SUMMARY
The tumor suppressor TP53 plays a crucial role in cancer biology, and the TP53 gene is the most mutated gene in human cancer. Trp53 knockout mouse models have been widely used in cancer etiology studies and in search for a cure of cancer with some limitations that other model organisms might help overcome. Via pronuclear microinjection of zinc finger nucleases (ZFNs), we created a Tp53 knockout rat that contains an 11-bp deletion in exon 3, resulting in a frameshift and premature terminations in the open reading frame. In cohorts of 25 homozygous (Tp53Δ11/Δ11), 37 heterozygous (Tp53Δ11/+) and 30 wild-type rats, the Tp53Δ11/Δ11 rats lived an average of 126 days before death or removal from study because of clinical signs of abnormality or formation of tumors. Half of Tp53Δ11/+ were removed from study by 1 year of age because of tumor formation. Both Tp53Δ11/+ and Tp53Δ11/Δ11 rats developed a wide spectrum of tumors, most commonly sarcomas. Interestingly, there was a strikingly high incidence of brain lesions, especially in Tp53Δ11/Δ11 animals. We believe that this mutant rat line will be useful in studying cancer types rarely observed in mice and in carcinogenicity assays for drug development.
INTRODUCTION
The TP53 protein is indispensible for genomic integrity, hence the name ‘the guardian of the genome’ (Lane, 1992). In humans, inherited monoallelic TP53 mutations lead to diverse early childhood cancers (Hollstein et al., 1991). Somatic mutations in the TP53 gene are responsible for the development of up to 50% of certain types of human cancer (Hollstein et al., 1991; Olivier et al., 2010). Analysis of the mutation spectrum of TP53, caused by various environmental factors, and the resulting tumor spectrum are valuable for elucidating cancer etiology (Olivier et al., 2010).
The TP53 protein is ubiquitously expressed throughout the body as an inactive transcription factor that is activated and stabilized by various cellular stresses (Lavin and Gueven, 2006). In the past three decades, various Trp53 loss-of-function mouse models have been developed (Clarke and Hollstein, 2003), including gene disruptions (Donehower et al., 1992; Jacks et al., 1994), humanization of the DNA binding domain (Luo et al., 2001) and a variety of point mutations (Petitjean et al., 2007), demonstrating good correlation between decreased TP53 activity and increased incidences of tumor formation. However, constitutively high expression of mutants that enhance TP53 activity is associated with increased tumor resistance and premature aging (Tyner et al., 2002), whereas an additional single copy of wild-type Trp53 gene under endogenous gene context (on a BAC) gained the transgenic animal tumor resistance without compromising longevity (García-Cao et al., 2002). These models have proven useful for studying TP53 protein function and its interaction network and have enabled many cancer studies that are not feasible to be carried out in humans. Yet, there are limitations of mouse models for certain types of cancers found in people with abnormal TP53 function, including breast cancer (Kim and Baek, 2010), brain and adrenocortical tumors (Olivier et al., 2003) and metastases (Bos et al., 2010; Francia et al., 2011).
The rat is a popular model system in biomedical research, with a wealth of information on its physiology, pathology and responses to drugs and other compounds. Tp53 knockout rats would be useful in both basic research and preclinical studies for drug development. As in the case of Trp53 knockout mice (MacDonald et al., 2004), a rat lacking functional TP53 could be used in the carcinogenicity bioassays required for each new drug, shortening the assay time from 2 years to several months (King-Herbert et al., 2010). Multiple efforts have been made to create Tp53 knockout rats. Tong and colleagues created a TP53-deficient Dark Agouti rat (referred to subsequently as the TP53-deficient DA rat) via homologous recombination in the rat embryonic stem cells (Tong et al., 2010). During the review process of this manuscript, the same group published phenotypic analysis of the TP53-deficient DA rats (Yan et al., 2012). Homozygous TP53-deficient DA rats lived no longer than 6 months and developed hemagiosarcomas and lymphomas. Heterozygous TP53-deficient DA rats survived to 12 months of age and exhibited a broader variety of sarcomas, including mammary carcinoma in about 20% of female rats. van Boxtel and colleagues recently reported phenotyping of a TP53-deficient rat model, containing a nonsense mutation in exon 6, C273X, created via ENU-mediated random mutagenesis (van Boxtel et al., 2011). The Tp53C273X/C273X rat, which was developed in a Wistar Han background (referred to subsequently as the TP53-deficient Wistar rat), expressed primarily sarcomas and survived until about 4 months of age, whereas the Tp53C273X/+ rats predominantly developed osteosarcoma, with onset of tumor occurring at around 43 weeks.
TRANSLATIONAL IMPACT.
Background
The most widely studied tumor suppressor is TP53, mutations in which cause impaired cell cycle arrest and increased susceptibility to cancer. Somatic loss-of-function mutations of the TP53 gene are present in more than 50% of human cancers. In addition, germline mutations, such as those identified in Li-Fraumeni syndrome, are associated with the development of certain types of cancer in children and young adults. Animal models of mutant TP53 are invaluable for basic cancer research, the development of cancer therapeutics and screening compounds for carcinogenicity potential. Mouse models have been used widely, despite limitations such as the underrepresentation of certain cancer types compared with humans. New models involving other species are needed to complement mouse studies and to advance the search for cures for cancer.
Results
Here, the authors describe a new rat model lacking TP53. Rats with one or both mutant Tp53 alleles developed cancer at a much earlier age than their wild-type littermates and, unlike other Tp53 mutant models, had diverse cancer types including brain neoplasia, lung, skin, mammary carcinomas and various sarcomas of muscle, bone and blood vessels. The spectrum of cancer types that develop in this new rat model is similar to that associated with Li-Fraumeni syndrome in humans.
Implications and future directions
This new rat model can be used for studying various cancers, including brain tumors, for evaluating new drug candidates and for screening compounds for their potential to induce cancer. Findings from this model, together with findings from other TP53-deficient rats, demonstrate that the spectrum of cancer types that develop depends on genetic background, underscoring the importance of developing new models in various strains and species to simulate the diverse genetics of humans. The growing availability of site-specific nucleases, such as the zinc-finger nucleases (ZFNs) used to create this model, has greatly expanded the number of species in which genome engineering is feasible, and will enable the creation of more animal models that closely resemble human cancers.
