Abstract
Background
We performed a genome-wide association study (GWAS) to identify common risk variants for schizophrenia.
Methods
The discovery scan included 1606 patients and 1794 controls from Ireland, using 6,212,339 directly genotyped or imputed single nucleotide polymorphisms (SNPs). A subset of this sample (270 cases and 860 controls) was subsequently included in the Psychiatric GWAS Consortium-schizophrenia GWAS meta-analysis.
Results
One hundred eight SNPs were taken forward for replication in an independent sample of 13,195 cases and 31,021 control subjects. The most significant associations in discovery, corrected for genomic inflation, were (rs204999, p combined = 1.34 × 10−9 and in combined samples (rs2523722 p combined = 2.88 × 10−16) mapped to the major histocompatibility complex (MHC) region. We imputed classical human leukocyte antigen (HLA) alleles at the locus; the most significant finding was with HLA-C*01:02. This association was distinct from the top SNP signal. The HLA alleles DRB1*03:01 and B*08:01 were protective, replicating a previous study.
Conclusions
This study provides further support for involvement of MHC class I molecules in schizophrenia. We found evidence of association with previously reported risk alleles at the TCF4, VRK2, and ZNF804A loci.
Keywords: CACNA1I, genetics, genome-wide association study, HLAC, major histocompatibility complex, polygene score, schizophrenia
Schizophrenia is a complex psychiatric disorder of substantial heritability (h2~.8) characterized by hallucinations, delusions, disordered thinking, and cognitive deficits (1). The disorder poses a significant public health problem, as it affects approximately .8% of the population, causes considerable morbidity, and reduces average life expectancy by 20 to 25 years (2). The onset of illness is typically in early adulthood, but despite current treatments, the evolution of symptoms, severity, and course of the disorder are variable.
Understanding the genetic component of schizophrenia has been a major focus in the field, with the hope that this will aid development of more effective diagnostics and therapeutics. Recently, a number of genome-wide association studies (GWAS) have made encouraging progress in identifying a spectrum of common and rare genetic risk variants that contribute to schizophrenia susceptibility (3–11). They have found evidence for a number of susceptibility loci, including the major histocompatibility complex (MHC) region on chromosome 6p21–6p22. Although the MHC has been implicated in multiple schizophrenia GWAS (3,6,12), the signals reported cover a broad region across the different studies from 27.2 megabase (Mb) to 32.7 Mb.
Here, we report a schizophrenia GWAS conducted as part of the Wellcome Trust Case Control Consortium 2 (WTCCC2) study of 15 complex disorders and traits. The discovery dataset was made up of an Irish cohort of 1606 affected and 1794 unaffected population control subjects. We performed a logistic regression case-control analysis (see Methods and Materials) and selected regions for follow-up with single nucleotide polymorphisms (SNPs) that surpassed a significance level of p < 10−4. Replication data consisted of a large independent set of 13,195 cases and 31,021 control subjects. Imputations based on the 1000 Genomes (13) reference panel yielded information at over six million SNPs after quality control (QC). In addition, we present evidence for association at loci that had met criteria for genome-wide association in previously reported schizophrenia and bipolar disorder GWAS. Finally, we imputed classical human leukocyte antigen (HLA) alleles and performed association analyses with these alleles to further define the MHC association seen in the SNP data.
Methods and Materials
Discovery Subjects
Discovery Sample Cases
The case sample of 1908 individuals was recruited through community mental services and inpatient units in the Republic of Ireland and Northern Ireland following similar research protocols and with local ethics approval. All participants were interviewed using a structured clinical interview (Structured Clinical Interview for DSM-IV [14]; Schedule for Affective Disorders and Schizophrenia [Lifetime Version] [15]; or Schedule for Clinical Assessment in Neuropsychiatry [16]). Diagnosis of a major psychotic disorder was made by the consensus lifetime best estimate method using DSM-IV criteria with all available information (interview, family or staff report, and chart review). All cases were over 18 years of age, were of Irish origin (being of Irish parents and having all four grandparents born in Ireland or the United Kingdom), and had been screened to exclude substance-induced psychotic disorder or psychosis due to a general medical condition. The final diagnostic determination was made by one of the four lead clinicians on the project (A.C., T.O.N., K.S.K., D.W.). The final analysis included samples from 1606 participants whose SNP data passed quality control filters, detailed in Table S1 in Supplement 1, and met DSM-IV criteria for schizophrenia (n = 1418), schizoaffective disorder (n = 182), or schizophreniform disorder (n = 6).
Discovery Sample Control Subjects
Control subjects (n = 2048) were ascertained with written informed consent from the Irish GeneBank and represented blood donors from the Irish Blood Transfusion Service recruited in the Republic of Ireland. They met the same ethnicity criteria as cases but were not specifically screened for psychiatric illness. Individuals taking regularly prescribed medication are excluded from blood donation in Ireland and donors are not financially remunerated. As the lifetime prevalence of schizophrenia is relatively low (<1%) and there was no obvious reason for individuals with schizophrenia to be overrepresented in the control subjects, the fraction of putative case individuals in the control collection was expected to be small. Following QC (Table S1 in Supplement 1), the control sample included 1794 participants.
Replication Subjects
The total replication sample included 13,195 cases and 31,021 control subjects. This was composed of nonoverlapping samples from the Psychiatric GWAS Consortium (PGC)-Schizophrenia consortium sample (9393 cases and 12,462 control subjects) and the SGENE+ consortium sample (1053 cases and 12,174 control subjects). It also included a WTCCC2 replication dataset (2749 cases from the United Kingdom and 6385 control subjects from the 1958 Birth Cohort and National Blood Service), genotyped on the Illumina Immunochip, which is referred to as the Immunochip replication sample.
Psychiatric GWAS Consortium Sample
The sample included individuals of European ancestry with a DSM-IV (or equivalent) diagnosis of schizophrenia or schizoaffective disorder. The full sample includes 9394 cases, which were collected with institutional review board approval at multiple centers in 17 individual cohorts representing 11 countries (Australia, Bulgaria, Denmark, Germany, Ireland, Netherlands, Norway, Portugal, Sweden, United Kingdom, and the United States of America). Control subjects consisted of 12,462 samples of European ancestry collected from the same countries. The PGC-Schizophrenia sample is described more fully elsewhere (9). A small number of Irish cases (n = 270) and control subjects (n = 860) overlapped with our discovery sample and were excluded from the replication analysis for this project. Due to the consortium nature of the PGC, the composite populations are genotyped on different platforms and are imputed to give a common set of SNPs. The SNPs for which we attempted replication are therefore not genotyped in all of the individual populations and are either made up of a subset of genotyped populations, with the remaining imputed, or are imputed in all populations.
SGENE+ Sample
Details of the SGENE+ sample have been published elsewhere (12). Part of the SGENE+ sample (identified in their paper as the SGENE-Bonn, Copenhagen, Munich, TOP3, and University of California, Los Angeles sites) had contributed to stage 1 of the PGC analysis and were excluded from the SGENE+ sample for replication purposes.
