ABSTRACT
Respiratory syncytial virus (RSV) is the most common viral cause of severe lower respiratory tract illness in infants and children. The virus replicates in polarized epithelial cells in the airway and, to a lesser extent, infects airway antigen-presenting cells, such as dendritic cells (DCs). RSV possesses a number of expressed genes that antagonize the effect of type I interferons and other related host factor pathways that inhibit replication efficiency. Virus infection alters host gene transcription and the translation of host transcripts through specific antagonism of the function of host proteins, through induction of RNA stress granules, and through induction of altered patterns of host gene expression. In healthy cells, microRNAs (miRNAs) regulate gene expression by targeting the noncoding region of mRNA molecules to cause silencing or degradation of transcripts. It is not known whether or not RSV infection alters the level of microRNAs in cells. We profiled the pattern of expression of host cell microRNAs in RSV-infected epithelial cells or DCs and found that RSV did alter microRNA expression but in a cell-type-specific manner. The studies showed that let-7b was upregulated in DCs, while let-7i and miR-30b were upregulated in epithelial cells in a process that required viral replication. Interestingly, we found that the RSV nonstructural genes NS1 and NS2 antagonized the upregulation of let-7i and miR-30b. RSV appears to manipulate host cell gene expression through regulation of expression of miRNAs related to the interferon response. The data suggest a new mechanism of virus-host cell interactions for paramyxoviruses.
IMPORTANCE
Respiratory syncytial virus (RSV) is the most common cause of serious lower respiratory tract illness in infants and children. The human innate immune response inhibits RSV replication early after inoculation, principally through the effect of substances called interferons. The virus, however, has developed several mechanisms for counteracting the host innate immune response. It is not known whether or not RSV infection alters the expression of host microRNAs, which are short RNA sequences that are posttranscriptional regulators. This paper shows that RSV does induce unique patterns of microRNA expression related to the NF-κB pathway or interferon pathways. The microRNA profiles differed depending on the cell type that was infected, airway cell or antigen-presenting cell. Interestingly, the virus appears to counteract the microRNA response by expressing nonstructural viral genes in the cell that reduce microRNA induction. The data suggest a new way in which paramyxoviruses regulate the host cell response to infection.
Introduction
Respiratory syncytial virus (RSV) is an enveloped negative-strand RNA virus in the family Paramyxoviridae (1). RSV is spread by large-particle aerosol droplets or direct contact, with infection initiating in the nasopharynx and then spreading to the lower respiratory tract (1). RSV mainly infects cells of the nasopharynx and lung but also can be detected in circulating mononuclear cells (2). RSV infects all age groups, but it rarely causes severe disease in otherwise healthy adults. In infants and the elderly, RSV infection can cause severe bronchiolitis or pneumonia (1). While mild disease does not require medical intervention, severe disease may require mechanical removal of secretions, humidified oxygen treatment, or mechanical ventilation (1). An effective RSV vaccine is not yet available.
MicroRNAs (miRNAs) are a class of noncoding RNAs with characteristic, complex secondary structures that are conserved evolutionarily in plants, invertebrates, and vertebrates. Target RNAs are identified through the 5′ 6- to 8-nucleotide seed sequence of the mature miRNAs (3). It has been hypothesized that each mature miRNA can recognize about 100 to 200 cellular transcripts (3). miRNAs act as part of the antiviral response in plants and invertebrates and exert their effects through a block in translation or direct degradation of target mRNAs (4). miRNAs also have antiviral responses in vertebrates. For example, the human microRNA hsa-miR-125a-5p interferes with expression of hepatitis B virus surface antigen to control viral replication (5).
miRNAs are categorized into families and clusters (6). Members of families of miRNAs have the same or similar seed sequences, and therefore, they may target the same genes. miRNAs are named through their family association. For example, members of the let-7 family of miRNAs have the nucleotide seed sequence 5′ UGAGGUAG 3′. Clusters of miRNAs are located in close proximity on chromosomes and can be regulated cotranscriptionally. Controlling expression of groups of miRNAs within clusters likely allows cells to regulate biological processes very quickly.
A small but growing body of literature indicates that mammals may respond with miRNAs as a part of the inflammatory response mounted against pathogens that includes factors such as interferons (IFNs). Beta interferon (IFN-β) treatment induced human miR-155 expression in cells in a Jun N-terminal protein kinase (JNK)-dependent fashion, possibly dependent upon tumor necrosis factor alpha (TNF-α) (7). Multiple human miRNAs are produced in response to IFN-β, with seed sequences that potentially target hepatitis C virus gene products and block replication of the virus in culture (8). Other human miRNAs are expressed differentially in resting versus activated T cells, a profile that has been hypothesized to promote HIV latency (9). RSV infection induces secretion of multiple proinflammatory cytokines, including, but not limited to, type I and type II IFNs, TNF-α, interleukin-12 (IL-12), and IL-6 (10–14). A recent publication has examined miRNA responses after RSV infection of human bronchial epithelial cells and found that several miRNAs were downregulated, including miRNA-221, which led to an increase in nerve growth factor (NGF) (15). Another group has recently published that RSV regulates miRNAs which affect viral replication (16).
RSV infection induces production of the type I IFNs IFN-α and -β (1, 17); however, induction is modest in comparison to that caused by many other RNA viruses (1, 10, 13, 18). RSV antagonizes IFN signaling by multiple mechanisms. RSV nonstructural proteins 1 and 2 (NS1 and NS2) block phosphorylation of interferon regulatory factor 3 (IRF-3) (2, 17). NS2, but not NS1, also blocks IFN signaling by decreasing expression of STAT2 (1, 19). The antiviral small RNA helicase retinoic acid-inducible gene I (RIG-I) binds the RSV genome and induces IFN-β production (1, 20). However, NS2 antagonizes IFN-β production through an interaction with RIG-I (3, 21). RSV N protein interacts with MDA5 and locates in close proximity to mitochondrial antiviral signaling protein (MAVS) in cytoplasmic granules termed viral inclusion bodies, thereby attenuating the IFN-β response (3, 22). Additionally, RSV G protein inhibits TLR3/4-mediated IFN-β production (4, 23).
