diversity of the modestly sized NINDS-II collection make it an efficient tool for conducting the first compound screen on the basis of the first fascin bioassay.
Fig. 2.
Molecular diversity of the NINDS-II compound collection screened in this study. (A) Relationship between a molecule and its scaffold, using tranilast as an example. The scaffold or cyclic system is obtained after iteratively removing the side chains and is identified by a five-character chemotype identifier code. The WSLAM scaffold is a singleton, i.e. tranilast is the only compound in the collection with this scaffold. (B) Scaffold-recovery curve. The x-axis represents the scaffolds, organized left-to-right from the most common (benzene) to the singletons, plotted as a cumulative fraction of the total (552, with all 66 acyclic systems considered a single scaffold). The y-axis shows the cumulative fraction of the 1040 compounds occupied by a given fraction of the scaffolds: ∼14% of the scaffolds contain 50% of the NINDS-II library (red lines), and 60% of the library is distributed over ∼24% of the scaffolds (green lines). Beyond this point, each scaffold is a singleton and the relationship becomes linear. (C) Common scaffolds in the NINDS-II library. 2D representations of cyclic systems found with a frequency ≤5; the number is under each structure. Highlighted are the pyridines (red), the quinolines (blue), and the scaffolds containing a four-ring sterol (green; 36 compounds). (D) Representation of chemical fingerprint analysis based on MACCS keys, using the antipsychotic drug droperidol as an example. Each of 166 possible substructural features is coded using a bit score: present at least once (1) or absent (0). Four of droperidol’s features (CN2O, fluorine, five-membered heterocycle and C3N) are shown with their positions in the MACCS keys bitstring. (E,F) 3D representations of the NINDS-II library in chemical space, obtained by principal component analysis (PCA) of the similarity matrix computed using MACCS keys and Tanimoto similarity. Each compound is shown as a sphere. The chemical space distribution of the compounds is extensive, with regional variation in population density. The 17 antipsychotic drugs (blue), are localized to a medium-sized sector of the space and interspersed among compounds with other activities. This is consistent with these drugs occupying 14 scaffolds and representing several different pharmacological mechanisms of action. (E) View centered on the vertical (PCA 2) axis. (F) The view obtained by rotation around the vertical axis. One antipsychotic drug is hidden behind another compound.
A bidirectional drug screen reveals diverse fascin-pathway enhancers and blockers
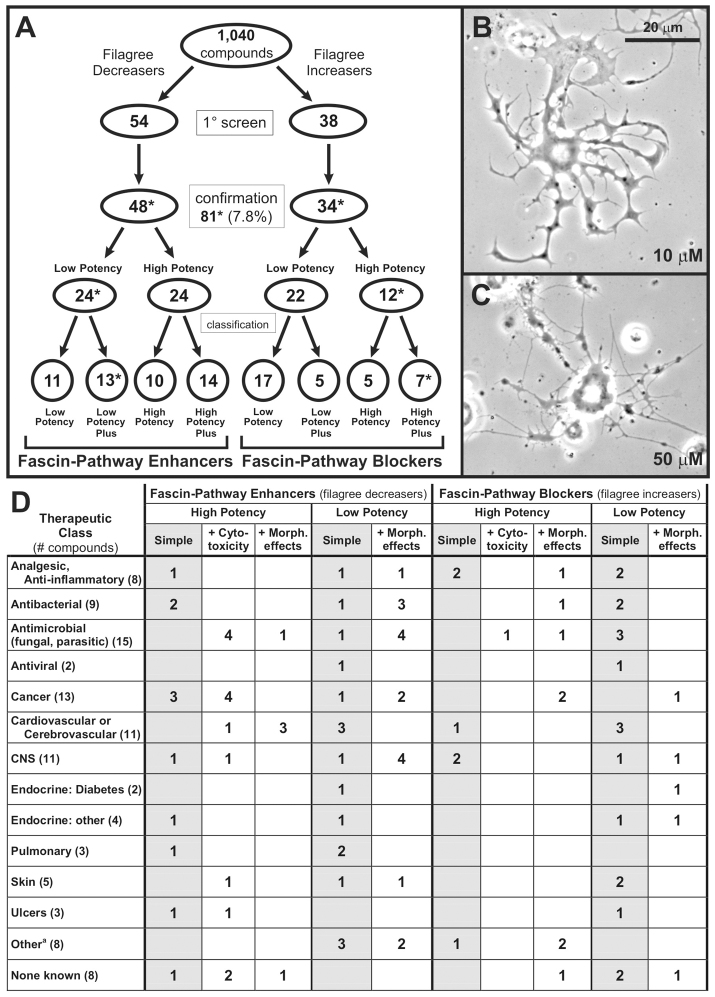
Of the 1040 compounds tested, 81 (7.8%) were active in the fascin bioassay at 10 and/or 50 μM concentrations, based on holistic scoring. We identified both drug-induced decreasers and increasers of the filagree phenotype (Figs 1, 3, 4; Table 1). Filagree decreasers (fascin-pathway enhancers; supplementary material Table S2) rescued the neurite-arbor shape defect of fascin-deficient mutant neurons, allowing them to extend neurites with normal trajectory and tapering (Fig. 1, compare F and G with A). Filagree increasers (fascin-pathway blockers; supplementary material Table S3) worsened the clockwise curvature and further disrupted neurite tapering, as seen in the fascin-null-mutant neurons (Fig. 1, compare H and I with C).
Fig. 3.
Summary of the fascin bioassay-based drug screen. (A) Compounds were screened by holistic scoring for their ability to modify the filagree phenotype, revealing two classes of activity, filagree decreasers (fascin-pathway enhancers) and filagree increasers (fascin-pathway blockers). One compound, cloxyquin, had dose-dependent activity (asterisk) and is counted in both groups. Confirmed active compounds were classified as ‘high potency’ (active at 10 and 50 μM) or ‘low potency’ (active at only 50 μM). Further classification distinguished between simple actives and those with cytotoxicity at the high dose or morphological effects (Plus). (B,C) Phase-contrast photomicrographs after 3 d.i.v. Cloxyquin increased the filagree phenotype at 10 μM (B), but decreased the filagree phenotype at 50 μM (C). (D) Therapeutic classes of filagree-modifying compounds, based on clinical and laboratory data. A compound may be in more than one therapeutic class. ‘Other’ includes compounds used for drug overdose, local anesthesia, gout, eye surgery, allergy, as homeopathic remedy or as insecticide. See supplementary material Tables S2 and S3 for information on chemical class, pharmacology and therapeutic use.
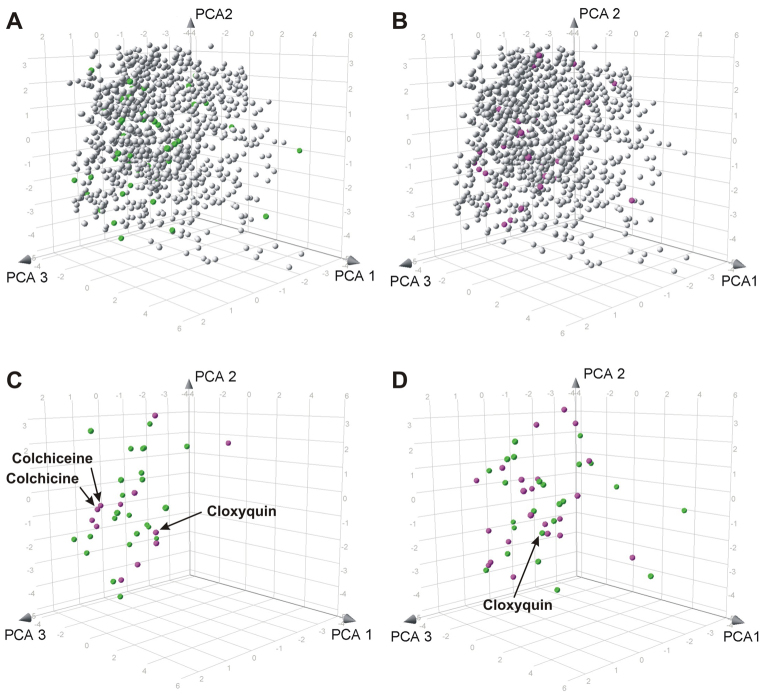
Fig. 4.
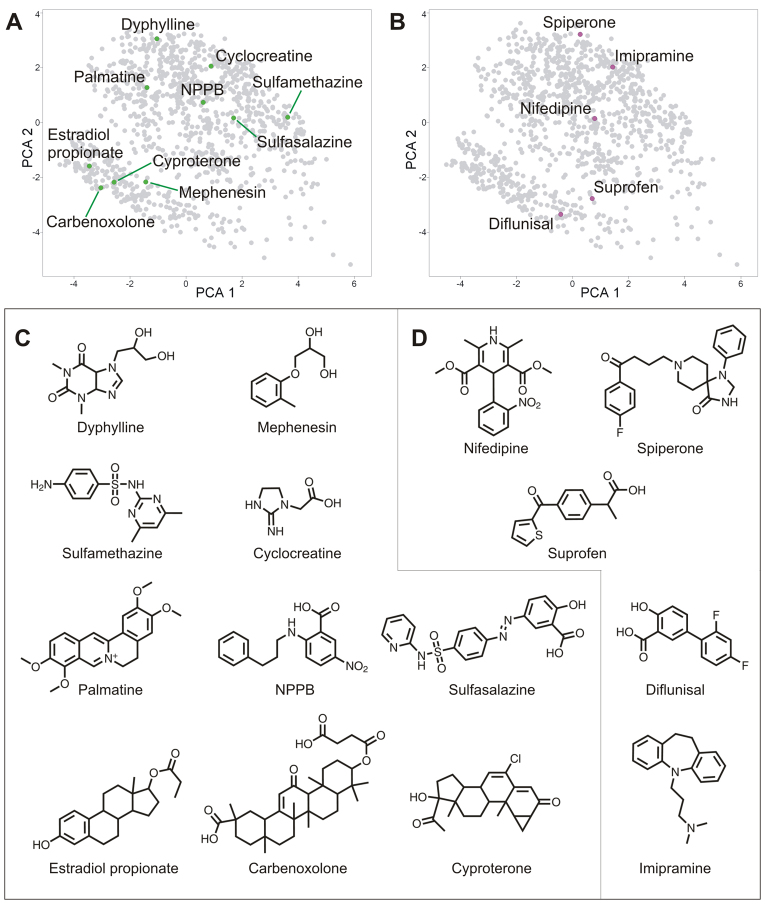
Chemical space distribution of compounds active in the fascin bioassay. 3D views of the chemical space occupied by NINDS-II, centered on the vertical (PCA 2) axis and highlighting different subsets of fascin-pathway modifiers. (A,B) Each class of active compounds is distributed throughout a large fraction of the chemical space occupied by the whole collection. Each compound is represented as a sphere with (A) the 48 fascin-pathway enhancers in green and (B) the 34 fascin-pathway blockers in magenta against the gray background of the inactive compounds. (C,D) Fascin-pathway modifiers of the same potency class, plotted in contrasting colors without the whole collection. Within each class, there is some intermingling of compounds with opposite activities, and some clustering of compounds with the same activity. (C) High-potency fascin-pathway enhancers (green; 24) and blockers (magenta; 12). (D) Low-potency fascin-pathway enhancers (green; 24) and blockers (magenta; 22). See supplementary material Fig. S2 for chemical structures of all active compounds.
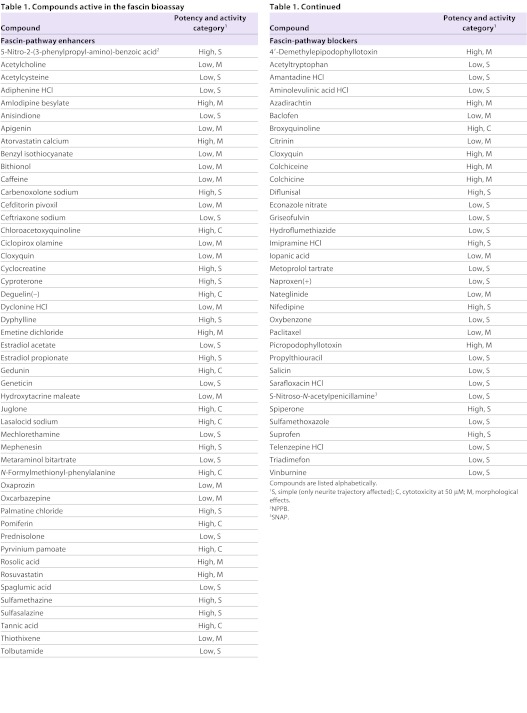
Table 1.