Here, we report the generation and initial characterization of a Tp53 knockout rat in the Sprague Dawley background using Zinc Finger Nuclease (ZFN) technology. ZFN technology has been shown to be a powerful tool for rat genome engineering, with high efficiency and incredible germline transmission rates (Cui et al., 2011; Geurts et al., 2009; Mashimo et al., 2010). Our results revealed a homozygous null rat, Tp53Δ11/Δ11, with complete loss of TP53 protein and a shortened disease-free lifespan due to early onset of cancer. The tumor spectrum in the null mutant rat included sarcomas and carcinomas, with a predominance of nervous system neoplasia. The Tp53Δ11/+ rats experienced a longer latency to tumorigenesis than Tp53Δ11/Δ11 rats and developed skin and endocrine cancer in addition to the cancer types recognized in the null model, demonstrating a broader spectrum than those reported in both TP53-deficient Wistar rats and the TP53-deficient DA rats. We believe our model will be a useful addition to the existing pool of rodent models for the mechanistic study of human cancers, development of anticancer drugs and for assessment of carcinogenicity of novel compounds.
RESULTS
ZFN validation
The TP53 ZFN target site is shown in supplementary material Fig. S1A. The target site is located in the 22-bp exon 3 of the gene. ZFN activity was validated by the presence of genome modifications with a mutation detection assay (see Materials and methods) in cells transfected with mRNA encoding for ZFN but not in cells transfected with a GFP-expressing plasmid as a negative control (supplementary material Fig. S1B).
Founder establishment
In vitro transcribed ZFN mRNA was used in pronuclear microinjection of single-cell embryos of Ntac:SD rats. The injected embryos were then transferred into pseudopregnant females. Twenty five founders were identified among 80 live births as described previously (Geurts et al., 2009; Mashimo et al., 2010) (supplementary material Fig. S2).
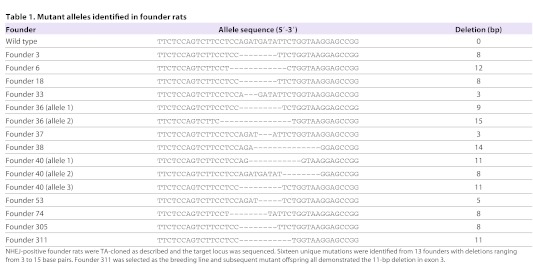
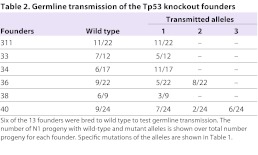
Sixteen distinct mutations were identified from 13 individual founders (Table 1). Two founders, 36 and 40, contained multiple mutant alleles and were mosaics, as we reported previously on other targets (Carbery et al., 2010; Cui, 2011). Six of the 13 founders were backcrossed to the wild-type Ntac:SD strain to obtain N1 generation: 311, 33, 34, 36, 38 and 40. All alleles transmitted through germline (Table 2). Homozygous animals were obtained by sibling matings between heterozygous littermates. Founder 311, carrying an allele with an 11-bp deletion was chosen to establish Tp53Δ11/+ and Tp53Δ11/Δ11 rat colonies.
Table 1.
Mutant alleles identified in founder rats
Table 2.
Germline transmission of the Tp53 knockout founders
Confirmation of lack of TP53 protein expression in Tp53Δ11/Δ11 rats
Various tissues from wild-type animals were harvested for western blot analysis, with little success because basal levels of TP53 are marginally detectable (An et al., 2004). Stress stimuli to the genome, such as DNA damage, are known to increase TP53 protein stability (Lavin and Gueven, 2006). Therefore, we derived primary cultures from tails of Tp53Δ11/Δ11 and wild-type rats and challenged the cultures with UV radiation before harvesting the cells for lysate preparation. TP53 was detected in wild-type culture but absent in the knockout culture, as demonstrated by western blot analysis (Fig. 1).
Fig. 1.
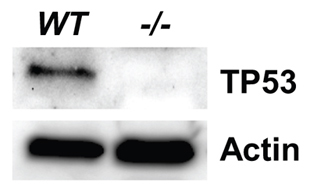
TP53 western blot of primary cultured tail cells from both wild-type (WT) and Tp53Δ11/Δ11 (−/−) rats. Actin was used as internal control for normalizing total input proteins. The monoclonal antibody employed for this blot targets an epitope near Ser20, which is located in the DNA binding domain of TP53.
Kaplan-Meier curve and tumor spectrum
It is well-documented that the lack of functional TP53 directly correlates with early and spontaneous tumorigenesis in both mice and humans. We conducted an aging study to assess tumor incidence and spectrum in cohorts of 30 wild-type, 37 Tp53Δ11/+ and 25 Tp53Δ11/Δ11 rats when housed under routine husbandry conditions. Animals were assessed for health twice daily and allowed to remain in the study unless they appeared to be moribund, lethargic and unable to obtain food and water, displayed labored breathing, or demonstrated deficiencies in nervous system function or visible or identifiable tumors. Animals demonstrating any of these criteria were no longer considered disease-free and were removed from the study for a comprehensive post-mortem examination.
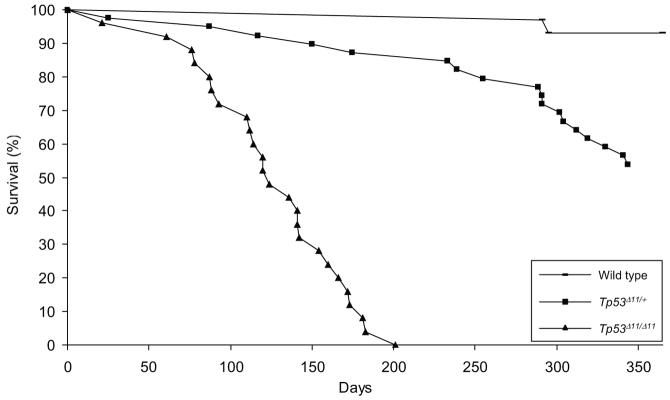
All wild-type rats were disease-free to the end of study, with the exception of two that were terminated upon recognition of subcutaneous masses. One rat had an undifferentiated subcutaneous sarcoma of the left flank, whereas the other had developed a benign mammary fibroadenoma. Both rats were just over 290 days old, and tumors of this nature have been reported as incidental findings in Sprague Dawley rats of this age (Ikezaki et al., 2011; Son and Gopinath, 2004). Tp53Δ11/+ rats developed neoplasia or diminished body condition as early as 25 days of age. By 1 year of age, 48.6% of the cohort had been removed from the study because of substantial decline in health or development of palpable tumors. The average disease-free lifespan of Tp53Δ11/+ rats was 252 days, and spanned from 25 to 375 days. A Kaplan-Meier curve is shown in Fig. 2. The Tp53Δ11/Δ11 rats displayed early onset of neoplasia, which shortened their survival. The average tumor-free lifespan for Tp53Δ11/Δ11 rats was 126 days, ranging from 21 days to 183 days.
