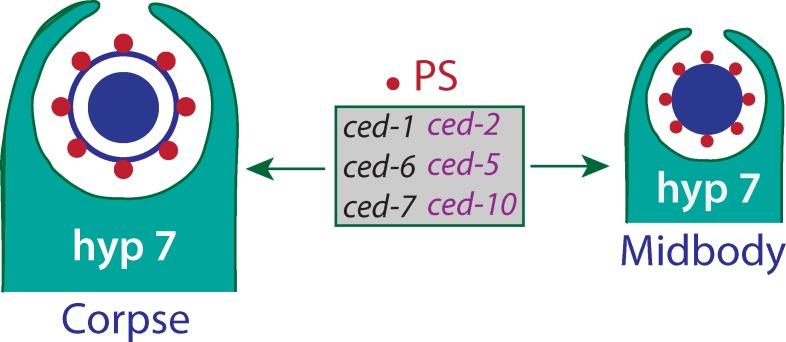
Independent of their role in apoptosis, cell engulfment proteins are essential for midbody internalization and degradation after cell division.
Abstract
Cell death genes are essential for apoptosis and other cellular events, but their nonapoptotic functions are not well understood. The midbody is an important cytokinetic structure required for daughter cell abscission, but its fate after cell division remains elusive in metazoans. In this paper, we show through live-imaging analysis that midbodies generated by Q cell divisions in Caenorhabditis elegans were released to the extracellular space after abscission and subsequently internalized and degraded by the phagocyte that digests apoptotic Q cell corpses. We further show that midbody degradation is defective in apoptotic cell engulfment mutants. Externalized phosphatidylserine (PS), an engulfment signal for corpse phagocytosis, exists on the outer surface of the midbody, and inhibiting PS signaling delayed midbody clearance. Thus, our findings uncover a novel function of cell death genes in midbody internalization and degradation after cell division.
Introduction
Midbody, a singular organelle formed between the two nascent daughter cells by the cleavage furrow ingression, constitutes a tightly packed, antiparallel microtubule bundle covered with electron-dense periphery material (Glotzer, 2009; Steigemann and Gerlich, 2009). A proposed function of the midbody is to provide an anchoring point for the final abscission, in which microtubules in the cytoplasmic bridge or membrane connecting the daughter cells are severed by microtubule-severing enzymes or by the endosomal sorting complex required for transport machinery (Yang et al., 2008; Elia et al., 2011). After cytokinesis, the midbody can be either shed to the extracellular space for degradation or asymmetrically retained by one daughter cell.
A recent study suggests that midbody degradation is an important cellular process and that defects of midbody clearance are associated with human lysosomal storage disorders (Pohl and Jentsch, 2009). Midbody degradation may also contribute to cell fate determination; stem cell markers, such as CD133, are present in midbodies, and daughter cells that remain as stem cells retain their midbodies, whereas those that differentiate degrade or shed the midbodies (Ettinger et al., 2011; Kuo et al., 2011; Schink and Stenmark, 2011). However, most of our current knowledge of the midbody was obtained from cultured cell lines, and the fate of the midbody, in live animals, remains largely unknown.
Programmed cell death, an integral component of metazoan development, selectively destroys a subpopulation of excessively produced cells in developing tissues or damaged cells for adult tissue maintenance (Conradt, 2009). Genetic studies in Caenorhabditis elegans uncovered that three sequential pathways specify the fate of cell death, activate the suicide program, and degrade the dying cells (Conradt and Xue, 2005; Conradt, 2009). Transcriptional regulatory networks first activate the expression of the death-inducing gene egl-1 in cells fated to die. EGL-1–mediated protein interaction cascade then activates a crucial caspase CED-3, which destroys subcellular structures and fragments nuclear DNA. Finally, the externalization of phosphatidylserine (PS) in the dying cell triggers two parallel signaling pathways (CED-1, CED-6, and CED-7; CED-2, CED-5, CED-10, and CED-12) in the phagocyte, which engulfs and degrades apoptotic cell corpses (Conradt and Xue, 2005; Conradt, 2009).
Cell death genes are also essential for various nonapoptotic cellular processes (Galluzzi et al., 2012; Hyman and Yuan, 2012). For example, a restricted and local modulation of caspase contributes to axon pruning and synapse elimination, which may refine mature neuronal circuits (Hyman and Yuan, 2012). During axon pruning, the phagocytic receptor, Draper/CED-1 functions inside glial cells to engulf pruned or injured axons and dendrites in Drosophila melanogaster and axons undergoing Wallerian degeneration (Awasaki et al., 2006; Hoopfer et al., 2006; MacDonald et al., 2006; Logan et al., 2012). Compared with the investigation of apoptotic function of cell death genes, our understanding of their nonapoptotic functions is limited.
This study, for the first time, followed the dynamic behavior of the midbody, generated by C. elegans Q neuroblast asymmetric division, by spinning-disk confocal microcopy. We showed that Q cell midbodies are released after abscission and engulfed and degraded by a phagocyte that also degrades Q cell corpse. We found that externalized PS exists on the outer surface of the midbody and is important for midbody clearance. We provided genetic evidence that midbody degradation in Q cell and other C. elegans lineages requires the function of apoptotic cell corpse engulfment genes but not caspase.
Results and discussion
Midbody release in C. elegans Q neuroblast lineage
We developed fluorescence markers and live-imaging techniques to study the fate of the midbody generated by asymmetric divisions in the C. elegans Q neuroblast lineage. Each of the two Q cells on the left (QL) or the right (QR) sides of L1 larvae undergoes four asymmetric divisions to create three distinct neurons and two apoptotic cells, producing four midbody remnants through these divisions (Fig. 1 A; Sulston and Horvitz, 1977; Ou and Vale, 2009; Ou et al., 2010; Li et al., 2012). To follow the dynamic behavior of Q cell midbody, we expressed GFP-tagged ZEN-4 (mammalian MKLP1 homologue), a widely used midbody marker across species (Kaitna et al., 2000; Glotzer, 2009) and mCherry-tagged plasma membrane and histone in Q cell. As expected, GFP::ZEN-4 accumulated in the midbody during cytokinesis (Video 1).