We hypothesized that RSV infection may upregulate cellular miRNA responses through an IFN-dependent mechanism. In this study, we identified miRNAs that were increased after RSV infection of primary dendritic cells (DCs) or normal human bronchial epithelial (NHBE) cells. Interestingly, the data revealed three modes of miRNA activation. In NHBE cells, let-7i and miR-30b were upregulated in a process that required viral replication. Inoculation of cells with RSV strains lacking NS1 or NS2 genes further upregulated let-7i and miR-30b. These data suggest that RSV NS1 and NS2 proteins normally antagonize the upregulation of miRNA expression in RSV-infected epithelial cells. In DCs, let-7b was upregulated. The induction of let-7b in DCs appeared to be enhanced by IFN in the inoculum, since inoculation of DCs with virus produced in IFN-deficient Vero cells exhibited reduced induction of let-7b.
RESULTS
After inoculation with RSV, let-7b was increased in MDDCs and let-7c, let-7i, and miR-30b were increased in NHBE cells.
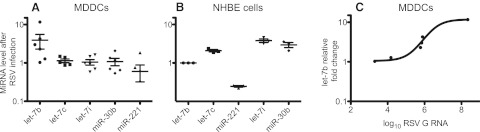
We hypothesized that RSV infection may upregulate specific miRNAs through induction of secretion of cytokines. We examined miRNA responses to RSV with miRNA microarrays using monocyte-derived dendritic cells (MDDCs) or primary NHBE cells following inoculation with RSV. The miRNA microarray assays were performed on two biological replicates of MDDCs and NHBE cells. We examined NHBE cells because bronchial epithelial cells are the primary targets for RSV infection in vivo. MDDCs were examined because they are productively infected by RSV and because they secrete cytokines that are likely to induce miRNA expression. The arrays suggested that several miRNAs were increased at least 2-fold or more in MDDCs after inoculation with RSV. We used quantitative PCR tests to measure the effect of RSV infection on candidate miRNAs with more accuracy. PCR validation has been performed on 8 biological replicates of MDDCs and 3 biological replicates of NHBE cells. Using the real-time PCR tests, we validated that let-7b was increased in MDDCs and let-7c, let-7i, and miR-30b were increased in NHBE cells following inoculation with RSV (Fig. 1). In MDDCs, let-7b was increased 2.7-fold, with a P value of 0.03 as measured by the Wilcoxon signed-rank test, comparing the actual mean to the theoretical mean of 1.0, or no change in let-7b (Fig. 1A). The other miRNAs tested all had P values greater than 0.2 in the real-time PCR validation assays. Two of the six MDDC samples tested did not exhibit increased let-7b. In our experience DCs exhibit a variable permissivity for infection with RSV. We tested for the presence of the RNA for the RSV G gene as a marker for viral infection and plotted those values against the level of the miRNA let-7b (Fig. 1C). The two samples that did not exhibit let-7b upregulation also did not have detectable RSV G RNA, suggesting that let-7b induction required productive infection by RSV in these cells.
FIG 1 .
let-7b is increased in MDDCs and let-7c, let-7i, and miR-30b are increased in NHBE cells after RSV inoculation. Cells were inoculated with RSV and harvested 48 h later. The microRNAs let-7b, let-7c, let-7i, miR-30b, and miR-221 were assayed in MDDCs (A) or NHBE cells (B) by real-time PCR. The relative fold change of the miRNAs was calculated by normalizing values against the GAPDH control in comparison to mock-infected cells. Data are plotted as means; error bars represent the standard errors of the means. Each point represents a biological replicate, and graphs are representative of three technical replicates. The RSV G RNA level also was assayed by real-time PCR in inoculated MDDCs and plotted as log10-transformed relative RNA level in comparison to mock-infected cells. The let-7b miRNA level was plotted against the RSV G RNA level (C).
In NHBE cells, miRNA activation was tested after inoculation with RSV in comparison to mock-infected cells (Fig. 1B). let-7b, which was the principal miRNA altered in DCs, was unchanged in NHBE cells. Three miRNAs, let-7c, let-7i, and miR-30b, increased in RSV-infected cells, while one, miR-221, decreased in response to RSV. Comparing each miRNA against the unchanged let-7b, all miRNAs tested were changed significantly in epithelial cells: let-7c had a P value of 0.0027, miR-221 had a P value of 0.0002, let-7i had a P value of 0.0023, and miR-30b had a P value of 0.0123. RSV infection and G detection did not vary between repetitions, and therefore, we were unable to correlate G expression with miRNA overexpression.
Activation of let-7i and miR-30b depended upon viral dose and viral replication.
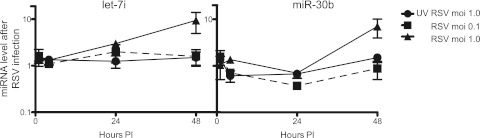
We chose to study further let-7i and miR-30b in NHBE cells, because they were induced most robustly. In order to begin to study the mechanism of miRNA induction after RSV infection, we determined the kinetics of induction and examined the effects of the inoculation viral dose and the presence of active replication in NHBE cells. NHBE cells were inoculated at a multiplicity of infection (MOI) of 0.1 or 1.0 with replication-competent virus or at an MOI of 1.0 with UV-inactivated virus. RNA was collected for miRNA analysis at 0, 1, 24, or 48 h postinoculation. Induction of both let-7i and miR-30b depended both on the dose of virus in the inoculum and on active viral replication (Fig. 2). let-7i increased at 24 and 48 h postinoculation, while miR-30b exhibited a lag of induction until 48 h postinoculation (Fig. 2). These data suggest that let-7i and miR-30b may be induced by different mechanisms. It was not possible to perform these experiments in MDDCs, because the number of cells needed for testing exceeded the number of DCs that are feasible to collect from individual donors using conventional high-volume peripheral blood collection.
FIG 2 .
Activation of let-7i or miR-30b depended on viral dose and active replication. NHBE cells were inoculated with live RSV that was not UV inactivated or with UV-inactivated RSV. RNAs were harvested 1, 24, or 48 h later and were assayed for miRNA expression by real-time PCR. The relative fold change of let-7i or miR-30b was calculated by normalizing values against the GAPDH control in comparison to mock-infected cells. Three technical replicates of three biological replicates were performed. Data are plotted as means; error bars represent the standard errors of the means. PI, postinoculation.
Activation of let-7b and let-7i correlated with levels of IFN-β in viral stocks.