Fig. 2.
Kaplan-Meier curve. Thirty wild-type (bar), 25 Tp53Δ11/Δ11 (triangle) and 37 Tp53Δ11/+ (square) rats were monitored for 365 days under conditions described in Materials and methods. Animals that displayed signs of declining health were euthanized, and a post-mortem examination performed. Deficiencies in functional TP53 significantly reduced the survival rate of the cohort. Neoplasia was present in all Tp53Δ11/Δ11 rats and in 89% of the Tp53Δ11/+ rats that were removed from study.
The spectrum of neoplasia in Trp53-deficient mutant mouse models is influenced by the genetic background of the mouse. Homozygous Trp53-null mice of various genetic backgrounds have a high frequency of lymphomas and sarcomas (Donehower et al., 1995). Trp53-null male mice on a 129/Sv background developed malignant testicular teratomas, which were not observed in Trp53-null male mice on a mixed C57Bl/6 and 129/Sv background (Donehower et al., 1995). A broader spectrum of sarcomatous tumors occurs in the Trp53 heterozygous mutant mice of C57BL/6 and 129/Sv genetic backgrounds, whereas mammary carcinoma is a predominant neoplasia that develops in heterozygous null mice on a BALB/c background (Kuperwasser et al., 2000).
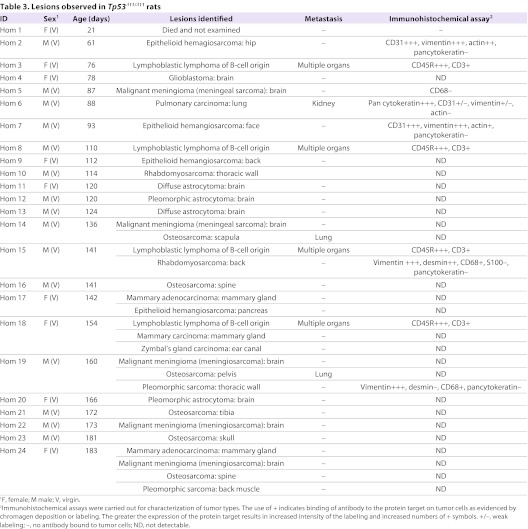
In our cohort of Tp53Δ11/Δ11 rats, the most common lesions were sarcomas, similar to the observations in Trp53 nullizygous mice and to findings in reports on ENU-induced TP53-deficient Wistar rats (van Boxtel et al., 2011) and TP53-deficient knockout DA rats (Yan et al., 2012). However, there are significant differences in tumor types between the three TP53-deficient rat models. In our Tp53Δ11/Δ11 cohort, all rats were diagnosed with neoplasia and 25% of the rats had multiple tumors. The most frequently observed tumor types were osteosarcoma (18%), malignant meningioma (15%), astrocytoma (12%), B-cell lymphoma (14%) and epithelioid hemangiosarcoma (14%) (Table 3). Representative photomicrographs of these tumors are shown in Fig. 3 and detailed descriptions of the tumors are provided in Materials and Methods. In striking contrast to the lack of cancers of the central nervous system (CNS) in the TP53-deficient Wistar or DA rats, 42% of our Tp53Δ11/Δ11 rats developed brain neoplasia. Additionally, our Tp53Δ11/Δ11 rats developed carcinomas of the mammary gland (n=3), Zymbal’s gland (n=1) and lung (n=1) before 6 months of age. Metastases were observed in two of the six rats with osteosarcoma and in the one rat with pulmonary carcinoma.
Table 3.
Lesions observed in Tp53Δ11/Δ11 rats
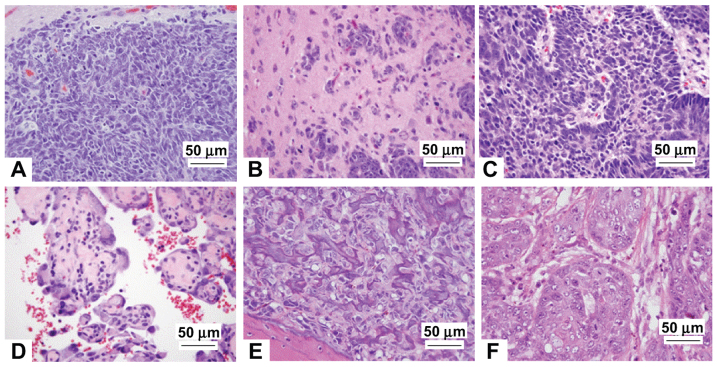
Fig. 3.
Examples of tumors represented in Tp53Δ11/Δ11 and Tp53Δ11/+ rats. The most common tumors of our TP53-deficient rats involved the CNS, blood vessels, bone and mammary gland: (A) meningeal sarcoma (CNS), (B) astrocytoma (CNS), (C) neuroblastoma (CNS), (D) epithelioid hemangiosarcoma (blood vessels), (E) osteosarcoma (bone) and (F) mammary carcinoma.
All but three Tp53Δ11/+ rats developed cancer. Of those tumor-bearing rats, three had multiple types of tumors and one rat had a metastatic neoplasm. The tumor spectrum of the Tp53Δ11/+ cohort was similar to that of Tp53Δ11/Δ11 rats. Of the 16 Tp53Δ11/+ rats with cancer, the most common tumors were of CNS origin, with eight rats bearing one of three types of nervous system neoplasia: olfactory neuroblastoma (n=3), astrocytoma (n=3) and malignant meningioma (n=2) (Table 4). Other tumors diagnosed in the Tp53Δ11/+ rats included squamous cell carcinoma, epithelioid hemangiosarcoma, adrenal cortical or medullary tumors and, with less frequency, osteosarcoma, subcutaneous myxosarcoma, Zymbal’s gland tumor, mammary fibroadenoma and an abdominal liposarcoma that metastasized to the lungs. Neoplasia observed in Tp53Δ11/+ but not Tp53Δ11/Δ11 rats included squamous cell carcinoma and neoplasia of the adrenal gland. The skin and endocrine tumors in combination with the hard and soft tissue sarcomas provided a spectrum of cancers similar to those of Li-Fraumeni syndrome in man (Olivier et al., 2010). Three Tp53Δ11/+ rats that died or were euthanized had no evidence of cancer, on the basis of gross and histopathologic review of tissues. Lesions observed were degenerative and common in aged SD rats, and included glomerulonephropathy, testicular degeneration and cardiac degeneration (data not shown).
Table 4.