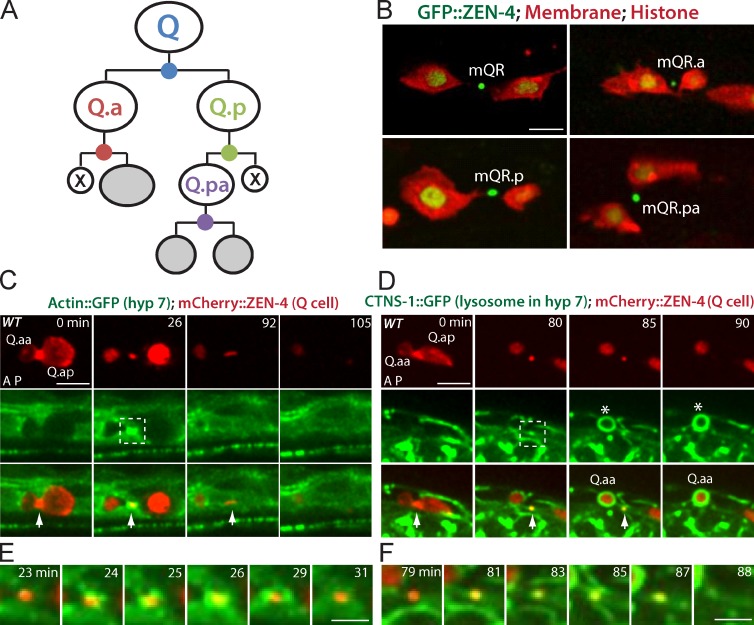
Figure 1.
Q cell midbody is released after cell division and degraded by the hyp7 cell. (A) C. elegans Q cells are bilaterally symmetric, and both QR (right) and QL (left) cells undergo three rounds of asymmetric division to generate three different neurons (gray) and two apoptotic cells (X). Four divisions by the Q, Q.a, Q.p and Q.pa cell make four midbodies (dots in blue, red, green, and purple). (B) Midbody release in the QR lineage with midbodies (ZEN-4/MKLP1 and GFP::ZEN-4) shown and the plasma membrane (mCherry with a myristoylation signal) and histone (his-24::mCherry) shown. Midbody name is adjacent. (C) Still images of the birth, engulfment, and degradation of the Q cell midbody in WT. (D) Lysosome recruitment from the hyp7 cell to the Q cell midbody. (C and D) mCherry labels the Q cell midbody in C and D (top), and GFP marks actin cytoskeleton (C) or lysosome (CTNS-1; D) in the hyp7 cell (middle). Arrows show midbodies, and asterisks in D show the apoptotic cells. The cell name is adjacent. Anterior of the cell is to the left. Time in minutes is given on the top of top row. The birth of the Q cell midbody was the end of cytokinesis of the mother cell (0 min in C and D). The degradation was the last frame showing the Q cell midbody (105 min in C and 90 min in D). (E and F) Enlarged time-lapse views of the dotted boxes (26 min in C and 80 min in D) in which GFP-actin (E) and lysosome (F) from the hyp7 cell were recruited onto the midbody. Bars: (C and D) 5 µm; (E and F) 2.5 µm.
We first examined the fate of the midbody after Q cell divisions by imaging at a single-cell resolution. Because the two daughter cells were well separated as a result of their migration, we were able to determine whether the midbody was associated with the neural progenitor, the apoptotic cell, or shed to extracellular space (Sulston and Horvitz, 1977; Ou and Vale, 2009). We found that GFP::ZEN-4 signals in the QR, QR.a, QR.p, or QR.pa were outside of the cell contour outlined by mCherry-tagged plasma membrane after division, indicating that the midbody remnants in the QR lineage were released to the extracellular space (Fig. 1 B and Video 1). To be different from the cell name, we used mQR, mQR.a, mQR.p, or mQR.pa to name related midbodies. For example, mQR (QR.a and QR.p) is generated by QR division.
The Q cell midbodies are internalized and degraded by the hyp7 cell
We examined midbody dynamics after release. We have recently found that the epithelial hyp7 cell in C. elegans is the phagocyte that internalizes apoptotic Q cell corpse (Li et al., 2012), so we examined the involvement of hyp7 cell in midbody engulfment. A hallmark for Q cell corpse internalization is the formation of an “actin halo” around the apoptotic cell corpse in the hyp7 cell (Li et al., 2012). Strikingly, we observed that GFP-tagged actin, specifically expressed in the hyp7 cell, also formed an actin halo around the Q.a midbody (mQ.a) at 37 ± 13 min (mean, SD; n = 23) after its birth (Fig. 1, C and E; and Video 2), indicating that mQ.a was internalized by the hyp7 cell.
We followed the behavior of Q cell midbody inside the hyp7 cell. After the apoptotic cell corpse is engulfed, the endocytic machinery in the phagocyte, such as RAB GTPases, mediates maturation events, culminating in lysosome delivery and the final digestion of cell corpse (Kinchen and Ravichandran, 2008). We examined whether such events also occur to Q cell midbodies. Indeed, live imaging revealed that GFP-tagged RAB-5 or RAB-7, an early or late endosome component, was recruited onto the mCherry-labeled mQ.a at 70 ± 20 min (n = 20) or 74 ± 22 min (n = 29) from the hyp7 cell after midbody birth (Videos 3 and 4). We further observed that the hyp7 cell lysosome, labeled by CTNS-1::GFP (Li et al., 2012), was recruited onto the Q cell midbody at 89 ± 18 min (n = 18) after its birth, and the CTNS-1::GFP aggregate lasted until the midbody was completely digested (Fig. 1, D and F; and Video 5). We further confirmed that the mQ.a was degraded in the hyp7 cell after internalization by using a different lysosome marker, NUC-1::mCherry (Video 6; Liu et al., 2012).