Compounds active in the fascin bioassay
Compounds with activity at 10 μM were considered ‘high potency’ whereas those with activity only at 50 μM were considered ‘low potency’. The 48 fascin-pathway enhancers were split equally between low- and high-potency compounds; the 34 fascin-pathway blockers included 22 low- and 12 high-potency compounds (Fig. 3A). Cloxyquin was the only compound with a bimodal effect, acting as a filagree increaser at 10 μM, but as a filagree decreaser at 50 μM (Fig. 3B,C). Within each potency class, we made a distinction between ‘simple’ actives (21 fascin-pathway enhancers, 22 fascin-pathway blockers) and those with other effects on the neurons beyond filagree modification (Fig. 3A,D; see below). The compounds of each activity group span a broad range of therapeutic indications (Fig. 3D; supplementary material Tables S2, S3).
When active compounds were re-tested for quantification of neurite curvature, we found that holistic scoring is relatively insensitive and very likely to underestimate true drug potencies. For example, the antifungal agent griseofulvin, initially identified as a low-potency fascin-pathway blocker (Fig. 1H), was active at both 10 μM (P=0.011) and 50 μM (P=0.0002), causing a dose-dependent increase in neurite curvature. Surprisingly, the mushroom body γ-neurons (γ-MB neurons) in the griseofulvin-treated cultures failed to show a significant change in neurite curvature at either 10 μM (P=0.13) or 50 μM (P=0.08). This also raises the question of whether any of the ten filagree-modifying compounds whose activity failed to replicate in confirmation tests (Fig. 3A) might have been too close to the detection threshold by holistic scoring, but nonetheless have some activity. This highlights the differential drug sensitivity of classes of neurons in the fascin-deficient brain. It is therefore possible that we did not detect drugs whose effects were restricted to neuron subtypes.
The 3D chemical-space distribution of the 48 fascin-pathway enhancers (filagree decreasers) and 34 fascin-pathway blockers (filagree increasers) are shown plotted against the background of the compound collection (Fig. 4, A and B, respectively). Each set of active compounds is quite diverse, as indicated by its distribution throughout the chemical space of the collection without obvious localization to any particular neighborhood. A notable exception is the pair of high-potency fascin-pathway blockers, colchicine and colchiceine (Fig. 4C), which are closely related alkaloids that differ only by a methyl group (Tanimoto similarity 0.956). When the fascin-pathway enhancers and blockers were plotted together in chemical space, many instances of intermingling between the two were seen among both high- and low-potency active compounds (Fig. 4, C and D, respectively). The chemical structures of the high-potency simple actives are shown in Fig. 5, along with 2D chemical-space plots. The ten simple high-potency fascin-pathway enhancers (Fig. 5C) and the five simple high-potency fascin-pathway blockers (Fig. 5D) each belong to a different scaffold.
Fig. 5.
Structures of high-potency simple fascin-pathway enhancers and blockers. (A,B) 2D chemical-space distributions of the high-potency simple filagree modifiers plotted against the background of the NINDS-II compound collection in gray. Each compound is represented as a circle; due to the projection, some compounds are hidden from view. (A) Ten fascin-pathway enhancers (green). (B) Five fascin-pathway blockers (magenta). (C,D) 2D chemical structures of the high-potency simple filagree modifiers. Within each class, there is considerable structural diversity. (C) Fascin-pathway enhancers. (D) Fascin-pathway blockers.
Consistent with this level of structural diversity, the active compounds represent diverse chemical classes and wide-ranging pharmacological mechanisms of action (supplementary material Tables S2, S3). For example, drugs that directly impact neurotransmitter systems were found among both fascin-pathway enhancers (acetylcholine, adiphenine HCl, mephenesin, metaraminol bitartrate, spaglumic acid) and fascin-pathway blockers (amantadine, baclofen, imipramine, spiperone). Several of the steroid hormones in the library were fascin-pathway enhancers (cyproterone, estradiol acetate, estradiol propionate, methylprednisolone). Among the fascin-pathway modifiers were two dozen antimicrobial agents, including antibacterial, antiviral, antifungal and anti-amoebic compounds.
Neither pharmacological nor chemical class reliably predict the direction of activity in the fascin bioassay. Of two sulfanilamide-type antibacterial agents, one (sulfamethoxazole) was a fascin-pathway blocker and the other (sulfamethazine) was a fascin-pathway enhancer. Four non-steroidal anti-inflammatory drugs that inhibit cyclooxygenase (COX) were fascin-pathway blockers (diflunisal, naproxen, salicin, suprofen), whereas other COX inhibitors, including diclofenac and ibuprofen, were inactive. The diversity of the active compounds suggests that multiple drug targets and mechanisms are operative, which is consistent with a screen based on a pathway that is regulated at multiple levels (Jayo and Parsons, 2010). These considerations led to further analysis of the chemical structures of active compounds and their inactive relatives.
Structure-activity relationships of fascin-pathway enhancers and blockers
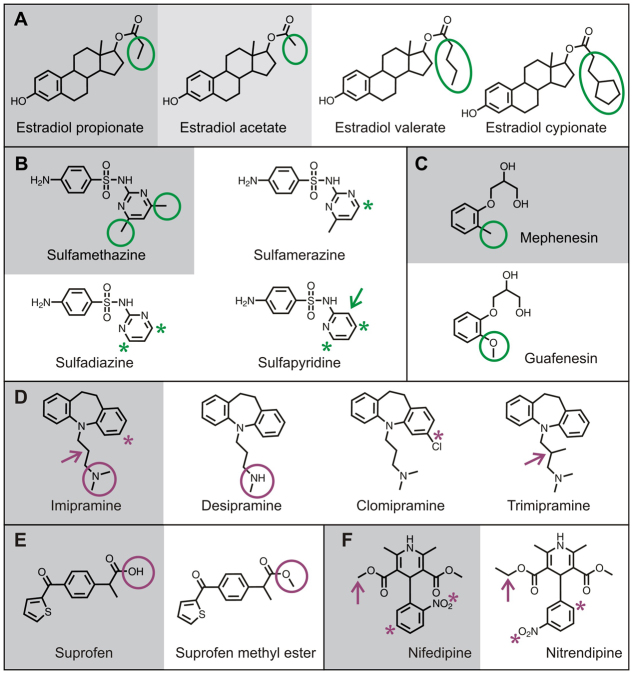
Due to the complexity of any cell-based assay, the activity of a compound can be affected by its transport, metabolism, binding to the molecular target(s) or a combination thereof. Nonetheless, small subsets of structurally similar compounds in the NINDS-II collection allowed us to conduct a preliminary analysis of structure-activity relationships (SARs) (Fig. 6) based on the concept of activity landscapes. Analogous to geographical landscapes, the latitude and longitude correspond to the position of a molecule in a 2D chemical space, and the altitude corresponds to its activity in a given assay. Because structurally similar compounds tend to have similar biological activities (Johnson and Maggiora, 1990), regions of active molecules in an activity landscape tend to resemble rolling hills. In some cases, however, similar molecules have significantly different activities, leading to cliff-like features. These activity cliffs contain significant information on SARs because they identify relatively small yet biologically relevant structural features associated with the dramatic changes in activity (Maggiora and Shanmugasundaram, 2011). We evaluated cases in which high-potency fascin-pathway enhancers (Fig. 6A–C) or blockers (Fig. 6D–F) were structurally very similar to low-potency or inactive compounds. By comparing the structures at the top of each cliff with those at its base, we could infer which structural features are necessary for activity. In some cases, the SAR hypotheses were also informed by the physical and/or chemical properties of the active and inactive compounds.
Fig. 6.
SAR analysis based on activity cliffs. (A–F) High-potency fascin-pathway modifiers (gray background) compared with structurally similar low-potency (light gray background) or inactive (white background) compounds. The highlighted substructures can be inferred to influence activity in the fascin bioassay. (A–C) Fascin-pathway enhancers (green highlights). (A) Estradiol propionate: the size of the side chain (oval) on the carboxy group on the sterol D ring is associated with activity, suggesting that in estradiol acetate the side chain is too small, whereas in estradiol valerate and cypionate, it is too large. (B) Sulfamethazine: activity is associated with dimethylation (circles) of the pyrimidine ring. Removal (asterisk) of one (sulfamerazine) or both (sulfadiazine and sulfapyridine) methyl groups is associated with loss of activity. (C) Mephenesin: replacement of the methyl group with a methoxy group in guaifenesin (circles) is associated with lack of activity. (D–F) Fascin-pathway blockers (magenta highlights). (D) Imipramine: in three other tricyclic compounds, single changes are associated with lack of activity. Desipramine is lacking a methyl group of the alkyl nitrogen (circles); clomipramine has a chloride on one of the aromatic rings (asterisks); trimipramine has a methyl substitution in the middle of the alkyl chain (arrows). (E) Suprofen: replacement of a hydrogen with a methyl group (circles) creates suprofen methyl ester, which is inactive. (F) Nifedipine: two changes, the position of the nitro group on the aromatic ring (asterisks) and addition of a methyl group (arrows), are associated with lack of activity of nitrendipine.
Comparing estradiol propionate with estradiol acetate (diminished activity) and two inactive estradiols (similarity to estradiol propionate ≥0.94), the key structural feature appears to be the substituent on the carboxy moiety (Fig. 6A, green ovals). Removing a methyl group lowers the activity, whereas lengthening the alkyl chain (estradiol valerate) or adding a terminal cyclopentane ring (estradiol cypionate) renders the compounds inactive. Because the steroid moiety remains unchanged, it can be inferred that the size of the carbon chain on the carboxy moiety is a crucial structural determinant of fascin-pathway enhancement.
For the high-potency fascin-pathway enhancer sulfamethazine, the focus is the pyrimidine ring, which is dimethylated in the active compound (Fig. 6B, green circles). Three inactive sulfonamides, sulfamerazine, sulfadiazine and sulfapyridine (with similarities to sulfamethazine of 0.98, 0.96 and 0.88, respectively), have reduced numbers of methyl substituents (Fig. 6B, asterisks). In sulfapyridine there is also a carbon atom replacing one of the nitrogens in the pyrimidine ring (Fig. 6B, green arrow). Because removing one or two methyl substituents renders the compound inactive, the importance of removing the ring nitrogen cannot be interpreted. Nevertheless, we can infer that dimethylation of the pyrimidine ring is important for activity. The high-potency fascin-pathway enhancer mephenesin was compared with the structurally related but inactive guaifenesin (similarity 0.87), which differs only by the replacement of the methyl substituent in the phenyl ring by a methoxy group (Fig. 6C, green circles). Thus, it can be inferred that activity is sensitive to substitution at that position of the phenyl ring.
The high-potency fascin-pathway blocker imipramine differs by a single change from each of three inactive tricyclic compounds, with similarities to imipramine of 0.93, 0.91 and 0.88 (Fig. 6D). Desipramine, an imipramine metabolite produced in the mammalian liver, lacks a methyl group on the alkyl nitrogen (Fig. 6D, circles). On the basis of experimentally determined dissociation constants, desipramine is a considerably stronger base than imipramine (Shalaeva et al., 2008). Hence, it has a harder time losing a proton to cross the plasma membrane and enter the neuron. This suggests that access to an intracellular target could be a factor controlling activity of tricyclic compounds in the fascin bioassay. For clomipramine, the single substitution of a chlorine atom on one aromatic ring (Fig. 6D, asterisks) eliminates activity. Lastly, trimipramine differs only by the presence of a methyl substitution on the alkyl chain (Fig. 6D, arrows). From these observations, it can be inferred that three distinct features are essential for imipramine activity.