Lesions observed in Tp53Δ11/+ rats and Founder 311
Fertility
Breeding of Tp53Δ11/Δ11 null mice is generally avoided because early onset of lymphoma occurs shortly after puberty and adults infrequently survive long enough to successfully raise litters (Donehower et al., 1992). However, mutant mice are fertile, and breeding heterozygous male and female mice is the standard approach for producing heterozygous mice for study (Donehower et al., 1992). To assess fecundity in our Tp53 knockout rat, we paired wild-type rats with Tp53Δ11/Δ11 rats of the opposite gender. All Tp53Δ11/Δ11 to wild-type pairings produced Tp53Δ11/+ offspring (Fig. 4) and litters were comparable in size and sex distribution.
Fig. 4.
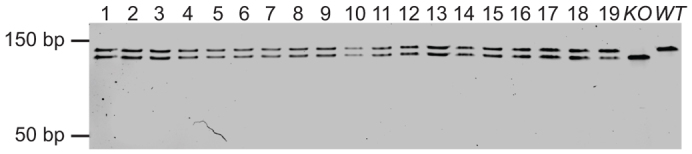
Mating of Tp53Δ11/Δ11 and wild-type rats resulted in expected heterozygosity in all offspring. PCR using primers flanking the mutation site generates a 131-bp amplicon in wild-type samples, a 120-bp amplicon in Tp53Δ11/Δ11 rats and both 13-bp and 120-bp amplicons in Tp53Δ11/+ rats. Pups 1–9 were offspring resulting from a mating of wild-type male with a Tp53Δ11/Δ11 female. Pups 10–19 were offspring resulting from the mating of a wild-type female and a Tp53Δ11/Δ11 male. KO, Tp53Δ11/Δ11 rat; WT, wild-type control.
DISCUSSION
ZFN-mediated KO rats
The successful creation of a Tp53 knockout rat model that develops cancer with an accelerated rate compared with wild-type rats is another example of general applicability of ZFN technology to animal model creation. A 100% germline transmission rate as well as high targeting efficiency and shorter timeline make it an attractive alternative to the conventional embryonic stem cell-based technology. Further, small deletions resulting from ZFN-mediated non-homologous end joining (NHEJ) events are likely to cause little disturbance of expression of neighboring genes, whereas the conventional knockout technology often relies on the insertion of a selection marker, with a higher chance of a neighboring effect. In this study, an 11-bp deletion in exon 3 leads to a lack of detectable TP53 protein expression in Tp53Δ11/Δ11 rats and cancer development in both Tp53Δ11/Δ11 and Tp53Δ11/+ rats.
Tp53 knockout rats for research
We believe that the addition of our Tp53 knockout rat to the current pool of TP53-deficient animals will provide a new lens through which we can view TP53 function. Similarly to Trp53 mutant mice and the TP53-deficient Wistar and DA rats, our model developed cancers at accelerated rates, and a large percentage of observed cancers were sarcomas. A unique feature about our model is that the cancers originated from diverse tissues, including brain, bone and epithelial organs. The expanded cancer spectrum in this model is most similar to those described in the Li-Fraumeni and Li-Fraumeni-like syndromes in man (Olivier et al., 2010). Li-Fraumeni patients have a range of inherited TP53 gene mutations and develop osteogenic and muscle sarcomas, breast cancer, brain cancer (predominantly glioblastoma in pediatric patients), lymphoma and adrenocortical tumors prior to the age of 45 years. Our Tp53 knockout rat might serve as a good model for Li-Fraumeni and Li-Fraumeni-like syndromes.
The spectrum of tumors that develops in the Trp53 knockout mouse is similar to that of spontaneous tumors in mice of the same genetic background (Jacks et al., 1994). Because rats are common rodent models for toxicity and carcinogenicity studies, historical databases of spontaneous lesions and tumors are available for SD, Wistar Han (Son et al., 2010) and DA rats (Deerberg, 1991). Interestingly, the spontaneous tumors of the parent stock and strains of rats are dissimilar to those that develop in Tp53 knockout rats of each respective background. In Wistar rats, the most common spontaneous tumor is lymphoma followed by mammary tumors, yet the TP53-deficient Wistar rats developed predominantly hemangiosarcoma and osteosarcoma. In the inbred DA rats, the most common spontaneous tumors are pituitary tumors, histiocytic sarcoma and endometrial carcinomas (Deerberg, 1991). However, the homozygous TP53-deficient DA rats develop liver hemangiosarcoma at a rate of about 80%, with most of the rest of the cohort developing lymphoma, whereas heterozygous TP53-deficient DA rat lesions were predominantly T cell lymphoma (55%), hemangiosarcoma (25%) and breast cancer (19%). In our study, the prevalence of neoplasia in the Tp53Δ11/Δ11 SD rats is essentially the reverse of that found in wild-type SD rats (Dinse et al., 2010; Ginkis, 2004). Osteosarcoma, brain neoplasia and hemangiosarcoma are less common in wild-type SD rats, but were the most frequent tumors in our SD Tp53 knockout model. These studies highlight the fact that differences in genetic background can significantly influence the development of tumors in the rat, and comparison of multiple models might help better understand cancer etiology in humans, who are genetically diverse. Our data also confirm the value of building new cancer models in search for those that better reflect human pathology.
In addition to the genetic differences between the Wistar, DA and SD rats, ENU mutagenesis can be another reason for the divergent tumor spectra observed between our Tp53 knockout rat and the TP53-deficient Wistar rat (van Boxtel et al., 2011). With ENU mutagenesis, it is necessary to introduce more than 1000 mutations per genome in order to identify mutations in the gene of interest (Wienholds et al., 2003). It is possible that other unidentified mutations in the genome of the TP53-deficient Wistar rat could influence its tumor spectrum.
Carcinogenicity assay
Chronic 2-year bioassay in both mice and rats for evaluation of the carcinogenic potential of drugs in development is well established. With the goals of decreasing the incidence of both false positives and false negatives as well as reducing the time the assay takes, many alternatives to the 2-year rodent cancer risk models have been evaluated, with varying results. Models such as the Trp53 knockout mouse and the Tg-rasH2 transgenic have been shown to be useful in some cancer bioassays (Cohen et al., 2001). Some working groups examining cancer hazard identification have recommended the use of multiple models and strains in order to best assess pharmaceutical compounds (King-Herbert and Thayer, 2006). Based on positive results seen through the use of the heterozygous Trp53 mouse in accurately determining compound toxicity, there is the potential for the heterozygous Tp53 knockout rat to be similarly useful. Comprehensive testing of a panel of chemicals and accompanying mechanistic research will need to be performed in order to determine the potential usefulness of the Tp53 knockout rat for screening of new pharmaceutical compounds.