We systematically analyzed the fate of midbodies after each division in Q cell lineages in wild-type (WT) animals. Midbody remnants in QL and QR lineages had identical fates, and the degradation of Q, Q.a, Q.p, and Q.pa midbody remnants followed the order of their birth (Fig. 2 A). QR midbody (mQR), the first ones to be created in the Q cell lineage, were all eliminated at the three-cell stage of Q cell development (i.e., after Q.ap, Q.paa, and Q.pap were born; Fig. 2, A and C). mQR.a and mQR.p are generated by the second round of asymmetric cell division. 92 and 80% of mQR.a and mQR.p, respectively, disappeared at the three-cell stage, and none of the mQR.a or mQR.p was visible at the three-neuron stage (i.e., when neurites of Q.ap, Q.paa and Q.pap were visible; Fig. 2, A and C). mQR.pa formed in the last Q cell division, and 72% of them were detectable at the three-cell stage, but only 13% remained at the three-neuron stage. The fate and degradation pattern of midbodies from the QL lineage is similar to the fate of midbodies from the QR lineage (Fig. 2, A and C; and Fig. S1 B).
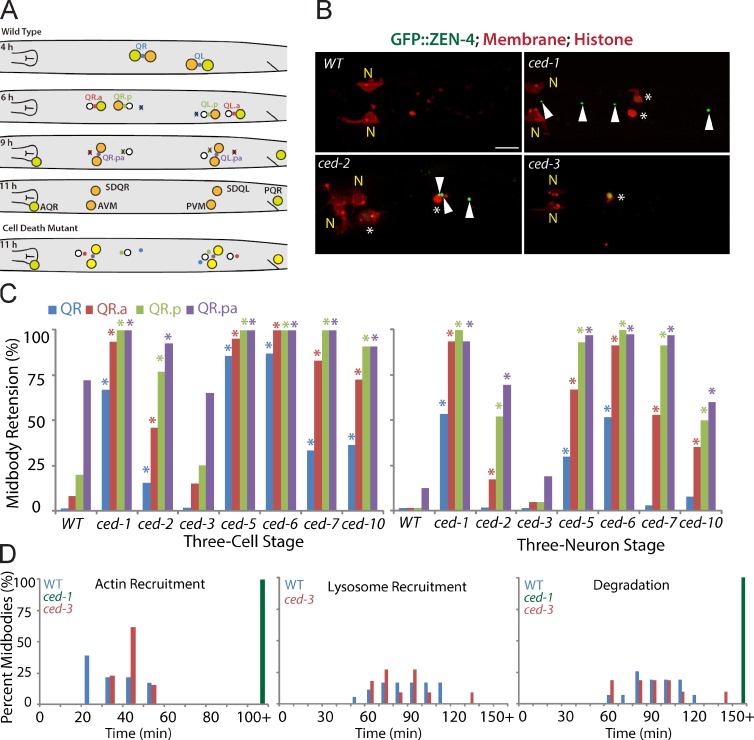
Figure 2.
Apoptotic cell engulfment genes are required for midbody degradation. (A) Cartoon shows the Q cell midbody fate. At 4 h after hatching, the Q midbody was generated by QR or QL division; mQ.a or mQ.p and mQ.pa were generated at 6 or 9 h (three-cell stage), respectively. mQ, mQ.a, mQ.p, or mQ.pa was degraded at 6, 9, or 11 h (three-neuron stage). (bottom) In cell death mutants, the midbody failed to degrade. PVM, posterior ventral microtubule. (B) Fluorescence images of Q cell midbody degradation in WT and cell death mutants. Midbodies (ZEN-4/MKLP1 and GFP::ZEN-4), plasma membrane (mCherry with a myristoylation signal), and histone (his-24::mCherry) are shown. N shows neurons (anterior ventral microtubule [AVM] and SDQR). Arrowheads show persistent midbodies in ced-1 and ced-2 mutants. Asterisks show cell corpses in ced-1 and ced-2 mutants and the nonapoptotic cell in the ced-3 mutant. Bar, 5 µm. (C) Quantifications of midbody degradation in the QR cell lineage in WT and mutants at the three-cell (left) or thee-neuron (right) developmental stage. *, P < 0.01, χ2 test (mutant paired with WT). For each data point, n = 11–45 from a single experiment. (D) Quantifications of the time for Q cell midbody engulfment (actin recruitment in hyp7 cell), lysosome recruitment, and degradation in WT, ced-1, and ced-3 mutants. n = 11–26 from a single experiment. Statistic analysis is shown in Fig. S3.