Immunochip Replication Sample
After quality control, this sample totaled 2749 cases collected in the United Kingdom and 6385 control subjects from 1958 Birth Cohort and National Blood Service cohorts. The control sample did not overlap with WTCCC1 control subjects included in the PGC sample. All cases were diagnosed with schizophrenia and were undergoing monitoring by the United Kingdom Clozaril Monitoring Service for clozapine therapy; full details on the control subjects are provided elsewhere (17). Samples were genotyped on the Illumina Immunochip platform at the Sanger Institute, United Kingdom. The Immunochip SNP chip was designed jointly by a number of groups, including the Wellcome Trust Case Control Consortium 2, and included 2400 SNPs selected from the top hits reported in individual schizophrenia GWAS (including from this study) and from the PGC meta-analysis of schizophrenia GWAS data. For the study presented here, the Immunochip data were used as a shallow replication cohort, where data at less than 100 SNPs were analyzed.
DNA Sample Preparation
For all participants in the discovery sample, DNA was extracted from whole blood using standard methods. Genomic DNA for all cases and control subjects was shipped to the Sanger Institute, United Kingdom, or the Broad Institute, United States. Similar procedures for DNA quality control were applied at both sites and are detailed elsewhere (3,17).
Genotype Calling
All discovery samples were genotyped using the Affymetrix 6.0 platform totaling 893,634 autosomal SNPs, either at the Affymetrix (Santa Clara, California) or Broad Institute (Cambridge, Massachusetts) laboratories. A subset of samples (n = 37) were genotyped at both sites and successfully identified as duplicates as part of the QC procedure. The final sample included 1606 cases (1302 were genotyped at Affymetrix) and 1794 control subjects (1001 were genotyped at Affymetrix). For all samples passing laboratory quality control, raw intensities (from the .CEL files) were renormalized within collections using CelQuantileNorm (http://outmodedbonsai.sourceforge.net/). These normalized intensities were used to call genotypes with an updated version of the Chiamo software (https://mathgen.stats.ox.ac.uk/genetics_software/chiamo/chiamo.html), adapted for Affymetrix 6.0 SNP data (17,18).
Quality Control
Full details of the quality control methods employed in the Wellcome Trust Case Control Consortium 2 have been published elsewhere (17,19) and additional details are provided in Supplemental Methods in Supplement 1. After applying all of the quality control criteria, from a total of 3956 samples, 566 samples were excluded, leaving 1606 cases and 1794 control subjects for further analysis (Table S1 in Supplement 1). After quality control of the SNP data (Supplemental Methods in Supplement 1), 696,951 directly assayed SNPs were available for analysis.
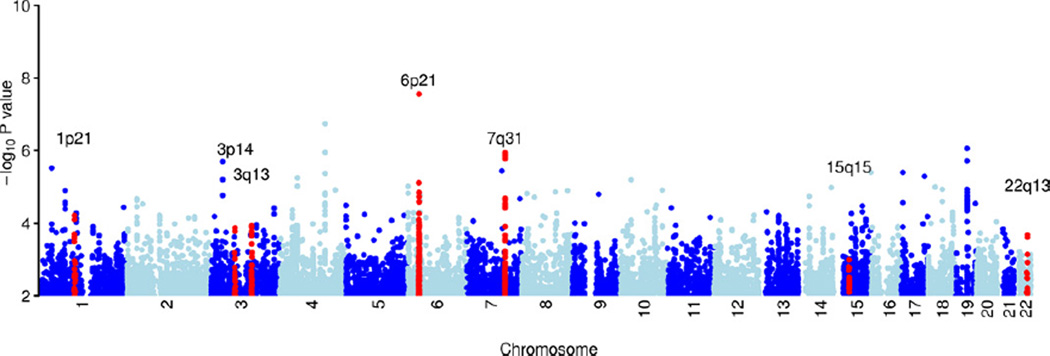
The genomic inflation statistic (λ) was 1.10 (Figure S1 in Supplement 1). This is slightly higher than would be ideal for a genome-wide study (20), and several methods were applied to try and understand any underlying structure in the data. Using a label corresponding to the genotyping center as a covariate gave a similar λ to just using the data that was genotyped at Affymetrix (λ = 1.095, λ = 1.09, respectively). Including the first two principal components as covariates also had no effect on reducing the value of λ (λ = 1.095). None of these methods substantially reduced the genomic inflation, suggesting there could be an underlying polygenic component to the inflation (21), as previous studies have found for schizophrenia (3). Therefore, no covariates were used for the following analyses unless explicitly described. Genomic control was not applied to the results of the primary analysis, but for completeness, Table S4 in Supplement 1 shows p values after applying genomic control at SNPs that were taken through to replication. Visual inspection of the Manhattan plot (Figure 1) suggested that the filtered data provide a good basis for association analyses.
Figure 1.
Plot of genome-wide association results after fitting the multiplicative model in SNPTEST. Genome-wide association results in the discovery data at 696,957 directly genotyped single nucleotide polymorphisms, each represented as a point on the plot. Chromosomes are colored dark blue and light blue alternatively, as labeled on the x axis. The −log 10 (p value) is shown on the y axis. Loci identified in this study as being potentially associated with schizophrenia (Table 2) are highlighted in red and labeled by chromosomal region.
Imputation
SNP Imputation
Haplotype phasing and imputation was performed using IMPUTE2 (22). Only samples that had passed the quality control filtering were imputed and before imputation SNPs were subjected to additional quality control to remove any that could adversely affect the quality of the imputation (Supplement 1). Imputation was performed using the HapMap2 and 3 and the 1000 Genomes reference panels; further details are in Supplement 1.
HLA Imputation
Human leukocyte antigen alleles were imputed using an updated version of HLA*IMP (23) (Supplement 1).
Statistical Methods
We report p values from 1-df Cochran-Armitage tests for trend as implemented in the software SNPTEST (https://mathgen.stats.ox.ac.uk/genetics_software/snptest/snptest.html). A parallel analysis using the Cochran-Armitage trend test in PLINK (24) gave near identical results (data not shown). We also performed 2-df genotypic tests to verify that none of our associations showed significant deviation from a multiplicative model and two marker logistic regressions to test for interactions between associated markers. Effect size (odds ratio [OR]) estimates represent per-allele increase of risk in a multiplicative model.
Polyscore Analyses
We performed a direct replication experiment of the polyscore analysis performed by the International Schizophrenia Consortium (ISC) (3). The principle, described in that paper, is that multiple genetic variants of very small effect (OR < 1.05) may have only a small chance of showing individual significance in a GWAS, but en masse they could contribute a substantial amount of risk. To be as close as possible to the original study, we chose the same 74,062 SNPs as in the original paper and used the same p value thresholds. Because of overlapping samples between this study and the International Schizophrenia Consortium, this analysis was restricted to a sample set of 1310 cases and 1023 control subjects.
Results
The top association in this GWAS was in the MHC region (rs204999, p = 2.77 × 10−8) (Figures 1 and 2). This SNP is 63 kilobase upstream from, and in weak linkage disequilibrium (1000 Genomes CEU data R2 = .23) with, the signal reported by Stefansson et al. (12) (rs3131296). After accounting for rs204999, rs3131296 does not add any further signal in our dataset (p = .816). The MHC was the only region in the discovery GWAS with SNPs that had p < 10−7. Association analysis was also carried out on the X chromosome, but following quality control, no SNPs were significant at p < 10−4 (Supplement 1).