In summary, we have created a Tp53 knockout rat that develops cancer within 4–5 months in homozygous animals and well under 1 year in heterozygous animals. The spectrum of cancers in Tp53Δ11/+ and Tp53Δ11/Δ11 rats includes brain, bone and endocrine sarcomas and carcinomas that allow new opportunities to study cancer pathogenesis, therapeutic interventions and carcinogenicity studies not previously possible with other rodent models. Our data also demonstrate the importance of using multiple models to better capture different aspects of human cancers and underline the value of new model development.
METHODS
Animals
Sprague Dawley rats (Ntac:SD) from Taconic Farms (Hudson, New York) were used for both microinjection and breeding. Microinjection of rat embryos was performed at Xenogen Biosciences (now part of Taconic Farms) as a service. The Tp53 ZFN mRNA mix was injected at 5 ng/μl final concentration.
The rat colonies were maintained at the University of Missouri, which operates under approved animal protocols overseen by its Institutional Animal Care and Use Committee (IACUC). Ntac: SD rats were housed in sterile, individually ventilated microisolator cages maintained on a 12 hour-12 hour light-dark cycle with ad libitum access to irradiated rodent diet (PicoLab 20; PMI Nutrition International, St Louis, MO) and acidified sterile water. Routine health monitoring of the colony was performed and revealed no evidence of infection with known serious pathogens including coronaviruses, Sendai virus, pneumonia virus of mice, rat theilovirus, reovirus 3, mouse adenoviruses 1 and 2, lymphocytic choriomeningitis virus, hantavirus, Mycoplasma pulmonis, CAR bacillus, Encephalitozoon cuniculi, Pneumocystis carinii, β-hemolytic streptococci, Streptococcus pneumoniae, Corynebacterium kutscheri, endoparasites and extoparasites.
Molecular biology and genotyping-related methods
ZFN validation and mRNA preparation
The ZFN expression plasmids were obtained from Sangamo Biosciences (Richmond, CA). To validate the efficacy of the ZFN pair, rat C6 cells were transfected with ZFN mRNA targeting. At 24 hours post transfection, genomic DNA was isolated from the cells using QuickExtract (Epicentre Biotechnologies, Madison, WI) and the locus of interest was amplified by PCR. Amplicons were analyzed for mutation by Surveyor Cel-1 enzymatic mutation detection assay (Transgenomics, Omaha, NE). The reaction results were analyzed using polyacrylamide gel electrophoresis (Bio-Rad, Hercules, CA), and visualized with ethidium bromide. mRNA was prepared from the constructs using MessageMax and Poly(A) polymerase tailing kits (Epicentre Biotechnology). The RNAs were purified, quantified, combined (1:1 ratio) and either transfected into tissue culture cells (for validation) or injected into single-cell embryos (for generating mutant animals).
Founder identification and Tp53Δ11/+ and Tp53Δ11/Δ11 genotyping
Tail or toe biopsies were used for genomic DNA extraction and analysis as described previously (Geurts et al., 2009). Primers flanking the mutation site (forward, 5′-GTGTGCTATTGG-GTGCCTCT-3′; reverse, 5′-ATTAGGTGACCTGTCGCTG-3′) amplified an amplicon of 326 bp, using 60°C for the annealing step, using JumpStart Taq ReadyMix (Sigma, St Louis, MO). The above PCR products were TA cloned following the manufacturer’s instruction (Invitrogen, Carlsbad, CA). Eight colonies per plate were picked and amplified by T7/T3 primers and sequenced. Sequencing was performed at ELIM Biopharmaceuticals (Hayward, CA). To genotype N1 and N2 generations, primers 50 bp forward (5′-TCTCCAAAGAACGGAAGGAA-3′) and 50 bp reverse (5′-CTACCAACCCACCAGCTCAC-3′) were used to amplify the target site in PCR reactions. Wild-type samples yielded an amplicon of 131 bp, Tp53Δ11/+ rat samples yielded amplicons of 131 bp and 120 bp, and Tp53Δ11/Δ11 rat samples yielded an amplicon of 120 bp.
Primary culture
Upon removal, tail tissue was immediately washed with ice-cold 1× PBS. Tissues were fragmented with a sterile scalpel into explants of approximately 25 mg in size and covered with a solution of Dispase II (Roche, Indianapolis, IN). Tissues were incubated at 37°C with constant shaking in Dispase II for 2 hours in six-well culture plates. Well contents were transferred to 15-ml conical tubes and centrifuged at 10,000 g for 2 minutes. The enzyme solution was decanted and washed with 1× PBS. Centrifugation was repeated and cell pellets were resuspended in 12 ml of Dulbecco’s modified Eagle’s medium/Nutrient Mixture F-12 Ham (Sigma D8437) containing 7.5% horse serum, 1.25% fetal bovine serum, 1× penicillin/streptomycin and 2 mM L-glutamine (liquid and sterile-filtered). Cells were transferred in 2 ml of medium to six-well cell culture plates (Corning, Corning, NY), and incubated at 37°C with 5% CO2 until explants yielded cell clusters. Cells were subcultured in 75 cm2 cell culture flasks (Corning) until approximately 90% confluence.
UV induction and western blots
Tail cells from wild-type and Tp53Δ11/Δ11 rats were cultured to passage 5 for western blot. Growth medium was removed from cells at approximately 90% confluence, and sterile PBS was used to wash the cells once. The cells were then exposed to ultraviolet light at 120,000 μJ cm−1 for 15 minutes in PBS, followed by recovering in growth medium at 37°C under 5% CO2 for 1 hour. The cells were then trypsinized, washed twice with PBS and then resuspended in 200 μl RIPA buffer (Sigma) containing 1× Protease Inhibitor Cocktail (Sigma). Cells were homogenized using a Dounce homogenizer and incubated on ice for 1 hour with 15 second vortexing every 10 minutes. Then, the homogenate was centrifuged at 14,000 g for 5 minutes at 4°C. The supernatant was then mixed with an equal volume of 2× Laemmli buffer (Sigma). Samples were denatured at 100°C for 5 minutes before loading. After denaturation, 20 μl of lysate was loaded onto a TGX 4–20% precast gel (Bio-Rad) and electrophoresed at 200 V for 25 minutes. The resolved gel was transferred to 0.45 μm nitrocellulose membrane (Bio-Rad) using a semi-dry transfer apparatus (Bio-Rad) for 30 minutes at 20 V. Transfer efficiency was analyzed using a Ponceau S (Sigma) stain. The membrane was then washed twice with 1× TBS-T for 5 minutes and blocked for 30 minutes in 5% milk. After blocking, the membrane was washed twice for 5 minutes in 1× TBS-T and incubated for 1 hour with primary antibody (#2524 at a 1:500 dilution in 5% milk solution; Cell Signaling Technology, Danvers, MA). After primary incubation, the membrane was washed twice in 1× TBS-T for 5 minutes and incubated with the secondary antibody (goat anti mouse 1:10,000; Jackson ImmunoResearch, West Grove, PA) for 30 minutes. The membrane was washed twice in 1× TBS-T and incubated for 2 minutes in Supersignal West Pico substrate (Thermo Scientific, Waltham, MA) and visualized on a ChemiDoc XRS+ (Bio-Rad).