In Fig. 6E, the high-potency fascin-pathway blocker suprofen is compared with its inactive methyl ester (similarity 0.82). Replacement of a hydrogen atom by a methyl group might disrupt activity because the acid moiety in suprofen is undoubtedly ionized in solution. Thus, the active form of suprofen is most probably an anion, whereas the inactive ester is uncharged. The high-potency fascin-pathway blocker nifedipine has two structural differences from its inactive analog nitrendipine (similarity 0.90). First, nifedipine has a methyl ester whereas nitrendipine has an ethyl ester (Fig. 6F, arrows). Second, the nitro group (Fig. 6F, asterisks) is in the ortho position in nifedipine, but in the meta position in nitrendipine. This change might affect the 3D conformation of nifedipine or indicate that a specific orientation of the nitro group is required for activity.
Other drug effects on cultured neurons
Roughly half of the actives had effects beyond modifying the filagree phenotype (Fig. 3A,D; Table 1; supplementary material Tables S2, S3), which we classified as cytotoxicity or morphological effects. Cytotoxicity was defined as apparent cell death without a neurite arbor (Fig. 7A,B) or, rarely, degeneration of neurites after formation of an arbor (data not shown). A subset of high-potency fascin-pathway modifiers caused overt cytotoxicity at the 50-μM concentration. Some inactive compounds also caused cytotoxicity. Distinctive morphological effects, the most common of which was reduced neurite outgrowth (Fig. 7E,F,H–K), were caused by both active and inactive compounds. Less common, but more dramatic, were changes in neuronal cell-body size or shape (Fig. 7C,D) and bizarre alterations in neurite morphology (Fig. 7C–E,G–K).
Fig. 7.
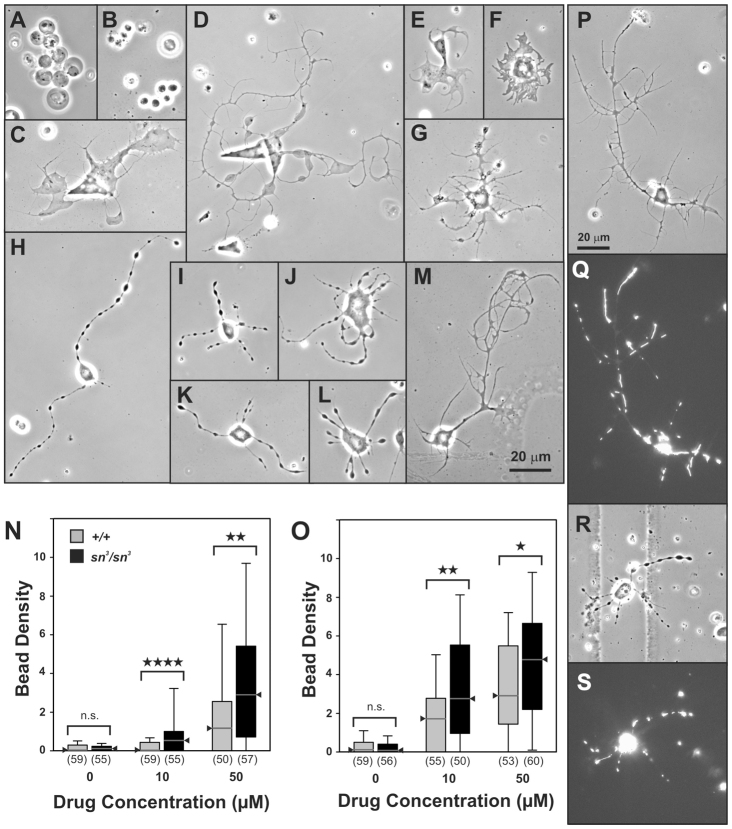
Neurotoxic morphological defects revealed in primary neuron culture. (A–M) Phase-contrast images (60×) of CNS neurons from sn3/sn3 (A–K) or wild-type (OreR-C; L–M) wandering third instar larvae, cultured for 3 d.i.v. with the following drugs (scale bar in M for images A–M): (A) tannic acid, 50 μM; (B) 3-3′-diindolylmethane; (C) ginkgolic acid, 50 μM; (D) tegaserod maleate, 50 μM; (E) azadirachtin, 50 μM; (F) 4’-demethylepipodopyllotoxin, 10 μM; (G) usnic acid, 10 μM; (H) atorvastatin, 50 μM; (I) rosuvastatin, 50 μM; (J) lovastatin, 50 μM; (K) pravastatin, 50 μM; (L) atorvastatin, 50 μM; (M) no-drug control. (N–O) The statin-induced BOS defect is modulated by genetic background. Box-plot distributions of the bead density (beads per 100 μm) along neurites of wild-type (+/+; gray) or fascin-deficient mutant (sn3/sn3; black) larval neurons cultured for 3 d.i.v. with pravastatin (N) or rosuvastatin (O) at the indicated concentrations. The median is indicated by the triangle. The top and bottom of the box represent the 75th and 25th percentiles, respectively; the top and bottom ‘whiskers’ represent the 90th and 10th percentiles, respectively. The number of neurons analyzed is indicated in parentheses below each group. At both concentrations, the mutant neurons are more sensitive than the wild-type neurons to the BOS effect of the statins (Mann-Whitney Rank Sum test). n.s., not significant; *P<0.05; **P<0.01; ****P<0.0001. (P–S) BOS beads contain mitochondria (scale bar in P for images P–S). Wild-type larval neurons expressing a GFP-tagged mitochondrial protein cultured for 3 d.i.v. and imaged live by phase-contrast (P,R) or fluorescence microscopy (Q,S). (Grid lines on the dish floor are visible behind the neuron in R.) In the no-drug control cultures (P,Q) GFP-tagged mitochondria are distributed as clusters or linear aggregates scattered throughout the neurite arbor, with a range of sizes and densities. In the atorvastatin-treated cultures (R,S), the GFP-tagged mitochondria are localized almost exclusively to the beads.
Within the NINDS-II library, 22% of the compounds are experimental (i.e. research reagents) rather than marketed pharmaceuticals or drugs that have been tested in clinical trials (supplementary material Table S1, Status column). Experimental compounds were over-represented in the cytotoxic-at-high-dose (54%) and morphological-effects (31%) groups, and were under-represented among the simple fascin-pathway modifiers (16%). It is not surprising that compounds with well-established toxicity, such as the sodium pump inhibitor ouabain and the protein synthesis inhibitors cycloheximide and anisomycin, had overt cytotoxic effects on cultured neurons. Similarly, the ionophore lasalocid, a high-potency fascin-pathway enhancer with cytotoxicity at 50 μM, is a known neurotoxin (Safran et al., 1996). Hence, much of the neurotoxicity among the active compounds was caused by molecules that are not drugs in the strict sense of the repurposing strategy. In many cases, synthetic chemistry can be used to modify such parent compounds to produce analogues that retain the desired activity but have reduced toxicity (e.g. Harrap, 1985).
Statin-induced BOS defect is modified by genetic background
Beads-on-a-string (BOS) is a striking morphological defect that was induced solely by the four statins in the library: atorvastatin, lovastatin, rosuvastatin and pravastatin (Fig. 7H–K). Wild-type neurons are also susceptible to the BOS defect when cultured with any of these four statins (Fig. 7L and data not shown). BOS has two components, ovoid intracellular nodules distributed along the length of the neurites and reduced neurite outgrowth (Fig. 7, compare L with M). These four statins differ in their potency of HMG-CoA reductase inhibition (White, 2002), and this paralleled their potency to induce BOS. Pravastatin and lovastatin induced BOS only at 50 μM, whereas rosuvastatin and atorvastatin did so at both 10 μM and 50 μM (Fig. 7N,O and data not shown). BOS was reversible when the culture media was replaced with drug-free media; the beads resolved and neurite outgrowth was markedly accelerated (data not shown). Using a transgenic GFP-tagged mitochondrial protein (Pilling et al., 2006), we found that the beads contained aggregations of mitochondria (Fig. 7P–S). The appearance of statin-induced beads is consistent with disrupted microtubule transport (Pilling et al., 2006), which would also contribute to reduced neurite outgrowth.
Because statin-induced myopathy is affected by genetic variants (Niemi, 2010), we tested the hypothesis that sensitivity of Drosophila neurons to statin-induced BOS is influenced by genotype. We compared the bead density of wild-type and fascin-deficient singed-mutant neurons in response to incubation with a low-potency (pravastatin) and a high-potency (rosuvastatin) statin. For both drugs at both 10 μM and 50 μM, fascin deficiency significantly enhanced the sensitivity of cultured neurons to statin-induced BOS (Fig. 7N,O).
DISCUSSION
The fascin pathway as a drug-discovery target
Motivated by the importance of fascin function in tumor invasion, as well as in brain development and plasticity, we designed and conducted a screen for pharmacological modifiers of the fascin pathway. By using primary cultured neurons in a cell-based assay, we avoided the disadvantages of immortalized cell lines and made use of the fact that dissociated mutant neurons can reveal cellular phenotypes with a magnitude far greater than that in the intact brain (Kraft et al., 2006; Liu et al., 2007; Chen and Herrup, 2008; Sawallisch et al., 2009). The filagree phenotype of fascin-deficient neurons, which is associated with a marked disruption of actin cytoskeleton organization (Kraft et al., 2006), causes a striking exaggeration of the intrinsic tendency of cultured neurites to turn clockwise (Tamada et al., 2010). This cell-based assay, with its easily observable morphological read-out, allowed us to cast a wide net, targeting the pathway without trying to predict a priori which molecule (fascin or one of its regulators or mediators) would be the best drug target for modulating fascin function. By choosing a well-characterized hypomorphic singed mutation, we were able to screen simultaneously for fascin-pathway blockers and enhancers in cells containing small amounts of wild-type fascin protein that could potentially serve as a drug target. Despite the limited sensitivity of holistic scoring, by screening a diverse collection we identified diverse high-potency compounds in both activity classes.
Fascin-pathway blockers identified in the Drosophila neuronal fascin bioassay are predicted to inhibit tumor invasion. Malignant glioblastoma represents a particularly challenging clinical problem because these cells migrate avidly along many routes beyond the boundaries of the main tumor mass. Chemotherapy with the new anti-proliferation drug temozolomide has increased patient survival, but only by months (Mangiola et al., 2010). It is likely that long-term survival will require a second drug to inhibit glioma dispersion (Giese et al., 2003; Salhia et al., 2006). Hence, we tested seven fascin-pathway blockers in a radial migration assay with two human glioblastoma multiforme cell lines, and found that three of the compounds caused dose-dependent inhibition of migration in both lines (L.L.R., W. S. McDonough, A.K. and M. E. Berens, unpublished results). This is encouraging preliminary cross-species evidence that a subset of compounds identified through the Drosophila fascin bioassay will have predictable effects on the phylogenetically conserved fascin pathway of mammalian cells. We note that, for both RET-dependent thyroid cancer (Das and Cagan, 2010) and fragile X syndrome (Bhogal and Jongens, 2010), Drosophila models have provided evidence of drug efficacy on the road to clinical trials (ClinicalTrials.gov database, NCT00514046 and NCT00965432, respectively).
The tricyclic antidepressant imipramine was a high-potency fascin-pathway blocker in our assay. An intriguing study of imipramine blue reported that this triphenylmethane derivative has anti-invasion properties against malignant glioma cells in vitro and in vivo (Munson et al., 2012). Although that study design was based on inhibition of NADPH oxidase by imipramine blue, glioma cells treated in vitro showed dramatic reorganization of their actin cytoskeleton, with a marked loss of actin bundle-based protrusions and extensions (Munson et al., 2012), consistent with the loss of fascin function (Vignjevic et al., 2006).
What can we infer about molecular mechanisms of action of the fascin-pathway modulators? Some possibilities can be proposed or ruled out based on the nature of the fascin bioassay. First, the molecular diversity of the fascin-pathway modulators argues strongly against a common target for either the enhancers or the blockers. Candidate targets, in addition to fascin per se, include CREB, CREBBP, Stat3, PKCα, Rab35, Rac and myosin X (Hashimoto et al., 2011).
Second, the ‘known’ pharmacology of the active compounds (supplementary material Tables S2, S3) might not be relevant, and this is implicit in the rationale of drug repurposing. For example, because the dissociated neuron cultures are low density by design, with limited cell-cell contact, the effects on neurite trajectory of neuroactive compounds like acetylcholine, adiphenine, baclofen and spiperone cannot be mediated through classical neurotransmission at functional synapses. Rather, their effects are better classified with the morphogenetic phenomena by which neurotransmitters influence brain development (Herlenius and Lagercrantz, 2001; Verney, 2003). Similarly, the antibiotic ceftriaxone, best known for blocking bacterial cell wall synthesis, has neuroprotective activity due to upregulation of glutamate transporter GT1 expression (Rothstein et al., 2005). However, this mechanism is unlikely to explain the activity of ceftriaxone in the fascin bioassay because GT1 is a glial protein, and our cell culture system is neuron-only.