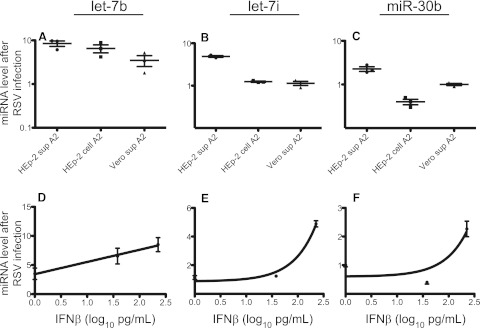
It was reported previously that let-7b and miR-30b were induced in cells treated with IFN-β (5, 8, 24, 25). RSV induces secretion of the type I interferons IFN-α and IFN-β (6, 10, 18, 26, 27). Therefore, we hypothesized that RSV may induce miRNAs through a mechanism involving type I IFNs. In order to test this hypothesis, we inoculated MDDCs or NHBE cells with (i) RSV collected from the supernatants of infected HEp-2 cells, (ii) cell-associated RSV from infected HEp-2 cells, or (iii) RSV collected from the supernatants of infected Vero cells. Vero cells are IFN deficient, and therefore, virus suspensions produced in these cells should not contain any IFN (7, 28, 29). IFN-β levels in viral preparations were assayed by enzyme-linked immunosorbent assay (ELISA). HEp-2 cell culture monolayer supernatants with RSV wild-type strain A2 contained 226 pg/ml IFN-β; the suspension of RSV strain A2 obtained from the cell-associated fraction of a HEp-2 cell culture contained 38 pg/ml IFN-β. RSV A2 suspension from Vero cell culture did not contain any detectable IFN-β (level of detection, 25 pg/ml IFN-β). At the time of inoculation, NHBE medium did not contain detectable levels of IFN-β in any of the biological replicates and rose to only 10 pg/ml in mock-treated cells. let-7b was assayed in separate cultures of MDDCs inoculated with each of the three viral preparations (Fig. 3A). Similarly, let-7i (Fig. 3B) and miR-30b (Fig. 3C) were assayed in NHBE cells inoculated with each of the three viral preparations. miRNA activation also was plotted against IFN-β levels to determine if miRNA expression correlated with the level of IFN (Fig. 3D to F). In MDDCs, let-7b was induced 8.5-fold in cells infected with RSV collected from HEp-2 cell supernatant (HEp-2 sup A2), 6.5-fold in cells infected with RSV collected from HEp-2 cell pellet fraction (HEp-2 cell A2), and 3.5-fold in cells infected with RSV collected from Vero cell supernatant (Vero sup A2) (Fig. 3A). The presence of IFN-β in viral preparations did correlate with an increase in let-7b induction. A comparison of the levels induced by HEp-2 sup A2 and Vero sup A2 yielded a P value of 0.017, while the comparison of HEp-2 cell A2 with Vero sup A2 yielded a P value of 0.015. When let-7b induction was plotted against log10-transformed IFN-β levels, a linear relationship between IFN-β levels and let-7b induction was observed with a significantly (P = 0.015) nonzero slope; however, the curve fit was not strong, with an r2 value of 0.6 (Fig. 3D). Furthermore, despite the absence of IFN in virus suspensions collected from Vero cells, induction of let-7b was reduced but not abrogated. These data suggest that let-7b induction is augmented by IFN during RSV infection but does not depend solely upon IFN.
FIG 3 .
Induction of let-7b or let-7i expression correlated with levels of IFN-β in viral inoculum. MDDCs or NHBE cells were inoculated with RSV collected from HEp-2 cell supernatants (HEp-2 sup A2), HEp-2 cell pellets (HEp-2 cell A2), or Vero cell supernatants (Vero sup A2). RNAs were harvested 48 h later and were assayed for let-7b, let-7i, or miR-30b expression by real-time PCR. The relative fold change of let-7b in MDDCs (A), let-7i in NHBE cells (B), or miR-30b in NHBE cells (C) was calculated by normalizing values against the GAPDH control in comparison to mock-infected cells. IFN-β in the viral stocks was assayed by ELISA. let-7b in MDDCs (D), let-7i in NHBE cells (E), or miR-30b in NHBE cells (F) was plotted against IFN-β.
Infection of NHBE cells with virus collected from HEp-2 cell pellet or Vero cell supernatant abrogated let-7i induction, indicating that RSV-associated let-7i induction may depend strictly upon IFN (Fig. 3B). When let-7i induction was plotted against the log10-transformed IFN-β levels, let-7i induction exhibited a nonlinear relationship to IFN-β levels, with a strong curve fit value (r2 = 0.96) (Fig. 3E). This finding confirmed a correlation of let-7i induction in NHBE cells with the presence of IFN-β. Other factors, however, could be contributing to let-7i induction, such as the presence of noninfectious viral particles. Conversely, infection of NHBE cells with virus collected from HEp-2 cell pellet caused a decrease in miR-30b that did not occur in NHBE cells infected with Vero cell supernatant (Fig. 3C). These data, in combination with plotting miR-30b induction against log10-transformed IFN-β levels, suggested that the level of miR-30b induction did not correlate with IFN-β levels (Fig. 3F).
Deletion of NS1 and NS2 augmented RSV activation of let-7i and miR-30b.
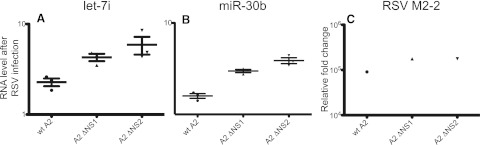
The ability of RSV NS1 and NS2 to antagonize type I IFN signaling is well established (8, 17, 19, 20). Comparison of the level of miRNA induction by RSV produced in HEp-2 cells with that in Vero cells indicated that let-7i induction is associated with the induction of IFN but that miR-30b levels are not. To further examine the association, we inoculated cells at an MOI of 0.5 with wild-type RSV A2 (A2), RSV A2 deleted for NS1 (RSV A2 ΔNS1), or RSV A2 deleted for NS2 (RSV A2 ΔNS2). IFN-β was assayed in the culture supernatant infected with A2, RSV A2 ΔNS1, and RSV A2 ΔNS2. Levels were 175, 429, and 398 pg/ml, respectively. Consistent with the above data indicating that let-7i induction correlated with IFN-β levels, infection of cells with virus deleted for IFN antagonists yielded even higher levels of induction of let-7i (Fig. 4A). When activation of let-7i was measured in cells inoculated with RSV A2 ΔNS1 (P = 0.011) or RSV A2 ΔNS2 (P = 0.05), expression was increased significantly with both deletion mutant viruses in comparison to that in cells infected with wild-type RSV. Expression of miR-30b was increased significantly in cells infected with RSV A2 ΔNS1 (P = 0.0.008) or RSV A2 ΔNS2 (P = 0.0016), compared to that in cells infected with wild-type RSV A2 (Fig. 4B). These data indicate that miR-30b induction does not correlate with IFN-β levels, and therefore, it is surprising that infection of NHBE cells with virus deleted for IFN antagonists also augmented miR-30b induction. The results suggest that miR-30b may be induced via an IFN-independent mechanism that also is antagonized by RSV NS1 and NS2 proteins. To confirm that wild-type RSV and the two deletion mutants replicated to approximately equal levels, the level of RSV M2-2 transcript was quantified by a real-time reverse transcription-PCR (RT-PCR) test. Equivalent levels of M2-2 RNA were detected in cells inoculated with wild-type RSV A2, RSV A2 ΔNS1, or RSV A2 ΔNS2 (Fig. 4C). It should be noted that NS1 and NS2 affect other cytokines and chemokines, and therefore, it remains possible that NS1 and NS2 may contribute to miRNA activation of other cytokines as well.