Euthanasia and end of life determination
Rats were monitored twice daily for any clinical signs of disease. Rats exhibiting abnormal gait, altered behavior, CNS signs, weight loss as determined by low body condition score (2 on a scale of 5), lethargy, labored breathing, unkempt coat or decreased alertness and responsiveness to environmental stimuli were removed from the colony. Rats were removed from the study if subtle behavioral changes were observed for more than 2 consecutive days, as signs often accelerated quickly if rats were left on study. Rats were also removed if a palpable mass was identified, even if the mass was determined by the clinical veterinarian to be small and not otherwise limiting normal behavior or activities. Using these criteria, most rats were in good physical condition at the time of removal. Lifespan in the context of this study is the time, in days, that rats displayed no abnormal clinical signs. Animal were euthanized by CO2 and subjected to a systematic necropsy exam.
Phenotype-related assays
Pathology
Lesions were recorded and the following tissues were collected for histopathologic examination: abdominal skin, adrenal gland, aorta, bone marrow (femoral), sternum, brain, cecum, cervical lymph node, colon, diaphragm, dorsal skin, oesophagus, eye, Harderian gland, heart, inner ear, kidney, lacrimal gland, liver, lung, mammary gland, mandibular salivary gland, mesenteric lymph node, muscle, nasal cavity, ovary (female), pancreas, parathyroid gland, parotid salivary gland, mammary gland, mesenteric lymph node, muscle, nasal cavity, ovary, pancreas, pituitary gland, prostate (male), sciatic nerve, skull, seminal vesicle (male); small intestine, spinal cord, spleen, stifle joint, stomach, thymus, thyroid gland, tongue, teeth, trachea, urinary bladder, uterus and Zymbal’s gland. Tumors and other abnormalities recognized during tissue collection were described, sampled and snap-frozen for genetic analysis. Soft tissues were immersed in 4% buffered paraformaldehyde for histological examination. Bones were fixed in 10% buffered formalin prior to treatment with acid for removal of calcium. Tissues were processed for paraffin embedment, blocked in wax and cut into 4-μm sections, which were stained with hematoxylin and eosin. Other staining methods used to characterize tissue lesions included Picrosirius Red for collagen, Alcian Blue Ph1.0 and 2.5 with Periodic acid Schiff for mucins, and Congo Red for amyloid.
Tumor characterization
Sarcomas were the most common category of neoplasms identified in our Tp53 knockout rats. Osteosarcomas were identified on the head, maxilla and vertebral column, and had the histologic appearance of pleomorphic spindle to polygonal cells, with scant to moderate amounts of amorphous eosinophilic stromal material with variable mineralization, which was interpreted to be osteoid. Rhabdomyosarcomas involving the epaxial muscles of the back, hind limb and intercostal muscles of the thoracic wall were composed of interlacing bundles of plump spindloid to globoid cells with oval to cigar-shaped nuclei with up to two prominent centrally located nucleoli and moderate amounts of eosinophilic cytoplasm, which often had striations typical for striated muscle. Numerous strap cells (elongated cells with cytoplasmic cross-striations) that are often multinucleated were evident throughout some of the masses. Most neoplastic cells had strong immunoreactivity for desmin and 40–50% of the cells, most notably strap cells, were immunoreactive for muscle actin. Hemangiosarcomas were most often observed as soft, often bloody masses in the subcutis, and also observed once in the pancreas of a Tp53Δ11/Δ11 rat. The endothelial tumors were locally invasive masses characterized by plump spindle to epithelioid cells that formed sheets and more often vasoform tubules or cystic spaces filled with red blood cells. Neoplastic endothelial cells were immunolabeled when probed with antibodies against CD31, or von Willebrand factor VIII and vimentin. The neoplastic adipocytes in an abdominal liposarcoma in a Tp53Δ11/+ rat had metastasized to the lung. Pleomorphic sarcomas characterized as a myxosarcoma contained interlacing bundles of spindle to polygonal, often multinucleated, cells in a mucinous matrix. Undifferentiated sarcomas were also in the subcutis, and were composed of sheets or bundles of pleomorphic spindle cells that were immunoreactive to vimentin, but lacked reactivity to a battery of antibody assays used to identify muscle, endothelial cells and nervous tissue (S100 protein). Multicentric lymphoma was recognized in splenic red pulp, bone marrow and vasculature of many parenchymal organs. Neoplastic lymphocytes were uniform, large lymphocytes of 8–10 μm in greatest diameter, with scant cytoplasm and nuclei with stippled chromatin. Neoplastic lymphocytes were labeled with antibodies to CD45R, identifying the cells as B lymphocytes.
Neoplasms of the CNS were often present in rats that displayed clinical signs of decreased activity, inappetance, head tilt or seizures. Primitive neuroectodermal tumors identified in the olfactory bulb region were classified as olfactory neuroblastomas. Olfactory neuroblastomas were highly cellular neoplasms that displaced and infiltrated the olfactory bulbs, adjacent cerebral neuropil and occasionally extended through the ethmoid plate to the nasal cavity. Neoplastic cells were pleomorphic to fusiform and were arranged in sheets and in pseudorossettes around capillaries or cores of eosinophilic fibrils. Individual cells varied from round to ovular in shape with large oval to carrot-shaped nuclei, and mitoses were common (averaged 10–20 per 400× field). Astrocytomas were diffusely infiltrating, highly cellular neoplasms composed of polygonal to spindle-shaped cells with moderate, lightly eosinophilic, fibrillar cytoplasm and indistinct borders. Multinucleated giant cells were occasionally observed in the pleomorphic astrocytomas. Meningeal sarcomas were expansile spindle cell masses within the meninges and invaded the adjacent neuropil of the cerebellum. Neoplastic cells formed streams and interlacing bundles amidst a very fine fibrovascular stroma and mitoses was common (20–40 per 400× field). A subset of meningeal sarcomas was probed with antibody to CD68 to determine whether the sarcomas were histiocytic in origin. A gliobastoma was recognized as a moderately pleomorphic, expansile mass of basophilic round cells in the pons. Neoplastic cells had indented, large, and occasionally bizarre nuclei with finely stippled chromatin, and vacuolated cytoplasm. Multinucleated cells were randomly distributed throughout the mass.