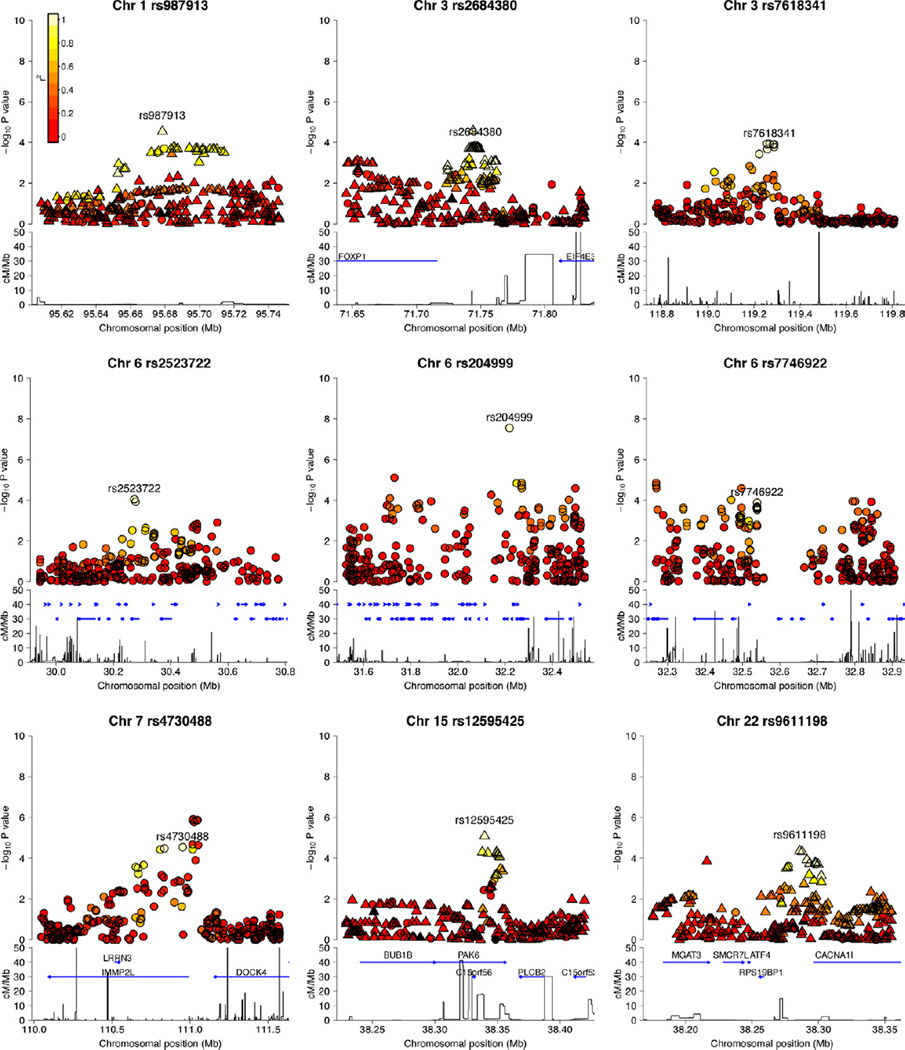
Figure 2.
Regional association plots of each of the loci described in Table 2. The −log 10 (p values) for the multiplicative model single nucleotide polymorphism (SNP) association in SNPTEST are shown on the upper part of the plot. Single nucleotide polymorphisms are colored based on their R2 with the labeled SNP, R2 calculated in the 58C control data. Where the SNP was chosen from the 1000 Genome imputation results, the plot includes the imputed SNPs. Circles represent the directly genotyped SNPs, and triangles represent the imputed SNPs. The bottom part of each plot shows the fine scale recombination rate estimated from individuals in the HapMap panel, and genes are marked by horizontal blue lines. Chr, chromosome; cM, centimorgan; Mb, megabase.
In total, we selected 79 regions for follow-up, where the most significant SNP had p < 10−4, and in the case of directly genotyped SNPs, the cluster plots met quality control standards. One or more SNPs were chosen for replication in each region, depending on the local pattern of linkage disequilibrium and the availability of SNPs in the replication data (108 SNPs in total). Seven SNPs spanning the association signal from 30.3 Mb to 34.1 Mb in the MHC region were included in the replication phase of the study. Of the other regions, 56 were identified from the genotyped scan (66 SNPs), 10 from the HapMap imputation (11 SNPs), and 12 from the 1000 Genomes imputation (24 SNPs). Replication was carried out in silico using the PGC and SGENE+ samples and SNPs were genotyped directly in the Immunochip replication samples, giving a total of 13,195 cases and 31,021 control subjects. Data were not available for all the SNPs on all samples due to differences in imputation strategies and genotyping platforms (see Methods and Materials).
Of note, a small proportion of the Irish cases in the discovery sample (n = 270) and Irish control subjects (n = 860) were included in the PGC-schizophrenia GWAS meta-analysis. We included these samples in the discovery phase for two reasons. First, this analysis had additional genotype information provided by 1000 Genomes imputation (6,212,339 directly genotyped and imputed SNPs) and analysis of the X chromosome when compared with the PGC analysis (1,252,901 autosomal SNPs). Second, existing data suggest genetic heterogeneity (e.g., at the MHC) may be important in schizophrenia etiology, indicating a value in reporting the results of the complete Irish sample. We performed a sensitivity analysis excluding these overlapping cases and control subjects from our discovery GWAS scan. This reduced the number of regions meeting criteria for follow-up (from 79 to 48) but did not materially affect the study outcome. We provide data on the SNP findings for the full and restricted discovery datasets at all loci selected for follow-up in Table S4 in Supplement 1. Expanding the sample to include additional cases with a broader psychotic disorder phenotype (n = 2049 cases, including mostly bipolar disorder cases) did not significantly alter the results.
Three of the seven SNPs tested in the MHC reached a p < 5 × 10−8 in a fixed effects meta-analysis across the four follow-up cohorts (p combined) (Table 1). These three SNPs showed evidence of association in two of the three replication datasets; no replication evidence (p < .05) at the MHC was found in the SGENE+ data (Figure S2 in Supplement 1). These MHC SNPs were the only replication results to meet standard significance criteria (p < .05) when correcting for the number of replication SNPs tested.
Table 1.