Third, blockers that target the residual wild-type fascin in the singed-mutant neurons most probably do not have the same mechanism as the migrastatin family. This is because Drosophila and mammalian fascin differ at amino acid position 474, which is essential for migrastatin binding (Chen et al., 2010). That site is on the surface of fascin (Jansen et al., 2011), a location with potential disadvantages for drug targeting. By contrast, high-resolution X-ray crystallographic studies identified internal pockets within the two actin-binding sites of fascin that, on the basis of glycerol and polyethylene glycol binding, could accommodate small-molecule blockers (Jansen et al., 2011).
Fourth, the fascin-pathway modulators with additional effects (Table 1) might represent distinct targets. For instance, 16 of the 18 fascin-pathway enhancers with other morphological effects caused reduced neurite outgrowth. Note that very small neurite arbors can manifest the filagree phenotype (Kraft et al., 2006). Therefore, our observations suggest that slowing neurite outgrowth compensates for fascin deficiency, allowing straighter neurite trajectory. Consistent with this, in vitro treatment of fascin-deficient neurons with a physiological hormone that boosts neurite outgrowth increases neurite curvature (Kraft et al., 2006). Although slowing the rate of neurite outgrowth might not be appropriate for therapeutic intervention, the observed relationship between neurite trajectory and outgrowth rate provides an important clue to fascin biology.
Repurposing and structure-activity relationships
Repurposing drug screens are done in the hope that the time to clinical efficacy trials will be considerably shorter than for a new molecular entity and, at least in theory, that retail prices will therefore be lower (Dueñas-González et al., 2008). Nonetheless, SAR investigation continues to be useful for probing the molecular requirements for activity. For example, the inferences from the activity cliffs (Fig. 6) represent SAR hypotheses that can be tested in the fascin bioassay using commercially available compounds that are similar to the original actives. Moreover, the actives can also be the starting point for structural modification for lead optimization, to enhance potency or to enhance blood-brain barrier penetration and CNS distribution (Denora et al., 2009).
Primary neuron culture for neurotoxicity screening in a genetic model organism
Insects lack the post-squalene sterol biosynthetic pathway and thus cannot synthesize cholesterol (Chapman, 1998). Hence, the BOS defect cannot be due to statin effects on cholesterol levels. HMG-CoA reductase inhibition also blocks the synthesis of isoprenoids that serve as membrane anchors for small GTPases such as Ras, Rho and Rac (Cordle et al., 2005; Wang et al., 2008). Isoprenyl membrane anchors are necessary for the activation of these signaling molecules, which serve essential roles in nervous system development and synaptic plasticity (Hall and Lalli, 2010; Samuel and Hynds, 2010; Ye and Carew, 2010). In mammals, statin neurotoxicity is manifest as reduced neurite outgrowth in vitro due to blockade of the isoprenoid pathway (Schulz et al., 2004) and, in the case of sympathetic neurons, specifically by inhibition of RhoA activation (Kim et al., 2009b). Thus, the BOS neurotoxic effect could involve the functional inhibition of small GTPases. We propose that BOS reflects disruption of microtubule-based transport, resulting in large aggregations of organelles, including mitochondria (Fig. 7P–S) and synaptic vesicles (data not shown). The synergistic effect of statin exposure and fascin deficiency highlights the importance of fascin for actin-myosin interactions (Ishikawa et al., 2003; Nagy et al., 2008; Tamada et al., 2010) that link actin and microtubule cytoskeletons (Cao et al., 2004).
BOS stands out from other neurotoxic effects (Fig. 7A–G) because of growing evidence that statins can cause significant, reversible neurocognitive or psychiatric side effects (King et al., 2003; Wagstaff et al., 2003; Golomb et al., 2004; Galatti et al., 2006; Tatley and Savage, 2007; Evans and Golomb, 2009). Recognition and reporting of these statin-caused adverse events, previously under-recognized and under-reported (Golomb et al., 2007), are likely to increase now that the FDA has added confusion and memory loss to statin warning labels (for more details, see http://www.fda.gov/Drugs/DrugSafety/ucm293101.htm). There is some urgency to address statin-induced cognitive side effects. In a randomized clinical trial, statin treatment disrupted learning in patients, who did not perceive that they had developed a cognitive deficit (Muldoon et al., 2004). In addition, there is a push from industry to approve statins for over-the-counter sales (Tinetti, 2008) and for use in children (de Ferranti and Ludwig, 2008). The latter is particularly worrisome because the developing brain might well be at higher risk for sides effect from statins.
There is presently no way to predict which patients will have statin-induced neurocognitive side effects. By contrast, statin-induced myopathy is associated with a single-nucleotide polymorphism in SLCO1B1 (MIM#604843), which encodes a hepatic statin transporter (Niemi, 2010). Our demonstration that fascin modulates BOS (Fig. 7N–O) suggests that Drosophila genetics and neuron culture could be applied systematically to identify candidate genes whose human orthologs control sensitivity to statin-induced cognitive side effects. More generally, tens of thousands of environmental compounds await testing for developmental neurotoxicity (Landrigan, 2010). Primary culture of developing Drosophila neurons could provide a starting point for neurotoxicity screening. There are many potential benefits for screening with an invertebrate in vitro model system (Coecke et al., 2007), especially one so well-suited to examining gene-X-environment interactions.
Brain development and malignant tumors: two sides of the same genetic coin
Fascin is not unique in being essential for brain development, yet also having pathological consequences when overexpressed or overactive in tumor cells. For example, the proto-oncogene MET (MIM#64860) encodes a receptor tyrosine kinase whose activation drives tumor progression and drug resistance (Stella et al., 2010). On the other hand, MET controls neuronal differentiation and is a risk gene for both schizophrenia (Cannon, 2010) and autism, with the autism-associated variants disrupting MET or reducing MET expression (Judson et al., 2011). Similarly, the group I PAK (p21-activated kinase) genes, which regulate cytoskeletal dynamics, are drivers of tumor invasion (Molli et al., 2009). However, they play crucial roles in brain development and synaptic plasticity, with cognitive and behavioral deficits resulting from PAK3 (MIM#300142) loss-of-function mutations (Kreis and Barnier, 2009).
Extrapolating further, one would predict that patients with developmental brain disorders due to loss-of-function mutations should be at lower risk for invasive cancers. A relevant case report describes a child with fragile X syndrome whose untreatable glioma had an indolent course for more than 8 years despite high-grade histopathology (Kalkunte et al., 2007). In fact, this biological duality might explain why patients with schizophrenia, a highly heritable, late-onset brain-development disorder (Karlsgodt et al., 2008), have lower rates of cancer despite engaging in high-risk behaviors such as smoking and drinking (Cannon, 2010). Thus, two seemingly unconnected medical conditions are linked by genes with dual roles in brain development and tumor biology. We propose that the bidirectional screen presented here represents a powerful new approach for simultaneous drug discovery for brain-development disorders and cancer.
METHODS
Drosophila culture and genetics
Fly stocks were maintained at room temperature on corn meal-yeast-agar medium obtained from the University of Arizona fly food facility. Experimental cultures were reared at 25°C and 60–80% relative humidity on corn flour-yeast-agar medium as previously described (Kraft et al., 2006). OreR-C was the wild-type laboratory strain. The sources of singed mutant stocks, the w(z) strain of w1118 (in a CantonS background), and the γ-MB driver/reporter stock (P[Gal4]201Y UAS-lacZ) were as described previously (Kraft et al., 1998; Kraft et al., 2006). The neuron-specific driver line elav-Gal4C155 (Lin and Goodman, 1994) was provided by Mani Ramaswami (then at University of Arizona, Tucson, AZ). The GAL4-responsive mitochondria-targeted GFP transgenic line w1118; P(UAS-mitoGFP) (Pilling et al., 2006) was provided by Konrad Zinsmaier (University of Arizona, Tucson, AZ). To identify mitochondria in living neurons, we generated elav-GAL4C155/w1118; P(UAS-mitoGFP)/+ progeny larvae.
γ-MB neurons with wild-type fascin/singed function were identified in vitro by crossing the γ-MB driver/reporter line to w(z) to generate w sn+/ w sn+; P[Gal4]201Y UAS-lacZ / +. To study the effect of fascin/singed mutation severity on the filagree phenotype of γ-MB neurons, we performed three crosses in parallel using the 201Y driver to produce female larval progeny of the desired genotypes. Crossing y w sn3/ Y; P[Gal4]201Y UAS-lacZ males to sn3/sn3 virgin females yielded y w sn3/sn3; P[Gal4]201Y UAS-lacZ / + daughters. In(1) dl-49, snX2/Y males were crossed either to y w sn3; P[Gal4]201Y UAS-lacZ females to produce y w sn3/snX2; P[Gal4]201Y UAS-lacZ/+ daughters, or to w; P[Gal4]201Y UAS-lacZ females to produce snX2/+; P[Gal4]201Y UAS-lacZ/+ daughters.
Chemical informatics
Scaffold analysis
The analysis of the scaffold distribution described in this work is based upon specific molecular chemotypes (Medina-Franco et al., 2006) called cyclic systems, originally defined by Xu and Johnson (Xu and Johnson, 2002). This approach is consistent with general guidelines for defining molecular scaffolds (Langdon et al., 2010). A cyclic system consists of a set of rings and the chains of atoms that link them to one another. To obtain the scaffold, all substituent groups are removed from the rings and the linkers except endocyclic carbonyls and imines. Heteroatoms are retained and all hydrogen atoms attached to them are considered side chains and therefore removed. The Meqi program (version 2.41), originally developed by Xu and Johnson (Xu and Johnson, 2002), was obtained from Mark Johnson (Pannanugget Consulting, Kalamazoo, MI) and used to determine the specific cyclic system (described by a five-character alphanumeric chemotype identifier) present in each compound in the NINDS-II library. The Scaffold Recovery Curve was generated as previously described (Lipkus et al., 2008; Medina-Franco et al., 2009).
Molecular similarity and library diversity
Molecular similarity refers to the relation between pairs of molecules, whereas molecular diversity is based on pair-wise relations of a population of molecules (e.g. a compound collection). Molecular fingerprints of each compound were used to measure all pair-wise similarities (Willett et al., 1998; Maggiora and Shanmugasundaram, 2011). In general, this approach involves designating the presence or absence of specific structural features, which can be considered molecular fragments. This information is encoded in ‘bit vectors’, each of whose components are assigned a value of ‘1’ if the fragment is present at least once in the molecule and ‘0’ if it is not. Thus, multiple occurrences of specific fragments are not explicitly counted. The work presented here employed MACCS key fingerprints (Anderson, 1984), the current version of which represents 166 molecular fragments and related structural data. MACCS Structural Keys software, now available through Accelrys, Inc. (San Diego, CA), was used to determine the fingerprints of all compounds in the library. The molecular fingerprints were then used to evaluate the Tanimoto similarities using the program Molecular Operating Environment (MOE, v.2010), available from the Chemical Computing Group, Inc. (Montreal, Quebec, Canada; www.chemcomp.com). The Tanimoto coefficient between two molecules, A and B, is given by T(A,B)=c/(a+b–c), where a is the number of fragments present in molecule A, b is the number of fragments present in molecule B and c is the number of fragments common to both A and B. Details of this procedure, which is widely used in chemical informatics applications, are available (Willett et al., 1998; Maggiora and Shanmugasundaram, 2011).