FIG 4 .
Infection with RSV mutant viruses lacking NS1 or NS2 genes induced more expression of let-7i or miR-30b than did infection with wild-type virus. NHBE cells were inoculated at an MOI of 0.5 with wild-type RSV (wt A2) or RSV with NS1 (A2 ΔNS1) or NS2 (A2 ΔNS2) deleted, and RNA was harvested 48 h later. let-7i (A) or miR-30b (B) was assayed by real-time PCR. RNA for the RSV gene M2-2 also was assayed by real-time PCR (C). Data have been confirmed with three biological replicates. Graphs are plotted as the relative fold change after GAPDH normalization in comparison to mock-infected cells. Data are plotted as means; error bars represent the standard errors of the means.
Expression of NF-κB superrepressor blocked RSV induction of miR-30b.
The findings above suggested that both let-7i and miR-30b were antagonized by NS1 and NS2 but that only let-7i was induced via an IFN-dependent mechanism. NS2 antagonizes RIG-I binding to IPS-1 (9, 21). RIG-I signaling leads to type I IFN activation but additionally leads to activation of NF-κB. Furthermore, it has been shown that the miR-30b promoter is bound by the transactivating NF-κB family member p65. We hypothesized that miR-30b might be upregulated through the NF-κB pathway after RSV infection.
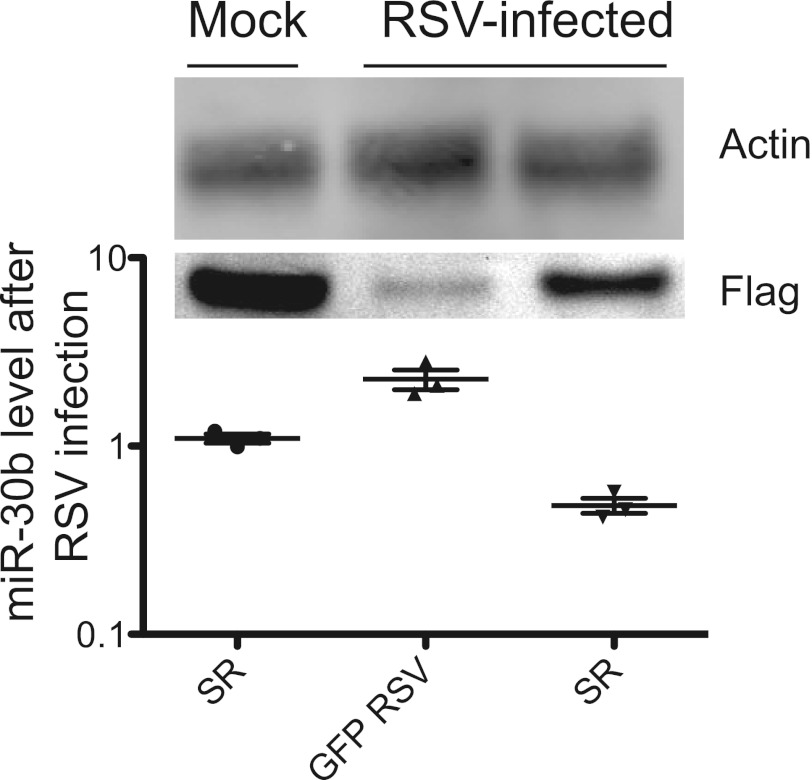
In order to test this hypothesis, we manipulated pathways regulating NF-κB transcription factors. The IκΒα protein inactivates NF-κB by masking the nuclear localization signals of NF-κB proteins and keeping them sequestered in an inactive state in the cytoplasm. The NF-κB superrepressor, when expressed, preferentially binds to the p65/p50 NF-κB transcription factor heterodimer but cannot be phosphorylated or degraded (10–14, 30, 31). Therefore, even in the presence of activating signals, the NF-κB superrepressor retains the NF-κB transcription factor in the cytosol and prevents NF-κB activation. We transiently transfected cells with a plasmid encoding the NF-κB superrepressor or a control plasmid encoding green fluorescent protein (GFP). Twenty-four hours posttransfection, cells were mock infected or inoculated with wild-type RSV A2 at an MOI of 1.0. RNA was harvested 48 h postinoculation and was assayed for miR-30b induction. The expression level of the superrepressor and equal loading were determined by immunoblotting (Fig. 5, top). Expression of the NF-κB superrepressor alone did not alter miR-30b levels, while RSV infection of GFP-expressing cells did induce miR-30b (Fig. 5, bottom). RSV infection of cells expressing NF-κB superrepressor resulted in a 2-fold decrease in miR-30b (P = 0.003). These data suggested that RSV likely induces miR-30b through the NF-κB pathway.
FIG 5 .
Transient expression of NF-κB superrepressor blocks miR-30b induction. NHBE cells were transfected with a plasmid expressing GFP or an NF-κB superrepressor (SR). Cells then were mock infected or inoculated with wild-type RSV A2 24 h later at an MOI of 1.0. Proteins and RNAs were harvested 48 h later. Expression of FLAG-tagged SR was confirmed by FLAG immunoblotting (top panel). miR-30b was assayed by real-time PCR (bottom graph). The graph is plotted as the relative fold change after GAPDH normalization in comparison to GFP-transfected, mock-infected cells. Data are plotted as means; error bars represent the standard errors of the means.