Epithelial tumors involved skin, mammary glands, the Zymbal’s gland and lungs. Squamous cell carcinomas primarily involved the skin, and neoplastic epithelial cells were locally invasive. Malignant mammary neoplasms (carcinoma and adenocarcinoma) were diagnosed in the Tp53Δ11/Δ11 rats and a benign mammary fibroadenoma was identified in a Tp53Δ11/+ rat. The frequency of mammary tumors was low, and the Tp53Δ11/Δ11 rats developed malignant mammary cancer; benign mammary cancer was identified in a Tp53Δ11/+ rat The Zymbal’s gland tumor formed an ulcerated mass on the side of the face. The unencapsulated mass formed multiple cystic cavities. The cellular portion of the mass contained sheets and nests of epithelioid cells with moderately light basophilic cytoplasm, round to oval finely stippled central nuclei and 1–4 prominent purple nucleoli. Cystic cavities were lined by keratinized squamous epithelial cells and cystic spaces were filled with keratin, neutrophils, necrotic neoplastic cells and a few scattered bacterial colonies of cocci. A pulmonary carcinoma with renal metastasis was identified in a Tp53Δ11/Δ11 rat. The pleomorphic appearance of the nests of large polygonal cells was consistent between lung and kidney, and cells immunostained with pancytokeratin, indicating epithelial origin.
Immunohistochemistry
Immunohistochemical assays were performed on lymphosarcomas for evaluation of lymphocytes lineage and on spindle cells tumors with poorly differentiated morphology for determination of tissue of origin. Sections for immunohistochemistry were probed with antibodies using standard procedures. Briefly, 4-μm sections were mounted on charged slides (Probe-on Plus, Fisher Scientific, Pittsburgh, PA). Wax was removed and sections were heated to 90°C in citrate buffer (10 mM, pH 6) for 20 minutes. Once cooled, sections were exposed to 3% hydrogen peroxide, rinsed and blocked with 5% bovine serum albumin. After a series of rinses, tissues were probed with one or more antibodies that recognized the following protein targets and the cells that express them: vimentin (mesenchymal cells) (1:100; sc7557, Santa Cruz Biotechnology, Santa Cruz, CA), pancytokeratin (AE1:AE3) (simple and stratified epithelial cells) (1:50; M3515, DAKO, Carpenteria, CA), CD31 (endothelial cells) (1:100; ab28364, Abcam, Cambridge, MA), CD45R (B lymphocytes) (1:100; 550301, BD Biosciences, San Diego, CA), CD68 (macrophages) (1:200; MCA341A, AbD Serotec, Raleigh, NC); factor VIII (endothelial cells) (1:800; A0082, DAKO), muscle actin (smooth muscle, skeletal muscle and myoepithelial cells) (1:400; M0635, DAKO), CD3 (T lymphocytes) (1:300; A0452, DAKO), and S100 (nervous and neuroendocrine cells) (1:600; Z3011, DAKO). After incubation of each antibody for 30 minutes, sections were rinsed and incubated with the appropriate secondary antibodies (DAKO). Bound antibodies were visualized with 3,3′-diaminobenzidine tetrahydrochloride (0.05% with 0.015% H2O2 in PBS). Sections were counterstained with Mayer’s hematoxylin, dehydrated, cleared and cover-slipped for microscopic examination. A summary of the assays used for each tumor and the interpretation of the immunostained sections is provided in Tables 3 and 4.
Supplementary Material
Acknowledgments
We thank Fyodor Urnov, Lei Zhang, Vivian Choi, George Katibah at Sangamo Biosciences for the Tp53 ZFNs, Dan Fisher for help with western blots, Diana Ji for suggestions on the manuscript, Lara Little, Christine McGregor, Christine Bethune and Beth Bauer for help managing the colony, and Qilong Ying for western blot suggestions. We also thank the IDEXX RADIL pathology staff, Shari Hamilton, Susan Caraker and Bettina Gentry and histology staff, Jill Hansen, Jan Adair and Bonita Cowan.
Footnotes
COMPETING INTERESTS
A.M., E.J.W. and X.C. are full-time employees of Sigma-Aldrich Corporation. C.L.B.-W. is a full-time employee of IDEXX RADIL.
AUTHOR CONTRIBUTIONS
E.J.W. and X.C. conceived and designed the experiments. A.M. and C.L.B.-W. performed the experiments. A.M., C.L.B.-W., C.L.F. and X.C. analyzed the data. A.M., C.L.B.-W., C.L.F., E.J.W. and X.C. wrote the paper.