Loci with Evidence for Association with Schizophrenia Identified in this Study
| Chromosome | rsID | Positiona | Candidate Locus |
Discovery |
p Value SGENE+ |
p Value PGC |
p Value Immunochip |
p Value Combined |
||||
| Risk Allele |
RAF Cases |
RAF Control |
p Value Discovery |
OR (95% CI) | ||||||||
| 1p21 | rs987913b | 95678503 | — | C | .405 | .357 | 2.92 × 10−5 | 1.24 (1.12–1.37) | NA | .0176 | NA | 1.48 × 10−4 |
| 3p14 | rs862689a | 71744029 | — | G | .412 | .364 | 2.68 × 10−5 | 1.24 (1.12–1.37) | NA | .0149 | .312e | 2.69 × 10−4 |
| 3p14 | rs2684380b | 71745822 | FOXP1 | T | .427 | .383 | 1.91 × 10−4 | 1.20 (1.09–1.33) | NA | .0332 | NA | 6.05 × 10−4 |
| 3q13 | rs7618341 | 119268497 | — | A | .599 | .552 | 1.17 × 10−4 | 1.20 (1.10–1.32) | .437 (−) | .0536 | .0196 | 2.65 × 10−4 |
| 6p21 | rs2523722 | 30273252 | MHC | G | .778 | .737 | 8.71 × 10−5 | 1.25 (1.12–1.39) | .736 | 8.74 × 10−9 | 6.08 × 10−7 | 1.47 × 10−16 |
| 6p21 | rs204999 | 32217957 | MHC | T | .695 | .631 | 2.77 × 10−8 | 1.33 (1.20–1.47) | .621 | .0442 | 1.93 × 10−7 | 5.37 × 10−10 |
| 6p21 | rs7746922 | 32538953 | MHC | G | .765 | .723 | 1.30 × 10−4 | 1.23 (1.10–1.36) | .197 | 2.82 × 10−5 | .0314 | 4.48 × 10−9 |
| 7q31 | rs4730488 | 110953401 | IMMP2L | C | .517 | .466 | 2.85 × 10−5 | 1.22 (1.11–1.34) | .0718 | .0287 | .152 | 2.09 × 10−5 |
| 15q15 | rs12595425b,c | 38350279 | PAK6 | G | .884 | .852 | 8.26 × 10−6 | 1.44 (1.23–1.68) | NA | .0116 | NA | 2.74 × 10−5 |
| 22q13 | rs9611198b | 38285819 | CACNA1I | C | .508 | .458 | 4.35 × 10−5 | 1.22 (1.11–1.34) | NA | 2.48 × 10−3 | NA | 7.56 × 10−6 |
| 22q13 | rs9607658d | 38287686 | CACNA1I | C | .513 | .463 | 5.60 × 10−5 | 1.21 (1.10–1.33) | .275 (−) | 7.19 × 10−3 | 6.33 × 10−3 | 1.11 × 10−5 |
The discovery, replication, and fixed effects meta-analysis results at SNPs for which there is evidence (one-sided p < .05) in the majority of the replication cohorts where data are available. All p values shown are two-sided, and (−) denotes that the direction of effect is opposite to that in the discovery analysis.
CI, confidence interval; NA, no data available; OR, odds ratio; PGC, Psychiatric GWAS Consortium; RAF, risk allele frequency; SNP, single nucleotide polymorphism.
National Center for Biotechnology Information human genome build 36 coordinates.
1000 Genomes imputed SNP.
SNP (rs12595425) not present in the replication data so replication measured at rs3784396 (R2 = .99).
HapMap imputed SNP.
Replication was attempted at a tag SNP in the Immunochip replication, rs2594113 (R2 = .97).
Six other regions showed some evidence for association in the replication dataset (Table 1). In each case, the direction of effect was consistent in the majority of replication datasets, although the combined p values were >5 × 10−8 (Table 1, Figure 2; Figure S2 in Supplement 1). These included two variants on chromosome 22, rs9611198 (p combined = 7.56 × 10−6) and rs9607658 (p combined = 1.11 × 10−5), at the calcium channel gene CACNA1I. This gene is of potential interest, as its protein product interacts with the product of CACNA1C, a susceptibility gene for both schizophrenia (25) and bipolar disorder (26). Other members of this calcium channel gene family have been implicated in both schizophrenia and bipolar disorder etiology (27) and childhood seizure disorder (28). We identified suggestive evidence for association at a locus that includes the gene IMMP2L (rs4730488) across three of the four datasets (p combined = 2.09 × 10−5). Disruption of this gene has been reported in sporadic and familial cases with Tourette syndrome (29,30) and in individuals with autistic spectrum disorder (31), and an IMMP2L-DOCK4 deletion has been identified in a family with dyslexia (32). Finally, we identified suggestive evidence for association with a SNP in the gene PAK6 (rs12595425, p combined = 2.74 × 10−5), a member of the group B family of p21-activated kinase serine/threonine kinases. PAK6 is highly expressed in the brain; disruption of this gene in mice is associated with deficits in learning and memory (33) and upregulation of the gene may be part of the response to traumatic brain injury (34).
In the schizophrenia GWAS literature to date, including the MHC region, SNPs at 13 independent loci have reached a significance threshold of p < 5 × 10−8. Results at these SNPs in the WTCCC2 genome-wide scan are shown in Table 2. Results from our study at additional loci reported for schizophrenia or bipolar disorder, as collated in the National Human Genome Research Institute catalogue of GWAS studies (http://www.genome.gov/26525384 [searched January 2012]), are included in Table S2 in Supplement 1.
Table 2.
Evidence in the Discovery Data at Loci Previously Reported to be Associated with Schizophrenia at p < 5 × 10−8
| Chr | rsID | Positiona | Locus of Interest |
Risk Allele |
Case RAF |
Control RAF |
p Valueb | OR | (95% CI) | Reference |
| 1 | rs1625579c | 98275522 | MIR137 | T | .83 | .82 | .035 | 1.12 | (.99–1.27) | (9) |
| 1 | rs10489202 | 166169703 | — | A | .25 | .23 | .027 | 1.11 | (1.00–1.24) | (10) |
| 2 | rs2312147d | 58076432 | VRK2 | C | .64 | .61 | 4.94 × 10−3 | 1.14 | (1.03–1.25) | (12) |
| 2 | rs1344706e | 185486673 | ZNF804A | T | .65 | .63 | 5.56 × 10−3 | 1.17 | (1.01–1.23) | (4) |
| 3 | rs6782299e | 182033396 | — | T | .74 | .73 | .148 | 1.07 | (.94–1.23) | (3) |
| 6 | rs1635d | 28335583 | NKAPL | C | .97 | .97 | .360 | 1.05 | (.79–1.41) | (11) |
| 6 | rs2142731 | 28358892 | PGBD1 | C | .003 | .002 | .099 | 1.92 | (.71–5.18) | (11) |
| 6 | rs3131296d | 32280971 | MHC | C | .85 | .82 | 1.88 × 10−4 | 1.26 | (1.11–1.44) | (12) |
| 6 | rs9272219 | 32710247 | MHC | G | .77 | .74 | .016 | 1.13 | (1.01–1.26) | (6) |
| 8 | rs10503253c | 4168252 | CSMD1 | A | .22 | .19 | 9.06 × 10−3 | 1.15 | (1.02 × 1.30) | (9) |
| 8 | rs16887244 | 38150502 | — | A | .77 | .75 | .084 | 1.08 | (.97–1.21) | (10) |
| 8 | rs7004633c | 89829427 | MMP16 | G | .18 | .16 | .062 | 1.11 | (.97–1.26) | (9) |
| 10 | rs11191580c | 104896201 | NT5C2 | T | .93 | .92 | .139 | 1.10 | (.92–1.32) | (9) |
| 11 | rs835784 | 44820394 | TSPAN18 | T | .48 | .47 | .256 | 1.03 | (.94–1.14) | (11) |
| 11 | rs12807809 | 124111495 | NRGN | T | .83 | .82 | .144 | 1.07 | (.94–1.22) | (12) |
| 18 | rs17594526e | 51209236 | TCF4 | A | .03 | .02 | 1.05 × 10−3 | 1.77 | (1.22–2.54) | (3) |
| 18 | rs9960767d | 51306000 | TCF4 | C | .05 | .04 | .080 | 1.18 | (.94–1.49) | (12) |
Chr, chromosome; CI, confidence interval; GWAS, genome-wide association studies; ISC, International Schizophrenia Consortium; OR, odds ratio; PGC, Psychiatric GWAS Consortium; RAF, risk allele frequency, SCZ, schizophrenia; SNP, single nucleotide polymorphism; WTCCC2, Wellcome Trust Case Control Consortium 2.