Apoptotic cell engulfment genes are required for midbody degradation
Given that the Q cell midbody is internalized and degraded by the hyp7 cell, the same phagocyte that digests apoptotic Q cell corpses, we asked whether cell death genes are essential for midbody clearance. Apoptosis is initiated by the activation of a caspase, C. elegans CED-3. Subsequently, two signaling pathways (CED-1, CED-6, and CED-7; CED-2, CED-5, and CED-12) activate a Rac GTPase (CED-10) that reorganizes the actin cytoskeleton to engulf apoptotic cell corpse (Conradt and Xue, 2005). We examined midbody degradation in mutants defective in the aforementioned pathways. In ced-1(e1735), ced-6(n2095), or ced-7(n2094) mutants, the clearance of all Q cell midbodies was significantly delayed; mQR normally disappeared at the three-cell stage, but >65% of mQR remained in ced-1 and ced-6 mutants, or 34% of mQR were retained in ced-7 worms, and about half of the mQR were still visible even at the three-neuron stage in ced-1 and ced-6 mutants. The mQR.a, mQR.p, and mQR.pa also showed significant defects in degradation defective in the ced-1 pathway (Fig. 2, B and C; and Fig. S1 A). All the midbody degradation in the QR lineage was similarly delayed in ced-2(n1994), ced-5(n1812), and ced-10(n3246) mutants (Fig. 2, B and C; and Fig. S1 A). However, midbody degradation in ced-3(n717) mutants was indistinguishable from that of the WT animals (Fig. 2, B and C). A similar impairment in midbody clearance of the QL lineage was observed in the cell corpse engulfment mutants but not ced-3 mutants (Fig. S1 B). Thus, apoptotic cell engulfment genes, but not the caspase, were required for Q cell midbody degradation.
We performed kinetic analysis of Q cell midbody clearance in ced-1 mutants to pinpoint which step was defective. Our time-lapse analysis (>150 min after midbody birth, n = 10) did not detect actin halo formation around the Q cell midbody in ced-1 worms, and the midbody did not degrade (Fig. 2 D; Fig. S2, A and B; and Fig. S3). In contrast, we found that actin halo formation, lysosome recruitment, and the final degradation of the midbody in ced-3 mutants were as normal as those in WT (Fig. 2 D; Fig. S2, A and B; and Fig. S3). Thus, the engulfment of the midbody by the phagocyte was defective in ced-1 but not ced-3 mutants.
PS functions as an engulfment signal for midbody internalization
During apoptosis, externalized PS can function as an engulfment signal to induce cell corpse phagocytosis (Mapes et al., 2012). We asked whether PS also exists on the outer surface of the midbody and acts as an “eat me” signal to trigger midbody engulfment. To test this idea, we used C. elegans heat shock promoters to express two PS-binding proteins, secreted Annexin V (AnxV; sAnxV)::GFP (Fig. 3 A) and secreted GFP (sGFP)::lactadherin C1 and C2 domains (sGFP::LactC1C2; Fig. 3 B; Mapes et al., 2012). Both markers were previously shown to recognize exposed PS (exPS) on apoptotic cell corpses in C. elegans with high sensitivity. We found that the sAnxV::GFP was associated with the mCherry::ZEN-4–tagged Q cell midbody (Fig. S2 C). We then performed time-lapse analysis of exPS dynamics on mQ.a. sAnxV::GFP or sGFP::LactC1C2 was first recruited onto the midbody 26 ± 11 min (n = 11) or 28 ± 9 min (n = 14) after midbody birth (Fig. 3, A–D; and Videos 7 and 8). The recruitment of the two exPS markers on the midbody was indistinguishable but earlier than actin halo formation during midbody engulfment (37 min after midbody birth; Fig. S3 D), suggesting that exPS could provide an engulfment signal for the hyp7 cell to internalize the midbody.
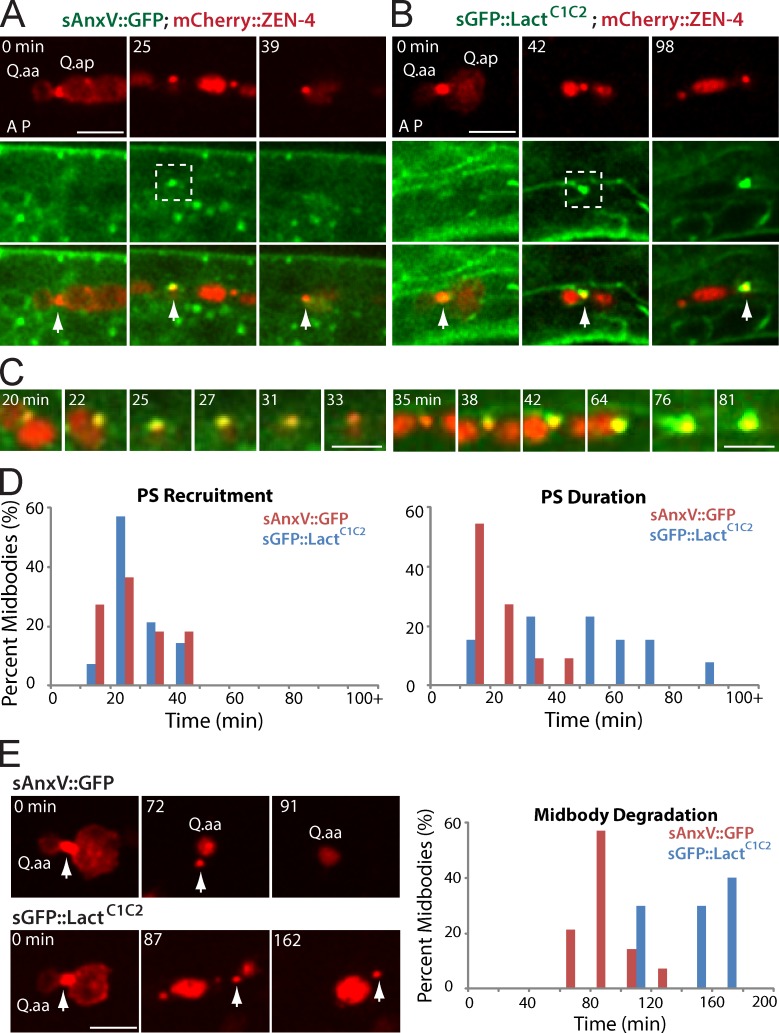
Figure 3.