DISCUSSION
This study represents the first mechanistic description of RSV-mediated overexpression of miRNAs. We have determined that RSV uses three modes to induce miRNA overexpression. In DCs, let-7b induction was enhanced by IFN-β. In NHBE cells, expression of let-7i depended on IFN-β. In NHBE cells, miR-30b was induced by an IFN-independent, NF-κB-dependent mechanism. The antiviral type I IFN response has been studied extensively; however, induction of miRNAs represents a new aspect of IFN effector function that has not been fully elucidated. In this work, we also demonstrated that let-7c was increased significantly in NHBE cells. We also saw a significant decrease in the expression of miR-221, consistent with previous work (15). In the paper that described the decrease of miR-221 after RSV infection, the authors did not describe an increase in miR-30b or let-7i despite using similar cells and the same virus strain. This group, however, examined responses at 24 h postinfection, whereas we focused on the 48-h time point. Our studies suggest that miR-30b and let-7i are at much higher levels at 48 h postinoculation, and therefore, the other group may not have detected the increase at 24 h.
We have determined that virally encoded NS1 and NS2 antagonized let-7i and miR-30b induction in NHBE cells, suggesting that the miRNA responses may play an essential role in the host defense against RSV. The use of several different mechanisms of induction of miRNAs, in addition to the presence of antagonists of miRNA induction within the viral genome, suggests that miRNA induction is an important feature of the host-cell interaction. RSV G is also a known regulator of type I IFN responses as well as IFN-λ. We have not yet evaluated the role of IFN-λ.
Dysregulation of miRNAs has been examined extensively in human cancers. There is a small but growing body of evidence suggesting that miRNAs also respond to the inflammatory response associated with infection by RNA viruses. Several miRNAs changed in the brains of mice infected with Venezuelan equine encephalitis virus (32). Furthermore, miRNAs that are modulated during influenza virus infection have been identified. let-7c, which was upregulated in influenza virus-infected epithelial cells as well as in our own studies with RSV, may inhibit influenza virus replication by directly targeting a viral gene product (33, 34). Infection of macaques with low- or high-pathogenicity avian influenza viruses suggested that infection with highly pathogenic viruses induced a specific miRNA signature within the lungs of infected animals (35). Studies of swine-origin low-pathogenicity influenza A viruses and high-pathogenicity avian influenza A viruses have suggested that the low- and high-pathogenicity influenza viruses induce distinct miRNA molecular fingerprints (36). These data suggest that miRNA expression profiles may reflect the severity of the disease or could drive the severity of disease. These data, in combination with a recent publication describing RSV-induced changes in nerve growth factor (NGF) levels through downregulation of miR-221, suggest that RSV infection also may exhibit a distinct miRNA fingerprint and that this profile may reflect the severity of disease (15).
We determined that let-7b is upregulated in DCs after inoculation with RSV but not in epithelial cells. Inoculation of DCs with virus collected from Vero cell supernatants that did not have detectable IFN-β exhibited reduced, but not completely absent, induction of let-7b. let-7b regulates IFN-β and is induced by IFN-β in macrophages (25), so it was not surprising that let-7b induction correlated with IFN levels. More specifically, STAT3 may regulate let-7b (25). Even in the absence of IFN-β, however, there is the potential for some residual let-7b upregulation. The let-7b promoter has a p53 binding site, so it could be regulated additionally through p53 (37), although other published data suggest that let-7b decreases in response to cellular stress dependent upon p53 (38). Since we saw an increase in let-7b, this scenario is unlikely. In another study, let-7d, let-7f, let-7a, and let-7b were found to be decreased by active NF-κB (39). Conversely, it has been suggested that let-7a-3 may be activated by NF-κB (40). let-7b is clustered with let-7a and therefore could be regulated transcriptionally with let-7a. Therefore, let-7b could be upregulated by both IFN-β and NF-κB after RSV inoculation.
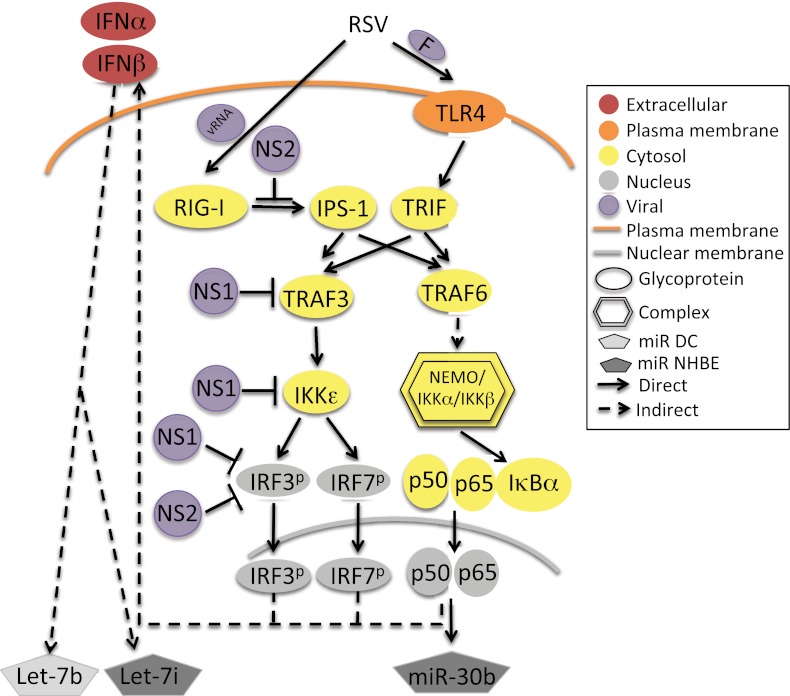
In NHBE cells, we determined that let-7i induction correlated with IFN-β and that miR-30b was induced via NF-κB. These findings are consistent with current knowledge, as RSV infection induces both IFN and NF-κB during RSV infection (10, 13, 18, 41). This activation could occur through RSV nucleic acid stimulation of RIG-I or RSV F stimulation of TLR4 (Fig. 6). While we have not categorically determined which of these routes is used by RSV, UV-inactivated virus did not activate let-7i or miR-30b. As RSV F protein is still present on UV-inactivated virion particles but the virus cannot replicate, these data might suggest a RIG-I-dependent mechanism. RSV genes NS1 and NS2 each antagonized both let-7i and miR-30b induction. It is known that NS1 and NS2 antagonize IFN and NF-κB signaling pathways at multiple points (Fig. 6). Since we have shown that let-7b and let-7i induction correlated with IFN-β levels, it was not surprising that both NS1 and NS2 antagonized induction, potentially through blocking IPS-1 binding to RIG-I, TRAF3 association with IPS-1, activation of IκB kinase epsilon (IKKε), and phosphorylation and activation of IRF-3 and IRF-7 (Fig. 6). miR-30b induction was blocked through expression of the NF-κB superrepressor and therefore may occur downstream of RIG-I/IPS-1 induction of RIP/FADD, TRAF6, NEMO, and the p50/p65 heterodimer (Fig. 6). In this scenario, NS2 should inhibit miR-30b induction by blocking RIG-I association with IPS-1. However, there is no known activity of NS1 that would antagonize miR-30b induction upstream of NF-κB activation. Therefore, NS1 may exhibit some activity that has not been discovered yet, or RSV also may activate NF-κB via a RIG-I-independent pathway. It has not been tested if RIG-I signaling by other mechanisms could lead to miR-30b induction. In the future, miR-30b induction will be tested following infection by other RNA viruses or stimulation with RIG-I and TLR4 agonists.