Principal component analysis of chemical-space distribution
Molecular similarity measures do not provide explicit information about the chemical space that molecules inhabit. To obtain a graphical representation of this space we created 3D plots using principal component analysis (PCA) (Jolliffe, 2002) based on the Tanimoto similarity matrix associated with the molecules in the NINDS-II collection. In the rectangular data matrix usually employed in PCA, the rows correspond to the objects under study and the columns to the descriptors, features or parameters used to describe the objects. By contrast, the similarity matrix for a set of n molecules is an n×n square symmetric array of Tanimoto similarity values where the rows and columns both correspond to the molecules under study. To obtain the principal components, the similarity matrix was treated as if it were a typical rectangular data matrix and the usual principal component procedure was applied. The columns were mean-centered, matrix multiplication was applied, and the product matrix was diagonalized yielding the eigenvectors and eigenvalues. The molecules (the objects in this study) were then plotted in the principal-component coordinate system using the first two or three eigenvectors or the principal components. Typically, the first three principal components represent most (65–75%) of the variance of the data. In this study, they represent ∼67% of the variance, whereas the first two represent ∼55% of the variance. The principal component data and plots were obtained using the Spotfire® dynamic data visualization program (v.9.1.2) from TIBCO Software, Inc. (Somerville, MA; spotfire.tibco.com).
Primary neuronal cell culture preparation and evaluation
Primary neuronal cell cultures from the central nervous system (CNS) of wandering third instar larvae were prepared as previously described (Kraft et al., 1998; Kraft et al., 2006) with minor modifications. This method generates pure-neuron cultures within several days of incubation. Enzymatic treatment of the dissected tissue prior to dissociation was done with a commercial blend of purified collagenase I, collagenase II and the neutral protease dispase (Liberase Blendzyme 1, #1988409; Roche, Indianapolis, IN) at a concentration of 72 μg/ml in Rinaldini’s saline. Schneider’s Insect Medium (#11720; Invitrogen, San Diego, CA) was supplemented with 10% fetal bovine serum of US origin (#SH30071; lot #APC20860; Hyclone, Logan, UT) and 50 μg/ml bovine insulin (#I-6634; Sigma, St Louis, MO). For experiments analyzed by random-neuron sampling and quantification of morphometric parameters, the floor of the culture dish was made with a photoetched (‘gridded’) glass cover slip (#1916-91818; Bellco, Vineland, NJ). For cultures to be evaluated by holistic scoring (see below), a round German #1 glass cover slip (#1943-10012; Bellco) was used. In either case, the resulting 8-mm-diameter wells were coated with Conconavalin A (#C-2010; Sigma) and laminin (#354232; BD Biosciences, Franklin Lakes, NJ).
Unless otherwise specified, CNS of female larvae were explanted by microdissection and the entire CNS was dissociated, with the cells distributed into six culture dishes, followed by incubation at 25°C for 3 days (70–80 hours). After this time, each culture dish typically contained 1500–2000 neurons bearing scorable neurite arbors. The phenotypic severity of the filagree defect of fascin-deficient singed-mutant neurons is similar whether they are from late larval or early pupal CNS (Kraft et al., 2006). We chose the larval stage for the screen because the CNS dissection is far easier. Neuronal cultures prepared from individual CNS regions (optic lobes, brain, ventral ganglion) of sn3/sn3 larvae had indistinguishable filagree severity (data not shown).
Although neurite curvature can be quantified using image-analysis software (Kraft et al., 2006; Narro et al., 2007), the method requires immunostaining, random sampling, image acquisition and image processing. Alternatively, classification of neurite-curvature phenotypes can be performed much more quickly by holistic scoring of all neurons in a culture well (generally 1500–2000 neurons distributed over a ∼50 mm2 area). Classification of wild-type (+/+), intermediate-filagree (sn3/sn3), and severe-filagree (snX2/Y) cultures using phase-contrast microscopy of live neurons is 100% accurate when performed by trained observers. Training was done using non-drug-treated cultures of known genotype after 3 d.i.v. Training consisted of (i) a learning phase, with repeated observations by phase-contrast microscopy of live cultured neurons with known genotypes (+/+, sn3/sn3, snX2/Y), exhibiting wild-type, moderate filagree, or severe filagree phenotypes, respectively; and (ii) a testing phase, using coded dishes or images of individual microscopic fields. Training continued until dishes were assigned the correct genotype with 100% accuracy, and individual fields with ∼90% accuracy (see Kraft et al., 2006).
To guide the selection of compound concentrations for screening, we tested the effect of the solvent dimethyl sulfoxide (DMSO, USP grade; #D-2438, Sigma) on neurite-arbor morphogenesis, selecting genetically marked γ-MB neurons (Kraft et al., 1998; Kraft et al., 2006) to minimize biological variation. There was no systematic concentration-dependent effect of DMSO on neurite curvature of fascin-deficient sn3/sn3 larval γ-neurons (supplementary material Fig. S1). However, at a concentration equivalent to a 100-μM drug treatment, DMSO significantly reduced neurite arbor size (data not shown). We therefore tested the compounds at 10 μM and 50 μM.
Screening the compound collection
The NINDS Custom Collection II (NINDS-II) library of 1040 biologically active compounds was purchased from Microsource Discovery Systems (Gaylordsville, CT). The collection was provided in 13 microplate-formatted storage racks, each with ten columns of eight polypropylene storage tubes sealed with a single non-separable CapStrip (#4415; Matrix Technology, Hudson, NH). Each compound was provided as a 10-mM solution in DMSO and uniquely identified by its rack number, row and column location. Each column of eight compounds was screened at the same time. The racks were stored at −80°C and each rack was removed only long enough to retrieve or replace a single column of compounds. Because the melting point for DMSO is 19°C, each column of tubes held together by a CapStrip was floated upright in a room-temperature water bath for 15 minutes to facilitate thawing the contents before use.
For each experiment, primary cultures were prepared from CNS of wild-type (OreR-C) and homozygous sn3 mutant larvae as described above, with each dissociated CNS preparation distributed into six 8-mm-diameter culture wells. Two of the sn3 mutant dishes served as no-drug controls for the baseline filagree phenotype. OreR-C cultures served as no-drug controls for wild-type neurite outgrowth and arbor morphology, and to verify that the culture medium and dish coating supported typical neuron survival and morphology. The cell suspension from each CNS was distributed equally into six culture dishes and the total volume in each dish was brought to 100 μl with culture medium. While the cells were allowed to settle and adhere for 2 hours at 25°C, eight compounds were each diluted in culture medium as needed for testing at final concentrations of 10 and 50 μM (final volume 1 ml). Liquid handling was facilitated by using an eight-channel electronic pipettor (Matrix) and barrier tips. Each dish of cells was supplemented with 900 μl of culture medium for final drug concentrations of 0, 10, 50 or 100 μM. The dishes were sealed with Parafilm-M® to minimize evaporation and then incubated with humidification at 25°C.
To minimize microbial contamination risk during the compound screening, we adopted a number of exceptional measures. A dissecting microscope and fiber-optic light source were moved into a dedicated tissue culture room that was decontaminated nightly with germicidal ultraviolet light. Dissections and dissociation were performed wearing sterile surgical gloves and a laboratory coat. The drug-containing racks were handled and the tubes thawed by a second person. The CapStrip was replaced after the column of drug vials was accessed.
After 3 d.i.v., the live cultured neurons were evaluated independently by two or three trained observers using phase-contrast microscopy on a Nikon Diaphot 300 inverted microscope using a 40× objective (numerical aperture, 0.7). Holistic scoring was performed by systematic scanning of each entire dish to assess the overall effect of each drug treatment on the severity of the moderate filagree phenotype of the sn3/sn3 mutant neurons. A compound at a given dose was scored as decreasing the filagree phenotype (fascin-pathway enhancer) or increasing the filagree phenotype (fascin-pathway blocker) when a clear preponderance of neurons in the dish were observed to exhibit an unambiguous shift of the phenotype in the same direction. If no change was evident or there was more than the usual variability in the filagree phenotype, the score was recorded as ‘no effect’. In addition to scoring the filagree phenotype, other effects on neuronal morphology were noted, again only when a significant majority of neurons in the culture displayed a consistent feature. An ‘other effect’ scoring scheme evolved over the course of the drug screening. A minimum of 3–5 minutes was required to examine each dish and additional time was necessary when an effect was noticed and its distribution throughout the dish was evaluated.
Scores for each compound at the low and high dose were recorded by each observer, after which the outcomes between observers were compared and discussed. In most cases, the scores were concordant. In cases of disagreement, the individuals would re-assess the cultures together and come to a consensus score. Drug-induced changes in filagree (increased or decreased severity) or in other aspects of neuronal morphology were documented by image acquisition of representative examples using a 60× oil-immersion objective (numerical aperture, 1.4) and a Hamamatsu ORCA285 digital camera with HCImage software (Hamamatsu Photonic Systems, Bridgewater, NJ).
Experiments were repeated for compounds causing filagree modification or other effects on neuron morphology so that the scores could be confirmed. In cases where the scores in the second test were not consistent with the original experiment, the compounds were repeated a third time and the majority score from the three experiments was taken as the definitive outcome. Compounds were also retested in instances when low neuron density or microbial contamination had compromised scoring. Once a coded compound was successfully evaluated, its name was revealed. A survey of published biomedical literature and publicly available databases was then conducted to collect pharmacology, toxicology and usage information.
Immunostaining and morphometric analysis of 2D neuron images
Statin compounds were re-tested on wild-type (OreR-C) neurons, on neurons expressing a mitochondria-targeted GFP and on singed-mutant neurons. Reciprocal drug wash-out experiments were performed on wild-type neurons in which the culture medium was replaced after 1 d.i.v. after a random sample of 25 neurons was imaged by phase-contrast microscopy. Those neurons were re-imaged live at 2 and 3 d.i.v. To quantify BOS severity, bead counting was based on phase-contrast images of live neurons. A ‘bead’ was defined as a phase-dark round or ovoid swelling within a neurite with a diameter greater than that of the flanking neurite on both sides. Because neurite expansions are often seen at branch points and neurite tips, potential beads at these locations were difficult to assess and therefore not counted.
Neurite length and curvature measurements were based on images of immunofluorescently labeled neurons. Fixation and immunostaining were performed after 3 d.i.v. as previously described (Kraft et al., 1998; Kraft et al., 2006). To visualize neuronal membranes with high resolution, the Drosophila Nervana 2 protein (Sun and Salvaterra, 1995) was labeled with a polyclonal goat anti-horseradish peroxidase antiserum (Sigma) at 1:500 and detected with an Alexa-Fluor®-488-conjugated donkey anti-goat antiserum (Invitrogen) at 1:500. To visualize β-galactosidase expression in genetically marked γ-MB neurons, a preabsorbed polyclonal rabbit anti-β-galactosidase antiserum (Cappel, West Chester, PA) was used at 1:5000 and detected with a Cy3-conjugated donkey anti-rabbit antiserum (Jackson ImmunoResearch, West Grove, PA) at 1:500. Labeled neurons were identified on the Nikon Diaphot 300 inverted microscope with the 60× oil-immersion objective using epifluorescence illumination. The Alexa Fluor 488 fluorescent signal was detected with filter cube Chroma #41001 (exciter 460-500 nm, dichroic 505 nm, band-pass emitter 510–560 nm) and the Cy3 signal with Nikon G-2A filter cube (exciter 510–560 nm, dichroic 580 nm, long-pass emitter 590 nm). Images were collected with the Hamamatsu ORCA285 digital camera and HCImage software.
Acquisition of images of γ-MB neurons and their generic neuron neighbors was performed as described (Kraft et al., 2006). The nearest-neighbor neurons constitute a random sample of the heterogeneous non-γ CNS neuron types in the culture. For quantitative study of neurons in non-genetically marked cultures, a rigorous method of random sampling was done by following the alphanumeric grid of the dish in a systematic manner, up and down a ‘staircase’, acquiring images of neurons along the ‘steps’ until at least 50 neurons were imaged.
Computer-assisted analysis of the fluorescent neuron images was performed using NeuronMetrics™ software, which converts each neurite arbor into a one-pixel-wide skeletal representation from which morphometric parameters, including length, are calculated (Narro et al., 2007). For each neuron, the number of beads per 100 μm (bead density) was calculated from the total bead count and total neurite length. The nonparametric Mann-Whitney rank sum test was used to compare the distributions of bead-density values between populations of the same genotype treated with different drug concentrations, and between populations with different genotypes treated with the same drug concentration. The statistical tests and graphical representation of the data were performed using SigmaStat and SigmaPlot (Systat Software, San Jose, CA). The neurite-arbor skeletons were also used for fully automated determination of mean neurite curvature for each neuron (Kraft et al., 2006). Distributions of neurite curvature were plotted as histograms with soft binning, to reduce the effect of discretization error, and a normal-distribution curve was plotted over the data. In soft binning, each value is considered the mean of a normal distribution, with the standard deviation being set to the bin size. We then distribute the value of that point to each of the bins in proportion to the value of the normal distribution at the bin centers. Comparisons of curvature distributions between groups were made using the Welch’s t-test (Myers and Well, 1991), which accounts for different variances and sample sizes.