FUNDING
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
SUPPLEMENTARY MATERIAL
Supplementary material for this article is available at http://dmm.biologists.org/lookup/suppl/doi:10.1242/dmm.009704/-/DC1
REFERENCES
- An W., Kim J., Roeder R. G. (2004). Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117, 735–748 [DOI] [PubMed] [Google Scholar]
- Bos P. D., Nguyen D. X., Massagué J. (2010). Modeling metastasis in the mouse. Curr. Opin. Pharmacol. 10, 571–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbery I. D., Ji D., Harrington A., Brown V., Weinstein E. J., Liaw L., Cui X. (2010). Targeted genome modification in mice using zinc finger nucleases. Genetics 186, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke A. R. H., Hollstein M. (2003). Mouse models with modified p53 sequences to study cancer and ageing. Cell Death Differ. 10, 443–450 [DOI] [PubMed] [Google Scholar]
- Cohen S. M., Robinson D., MacDonald J. (2001). Alternative models for carcinogenicity testing. Toxicol. Sci. 64, 14–19 [DOI] [PubMed] [Google Scholar]
- Cui X., Ji D., Fisher D. A., Wu Y., Briner D. M., Weinstein E. J. (2011). Targeted integration in rat and mouse embryos with zinc-finger nucleases. Nat. Biotechnol. 29, 64–67 [DOI] [PubMed] [Google Scholar]
- Deerberg F. (1991). Age-associated versus husbandry-related pathology of aging rats. Neurobiol. Aging 12, 659–662 [DOI] [PubMed] [Google Scholar]
- Dinse G. E., Peddada S. D., Harris S. F., Elmore S. A. (2010). Comparison of NTP historical control tumor incidence rates in female Harlan Sprague-Dawley and Fischer 344/N rats. Toxicol. Pathol. 38, 765–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower L. A., Harvey M., Slagle B. L., McArthur M. J., Montgomery C. A., Jr, Butel J. S., Bradley A. (1992). Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356, 215–221 [DOI] [PubMed] [Google Scholar]
- Donehower L. A., Harvey M., Vogel H., McArthur M. J., Montgomery C. A., Jr, Park S. H., Thompson T., Ford R. J., Bradley A. (1995). Effects of genetic background on tumorigenesis in p53-deficient mice. Mol. Carcinogen. 14, 16–22 [DOI] [PubMed] [Google Scholar]
- Francia G., Cruz-Munoz W., Man S., Xu P., Kerbel R. S. (2011). Mouse models of advanced spontaneous metastasis for experimental therapeutics. Nat. Rev. Cancer 11, 135–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Cao I., García-Cao M., Martín-Caballero J., Criado L. M., Klatt P., Flores J. M., Weill J. C., Blasco M. A., Serrano M. (2002). “Super p53” mice exhibit enhanced DNA damage response, are tumor resistant and age normally. EMBO J. 21, 6225–6235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts A. M., Cost G. J., Freyvert Y., Zeitler B., Miller J. C., Choi V. M., Jenkins S. S., Wood A., Cui X., Meng X., et al. (2009). Knockout rats via embryo microinjection of zinc-finger nucleases. Science 325, 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giknis M. L. A., Clifford C. B. (2004). Compilation of spontaneous neoplastic lesions and survival in Crl:CD (SD) rats from control groups. Charles River Laboratories. http://www.criver.com/SiteCollectionDocuments/rm_rm_r_lesions_survival_crlcd_sd_rats.pdf
- Hollstein M., Sidransky D., Vogelstein B., Harris C. C. (1991). p53 mutations in human cancers. Science 253, 49–53 [DOI] [PubMed] [Google Scholar]
- Ikezaki S., Takagi M., Tamura K. (2011). Natural occurrence of neoplastic lesions in young sprague-dawley rats. J. Toxicol. Pathol. 24, 37–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Remington L., Williams B. O., Schmitt E. M., Halachmi S., Bronson R. T., Weinberg R. A. (1994). Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 4, 1–7 [DOI] [PubMed] [Google Scholar]
- Kim I. S. V., Baek S. H. (2010). Mouse models for breast cancer metastasis. Biochem. Biophys. Res. Commun. 394, 443–447 [DOI] [PubMed] [Google Scholar]
- King-Herbert A., Thayer K. (2006). NTP workshop: animal models for the NTP rodent cancer bioassay: stocks and strains – should we switch? Toxicol. Pathol. 34, 802–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King-Herbert A. P., Sills R. C., Bucher J. R. (2010). Commentary: update on animal models for NTP studies. Toxicol. Pathol. 38, 180–181 [DOI] [PubMed] [Google Scholar]
- Kuperwasser C., Hurlbut G. D., Kittrell F. S., Dickensn E. S., Laucirica R., Medina D., Nader S. P., Jerry J. D. (2000). Development of spontaneous mammary tumors in BALC/c p53 heterozygous mice. Am. J. Pathol. 157, 2151–2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D. P. (1992). Cancer. p53, guardian of the genome. Nature 358, 15–16 [DOI] [PubMed] [Google Scholar]
- Lavin M. F., Gueven N. (2006). The complexity of p53 stabilization and activation. Cell Death Differ. 13, 941–950 [DOI] [PubMed] [Google Scholar]
- Luo J. L., Yang Q., Tong W. M., Hergenhahn M., Wang Z. Q., Hollstein M. (2001). Knock-in mice with a chimeric human/murine p53 gene develop normally and show wild-type p53 responses to DNA damaging agents: a new biomedical research tool. Oncogene 20, 320–328 [DOI] [PubMed] [Google Scholar]
- MacDonald J., French J. E., Gerson R. J., Goodman J., Inoue T., Jacobs A., Kasper P., Keller D., Lavin A., Long G., et al. (2004). The utility of genetically modified mouse assays for identifying human carcinogens: a basic understanding and path forward. Toxicol. Sci. 77, 188–194 [DOI] [PubMed] [Google Scholar]
- Mashimo T., Takizawa A., Voigt B., Yoshimi K., Hiai H., Kuramoto T., Serikawa T. (2010). Generation of knockout rats with X-linked severe combined immunodeficiency (X-SCID) using zinc-finger nucleases. PLoS ONE 5, e8870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier M., Goldgar D. E., Sodha N., Ohgaki H., Kleihues P., Hainaut P., Eeles R. A. (2003). Li-Fraumeni and related syndromes: correlation between tumor type, family structure, and TP53 genotype. Cancer Res. 63, 6643–6650 [PubMed] [Google Scholar]
- Olivier M., Hollstein M., Hainaut P. (2010). TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb. Perspect. Biol. 2, a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitjean A., Mathe E., Kato S., Ishioka C., Tavtigian S. V., Hainaut P., Olivier M. (2007). Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum. Mutat. 28, 622–629 [DOI] [PubMed] [Google Scholar]
- Son W.-C., Gopinath C. (2004). Early occurrence of spontaneous tumors in CD-1 mice and Sprague-Dawley rats. Toxicol. Pathol. 32, 371–374 [DOI] [PubMed] [Google Scholar]
- Son W. C., Bell D., Taylor I., Mowat V. (2010). Profile of early occurring spontaneous tumors in Han Wistar rats. Toxicol. Pathol. 38, 292–296 [DOI] [PubMed] [Google Scholar]
- Tong C., Li P., Wu N. L., Yan Y., Ying Q. L. (2010). Production of p53 gene knockout rats by homologous recombination in embryonic stem cells. Nature 467, 211–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyner S. D., Venkatachalam S., Choi J., Jones S., Ghebranious N., Igelmann H., Lu X., Soron G., Cooper B., Brayton C., et al. (2002). p53 mutant mice that display early ageing-associated phenotypes. Nature 415, 45–53 [DOI] [PubMed] [Google Scholar]
- van Boxtel R., Kuiper R. V., Toonen P. W., van Heesch S., Hermsen R., de Bruin A., Cuppen E. (2011). Homozygous and heterozygous p53 knockout rats develop metastasizing sarcomas with high frequency. Am. J. Pathol. 179, 1616–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E., van Eeden F., Kosters M., Mudde J., Plasterk R. H., Cuppen E. (2003). Efficient target-selected mutagenesis in zebrafish. Genome Res. 13, 2700–2707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H. X., Wu H. P., Ashton C., Tong C., Ying Q. L. (2012). Rats deficient for p53 are susceptible to spontaneous and carcinogen-induced tumorigenesis. Carcinogenesis 33, 2001–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.