FIG 6 .
Model of RSV-mediated activation of let-7b, let-7i, and miR-30b. RSV activates the small RNA helicase RIG-I through an interaction with viral RNA and TLR4 through binding of virion protein F. RIG-I binds to the cellular adaptor protein IPS-1, which is antagonized by RSV NS2. TLR4 interacts with the cellular adaptor protein TRIF. Both IPS-1 and TRIF interact with cellular adaptor proteins TRAF3 and TRAF6. RSV NS1 blocks TRAF3 interaction with IPS-1. TRAF3 binding to IPS-1 leads to activation of the IΚKε kinase, which phosphorylates IRF-3 and -7. NS1 blocks activation of IΚKε, and both NS1 and NS2 block phosphorylation of IRFs. IRFs initiate expression of the type I IFNs IFN-α and IFN-β, which leads to downstream activation of let-7b and let-7i. TRAF6 interaction with IPS-1 or TRIF leads to downstream activation of the NEMO-IΚKα-IΚKβ kinase complex, which phosphorylates IκBα bound to the cytosolic p50-p65 NF-κB heterodimer. Phosphorylated IκBα is degraded by the proteasome, which leads to the release of the p50-p65 heterodimer and exposes their nuclear localization sequences. The heterodimer is transported to the nucleus and initiates transcriptional activation of NF-κB target genes, which include miR-30b as well as type I IFNs.
In this study, we identified three different members of the let-7 family that are induced by inoculation with RSV. Members of the let-7 family share the same seed sequence, and therefore, they should target the same cellular transcripts for degradation or block in translation. let-7c increases in response to infection with another important respiratory virus, influenza virus (33). The let-7 family has an extensive list of experimentally validated direct targets, including SOCS4, caspase-3, p27, Lin28, TRIM71, IL-13, TLR4, RAS, c-Myc, HMGA2, IL-10, and IL-6 (42). Several of the experimentally confirmed targets could impact RSV replication or the disease process. An experimentally confirmed target of let-7 is IL-13. It was found that during allergic inflammation in mice, let-7 was induced (43). Surprisingly, however, when let-7a is inhibited in mice during allergic airway inflammation, there was a decrease in secretion of proinflammatory cytokines (44). In animal models, IL-13 appears to enhance the severity of disease (45, 46). In children with RSV, increased levels of serum IL-13 were detected (47). This finding could indicate that RSV induces let-7 and enhances secretion of proinflammatory cytokines, thus enhancing the severity of disease.
Members of the let-7 family also target IL-6 (39). RSV induces IL-6 expression and secretion in macrophages (48). Data from our laboratory also indicated that neonatal cells inoculated with RSV secreted more IL-6 than did adult cells (14). let-7 regulation could be a mechanism of IL-6 secretion during RSV infection. let-7 also targets TLR4 (49). Polymorphisms in the TLR4 gene are associated with enhanced risk for severe RSV disease (50). TLR4 is stimulated by the RSV F protein (51), and TLR4-deficient mice who were infected exhibited enhanced disease (51, 52). let-7 overexpression may enhance the severity of disease by blocking expression of TLR4.
The functions of miR-30b and the miR-30 family have been less extensively studied. Targets of miR-30b have been experimentally confirmed only by identifying target sequences in their 3′ untranslated regions (UTRs). Genes confirmed by more rigorous methods to be targeted by other members of the miR-30 family include RUNX2, beclin-1, and GNAI2 (42).
Another potential effect of miRNA induction is degradation of viral gene products, which would result in inhibition of viral replication. Several groups have suggested that miRNAs may target viral mRNAs directly as a new antiviral immune mechanism (33, 34, 53, 54).
In summary, miRNAs appear to play an important role in the host response to RSV infection. Multiple miRNAs are induced by infection, in a cell-type-specific fashion. The particular miRNAs induced are known to relate to primary pathways in the host innate immune response involving classical regulators such as NF-κB and the type I IFNs. Thus, it is unlikely that miRNAs represent a completely new and independent antiviral program but rather that they appear to contribute to regulation of the overall innate response to infection. The fact that two RSV nonstructural genes inhibit the induction of miRNAs reveals how complex and multilayered is the interaction between paramyxoviruses and the innate response in epithelial and immune system cells.
MATERIALS AND METHODS
Cell lines and virus.
Vero cells were maintained in Eagle’s minimum essential medium (Mediatech) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. Normal human bronchial epithelial (NHBE) cells were obtained from Lonza and propagated in bronchial epithelial growth medium (BEGM) with retinoic acid, as directed by the manufacturer. Live RSV was UV inactivated with a 115-V VWR cross-linker for 15 min. UV inactivation was confirmed by plaque assay. Live RSV strains lacking the NS1 or NS2 genes that had been generated by reverse genetics techniques were kindly provided by MedImmune. The RSV wild-type strain A2 and the NS1 and NS2 gene-deletion RSV mutants were expanded in Vero cell monolayer cultures. Virus was isolated from infected cell monolayers after scraping, pelleting, three successive freeze-thaw cycles, and resuspension in fresh medium. RSV was purified further with two rounds of centrifugation at 2,500 × g for 10 min at 4°C and then filtration through a 45-µm filter. For mock infections, uninfected Vero cell culture monolayers were harvested and treated as described above.
Generation of MDDCs.