National Center for Biotechnology Information human genome build 36 coordinates.
p value is one sided in the direction of risk in the original study; the direction of effect is the same as in the original study for all SNPs.
The WTCCC2-SCZ data contributed to the PGC GWAS meta-analysis.
SNP imputed in WTCCC2 data.
There was some overlap between the WTCCC2-SCZ sample and the O’Donovan et al. (4) and ISC studies. These cases and control subjects are excluded from the WTCCC2-SCZ sample for the purposes of the current analysis.
The strongest replication of currently known loci was found on chromosome 6p21 (SNP imputed, rs3131296, one-sided p = 1.88 × 10−4) at the MHC locus. A number of schizophrenia GWAS studies (3,6,12) have reported association at this locus; in our data, there was varying support for the SNPs previously reported (Table S3 in Supplement 1). Analysis of the MHC in our dataset is discussed more fully below. Outside the MHC, we found evidence of association at 6 of the 13 previously identified risk alleles, where the one-sided p < .05 is in the same direction of effect as the original study. The three most significant findings mapped to the genes TCF4, ZNF804A, and VRK2 (Table 2).
The MHC is a highly polymorphic and complex region of the genome, containing many genes. To date, attempts to dissect the schizophrenia association signal at the MHC have been inconclusive (3,6,12). In this study, three MHC SNPs each reached a fixed effect p combined < 5 × 10−8 (rs204999, rs2523722, rs7746922). In the discovery cohort, conditioning on the most significantly associated SNP (rs204999) substantially reduced the evidence of association at the other replicated MHC SNPs (rs2523722 p = .071 and rs7746922 p = .250). However, rs204999 did not account for all evidence of SNP association at the locus. A secondary signal was seen at rs2499690 and rs3103431 (Figure S3 in Supplement 1), although there was not strong replication evidence for this second signal.
Previous HLA analyses, including those reported from large collaborative GWAS efforts (representing most of our follow-up sample), have been equivocal, which likely reflects known heterogeneity at the locus and the contribution of multiple different populations to these studies. For this reason, we attempted to identify whether a specific classical allele increased risk in our sample and whether this could be replicated in a large, new sample collection from the United Kingdom (the Immunochip replication sample). To investigate whether any of the classical human leukocyte antigen alleles could be driving the SNP signal at the MHC, classical HLA alleles were imputed using an updated version of the HLA*IMP algorithm (23,35). Human leukocyte antigen alleles were imputed to a four-digit accuracy at HLA-A, HLA-B, HLA-C, HLA-DQA1, HLA-DQB1, and HLA-DRB1 (see Supplemental Methods in Supplement 1).
An association test for each allele was performed in the Irish sample (Table S5 in Supplement 1), and HLA-C*01:02 showed the strongest evidence of association with schizophrenia (p = 3.83 × 10−4), where presence of HLA-C*01:02 was associated with increased risk of schizophrenia (OR [95%CI] = 1.65 [1.25–2.18]). HLA-C*01:02 was found at a frequency of 4.1% in cases and 2.5% in control subjects and was imputed with high specificity and sensitivity (see Supplemental Methods in Supplement 1). This allele was also associated with risk of schizophrenia in the Immunochip replication data, where HLA imputation was available for 2690 cases and 6279 control subjects (p = .045, OR [95%CI] = 1.19 [1.01–1.41]).
When conditioning on HLA-C*01:02, the top SNP from the discovery scan, rs204999, was still the most significant SNP (p = 2.51 × 10−7), and similarly, when conditioning on rs204999, HLA-C*01:02 was still significant (p = 2.82 × 10−3). This suggests that both signals may be important in this dataset.
Stefansson et al. (12) found that SNP haplotypes tagging DRB1*03 and HLA-B*08 were protective for schizophrenia. In this study, we replicated these findings at the imputed HLA alleles directly rather than at tagging SNP haplotypes: HLA-B*08:01 p = .024 and DRB1*03:01 p = .068 (one-sided p values shown). Both of these HLA alleles were imputed with high specificity and sensitivity (see Supplemental Methods in Supplement 1). DRB1*03:01 and B*08:01 were in high linkage disequilibrium (LD) (R2 = .61) and from these data it appears that these two HLA alleles were tagging the same signal.
Taking the large set of autosomal score alleles (n = 74,062) from the ISC dataset, we investigated the aggregate risk scores for individuals in the WTCCC2 schizophrenia sample. We examined increasingly liberal p value thresholds (PT) from PT < .01 to PT < .7. We identified higher mean score in cases compared with control subjects in the WTCCC2 dataset. The ISC-derived score was highly associated with disease at PT < .5, as has previously been reported in other schizophrenia datasets (PGC) (36). In our sample, the most significant PT finding was at PT < .3 (p < 10−15) (Figure S4 in Supplement 1).
Discussion
We report a schizophrenia GWAS in a sample of 1606 cases and 1794 control subjects. We found strong support for association at three SNPs in the MHC, including the lead SNP in the discovery data rs204999. Multiple SNPs in the MHC region have been identified in previous schizophrenia GWAS (3,6,12). Conditional analysis of the associated SNPs suggests that in this data the signal is being driven by association with rs204999, although there is evidence of association with other SNPs at the locus. Previous work to pinpoint HLA alleles that could be responsible for the signal in the MHC have had varied success. Comparison of association with HLA alleles across studies is complicated by differences in the methodology used to call alleles, as well as differences in the populations being studied. In this study, the strongest HLA allele association was found with HLA-C*01:02, and this was separate from the top SNP signal. We also replicated association with two protective HLA alleles, HLA-B*08:01 and DRB1*03:01, which had previously been identified by Stefansson et al. (5). Our analysis suggests that these alleles tag the same association signal.
There is growing awareness of the important role that MHC genes play in nervous system development and function (37). Major histocompatibility complex class I molecules are expressed in the brain and function both presynaptically and postsynaptically in forming and regulating cortical connectivity across brain circuitry (38). Our study identifies risk and protective HLA alleles for schizophrenia, which may be particularly relevant because of autoimmune or infectious risk factors in this population.
Because of extensive linkage disequilibrium, localizing signals within the MHC is extremely challenging, as evident from Figure 2. The SNP rs204999 is in strong LD with seven other SNPs (R2 = .66 –.68; rs3130284, rs3134608, rs3130347, rs3134945, rs3096697, rs3134947, rs3132965), which span a region from 32.22 Mb to 32.26 Mb containing PPT2, PRRT1, EGFL8, AGPAT1, and RNF5. None of these genes feature in the pre-GWAS schizophrenia genetics literature and there is no strong in silico information available to suggest a functional role for rs204999 in the regulation of these genes or others. Single nucleotide polymorphism rs2523722 is at 30.27 Mb on chromosome 6 and is located in an intron of TRIM26. This variant is in complete LD (R2 = 1) with 11 other SNPs within a 10 kilobase region, all of which map to TRIM26. Nine of these SNPs are intronic, one (rs2523721) is a coding silent SNP, and one (rs971570) is located in the 5’ untranslated region. There is no prior evidence of association between TRIM26 and schizophrenia and the gene is a member of a tripartite motif family of unknown function.