The recruitment and effect of PS markers to the Q cell midbody. (A and B) Still images of sAnxV::GFP (A) and sGFP::LactC1C2 (B) on the Q cell midbody. mCherry labels the Q cell midbody/ZEN-4 (top), and GFP marks PS (middle). Arrows show the midbody. A, anterior; P, posterior. (C) Enlarged time-lapse views of the dotted boxes (left, 25 min; right, 80 min) in which sAnxV::GFP or sGFP::LactC1C2 was recruited onto the midbody. (D and E) Quantifications of the time for PS recruitment (D) and duration (E) on the Q cell midbody. n = 10–15 from a single experiment. (E) Still images (left) show midbody degradation in the presence of sAnxV::GFP (top) or sGFP::LactC1C2 (bottom). sGFP::LactC1C2 overexpression caused a delay of midbody degradation. Arrows show the midbody. Quantifications are given on the right. Statistic analysis is shown in Fig. S3. Bars: (A and B) 5 µm; (C) 2.5 µm.
We validated the specificity of sAnxV::GFP in C. elegans larvae. For this experiment, we deleted the secretion signal from sAnxV::GFP. We found that AnxV::GFP is largely diffusive, whereas sAnxV::GFP forms puncta (Video 9). Our time-lapse analysis showed that AnxV::GFP was not associated with the Q.a midbody within 1 h since its birth (n = 13; Video 9), suggesting that sAnxV::GFP reliably marks exPS in C. elegans larvae.
We examined the importance of exPS signaling in midbody degradation. In mammalian cells, pretreatment of macrophages with AnxV masks PS and blocks apoptotic cell internalization (Callahan et al., 2003). An expression of sGFP::LactC1C2, but not sAnxV::GFP, inhibits PS signaling and causes a delay of apoptotic cell clearance in C. elegans (Mapes et al., 2012). We measured the duration of sAnxV::GFP and sGFP::LactC1C2 on the midbody. sAnxV::GFP only stained the midbody for 21 ± 10 min (n = 11) after its first appearance, whereas sGFP::LactC1C2 stayed on the midbody for 52 ± 24 min (n = 13; Fig. 3, C and E; and Fig. S3 E). We compared the lifetime of the midbody in the presence of sAnxV::GFP or sGFP::LactC1C2. The midbody was degraded 91 ± 16 min (n = 14) after its birth in the presence of sAnxV::GFP, similar to the WT midbody lifetime (97 min). However, the expression of sGFP::LactC1C2 significantly prolonged midbody lifetime to 145 ± 25 min (n = 10; Fig. 3 E and Fig. S3 C). The association of sGFP::LactC1C2 with exPS on the midbody may block the interaction of exPS with its receptor, inhibiting the downstream signal transduction for midbody internalization. Thus, exPS presence on the midbody is important for midbody degradation.
The requirement of cell death genes for midbody degradation in other C. elegans lineages
To further explore whether cell death genes are required for midbody clearance in other C. elegans lineages, we examined midbody degradation in embryos. Most embryonic cell divisions are completed within 6 h after fertilization, and a C. elegans hermaphrodite generates 671 cells (Sulston et al., 1983) by 670 cell divisions. The rapid generation and large amount of embryonic midbody remnants precluded an accurate analysis of midbody degradation in embryos. We thus counted the number of embryonic midbodies in C. elegans L1 young hatchlings (0–4 h after hatching) because the postembryonic cells do not divide until 4 h after hatching. Using a strain expressing ZEN-4::GFP under its endogenous promoter to track all C. elegans midbody remnants (Kaitna et al., 2000), we found that only 6 ± 3 (n = 10) out of 670 embryonic midbody remnants were visible in L1 young hatchlings, ranging from 2 to 11 (Fig. 4, A and C). In contrast, embryonic midbody clearance was defective in ced-1 mutants, which contained 33 ± 7 (n = 10) midbody remnants in young hatchlings, ranging from 23 to 47 (Fig. 4, A and C). Like that of the Q lineage, embryonic midbody clearance was normal in ced-3 mutants (Fig. 4, A and C).
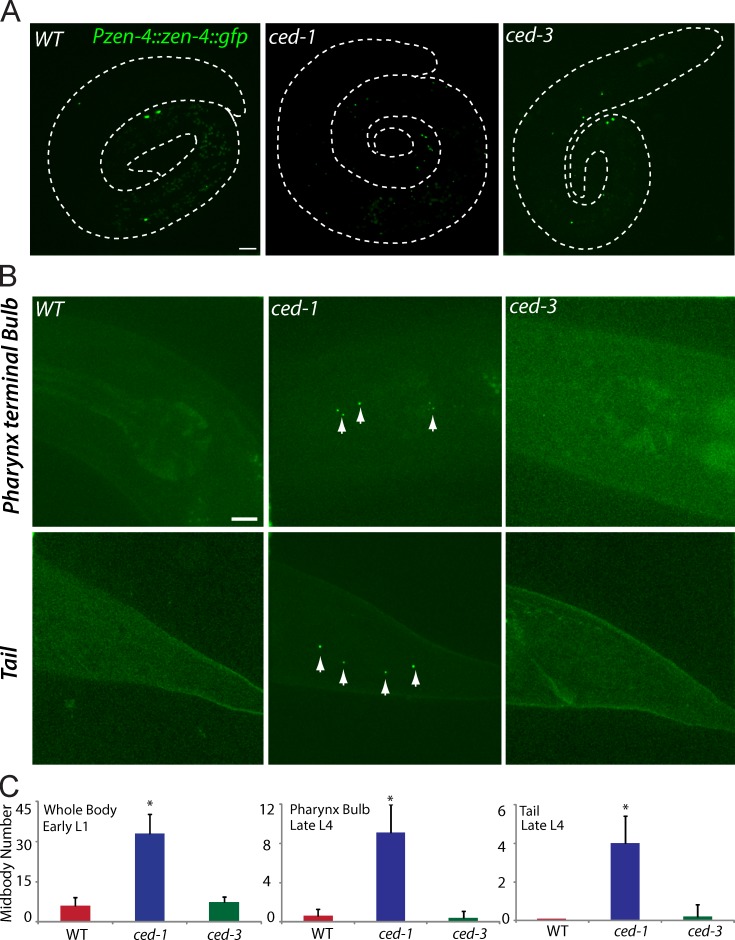
Figure 4.