Supplementary Material
Acknowledgments
The authors thank Mindy Escobar, Benjamin Reddoch, Michelle O’Shea, David Andrew and Katie Olson for assistance in the early stages of the drug screen. We are also grateful to Anna Burns and Lynne Oland for expert oversight of the Department of Neuroscience tissue culture facility, to Konrad Zinsmaier and Mani Ramaswami for Drosophila stocks, to Andrew Predoehl for help running the neurite-curvature software, to Nina Ossanna and Laurence Hurley for advice about drug discovery, and to Mark Johnson (Pannanugget Consulting) for providing the Meqi program.
Footnotes
FUNDING
The project was funded by awards to L.L.R. from the National Institutes of Health [grant number NS055774], to L.L.R. and G.M.M. from the Center for Insect Science (University of Arizona), and to J.L.M.-F. from the State of Florida and the Menopause & Women’s Health Research Center (Port St Lucie, FL). C.B. was funded by NIGMS (Minority Access to Research Careers).
COMPETING INTERESTS
G.M.M. was previously employed by Upjohn and Pharmacia.
AUTHOR CONTRIBUTIONS
L.L.R. conceived and developed the experimental concepts and directed the project; L.L.R. and R.K. designed experiments; R.K., A.K., M.L.O. and C.B. performed laboratory experiments; R.K., K.B., G.M.M., J.M.-F., F.L.-V., M.L.O., C.B. and L.L.R. contributed data analysis; and L.L.R., R.K., K.B., G.M.M., F.L.-V. and J.M.-F. prepared and edited the manuscript and figures.
SUPPLEMENTARY MATERIAL
Supplementary material for this article is available at http://dmm.biologists.org/lookup/suppl/doi:10.1242/dmm.008243/-/DC1
REFERENCES
- Aiken C. T., Tobin A. J., Schweitzer E. S. (2004). A cell-based screen for drugs to treat Huntington’s disease. Neurobiol. Dis. 16, 546–555 [DOI] [PubMed] [Google Scholar]
- Al-Alwan M., Olabi S., Ghebeh H., Barhoush E., Tulbah A., Al-Tweigeri T., Ajarim D., Adra C. (2011). Fascin is a key regulator of breast cancer invasion that acts via the modification of metastasis-associated molecules. PLoS ONE 6, e27339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S. (1984). Graphical representation of molecules and substructure-search queries in MACCSTM. J. Mol. Graph. 2, 83–90 [Google Scholar]
- Anilkumar N., Parsons M., Monk R., Ng T., Adams J. C. (2003). Interaction of fascin and protein kinase Cα: a novel intersection in cell adhesion and motility. EMBO J. 22, 5390–5402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aratyn Y. S., Schaus T. E., Taylor E. W., Borisy G. G. (2007). Intrinsic dynamic behavior of fascin in filopodia. Mol. Biol. Cell 18, 3928–3940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburn T. T., Thor K. B. (2004). Drug repositioning: identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 3, 673–683 [DOI] [PubMed] [Google Scholar]
- Bemis G. W., Murcko M. A. (1996). The properties of known drugs. 1. Molecular frameworks. J. Med. Chem. 39, 2887–2893 [DOI] [PubMed] [Google Scholar]
- Bender H. A. (1960). Studies on the expression of various singed alleles in Drosophila melanogaster. Genetics 45, 867–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhogal B., Jongens T. A. (2010). Fragile X syndrome and model organisms: identifying potential routes of therapeutic intervention. Dis. Model. Mech. 3, 693–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc F. V., Tully T. (2009). Fruit flies and intellectual disability. Fly (Austin) 3, 91–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan J., Edwards R., Matsudaira P., Otto J., Wulfkühle J. (1993). Fascin, an echinoid actin-bundling protein, is a homolog of the Drosophila singed gene product. Proc. Natl. Acad. Sci. USA 90, 9115–9119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke S. N., Barnes C. A. (2006). Neural plasticity in the ageing brain. Nat. Rev. Neurosci. 7, 30–40 [DOI] [PubMed] [Google Scholar]
- Cannon T. D. (2010). Candidate gene studies in the GWAS era: the MET proto-oncogene, neurocognition, and schizophrenia. Am. J. Psychiatry 167, 369–372 [DOI] [PubMed] [Google Scholar]
- Cant K., Knowles B. A., Mooseker M. S., Cooley L. (1994). Drosophila singed, a fascin homolog, is required for actin bundle formation during oogenesis and bristle extension. J. Cell Biol. 125, 369–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao T. T., Chang W., Masters S. E., Mooseker M. S. (2004). Myosin-Va binds to and mechanochemically couples microtubules to actin filaments. Mol. Biol. Cell 15, 151–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman R. F. (1998). Nutrition. In The Insects–Structure and Function, pp. 69–93 Cambridge: Cambridge University Press [Google Scholar]
- Chen J., Herrup K. (2008). Selective vulnerability of neurons in primary cultures and in neurodegenerative diseases. Rev. Neurosci. 19, 317–326 [DOI] [PubMed] [Google Scholar]
- Chen L., Yang S., Jakoncic J., Zhang J. J., Huang X. Y. (2010). Migrastatin analogues target fascin to block tumour metastasis. Nature 464, 1062–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coecke S., Goldberg A. M., Allen S., Buzanska L., Calamandrei G., Crofton K., Hareng L., Hartung T., Knaut H., Honegger P., et al. (2007). Workgroup report: incorporating in vitro alternative methods for developmental neurotoxicity into international hazard and risk assessment strategies. Environ. Health Perspect. 115, 924–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordle A., Koenigsknecht-Talboo J., Wilkinson B., Limpert A., Landreth G. (2005). Mechanisms of statin-mediated inhibition of small G-protein function. J. Biol. Chem. 280, 34202–34209 [DOI] [PubMed] [Google Scholar]
- Das T., Cagan R. (2010). Drosophila as a novel therapeutic discovery tool for thyroid cancer. Thyroid 20, 689–695 [DOI] [PubMed] [Google Scholar]
- De Arcangelis A., Georges-Labouesse E., Adams J. C. (2004). Expression of fascin-1, the gene encoding the actin-bundling protein fascin-1, during mouse embryogenesis. Gene Expr. Patterns 4, 637–643 [DOI] [PubMed] [Google Scholar]
- de Ferranti S., Ludwig D. S. (2008). Storm over statins – the controversy surrounding pharmacologic treatment of children. N. Engl. J. Med. 359, 1309–1312 [DOI] [PubMed] [Google Scholar]
- Deinhardt K., Kim T., Spellman D. S., Mains R. E., Eipper B. A., Neubert T. A., Chao M. V., Hempstead B. L. (2011). Neuronal growth cone retraction relies on proneurotrophin receptor signaling through Rac. Sci. Signal. 4, ra82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denora N., Trapani A., Laquintana V., Lopedota A., Trapani G. (2009). Recent advances in medicinal chemistry and pharmaceutical technology – strategies for drug delivery to the brain. Curr. Top. Med. Chem. 9, 182–196 [DOI] [PubMed] [Google Scholar]
- Desai U. A., Pallos J., Ma A. A., Stockwell B. R., Thompson L. M., Marsh J. L., Diamond M. I. (2006). Biologically active molecules that reduce polyglutamine aggregation and toxicity. Hum. Mol. Genet. 15, 2114–2124 [DOI] [PubMed] [Google Scholar]
- Dueñas-González A., García-López P., Herrera L. A., Medina-Franco J. L., González-Fierro A., Candelaria M. (2008). The prince and the pauper. A tale of anticancer targeted agents. Mol. Cancer 7, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ess K. C. (2006). The neurobiology of tuberous sclerosis complex. Semin. Pediatr. Neurol. 13, 37–42 [DOI] [PubMed] [Google Scholar]
- Evans M. A., Golomb B. A. (2009). Statin-associated adverse cognitive effects: survey results from 171 patients. Pharmacotherapy 29, 800–811 [DOI] [PubMed] [Google Scholar]
- Fu H., Wen J. F., Hu Z. L., Luo G. Q., Ren H. Z. (2009). Knockdown of fascin1 expression suppresses the proliferation and metastasis of gastric cancer cells. Pathology 41, 655–660 [DOI] [PubMed] [Google Scholar]
- Galatti L., Polimeni G., Salvo F., Romani M., Sessa A., Spina E. (2006). Short-term memory loss associated with rosuvastatin. Pharmacotherapy 26, 1190–1192 [DOI] [PubMed] [Google Scholar]
- Gan B., Sahin E., Jiang S., Sanchez-Aguilera A., Scott K. L., Chin L., Williams D. A., Kwiatkowski D. J., DePinho R. A. (2008). mTORC1-dependent and -independent regulation of stem cell renewal, differentiation, and mobilization. Proc. Natl. Acad. Sci. USA 105, 19384–19389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giese A., Bjerkvig R., Berens M. E., Westphal M. (2003). Cost of migration: invasion of malignant gliomas and implications for treatment. J. Clin. Oncol. 21, 1624–1636 [DOI] [PubMed] [Google Scholar]
- Golomb B. A., Kane T., Dimsdale J. E. (2004). Severe irritability associated with statin cholesterol-lowering drugs. QJM 97, 229–235 [DOI] [PubMed] [Google Scholar]
- Golomb B. A., McGraw J. J., Evans M. A., Dimsdale J. E. (2007). Physician response to patient reports of adverse drug effects: implications for patient-targeted adverse effect surveillance. Drug Saf. 30, 669–675 [DOI] [PubMed] [Google Scholar]
- Greenspan R. J., Dierick H. A. (2004). ‘Am not I a fly like thee?’ From genes in fruit flies to behavior in humans. Hum. Mol. Genet. 13 Suppl. 2, R267–R273 [DOI] [PubMed] [Google Scholar]
- Guild G. M., Connelly P. S., Ruggiero L., Vranich K. A., Tilney L. G. (2003). Long continuous actin bundles in Drosophila bristles are constructed by overlapping short filaments. J. Cell Biol. 162, 1069–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunal A., Onguru O., Safali M., Beyzadeoglu M. (2008). Fascin expression in glial tumors and its prognostic significance in glioblastomas. Neuropathology 28, 382–386 [DOI] [PubMed] [Google Scholar]
- Hall A., Lalli G. (2010). Rho and Ras GTPases in axon growth, guidance, and branching. Cold Spring Harb. Perspect. Biol. 2, 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrap K. R. (1985). Preclinical studies identifying carboplatin as a viable cisplatin alternative. Cancer Treat Rev. 12 Suppl. A, 21–33 [DOI] [PubMed] [Google Scholar]
- Hashimoto Y., Parsons M., Adams J. C. (2007). Dual actin-bundling and protein kinase C-binding activities of fascin regulate carcinoma cell migration downstream of Rac and contribute to metastasis. Mol. Biol. Cell 18, 4591–4602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y., Kim D. J., Adams J. C. (2011). The roles of fascins in health and disease. J. Pathol. 224, 289–300 [DOI] [PubMed] [Google Scholar]
- Heemskerk J. (2005). Screening existing drugs for neurodegeneration: The National Institute of Neurologic Disorders and Stroke (NINDS) model. Retina 25, S56–S57 [DOI] [PubMed] [Google Scholar]
- Herlenius E., Lagercrantz H. (2001). Neurotransmitters and neuromodulators during early human development. Early Hum. Dev. 65, 21–37 [DOI] [PubMed] [Google Scholar]
- Hwang J. H., Smith C. A., Salhia B., Rutka J. T. (2008). The role of fascin in the migration and invasiveness of malignant glioma cells. Neoplasia 10, 149–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inlow J. K., Restifo L. L. (2004). Molecular and comparative genetics of mental retardation. Genetics 166, 835–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa R., Sakamoto T., Ando T., Higashi-Fujime S., Kohama K. (2003). Polarized actin bundles formed by human fascin-1: their sliding and disassembly on myosin II and myosin V in vitro. J. Neurochem. 87, 676–685 [DOI] [PubMed] [Google Scholar]