We performed a polygene score analysis using the large set of autosomal score alleles identified in the previous ISC schizophrenia study. The proportion of variance explained (estimated by Nagelkerke’s R2) in the original analysis was 3% to 4% and was 6% in this sample, adding support to the hypothesis that a substantial fraction of schizophrenia heritability is accounted for by a polygenic component. In this study, we attempted replication at 79 regions and identified nominal significant evidence of association (p < .05) with the same risk allele at 17 regions; this is significantly more than expected by chance (p = 3.09 × 10−7). Larger studies are required to confirm which of these regions harbor schizophrenia susceptibility variants. We identified suggestive evidence of association at loci that could implicate three novel genes in schizophrenia susceptibility (IMMP2L, PAK6, and CACNA1I). Two of these loci were identified from imputation based on reference panels from the 1000 Genomes data, suggesting the inclusion of the imputation data was a useful addition to this study. Based on the experience for other common disorders, further investigation of these loci in a larger meta-analysis of GWAS datasets may be warranted.
Supplementary Material
Acknowledgments
The authors sincerely thank all patients who contributed to this study and all staff who facilitated their involvement. Funding for this study was provided by the Wellcome Trust Case Control Consortium 2 project (085475/B/08/Z and 085475/Z/08/Z), the Wellcome Trust (072894/Z/03/Z, 090532/Z/09/Z and 075491/Z/04/B), NIMH grants (MH 41953 and MH083094) and Science Foundation Ireland (08/IN.1/B1916). P. Donnelly was supported in part by a Wolfson-Royal Society Merit Award. We also thank S. Bertrand, J. Bryant, S.L. Clark, J.S. Conquer, T. Dibling, J.C. Eldred, S. Gamble, C. Hind, A. Wilk, C.R. Stribling and S. Taylor of the Wellcome Trust Sanger Institute’s Sample and Genotyping Facilities for technical assistance. We thank S. Leslie for support with the HLA imputation analysis. We acknowledge use of the Irish GeneBank sample, the British 1958 Birth Cohort DNA collection funded by the Medical Research Council (grant G0000934) and the Wellcome Trust (grant 068545/Z/02) and of the UK National Blood Service controls funded by the Wellcome Trust.
Appendix A
The authors of this paper are: Amy Strange1,*, Brien P Riley2,*, Chris C A Spencer1, Derek W Morris3, Matti Pirinen1, Colm T O’Dushlaine3, Zhan Su1, Brion S Maher2, Colin Freeman1, Paul Cormican3, Celine Bellenguez1, Elaine M Kenny3, Gavin Band1, Brandon Wormley2, Gary Donohoe3, Alexander Dilthey4, Loukas Moutsianas1,4, Emma Quinn3, Sarah Edkins5, Roisin Judge3, Kim Coleman3, Sarah Hunt5, Daniela Tropea3, Siobhan Roche6, Liz Cummings3, Eric Kelleher3, Patrick McKeon6, Ted Dinan7, Colm McDonald8, Kieran C Murphy9, Eadbhard O’Callaghan10,†, Francis A O’Neill11, John L Waddington12, Dermot Walsh13, Eleni Giannoulatou1, Cordelia Langford5, Panos Deloukas5, Emma Gray5, Serge Dronov5, Simon Potter5, Richard Pearson1, Damjan Vukcevic1, Avazeh Tashakkori-Ghanbaria5, Jenefer M Blackwell14,15, Elvira Bramon16, Matthew A Brown17, Juan P Casas18, Audrey Duncanson19, Janusz Jankowski20, Hugh S Markus21, Christopher G Mathew22, Colin NA Palmer23, Robert Plomin24, Anna Rautanen1, Stephen J Sawcer25, Richard C Trembath22, Ananth C Viswanathan26, Nicholas W Wood27, Jennifer Stone28, Ed Scolnick28,, Shaun Purcell28, Pamela Sklar28, SGENE+ Consortium, Stephan Ripke29, James Walters29, Michael J Owen29, Michael C O’Donovan29, Schizophrenia Working Group of the Psychiatric GWAS Consortium, Leena Peltonen5,†, Gil McVean4, Ken S Kendler2, Michael Gill3, Peter Donnelly1,^, Aiden Corvin3,^
*Authors contributed equally to this work
^Authors jointly directed this work
†Deceased
1Wellcome Trust Centre for Human Genetics, Oxford, UK; 2Depts of Psychiatry and Human Genetics, Virginia Institute of Psychiatric and Behavioral Genetics, Virginia Commonwealth University, Richmond, VA, USA; 3Dept of Psychiatry & Neuropsychiatric Genetics Group, Institute of Molecular Medicine, Trinity College Dublin, Dublin, Ireland; 4Dept Statistics, University of Oxford, Oxford, UK; 5Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Cambridge, UK; 6St Patrick’s University Hospital, James St., Dublin, Ireland; 7Dept of Psychiatry, University College Cork, Cork, Ireland; 8Dept of Psychiatry, National University of Ireland, Galway, Galway, Ireland; 9Dept of Psychiatry, RCSI Education and Research Centre, Beaumont Hospital, Dublin, Ireland; 10DETECT Early Intervention in Psychosis Services, Dun Laoghaire, Co. Dublin, Ireland; 11Dept of Psychiatry, Queen’s University, Belfast, Northern Ireland, UK; 2Dept Statistics, University of Oxford, Oxford, UK; 12Molecular and Cellular Therapeutics, Royal College of Surgeons in Ireland, Dublin, Ireland; 13Health Research Board, Dublin 2, Ireland; 14Telethon Institute for Child Health Research, Centre for Child Health Research, University of Western Australia, Subiaco, Western Australia; 15Cambridge Institute for Medical Research, University of Cambridge School of Clinical Medicine, Cambridge, UK; 16Department of Psychosis Studies, NIHR Biomedical Research Centre for Mental Health at the Institute of Psychiatry, King’s College London and The South London and Maudsley NHS Foundation Trust, Denmark Hill, London, UK; 17University of Queensland Diamantina Institute, Immunology and Metabolic Medicine, Princess Alexandra Hospital, University of Queensland, Brisbane, Queensland, Australia; 18Dept Epidemiology and Population Health, London School of Hygiene and Tropical Medicine, London and Dept Epidemiology and Public Health, University College London, UK; 19Molecular and Physiological Sciences, The Wellcome Trust, London, UK; 20Centre for Digestive Diseases, Queen Mary University of London, London, UK and Digestive Diseases Centre, Leicester Royal Infirmary, Leicester, UK and Department of Clinical Pharmacology, Old Road Campus, University of Oxford, Oxford, UK; 21Clinical Neurosciences, St George’s University of London, London, UK; 22King’s College London Dept Medical and Molecular Genetics, School of Medicine, Guy’s Hospital, London, UK; 23Biomedical Research Centre, Ninewells Hospital and Medical School, Dundee, UK; 24King’s College London Social, Genetic and Developmental Psychiatry Centre, Institute of Psychiatry, Denmark Hill, London, UK; 25University of Cambridge Dept Clinical Neurosciences, Addenbrooke’s Hospital, Cambridge, UK; 26NIHR Biomedical Research Centre for Ophthalmology, Moorfields Eye Hospital NHS Foundation Trust and UCL Institute of Ophthalmology, London, UK; 27Dept Molecular Neuroscience, Institute of Neurology, Queen Square, London, UK; 28Broad Institute and Center for Human Genetics Research of Massachusetts General Hospital, Boston, MA, USA; 29MRC Centre for Neuropsychiatric Genetics and Genomics, School of Medicine, Cardiff University, Heath Park, Cardiff, UK.