Midbody degradation in other cell lineages. (A, left) Midbodies from C. elegans embryonic lineages normally degrade when L1 larvae hatch. Persistent midbodies remain in ced-1 (middle) but not in ced-3 mutants (right). Midbodies were visualized by Pzen-4::zen-4::gfp. Dotted lines show the C. elegans periphery. (B) Midbody degradation in the pharynx terminal bulb (top) and tail (bottom) of the late L4 animals in WT (left), ced-1 (middle), and ced-3 (right) mutant. Arrows show the persistent midbodies in ced-1 mutants. (C) Quantifications of midbody degradation in embryonic (left) and postembryonic (pharynx terminal bulb [middle] or tail [right]) lineages. *, P < 0.01, the Student’s t test. Error bars represent SDs. Bars, 5 µm.
We further quantified midbody number in the pharynx terminal bulb and the tail of WT L4 larvae and that in cell death mutants. Zero to one midbody was visible around the pharynx terminal bulb or tail in the WT or ced-3 mutants, but four to nine midbody remnants were detected in ced-1 mutants (n = 10 for each measurement; Fig. 4, B and C). Thus, the requirement of the apoptotic cell engulfment genes in midbody clearance appears to be a general mechanism in C. elegans embryonic and postembryonic lineages.
Conclusion
In summary, our findings show that the midbody generated in divisions by cells of the Q cell lineage in C. elegans is released after abscission and appears to use the same molecular and cellular mechanisms—namely the same “eat me” signal and the phagocyte—as those for an apoptotic cell for engulfment and degradation (Fig. 5). Although cell death genes are involved in midbody degradation, each of them may contribute to this process differently. For example, the defects of midbody clearance in ced-7, ced-2, and ced-10 mutants were less severe than ced-1, ced-6, and ced-5 mutants (Fig. 2 C and Fig. S1 B); TTR-52, a PS-binding bridging molecule, critical for apoptotic cell corpse degradation, was not essential for midbody clearance (unpublished data; Mapes et al., 2012); and ced-1 and ced-2 function in two parallel pathways in cell corpse clearance, but ced-1; ced-2 double mutants did not enhance the ced-1 single mutant phenotype in midbody degradation (Fig. S3 F). Such differences might be caused by the different sizes or other properties between cell corpses and midbodies. The diameter of cell corpses is ∼3 µm, whereas the diameter of the midbody is only ∼0.5 µm (Fig. 1, B–D; Mullins and Biesele, 1977). The smaller size of the midbody may make it an easier target to be engulfed, even without PS bridging molecules or with less help from other factors.
Figure 5.
Apoptotic mimicry in midbody degradation. An apoptotic cell externalizes phosphatidylserine (PS) as an engulfment signal to activate two signaling pathways (CED-1, -6, and -7; CED-2, -5, and -10) for apoptotic cell corpse internalization. The same PS signal, transduction pathways, and phagocyte are used for midbody engulfment and degradation.
Our finding of Q cell midbody release is consistent with the notion that the midbody is shed or degraded in the differentiating cells (Ettinger et al., 2011; Kuo et al., 2011; Schink and Stenmark, 2011) because Q cells differentiate into neurons after three rounds of division. Interestingly, the midbody formed by the C. elegans first embryonic cell division is retained by the P1 cell (Oegema, K., and R. Green, personal communication; Raich et al., 1998), suggesting that it may not use apoptotic mimicry for degradation. Midbody remnants can be degraded by autophagy in mammalian cells (Pohl and Jentsch, 2009), so it is possible that distinct mechanisms are used to degrade midbodies, depending on specific cell types or development stages.
Nevertheless, our finding expands the nonapoptotic functional repertoire of cell death genes. A range of cellular processes require the nonapoptotic function of cell death genes (Galluzzi et al., 2012; Hyman and Yuan, 2012). For example, during axon pruning or neural injury, Draper/CED-1 and CED-6 are required for the degradation of axon debris (Awasaki et al., 2006; Hoopfer et al., 2006; MacDonald et al., 2006), whereas the entry of vaccinia virus into the host cell critically depends on the exPS on the viral membrane (Mercer and Helenius, 2008). However, the engulfment signal from the damaged axons is largely unknown, and whether cell death genes are involved in viral entry was not examined. Our study has identified both PS signaling and apoptotic cell engulfment genes as essential components for midbody internalization, suggesting that a complete apoptotic mimicry functions in this process. The same genes may also play a broader role in engulfing other exPS-containing cellular debris; many exPS-positive particles are detectable in our live-imaging analysis of C. elegans larva (Videos 7 and 8).
Although vertebrates carrying mutations of cell death genes exhibit severe development disorders (Schafer and Kornbluth, 2006), apoptotic cell corpse engulfment genes in C. elegans are not essential for the viability of this organism (Conradt and Xue, 2005). Thus, the functional importance of midbody clearance in C. elegans remains unclear. It is, however, worth pointing out that midbody degradation was known to be associated with human lysosomal storage disorders or mammalian cell fate determination (Pohl and Jentsch, 2009; Ettinger et al., 2011; Kuo et al., 2011; Schink and Stenmark, 2011).