- Ito T., Handa H. (2012). Deciphering the mystery of thalidomide teratogenicity. Congenit. Anom. (Kyoto) 52, 1–7 [DOI] [PubMed] [Google Scholar]
- Jansen S., Collins A., Yang C., Rebowski G., Svitkina T., Dominguez R. (2011). Mechanism of actin filament bundling by fascin. J. Biol. Chem. 286, 30087–30096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jawhari A. U., Buda A., Jenkins M., Shehzad K., Sarraf C., Noda M., Farthing M. J., Pignatelli M., Adams J. C. (2003). Fascin, an actin-bundling protein, modulates colonic epithelial cell invasiveness and differentiation in vitro. Am. J. Pathol. 162, 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayo A., Parsons M. (2010). Fascin: a key regulator of cytoskeletal dynamics. Int. J. Biochem. Cell Biol. 42, 1614–1617 [DOI] [PubMed] [Google Scholar]
- Johnson M. A., Maggiora G. A. (eds) (1990). Concepts and Applications of Molecular Similarity, pp. 393 New York, NY: Wiley-Interscience [Google Scholar]
- Johnston M. V. (2004). Clinical disorders of brain plasticity. Brain Dev. 26, 73–80 [DOI] [PubMed] [Google Scholar]
- Jolliffe I. T. (2002). Principal Component Analysis. New York, NY: Springer Series in Statistics [Google Scholar]
- Judson M. C., Eagleson K. L., Levitt P. (2011). A new synaptic player leading to autism risk: Met receptor tyrosine kinase. J. Neurodev. Disord. 3, 282–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkunte R., Macarthur D., Morton R. (2007). Glioblastoma in a boy with fragile X: an unusual case of neuroprotection. Arch. Dis. Child. 92, 795–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsgodt K. H., Sun D., Jimenez A. M., Lutkenhoff E. S., Willhite R., van Erp T. G., Cannon T. D. (2008). Developmental disruptions in neural connectivity in the pathophysiology of schizophrenia. Dev. Psychopathol. 20, 1297–1327 [DOI] [PubMed] [Google Scholar]
- Kim M. Y., Oskarsson T., Acharyya S., Nguyen D. X., Zhang X. H., Norton L., Massagué J. (2009a). Tumor self-seeding by circulating cancer cells. Cell 139, 1315–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W. Y., Gonsiorek E. A., Barnhart C., Davare M. A., Engebose A. J., Lauridsen H., Bruun D., Lesiak A., Wayman G., Bucelli R., et al. (2009b). Statins decrease dendritic arborization in rat sympathetic neurons by blocking RhoA activation. J. Neurochem. 108, 1057–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. J., Choi I. J., Cheong T. C., Lee S. J., Lotan R., Park S. H., Chun K. H. (2010). Galectin-3 increases gastric cancer cell motility by up-regulating fascin-1 expression. Gastroenterology 138, 1035–1045, e1–e2 [DOI] [PubMed] [Google Scholar]
- King D. S., Wilburn A. J., Wofford M. R., Harrell T. K., Lindley B. J., Jones D. W. (2003). Cognitive impairment associated with atorvastatin and simvastatin. Pharmacotherapy 23, 1663–1667 [DOI] [PubMed] [Google Scholar]
- Kraft R., Levine R. B., Restifo L. L. (1998). The steroid hormone 20-hydroxyecdysone enhances neurite growth of Drosophila mushroom body neurons isolated during metamorphosis. J. Neurosci. 18, 8886–8899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft R., Escobar M. M., Narro M. L., Kurtis J. L., Efrat A., Barnard K., Restifo L. L. (2006). Phenotypes of Drosophila brain neurons in primary culture reveal a role for fascin in neurite shape and trajectory. J. Neurosci. 26, 8734–8747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreis P., Barnier J. V. (2009). PAK signalling in neuronal physiology. Cell. Signal. 21, 384–393 [DOI] [PubMed] [Google Scholar]
- Kureishy N., Sapountzi V., Prag S., Anilkumar N., Adams J. C. (2002). Fascins, and their roles in cell structure and function. Bioessays 24, 350–361 [DOI] [PubMed] [Google Scholar]
- Landrigan P. J. (2010). What causes autism? Exploring the environmental contribution. Curr. Opin. Pediatr. 22, 219–225 [DOI] [PubMed] [Google Scholar]
- Langdon S. R., Ertl P., Brown N. (2010). Bioisosteric replacement and scaffold hopping in lead generation and optimization. Mol. Inf. 29, 366–385 [DOI] [PubMed] [Google Scholar]
- Li A., Dawson J. C., Forero-Vargas M., Spence H. J., Yu X., König I., Anderson K., Machesky L. M. (2010a). The actin-bundling protein fascin stabilizes actin in invadopodia and potentiates protrusive invasion. Curr. Biol. 20, 339–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Mauric V., Zheng J. F., Kang S. U., Patil S., Höger H., Lubec G. (2010b). Olfactory bulb proteins linked to olfactory memory in C57BL/6J mice. Amino Acids 39, 871–886 [DOI] [PubMed] [Google Scholar]
- Lin D. M., Goodman C. S. (1994). Ectopic and increased expression of Fasciclin II alters motoneuron growth cone guidance. Neuron 13, 507–523 [DOI] [PubMed] [Google Scholar]
- Lipkus A. H., Yuan Q., Lucas K. A., Funk S. A., Bartelt W. F., 3rd, Schenck R. J., Trippe A. J. (2008). Structural diversity of organic chemistry. A scaffold analysis of the CAS Registry. J. Org. Chem. 73, 4443–4451 [DOI] [PubMed] [Google Scholar]
- Liu A. C., Welsh D. K., Ko C. H., Tran H. G., Zhang E. E., Priest A. A., Buhr E. D., Singer O., Meeker K., Verma I. M., et al. (2007). Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell 129, 605–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky L. M., Li A. (2010). Fascin: Invasive filopodia promoting metastasis. Commun. Integr. Biol. 3, 263–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggiora G. M., Shanmugasundaram V. (2011). Molecular similarity measures. In Chemoinformatics and Computational Chemical Biology (ed. Bajorath J.), pp. 39–100 New York: Humana Press, Springer [Google Scholar]
- Mangiola A., Anile C., Pompucci A., Capone G., Rigante L., De Bonis P. (2010). Glioblastoma therapy: going beyond Hercules Columns. Expert Rev. Neurother. 10, 507–514 [DOI] [PubMed] [Google Scholar]
- Marín-Vicente C., Guerrero-Valero M., Nielsen M. L., Savitski M. M., Gómez-Fernández J. C., Zubarev R. A., Corbalán-García S. (2011). ATP enhances neuronal differentiation of PC12 cells by activating PKCα interactions with cytoskeletal proteins. J. Proteome Res. 10, 529–540 [DOI] [PubMed] [Google Scholar]
- Medina-Franco J. L., Petit J., Maggiora G. M. (2006). Hierarchical strategy for identifying active chemotype classes in compound databases. Chem. Biol. Drug Des. 67, 395–408 [DOI] [PubMed] [Google Scholar]
- Medina-Franco J. L., Martínez-Mayorga K., Bender A., Scior T. (2009). Scaffold diversity analysis of compound data sets using an entropy-based measure. QSAR Comb. Sci. 28, 1551–1560 [Google Scholar]
- Megiorni F., Indovina P., Mora B., Mazzilli M. C. (2005). Minor expression of fascin-1 gene (FSCN1) in NTera2 cells depleted of CREB-binding protein. Neurosci. Lett. 381, 169–174 [DOI] [PubMed] [Google Scholar]
- Meller R. (2009). The role of the ubiquitin proteasome system in ischemia and ischemic tolerance. Neuroscientist 15, 243–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles W. O., Dyson N. J., Walker J. A. (2011). Modeling tumor invasion and metastasis in Drosophila. Dis. Model. Mech. 4, 753–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molli P. R., Li D. Q., Murray B. W., Rayala S. K., Kumar R. (2009). PAK signaling in oncogenesis. Oncogene 28, 2545–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muldoon M. F., Ryan C. M., Sereika S. M., Flory J. D., Manuck S. B. (2004). Randomized trial of the effects of simvastatin on cognitive functioning in hypercholesterolemic adults. Am. J. Med. 117, 823–829 [DOI] [PubMed] [Google Scholar]
- Munson J. M., Fried L., Rowson S. A., Bonner M. Y., Karumbaiah L., Diaz B., Courtneidge S. A., Knaus U. G., Brat D. J., Arbiser J. L., et al. (2012). Anti-invasive adjuvant therapy with imipramine blue enhances chemotherapeutic efficacy against glioma. Sci. Transl. Med. 4, 127ra36. [DOI] [PubMed] [Google Scholar]
- Myers J. L., Well A. D. (1991). Research Design and Statistical Analysis. New York: HarperCollins [Google Scholar]
- Nagy S., Ricca B. L., Norstrom M. F., Courson D. S., Brawley C. M., Smithback P. A., Rock R. S. (2008). A myosin motor that selects bundled actin for motility. Proc. Natl. Acad. Sci. USA 105, 9616–9620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narro M. L., Yang F., Kraft R., Wenk C., Efrat A., Restifo L. L. (2007). NeuronMetrics: software for semi-automated processing of cultured neuron images. Brain Res. 1138, 57–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemi M. (2010). Transporter pharmacogenetics and statin toxicity. Clin. Pharmacol. Ther. 87, 130–133 [DOI] [PubMed] [Google Scholar]
- Ono S., Yamakita Y., Yamashiro S., Matsudaira P. T., Gnarra J. R., Obinata T., Matsumura F. (1997). Identification of an actin binding region and a protein kinase C phosphorylation site on human fascin. J. Biol. Chem. 272, 2527–2533 [DOI] [PubMed] [Google Scholar]
- Opii W. O., Joshi G., Head E., Milgram N. W., Muggenburg B. A., Klein J. B., Pierce W. M., Cotman C. W., Butterfield D. A. (2008). Proteomic identification of brain proteins in the canine model of human aging following a long-term treatment with antioxidants and a program of behavioral enrichment: relevance to Alzheimer’s disease. Neurobiol. Aging 29, 51–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbo A., Facon T., Sonneveld P., Bladè J., Offidani M., Gay F., Moreau P., Waage A., Spencer A., Ludwig H., et al. (2008). Thalidomide for treatment of multiple myeloma: 10 years later. Blood 111, 3968–3977 [DOI] [PubMed] [Google Scholar]
- Paterson J., O’Hare K. (1991). Structure and transcription of the singed locus of Drosophila melanogaster. Genetics 129, 1073–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peraud A., Mondal S., Hawkins C., Mastronardi M., Bailey K., Rutka J. T. (2003). Expression of fascin, an actin-bundling protein, in astrocytomas of varying grades. Brain Tumor Pathol. 20, 53–58 [DOI] [PubMed] [Google Scholar]
- Piccioni F., Roman B. R., Fischbeck K. H., Taylor J. P. (2004). A screen for drugs that protect against the cytotoxicity of polyglutamine-expanded androgen receptor. Hum. Mol. Genet. 13, 437–446 [DOI] [PubMed] [Google Scholar]
- Pilling A. D., Horiuchi D., Lively C. M., Saxton W. M. (2006). Kinesin-1 and Dynein are the primary motors for fast transport of mitochondria in Drosophila motor axons. Mol. Biol. Cell 17, 2057–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelfsema J. H., Peters D. J. (2007). Rubinstein-Taybi syndrome: clinical and molecular overview. Expert Rev. Mol. Med. 9, 1–16 [DOI] [PubMed] [Google Scholar]
- Roma A. A., Prayson R. A. (2005). Fascin expression in 90 patients with glioblastoma multiforme. Ann. Diagn. Pathol. 9, 307–311 [DOI] [PubMed] [Google Scholar]
- Rothstein J. D., Patel S., Regan M. R., Haenggeli C., Huang Y. H., Bergles D. E., Jin L., Dykes Hoberg M., Vidensky S., Chung D. S., et al. (2005). Beta-lactam antibiotics offer neuroprotection by increasing glutamate transporter expression. Nature 433, 73–77 [DOI] [PubMed] [Google Scholar]