Membership of Wellcome Trust Case Control Consortium 2:
Management Committee: Peter Donnelly (Chair)1,2, Ines Barroso (Deputy Chair)3, Jenefer M Blackwell4, 5, Elvira Bramon6, Matthew A Brown7, Juan P Casas8, Aiden Corvin9, Panos Deloukas3, Audrey Duncanson10, Janusz Jankowski11, Hugh S Markus12, Christopher G Mathew13, Colin NA Palmer14, Robert Plomin15, Anna Rautanen1, Stephen J Sawcer16, Richard C Trembath13, Ananth C Viswanathan17, Nicholas W Wood18
Data and Analysis Group: Chris C A Spencer1, Gavin Band1, Céline Bellenguez1, Alexander Dilthey2, Colin Freeman1, Garrett Hellenthal1, Eleni Giannoulatou1, Gil McVean2, Loukas Moutsianas2, Matti Pirinen1, Richard Pearson1, Amy Strange1, Zhan Su1, Avazeh Tashakkori-Ghanbaria3, Damjan Vukcevic1, Peter Donnelly1,2
DNA, Genotyping, Data QC and Informatics Group: Cordelia Langford3, Sarah E Hunt3, Sarah Edkins3, Rhian Gwilliam3, Hannah Blackburn3, Suzannah J Bumpstead3, Serge Dronov3, Matthew Gillman3, Emma Gray3, Naomi Hammond3, Alagurevathi Jayakumar3, Owen T McCann3, Jennifer Liddle3, Simon C Potter3, Radhi Ravindrarajah3, Michelle Ricketts3, Matthew Waller3, Paul Weston3, Sara Widaa3, Pamela Whittaker3, Ines Barroso3, Panos Deloukas3
Publications Committee: Christopher G Mathew (Chair)13, Jenefer M Blackwell4,5, Matthew A Brown7, Aiden Corvin9, Chris C A Spencer1
Membership of the Irish Schizophrenia Genomics Consortium:
Brien P Riley18, Derek W Morris9, Colm T O’Dushlaine9, Brion S Maher18, Paul Cormican9, Elaine M Kenny9, Brandon Wormley18, Gary Donohoe9, Emma Quinn9, Roisin Judge9, Kim Coleman9, Daniela Tropea9, Siobhan Roche19, Liz Cummings9, Eric Kelleher9, Patrick McKeon19, Ted Dinan20, Colm McDonald21, Kieran C Murphy22, Eadbhard O’Callaghan23, Francis A O’Neill24, John L Waddington22, Dermot Walsh25, Jennifer Stone26, Ed Scolnick26,, Shaun Purcell26, Pamela Sklar26, SGENE+ Consortium, Stephan Ripke26, James Walters27, Michael J Owen27, Michael C O’Donovan27, Schizophrenia Working Group of the Psychiatric GWAS Consortium, Ken S Kendler18, Michael Gill9, Aiden Corvin9
Membership lists of SGENE+ Consortium and Schizophrenia Working Group of the Psychiatric GWAS Consortium are provided in the Supplement.
1 Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford, UK; 2 Dept Statistics, University of Oxford, Oxford, UK; 3 Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Cambridge, UK; 4 Telethon Institute for Child Health Research, Centre for Child Health Research, University of Western Australia, Subiaco, Western Australia; 5 Cambridge Institute for Medical Research, University of Cambridge School of Clinical Medicine, Cambridge, UK; 6 Department of Psychosis Studies, NIHR Biomedical Research Centre for Mental Health at the Institute of Psychiatry, King’s College London and The South London and Maudsley NHS Foundation Trust, Denmark Hill, London, UK; 7 University of Queensland Diamantina Institute, Brisbane, Queensland, Australia; 8 Dept Epidemiology and Population Health, London School of Hygiene and Tropical Medicine, London, and Dept Epidemiology and Public Health, University College London, UK; 9 Neuropsychiatric Genetics Research Group, Institute of Molecular Medicine, Trinity College Dublin, Dublin, Ireland; 10 Molecular and Physiological Sciences, The Wellcome Trust, London, UK; 11 Department of Oncology, Old Road Campus, University of Oxford, Oxford, UK, Digestive Diseases Centre, Leicester Royal Infirmary, Leicester, UK and Centre for Digestive Diseases, Queen Mary University of London, London, UK; 12 Clinical Neurosciences, St George’s University of London, London, UK; 13 King’s College London Dept Medical and Molecular Genetics, King’s Health Partners, Guy’s Hospital, London, UK; 14 Biomedical Research Centre, Ninewells Hospital and Medical School, Dundee, UK; 15 King’s College London Social, Genetic and Developmental Psychiatry Centre, Institute of Psychiatry, Denmark Hill, London, UK; 16 University of Cambridge Dept Clinical Neurosciences, Addenbrooke’s Hospital, Cambridge, UK; 17 NIHR Biomedical Research Centre for Ophthalmology, Moorfields Eye Hospital NHS Foundation Trust and UCL Institute of Ophthalmology, London, UK; 18 Dept Molecular Neuroscience, Institute of Neurology, Queen Square, London, UK, and Depts of Psychiatry and Human Genetics, Virginia Institute of Psychiatric and Behavioral Genetics, Virginia Commonwealth University, Richmond, VA, USA; 19 St Patrick’s University Hospital, James St., Dublin, Ireland; 20 Dept of Psychiatry, University College Cork, Cork, Ireland; 21 Dept of Psychiatry, National University of Ireland, Galway, Galway, Ireland; 22 Dept of Psychiatry, RCSI Education and Research Centre, Beaumont Hospital, Dublin, Ireland; 23 DETECT Early Intervention in Psychosis Services, Dun Laoghaire, Co. Dublin, Ireland; 24 Dept of Psychiatry, Queen’s University, Belfast, Northern Ireland, UK; 25 Health Research Board, Dublin, Ireland; 26 Broad Institute and Center for Human Genetics Research of Massachusetts General Hospital, Boston, MA, USA; 27 MRC Centre for Neuropsychiatric Genetics and Genomics, School of Medicine, Cardiff University, Heath Park, Cardiff, UK.
Footnotes
The authors report no biomedical financial interests or potential conflicts of interest.
Supplementary material cited in this article is available online.