Materials and methods
C. elegans strains, genetics, and DNA manipulations
We raised C. elegans strains on nematode growth media plate seeded with the Escherichia coli strain OP50 at 20°C. Tables S1 and S2 show the information of strain genotype, primers, and plasmids. To make transgenic C. elegans by germline transformation, 20–50 ng/µl DNA and a unc-76–rescuing plasmid were microinjected into unc-76(e911) animals, and γ-ray irradiation was used to integrate transgenes into the chromosome.
Live-cell imaging
We anesthetized C. elegans L1 larva with 0.1 mmol/liter levamisole in M9 buffer and mounted them on 3% agarose pads at 20°C. Our imaging system consists of a microscope (Axio Observer.Z1; Carl Zeiss) equipped with a 100×, 1.45 NA objective, an electron multiplying charge-coupled device camera (iXon+ series DU-897D-C00-#BV-500; Andor Technology), and the 488- and 568-nm lines of a continuous wave Center for Devices and Radiological Health universal serial bus laser system (Sapphire; Coherent, Inc.) attached to a spinning-disk confocal scan head (CSU-X1; Yokogawa Corporation of America). Time-lapse images were acquired with an exposure time of 300 msec every 30–60 s by Focus Image software (developed by X. Zhang, Institute of Biophysics, Chinese Academy of Sciences, Beijing, China). Images were processed by ImageJ software (National Institutes of Health).
We quantified the midbody persistence at the three-cell or three-neuron stage of Q neuroblast development. Each stage was defined in the Results and discussion section. Because the midbody is immotile and Q cells migrate after asymmetric divisions, we determined the midbody of Q, Q.a, Q.p, and Q.ap based on the distance from the birthplace. For example, mQR is the most posterior, and mQR.ap is the most anterior in the QR lineage. mQR.a is more anterior than mQR.p.
Heat shock treatment
Transgenic L1 larvae (0–2 h after hatching) were placed at 33°C for 45 min followed by a recovery of 2–3 h at 20°C.
Statistical analysis
We used the Student’s t test and χ2 analysis to examine statistical differences of the midbody degradation, actin and lysosome recruitment, and PS recruitment and duration in WT and different genetic backgrounds as indicated in the figure legends.
Online supplemental material
Fig. S1shows Q cell midbody degradation in cell death mutants. Fig. S2 shows actin, lysosome, and PS recruitment on the midbody. Fig. S3 shows statistic analysis of Q cell midbody degradation in WT and different genetic backgrounds. Table S1 shows C. elegans strains used in this study. Table S2 shows primers and plasmids for C. elegans transgenesis. Video 1 shows the birth and release of QR.a and QR.p midbodies. Video 2 shows the engulfment of the Q.a cell midbody by the hyp7 cell. Videos 3–8 show the recruitment RAB-5 (Video 3), RAB-7 (Video 4), CTNS-1 (Video 5), NUC-1 (Video 6), sAnxV::GFP (Video 7), and sGFP::LactC1C2 (Video 8) onto the Q.a midbody. Video 9 shows the dynamics of AnxV::GFP in the C. elegans larva. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201209050/DC1.
Supplementary Material
Acknowledgments
We thank Dr. X. Wang (National Institute of Biological Sciences, Beijing, China), C. Yang (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China), R. Vale (University of California, San Francisco, San Francisco, CA), D. Xue (University of Colorado Boulder, Boulder, CO), and the Caenorhabditis Genetics Center for reagents and strains and Drs. P. Liu, W. Zhong, G. Garriga, Z. Bao, K. Shen, and K. Oegema for helpful discussion.
This work was supported by the National Basic Research Program of China to W. Li and G. Ou (973 Program, 2012CB966800, 2012CB945002, and 2013CB945600), the National Natural Science Foundation of China to W. Li, Y. Yang, Y. Chai, and G. Ou (31101002, 31100972, 31171295, 31190063, 31201009, and 31222035), and the Junior Thousand Talents Program of China to G. Ou.
Footnotes
Abbreviations used in this paper:
- AnxV
- Annexin V
- exPS
- exposed PS
- PS
- phosphatidylserine
- sAnxV
- secreted AnxV
- sGFP
- secreted GFP
- WT
- wild type
References
- Awasaki T., Tatsumi R., Takahashi K., Arai K., Nakanishi Y., Ueda R., Ito K. 2006. Essential role of the apoptotic cell engulfment genes draper and ced-6 in programmed axon pruning during Drosophila metamorphosis. Neuron. 50:855–867 10.1016/j.neuron.2006.04.027 [DOI] [PubMed] [Google Scholar]
- Callahan M.K., Halleck M.S., Krahling S., Henderson A.J., Williamson P., Schlegel R.A. 2003. Phosphatidylserine expression and phagocytosis of apoptotic thymocytes during differentiation of monocytic cells. J. Leukoc. Biol. 74:846–856 10.1189/jlb.0902433 [DOI] [PubMed] [Google Scholar]