Human peripheral blood was collected with the approval of the Vanderbilt University Institutional Review Board. Sixty milliliters of peripheral blood was collected from healthy adult donors. Peripheral blood mononuclear cells (PBMCs) were isolated using Ficoll (Sigma; Histopaque 1077). Monocytes were enriched using CD14 microbeads and a QuadroMACS magnet (Miltenyi). Purified CD14+ monocytes were cultured in the presence of recombinant human granulocyte-macrophage colony-stimulating factor (GM-CSF) (800 U/ml) and recombinant human IL-4 (500 U/ml; PeproTech) for 6 days to generate MDDCs prior to virus inoculation.
miRNA array.
NHBE cells or MDDCs were inoculated with RSV wild-type strain A2 at an MOI of 1.0. Total RNA with preserved miRNAs was harvested at 48 h after inoculation using an miRNA isolation kit, as directed by the manufacturer (Exiqon). RNA concentration was measured on a NanoDrop ND-1000 spectrophotometer, and integrity was measured with an Agilent 2100 bioanalyzer. MicroRNA responses to RSV infection were assayed with the Exiqon miRCURY LNA microRNA array at the Vanderbilt University Genomic Science Resource core using miRXPLORE (Miltenyi).
miRNA isolation and detection.
RSV- or mock-infected cells were harvested at the indicated time points after inoculation. MicroRNAs were isolated from infected cell monolayers using the mirVana miRNA isolation kit (Ambion), as directed by the manufacturer. Both small and large RNAs were collected during isolation in order to retain cellular glyceraldehyde-3-phosphate dehydrogenase (GAPDH) RNA for normalization of data to an unaffected housekeeping gene. RNA concentration was measured with a NanoDrop spectrophotometer. MicroRNAs were detected by TaqMan real-time PCR using miRNA-specific primer probe sets for let-7b, let-7i, or miR-30b (Applied Biosystems). MicroRNAs were expanded by reverse transcription from 50 ng RNA with the TaqMan microRNA RT kit and sequence-specific primers (Applied Biosystems), as directed by the manufacturer. Copy DNA for the cellular control gene GAPDH was synthesized from 50 ng of RNA using a high-capacity cDNA reverse transcription kit (Applied Biosystems), as directed by the manufacturer. Individual gene products were detected by real-time PCR performed in a SmartCycler II (Cepheid) thermocycler using TaqMan gene expression assays and TaqMan universal PCR master mix (Applied Biosystems), as directed by the manufacturer. Expression assays used included GAPDH (4352934E), let-7b, let-7i, and miR-30b (Applied Biosystems). Custom TaqMan assays also were designed for detection of RSV M2-2 and G RNAs (Applied Biosystems Assays on Demand). Statistics were calculated using the Wilcoxon rank-sum test.
IFN-β ELISA.
IFN-β was assayed in viral preparations by ELISA (R&D Systems), as directed by the manufacturer. Plates were read using a SpectraMax M5 plate reader (Molecular Devices).
Transfection of NHBE cells.
NHBE cells were transiently transfected using an Amaxa Nucleofector device and Amaxa NHBE transfection solution (Lonza) as directed by the manufacturer. Briefly, cells were passaged 2 days before transfection. On the day of transfection, cells were trypsinized, trypsin was inactivated, and 5 × 105 cells were resuspended in transfection reagent spiked with supplement and 2 µg plasmid DNA. Cells were transfected with pmaxGFP as plasmid control or pCDNA3.1+ with the NF-κB superrepressor. Cells were then inoculated with RSV at 16 h posttransfection.
Immunoblotting to demonstrate gene expression.
Protein expression of the NF-κB superrepressor (an IκBα mutant that prevents NF-κB activation) was confirmed by immunoblotting using primary antibodies directed against the FLAG M2 epitope tag (Sigma). Equal loading of the blots was confirmed by actin. Cells were disrupted in lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 1% Triton X-100, pH 8.0) containing 0.5% (vol/vol) protease inhibitor cocktail (Sigma) and 1.0% (vol/vol) phosphatase inhibitors (Sigma). Lysates were run on 4 to 12% NuPAGE Bis-Tris gels (Invitrogen) and transferred to polyvinylidene difluoride (PVDF) membranes with an iBlot dry-blotting system (Invitrogen). Membranes were blocked with 50% Odyssey blocking buffer (Li-Cor) diluted in phosphate-buffered saline (PBS). Membranes were incubated in primary antibodies overnight at 4°C, washed, and then incubated with Li-Cor anti-mouse IRDye 800CW secondary antibody for 1 h at room temperature. Protein bands were detected and quantified using the Odyssey infrared imaging system (Li-Cor Biosciences).
ACKNOWLEDGMENTS
This work was supported by a grant from the March of Dimes, NIH grant R01 GM094198, and a Vanderbilt Ingram Cancer Center pilot project grant supported by NIH P30 CA068485 (Cancer Center Support Grant).
We thank Dean Ballard for his generous donation of the NF-κB superrepressor plasmid. We thank MedImmune for their generous donation of RSV NS1- and NS2-deletion viruses.
Footnotes
Citation Thornburg NJ, Hayward SL, Crowe JE, Jr. 2012. Respiratory syncytial virus regulates human microRNAs by using mechanisms involving beta interferon and NF-κB. mBio 3(6):e00220-12. doi:10.1128/mBio.00220-12.