References
- 1.Cardno AG, Gottesman II. Twin studies of schizophrenia: From bow-and-arrow concordances to star wars Mx and functional genomics. Am J Med Genet. 2000;97:12–17. [PubMed] [Google Scholar]
- 2.Tiihonen J, Lonnqvist J, Wahlbeck K, Klaukka T, Niskanen L, Tanskanen A, Haukka J. 11-year follow-up of mortality in patients with schizophrenia: A population-based cohort study (FIN11 study) Lancet. 2009;374:620–627. doi: 10.1016/S0140-6736(09)60742-X. [DOI] [PubMed] [Google Scholar]
- 3.International Schizophrenia Consortium. Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Donovan MC, Craddock N, Norton N, Williams H, Peirce T, Moskvina V, et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008;40:1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- 5.Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi J, Levinson DF, Duan J, Sanders AR, Zheng Y, Pe’er I, et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460:753–757. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Athanasiu L, Mattingsdal M, Kahler AK, Brown A, Gustafsson O, Agartz I, et al. Gene variants associated with schizophrenia in a Norwegian genome-wide study are replicated in a large European cohort. J Psychiatr Res. 2010;44:748–753. doi: 10.1016/j.jpsychires.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ripke S, Sanders AR, Kendler KS, Levinson DF, Sklar P, Holmans PA, et al. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi Y, Li Z, Xu Q, Wang T, Li T, Shen J, et al. Common variants on 8p12 and 1q24.2 confer risk of schizophrenia. Nat Genet. 2011;43:1224–1227. doi: 10.1038/ng.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yue WH, Wang HF, Sun LD, Tang FL, Liu ZH, Zhang HX, et al. Genome-wide association study identifies a susceptibility locus for schizophrenia in Han Chinese at 11p11.2. Nat Genet. 2011;43:1228–1231. doi: 10.1038/ng.979. [DOI] [PubMed] [Google Scholar]
- 12.Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.First MB, Spitzer RL, Robert L, Gibbon M, Williams JBW. Structured Clinical Interview for DSM-IV-TR Axis I Disorders, Research Version, Patient Edition. New York: Biometrics Research, New York State Psychiatric Institute; 2002. [Google Scholar]
- 15.Endicott J, Spitzer RL. A diagnostic interview: The schedule for affective disorders and schizophrenia. Arch Gen Psychiatry. 1978;35:837–844. doi: 10.1001/archpsyc.1978.01770310043002. [DOI] [PubMed] [Google Scholar]
- 16.World Health Organization. Schedules for Clinical Assessment in Neuropsychiatry (Version 1 0) Geneva: WHO; 1992. [Google Scholar]
- 17.UK IBD Genetics Consortium. Barrett JC, Lee JC, Lees CW, Prescott NJ, Anderson CA, et al. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat Genet. 2009;41:1330–1334. doi: 10.1038/ng.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bellenguez C, Strange A, Freeman C, Donnelly P, Spencer CC Wellcome Trust Case Control Consortium. A robust clustering algorithm for identifying problematic samples in genome-wide association studies. Bioinformatics. 2012;28:134–135. doi: 10.1093/bioinformatics/btr599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Price AL, Zaitlen NA, Reich D, Patterson N. New approaches to population stratification in genome-wide association studies. Nat Rev Genet. 2010;11:459–463. doi: 10.1038/nrg2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J, Weedon MN, Purcell S, Lettre G, Estrada K, Willer CJ, et al. Genomic inflation factors under polygenic inheritance. Eur J Hum Genet. 2011;19:807–812. doi: 10.1038/ejhg.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000529. e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dilthey AT, Moutsianas L, Leslie S, McVean G. HLA*IMP--an integrated framework for imputing classical HLA alleles from SNP genotypes. Bioinformatics. 2011;27:968–972. doi: 10.1093/bioinformatics/btr061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Green EK, Grozeva D, Jones I, Jones L, Kirov G, Caesar S, et al. The bipolar disorder risk allele at CACNA1C also confers risk of recurrent major depression and of schizophrenia. Mol Psychiatry. 2010;15:1016–1022. doi: 10.1038/mp.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferreira MA, O’Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L, et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008;40:1056–1058. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Curtis D, Vine AE, McQuillin A, Bass NJ, Pereira A, Kandaswamy R, et al. Case-case genome-wide association analysis shows markers differentially associated with schizophrenia and bipolar disorder and implicates calcium channel genes. Psychiatr Genet. 2011;21:1–4. doi: 10.1097/YPG.0b013e3283413382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y, Lu J, Pan H, Zhang Y, Wu H, Xu K, et al. Association between genetic variation of CACNA1H and childhood absence epilepsy. Ann Neurol. 2003;54:239–243. doi: 10.1002/ana.10607. [DOI] [PubMed] [Google Scholar]
- 29.Patel C, Cooper-Charles L, McMullan DJ, Walker JM, Davison V, Morton J. Translocation breakpoint at 7q31 associated with tics: Further evidence for IMMP2L as a candidate gene for Tourette syndrome. Eur J Hum Genet. 2011;19:634–639. doi: 10.1038/ejhg.2010.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petek E, Windpassinger C, Vincent JB, Cheung J, Boright AP, Scherer SW, et al. Disruption of a novel gene (IMMP2L) by a breakpoint in 7q31 associated with Tourette syndrome. Am J Hum Genet. 2001;68:848–858. doi: 10.1086/319523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maestrini E, Pagnamenta AT, Lamb JA, Bacchelli E, Sykes NH, Sousa I, et al. High-density SNP association study and copy number variation analysis of the AUTS1 and AUTS5 loci implicate the IMMP2L-DOCK4 gene region in autism susceptibility. Mol Psychiatry. 2010;15:954–968. doi: 10.1038/mp.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pagnamenta AT, Bacchelli E, de Jonge MV, Mirza G, Scerri TS, Minopoli F, et al. Characterization of a family with rare deletions in CNTNAP5 and DOCK4 suggests novel risk loci for autism and dyslexia. Biol Psychiatry. 2010;68:320–328. doi: 10.1016/j.biopsych.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nekrasova T, Jobes ML, Ting JH, Wagner GC, Minden A. Targeted disruption of the Pak5 and Pak6 genes in mice leads to deficits in learning and locomotion. Dev Biol. 2008;322:95–108. doi: 10.1016/j.ydbio.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 34.Zhao W, Yang J, Shi W, Wu X, Shao B, Wu Q, et al. Upregulation of p21-activated Kinase 6 in rat brain cortex after traumatic brain injury. J Mol Histol. 2011;42:195–203. doi: 10.1007/s10735-011-9324-8. [DOI] [PubMed] [Google Scholar]
- 35.Leslie S, Donnelly P, McVean G. A statistical method for predicting classical HLA alleles from SNP data. Am J Hum Genet. 2008;82:48–56. doi: 10.1016/j.ajhg.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ripke S, Sanders AR, Kendler KS, Levinson DF, Sklar P, Holmans PA, et al. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shatz CJ. MHC class I: An unexpected role in neuronal plasticity. Neuron. 2009;64:40–45. doi: 10.1016/j.neuron.2009.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Needleman LA, Liu XB, El-Sabeawy F, Jones EG, McAllister AK. MHC class I molecules are present both pre- and postsynaptically in the visual cortex during postnatal development and in adulthood. Proc Natl Acad Sci U S A. 2011;107:16999–17004. doi: 10.1073/pnas.1006087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.