- Conradt B. 2009. Genetic control of programmed cell death during animal development. Annu. Rev. Genet. 43:493–523 10.1146/annurev.genet.42.110807.091533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conradt B., Xue D. 2005. Programmed cell death. WormBook. 10.1895/wormbook.1.34.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia N., Sougrat R., Spurlin T.A., Hurley J.H., Lippincott-Schwartz J. 2011. Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc. Natl. Acad. Sci. USA. 108:4846–4851 10.1073/pnas.1102714108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettinger A.W., Wilsch-Bräuninger M., Marzesco A.M., Bickle M., Lohmann A., Maliga Z., Karbanová J., Corbeil D., Hyman A.A., Huttner W.B. 2011. Proliferating versus differentiating stem and cancer cells exhibit distinct midbody-release behaviour. Nat Commun. 2:503 10.1038/ncomms1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L., Kepp O., Trojel-Hansen C., Kroemer G. 2012. Non-apoptotic functions of apoptosis-regulatory proteins. EMBO Rep. 13:322–330 10.1038/embor.2012.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M. 2009. The 3Ms of central spindle assembly: microtubules, motors and MAPs. Nat. Rev. Mol. Cell Biol. 10:9–20 10.1038/nrm2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoopfer E.D., McLaughlin T., Watts R.J., Schuldiner O., O’Leary D.D., Luo L. 2006. Wlds protection distinguishes axon degeneration following injury from naturally occurring developmental pruning. Neuron. 50:883–895 10.1016/j.neuron.2006.05.013 [DOI] [PubMed] [Google Scholar]
- Hyman B.T., Yuan J. 2012. Apoptotic and non-apoptotic roles of caspases in neuronal physiology and pathophysiology. Nat. Rev. Neurosci. 13:395–406 10.1038/nrn3228 [DOI] [PubMed] [Google Scholar]
- Kaitna S., Mendoza M., Jantsch-Plunger V., Glotzer M. 2000. Incenp and an aurora-like kinase form a complex essential for chromosome segregation and efficient completion of cytokinesis. Curr. Biol. 10:1172–1181 10.1016/S0960-9822(00)00721-1 [DOI] [PubMed] [Google Scholar]
- Kinchen J.M., Ravichandran K.S. 2008. Phagosome maturation: going through the acid test. Nat. Rev. Mol. Cell Biol. 9:781–795 10.1038/nrm2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo T.C., Chen C.T., Baron D., Onder T.T., Loewer S., Almeida S., Weismann C.M., Xu P., Houghton J.M., Gao F.B., et al. 2011. Midbody accumulation through evasion of autophagy contributes to cellular reprogramming and tumorigenicity. Nat. Cell Biol. 13:1214–1223 10.1038/ncb2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Zou W., Yang Y., Chai Y., Chen B., Cheng S., Tian D., Wang X., Vale R.D., Ou G. 2012. Autophagy genes function sequentially to promote apoptotic cell corpse degradation in the engulfing cell. J. Cell Biol. 197:27–35 10.1083/jcb.201111053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Du H., Rutkowski R., Gartner A., Wang X. 2012. LAAT-1 is the lysosomal lysine/arginine transporter that maintains amino acid homeostasis. Science. 337:351–354 10.1126/science.1220281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan M.A., Hackett R., Doherty J., Sheehan A., Speese S.D., Freeman M.R. 2012. Negative regulation of glial engulfment activity by Draper terminates glial responses to axon injury. Nat. Neurosci. 15:722–730 10.1038/nn.3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald J.M., Beach M.G., Porpiglia E., Sheehan A.E., Watts R.J., Freeman M.R. 2006. The Drosophila cell corpse engulfment receptor Draper mediates glial clearance of severed axons. Neuron. 50:869–881 10.1016/j.neuron.2006.04.028 [DOI] [PubMed] [Google Scholar]
- Mapes J., Chen Y.Z., Kim A., Mitani S., Kang B.H., Xue D. 2012. CED-1, CED-7, and TTR-52 regulate surface phosphatidylserine expression on apoptotic and phagocytic cells. Curr. Biol. 22:1267–1275 10.1016/j.cub.2012.05.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer J., Helenius A. 2008. Vaccinia virus uses macropinocytosis and apoptotic mimicry to enter host cells. Science. 320:531–535 10.1126/science.1155164 [DOI] [PubMed] [Google Scholar]
- Mullins J.M., Biesele J.J. 1977. Terminal phase of cytokinesis in D-98s cells. J. Cell Biol. 73:672–684 10.1083/jcb.73.3.672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou G., Vale R.D. 2009. Molecular signatures of cell migration in C. elegans Q neuroblasts. J. Cell Biol. 185:77–85 10.1083/jcb.200812077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou G., Stuurman N., D’Ambrosio M., Vale R.D. 2010. Polarized myosin produces unequal-size daughters during asymmetric cell division. Science. 330:677–680 10.1126/science.1196112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl C., Jentsch S. 2009. Midbody ring disposal by autophagy is a post-abscission event of cytokinesis. Nat. Cell Biol. 11:65–70 10.1038/ncb1813 [DOI] [PubMed] [Google Scholar]
- Raich W.B., Moran A.N., Rothman J.H., Hardin J. 1998. Cytokinesis and midzone microtubule organization in Caenorhabditis elegans require the kinesin-like protein ZEN-4. Mol. Biol. Cell. 9:2037–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer Z.T., Kornbluth S. 2006. The apoptosome: physiological, developmental, and pathological modes of regulation. Dev. Cell. 10:549–561 10.1016/j.devcel.2006.04.008 [DOI] [PubMed] [Google Scholar]
- Schink K.O., Stenmark H. 2011. Cell differentiation: midbody remnants - junk or fate factors? Curr. Biol. 21:R958–R960 10.1016/j.cub.2011.10.035 [DOI] [PubMed] [Google Scholar]
- Steigemann P., Gerlich D.W. 2009. Cytokinetic abscission: cellular dynamics at the midbody. Trends Cell Biol. 19:606–616 10.1016/j.tcb.2009.07.008 [DOI] [PubMed] [Google Scholar]
- Sulston J.E., Horvitz H.R. 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56:110–156 10.1016/0012-1606(77)90158-0 [DOI] [PubMed] [Google Scholar]
- Sulston J.E., Schierenberg E., White J.G., Thomson J.N. 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100:64–119 10.1016/0012-1606(83)90201-4 [DOI] [PubMed] [Google Scholar]
- Yang D., Rismanchi N., Renvoisé B., Lippincott-Schwartz J., Blackstone C., Hurley J.H. 2008. Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Nat. Struct. Mol. Biol. 15:1278–1286 10.1038/nsmb.1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.