- Ryu M. J., Kim D., Kang U. B., Kim J., Shin H. S., Lee C., Yu M. H. (2007). Proteomic analysis of gamma-butyrolactone-treated mouse thalamus reveals dysregulated proteins upon absence seizure. J. Neurochem. 102, 646–656 [DOI] [PubMed] [Google Scholar]
- Ryu M. J., Lee C., Kim J., Shin H. S., Yu M. H. (2008). Proteomic analysis of stargazer mutant mouse neuronal proteins involved in absence seizure. J. Neurochem. 104, 1260–1270 [DOI] [PubMed] [Google Scholar]
- Safran N., Haring R., Gurwitz D., Shainberg A., Halili I., Levy A., Bogin E., Shahar A. (1996). Selective neurotoxicity induced by the ionophore lasalocid in rat dissociated cerebral cultures, involvement of the NMDA receptor/channel. Neurotoxicology 17, 883–895 [PubMed] [Google Scholar]
- Salhia B., Tran N. L., Symons M., Winkles J. A., Rutka J. T., Berens M. E. (2006). Molecular pathways triggering glioma cell invasion. Expert Rev. Mol. Diagn. 6, 613–626 [DOI] [PubMed] [Google Scholar]
- Samuel F., Hynds D. L. (2010). RHO GTPase signaling for axon extension: is prenylation important? Mol. Neurobiol. 42, 133–142 [DOI] [PubMed] [Google Scholar]
- Sawallisch C., Berhörster K., Disanza A., Mantoani S., Kintscher M., Stoenica L., Dityatev A., Sieber S., Kindler S., Morellini F., et al. (2009). The insulin receptor substrate of 53 kDa (IRSp53) limits hippocampal synaptic plasticity. J. Biol. Chem. 284, 9225–9236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz J. G., Bösel J., Stoeckel M., Megow D., Dirnagl U., Endres M. (2004). HMG-CoA reductase inhibition causes neurite loss by interfering with geranylgeranylpyrophosphate synthesis. J. Neurochem. 89, 24–32 [DOI] [PubMed] [Google Scholar]
- Sedeh R. S., Fedorov A. A., Fedorov E. V., Ono S., Matsumura F., Almo S. C., Bathe M. (2010). Structure, evolutionary conservation, and conformational dynamics of Homo sapiens fascin-1, an F-actin crosslinking protein. J. Mol. Biol. 400, 589–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalaeva M., Kenseth J., Lombardo F., Bastin A. (2008). Measurement of dissociation constants (pKa values) of organic compounds by multiplexed capillary electrophoresis using aqueous and cosolvent buffers. J. Pharm. Sci. 97, 2581–2606 [DOI] [PubMed] [Google Scholar]
- Shan D., Chen L., Njardarson J. T., Gaul C., Ma X., Danishefsky S. J., Huang X. Y. (2005). Synthetic analogues of migrastatin that inhibit mammary tumor metastasis in mice. Proc. Natl. Acad. Sci. USA 102, 3772–3776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J. H., Guedj F., Delabar J. M., Lubec G. (2007). Dysregulation of growth factor receptor-bound protein 2 and fascin in hippocampus of mice polytransgenic for chromosome 21 structures. Hippocampus 17, 1180–1192 [DOI] [PubMed] [Google Scholar]
- Shin J. B., Longo-Guess C. M., Gagnon L. H., Saylor K. W., Dumont R. A., Spinelli K. J., Pagana J. M., Wilmarth P. A., David L. L., Gillespie P. G., et al. (2010). The R109H variant of fascin-2, a developmentally regulated actin crosslinker in hair-cell stereocilia, underlies early-onset hearing loss of DBA/2J mice. J. Neurosci. 30, 9683–9694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N., Guha R., Giulianotti M. A., Pinilla C., Houghten R. A., Medina-Franco J. L. (2009). Chemoinformatic analysis of combinatorial libraries, drugs, natural products, and molecular libraries small molecule repository. J. Chem. Inf. Model. 49, 1010–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaugenhaupt S. A., Mull J., Leyne M., Cuajungco M. P., Gill S. P., Hims M. M., Quintero F., Axelrod F. B., Gusella J. F. (2004). Rescue of a human mRNA splicing defect by the plant cytokinin kinetin. Hum. Mol. Genet. 13, 429–436 [DOI] [PubMed] [Google Scholar]
- Sleeman J., Steeg P. S. (2010). Cancer metastasis as a therapeutic target. Eur. J. Cancer 46, 1177–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporn M. B. (1996). The war on cancer. Lancet 347, 1377–1381 [DOI] [PubMed] [Google Scholar]
- Stavrovskaya I. G., Narayanan M. V., Zhang W., Krasnikov B. F., Heemskerk J., Young S. S., Blass J. P., Brown A. M., Beal M. F., Friedlander R. M., et al. (2004). Clinically approved heterocyclics act on a mitochondrial target and reduce stroke-induced pathology. J. Exp. Med. 200, 211–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stella G. M., Benvenuti S., Comoglio P. M. (2010). Targeting the MET oncogene in cancer and metastases. Expert Opin. Investig. Drugs 19, 1381–1394 [DOI] [PubMed] [Google Scholar]
- Sun B., Salvaterra P. M. (1995). Characterization of nervana, a Drosophila melanogaster neuron-specific glycoprotein antigen recognized by anti-horseradish peroxidase antibodies. J. Neurochem. 65, 434–443 [DOI] [PubMed] [Google Scholar]
- Svitkina T. M., Bulanova E. A., Chaga O. Y., Vignjevic D. M., Kojima S., Vasiliev J. M., Borisy G. G. (2003). Mechanism of filopodia initiation by reorganization of a dendritic network. J. Cell Biol. 160, 409–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamada A., Kawase S., Murakami F., Kamiguchi H. (2010). Autonomous right-screw rotation of growth cone filopodia drives neurite turning. J. Cell Biol. 188, 429–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z. L., Zhang X. J., Yang S. L., Mu Y. L., Cui W. T., Ao H., Li K. (2010). The chromosomal localization, expression pattern and polymorphism analysis of porcine FSCN1 gene differently expressed from LongSAGE library. Mol. Biol. Rep. 37, 2361–2367 [DOI] [PubMed] [Google Scholar]
- Tatley M., Savage R. (2007). Psychiatric adverse reactions with statins, fibrates and ezetimibe: implications for the use of lipid-lowering agents. Drug Saf. 30, 195–201 [DOI] [PubMed] [Google Scholar]
- Tilney L. G., Tilney M. S., Guild G. M. (1995). F actin bundles in Drosophila bristles. I. Two filament cross-links are involved in bundling. J. Cell Biol. 130, 629–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinetti M. E. (2008). Over-the-counter sales of statins and other drugs for asymptomatic conditions. N. Engl. J. Med. 358, 2728–2732 [DOI] [PubMed] [Google Scholar]
- Traynor B. J., Bruijn L., Conwit R., Beal F., O’Neill G., Fagan S. C., Cudkowicz M. E. (2006). Neuroprotective agents for clinical trials in ALS: a systematic assessment. Neurology 67, 20–27 [DOI] [PubMed] [Google Scholar]
- Tseng Y., Kole T. P., Lee J. S., Fedorov E., Almo S. C., Schafer B. W., Wirtz D. (2005). How actin crosslinking and bundling proteins cooperate to generate an enhanced cell mechanical response. Biochem. Biophys. Res. Commun. 334, 183–192 [DOI] [PubMed] [Google Scholar]
- Tubb B. E., Bardien-Kruger S., Kashork C. D., Shaffer L. G., Ramagli L. S., Xu J., Siciliano M. J., Bryan J. (2000). Characterization of human retinal fascin gene (FSCN2) at 17q25: close physical linkage of fascin and cytoplasmic actin genes. Genomics 65, 146–156 [DOI] [PubMed] [Google Scholar]
- Tubb B., Mulholland D. J., Vogl W., Lan Z. J., Niederberger C., Cooney A., Bryan J. (2002). Testis fascin (FSCN3): a novel paralog of the actin-bundling protein fascin expressed specifically in the elongate spermatid head. Exp. Cell Res. 275, 92–109 [DOI] [PubMed] [Google Scholar]
- Verney C. (2003). Phenotypic expression of monoamines and GABA in the early development of human telencephalon, transient or not transient. J. Chem. Neuroanat. 26, 283–292 [DOI] [PubMed] [Google Scholar]
- Vignjevic D., Kojima S., Aratyn Y., Danciu O., Svitkina T., Borisy G. G. (2006). Role of fascin in filopodial protrusion. J. Cell Biol. 174, 863–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent A. M., Backus C., Taubman A. A., Feldman E. L. (2005). Identification of candidate drugs for the treatment of ALS. Amyotroph. Lateral Scler. Other Motor Neuron Disord. 6, 29–36 [DOI] [PubMed] [Google Scholar]
- Wagstaff L. R., Mitton M. W., Arvik B. M., Doraiswamy P. M. (2003). Statin-associated memory loss: analysis of 60 case reports and review of the literature. Pharmacotherapy 23, 871–880 [DOI] [PubMed] [Google Scholar]
- Wang J., Gines S., MacDonald M. E., Gusella J. F. (2005a). Reversal of a full-length mutant huntingtin neuronal cell phenotype by chemical inhibitors of polyglutamine-mediated aggregation. BMC Neurosci. 6, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Duan W., Igarashi S., Morita H., Nakamura M., Ross C. A. (2005b). Compounds blocking mutant huntingtin toxicity identified using a Huntington’s disease neuronal cell model. Neurobiol. Dis. 20, 500–508 [DOI] [PubMed] [Google Scholar]
- Wang C. Y., Liu P. Y., Liao J. K. (2008). Pleiotropic effects of statin therapy: molecular mechanisms and clinical results. Trends Mol. Med. 14, 37–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White C. M. (2002). A review of the pharmacologic and pharmacokinetic aspects of rosuvastatin. J. Clin. Pharmacol. 42, 963–970 [PubMed] [Google Scholar]
- Willett P., Barnard J. M., Downs G. M. (1998). Chemical similarity searching. J. Chem. Inf. Comput. Sci. 38, 983–996 [Google Scholar]
- Wishart D. S., Knox C., Guo A. C., Cheng D., Shrivastava S., Tzur D., Gautam B., Hassanali M. (2008). DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 36, D901–D906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulfkühle J. D., Petersen N. S., Otto J. J. (1998). Changes in the F-actin cytoskeleton during neurosensory bristle development in Drosophila: the role of singed and forked proteins. Cell Motil. Cytoskeleton 40, 119–132 [DOI] [PubMed] [Google Scholar]
- Xu Y. J., Johnson M. (2002). Using molecular equivalence numbers to visually explore structural features that distinguish chemical libraries. J. Chem. Inf. Comput. Sci. 42, 912–926 [DOI] [PubMed] [Google Scholar]
- Yamakita Y., Matsumura F., Yamashiro S. (2009). Fascin1 is dispensable for mouse development but is favorable for neonatal survival. Cell Motil. Cytoskeleton 66, 524–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro S., Yamakita Y., Ono S., Matsumura F. (1998). Fascin, an actin-bundling protein, induces membrane protrusions and increases cell motility of epithelial cells. Mol. Biol. Cell 9, 993–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X., Carew T. J. (2010). Small G protein signaling in neuronal plasticity and memory formation: the specific role of ras family proteins. Neuron 68, 340–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder B. J., Tso E., Skacel M., Pettay J., Tarr S., Budd T., Tubbs R. R., Adams J. C., Hicks D. G. (2005). The expression of fascin, an actin-bundling motility protein, correlates with hormone receptor-negative breast cancer and a more aggressive clinical course. Clin. Cancer Res. 11, 186–192 [PubMed] [Google Scholar]
- Zanet J., Stramer B., Millard T., Martin P., Payre F., Plaza S. (2009). Fascin is required for blood cell migration during Drosophila embryogenesis. Development 136, 2557–2565 [DOI] [PubMed] [Google Scholar]
- Zhang F. R., Tao L. H., Shen Z. Y., Lv Z., Xu L. Y., Li E. M. (2008). Fascin expression in human embryonic, fetal, and normal adult tissue. J. Histochem. Cytochem. 56, 193–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zins G. R. (1988). The history of the development of minoxidil. Clin. Dermatol. 6, 132–147 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.