REFERENCES
- 1. Collins PL, Crowe JE., Jr. 2006. Respiratory syncytial virus and metapneumovirus, p 1449–1496 In Knipe DM, Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2. Domurat F, Roberts NJ, Walsh EE, Dagan R. 1985. Respiratory syncytial virus infection of human mononuclear leukocytes in vitro and in vivo. J. Infect. Dis. 152:895–902 [DOI] [PubMed] [Google Scholar]
- 3. Sarnow P, Jopling CL, Norman KL, Schütz S, Wehner KA. 2006. MicroRNAs: expression, avoidance and subversion by vertebrate viruses. Nat. Rev. Microbiol. 4:651–659 [DOI] [PubMed] [Google Scholar]
- 4. Cullen BR. 2006. Is RNA interference involved in intrinsic antiviral immunity in mammals? Nat. Immunol. 7:563–567 [DOI] [PubMed] [Google Scholar]
- 5. Potenza N, et al. 2011. Human microRNA hsa-miR-125a-5p interferes with expression of hepatitis B virus surface antigen. Nucleic Acids Res. 39:5157–5163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Roush S, Slack FJ. 2008. The let-7 family of microRNAs. Trends Cell Biol. 18:505–516 [DOI] [PubMed] [Google Scholar]
- 7. O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. 2007. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. U. S. A. 104:1604–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pedersen IM, et al. 2007. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature 449:919–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huang J, et al. 2007. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat. Med. 13:1241–1247 [DOI] [PubMed] [Google Scholar]
- 10. Garofalo R, et al. 1996. Respiratory syncytial virus infection of human respiratory epithelial cells up-regulates class I MHC expression through the induction of IFN-beta and IL-1 alpha. J. Immunol. 157:2506–2513 [PubMed] [Google Scholar]
- 11. König B, Streckert HJ, Krusat T, König W. 1996. Respiratory syncytial virus G-protein modulates cytokine release from human peripheral blood mononuclear cells. J. Leukoc. Biol. 59:403–406 [DOI] [PubMed] [Google Scholar]
- 12. Krishnan S, Craven M, Welliver RC, Ahmad N, Halonen M. 2003. Differences in participation of innate and adaptive immunity to respiratory syncytial virus in adults and neonates. J. Infect. Dis. 188:433–439 [DOI] [PubMed] [Google Scholar]
- 13. Lee FE, et al. 2007. Human infant respiratory syncytial virus (RSV)-specific type 1 and 2 cytokine responses ex vivo during primary RSV infection. J. Infect. Dis. 195:1779–1788 [DOI] [PubMed] [Google Scholar]
- 14. Thornburg NJ, Shepherd B, Crowe JE., Jr. 2010. Transforming growth factor beta is a major regulator of human neonatal immune responses following respiratory syncytial virus infection. J. Virol. 84:12895–12902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Othumpangat S, Walton C, Piedimonte G. 2012. MicroRNA-221 modulates RSV replication in human bronchial epithelium by targeting NGF expression. PLoS One 7:e30030 http://dx.doi.org/10.1371/journal.pone.0030030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bakre A, et al. 2012. Respiratory syncytial virus modifies microRNAs regulating host genes that affect virus replication. J. Gen. Virol. 93:2346–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Spann KM, Tran KC, Collins PL. 2005. Effects of nonstructural proteins NS1 and NS2 of human respiratory syncytial virus on interferon regulatory factor 3, NF-kappaB, and proinflammatory cytokines. J. Virol. 79:5353–5362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jewell NA, et al. 2007. Differential type I interferon induction by respiratory syncytial virus and influenza A virus in vivo. J. Virol. 81:9790–9800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lo MS, Brazas RM, Holtzman MJ. 2005. Respiratory syncytial virus nonstructural proteins NS1 and NS2 mediate inhibition of Stat2 expression and alpha/beta interferon responsiveness. J. Virol. 79:9315–9319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Groskreutz DJ, et al. 2006. Respiratory syncytial virus induces TLR3 protein and protein kinase R, leading to increased double-stranded RNA responsiveness in airway epithelial cells. J. Immunol. 176:1733–1740 [DOI] [PubMed] [Google Scholar]
- 21. Ling Z, Tran KC, Teng MN. 2009. Human respiratory syncytial virus nonstructural protein NS2 antagonizes the activation of beta interferon transcription by interacting with RIG-I. J. Virol. 83:3734–3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lifland AW, et al. 2012. Human respiratory syncytial virus nucleoprotein and inclusion bodies antagonize the innate immune response mediated by MDA5 and MAVS. J. Virol. 86:8245–8258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shingai M, et al. 2008. Soluble G protein of respiratory syncytial virus inhibits Toll-like receptor 3/4-mediated IFN-beta induction. Int. Immunol. 20:1169–1180 [DOI] [PubMed] [Google Scholar]
- 24. Scagnolari C, et al. 2010. Differential expression of interferon-induced microRNAs in patients with chronic hepatitis C virus infection treated with pegylated interferon alpha. Virol. J. 7:311 http://dx.doi.org/10.1186/1743-422X-7-311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Witwer KW, Sisk JM, Gama L, Clements JE. 2010. MicroRNA regulation of IFN-beta protein expression: rapid and sensitive modulation of the innate immune response. J. Immunol. 184:2369–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. de Graaff PM, et al. 2005. Respiratory syncytial virus infection of monocyte-derived dendritic cells decreases their capacity to activate CD4 T cells. J. Immunol. 175:5904–5911 [DOI] [PubMed] [Google Scholar]
- 27. Stewart MJ, Kulkarni SB, Meusel TR, Imani F. 2006. C-Jun N-terminal kinase negatively regulates dsRNA and RSV induction of tumor necrosis factor-alpha transcription in human epithelial cells. J. Interferon Cytokine Res. 26:521–533 [DOI] [PubMed] [Google Scholar]
- 28. Colamonici OR, Porterfield B, Domanski P, Constantinescu S, Pfeffer LM. 1994. Complementation of the interferon alpha response in resistant cells by expression of the cloned subunit of the interferon alpha receptor. A central role of this subunit in interferon alpha signaling. J. Biol. Chem. 269:9598–9602 [PubMed] [Google Scholar]
- 29. Desmyter J, Melnick JL, Rawls WE. 1968. Defectiveness of interferon production and of rubella virus interference in a line of African green monkey kidney cells (Vero). J. Virol. 2:955–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brockman JA, et al. 1995. Coupling of a signal response domain in IκBα to multiple pathways for NF-κB activation. Mol. Cell. Biol. 15:2809–2818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang CY, Mayo MW, Baldwin AS. 1996. TNF- and cancer therapy-induced apoptosis: potentiation by inhibition of NF-kappaB. Science 274:784–787 [DOI] [PubMed] [Google Scholar]
- 32. Bhomia M, et al. 2010. Analysis of microRNAs induced by Venezuelan equine encephalitis virus infection in mouse brain. Biochem. Biophys. Res. Commun. 395:11–16 [DOI] [PubMed] [Google Scholar]
- 33. Ma YJ, et al. 2012. Cellular microRNA let-7c inhibits M1 protein expression of the H1N1 influenza A virus in infected human lung epithelial cells. J. Cell. Mol. Med. 16:2539–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Song L, Liu H, Gao S, Jiang W, Huang W. 2010. Cellular microRNAs inhibit replication of the H1N1 influenza A virus in infected cells. J. Virol. 84:8849–8860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li Y, et al. 2011. Differential microRNA expression and virulence of avian, 1918 reassortant, and reconstructed 1918 influenza A viruses. Virology 421:105–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Loveday EK, Svinti V, Diederich S, Pasick J, Jean F. 2012. Temporal- and strain-specific host microRNA molecular signatures associated with swine-origin H1N1 and avian-origin H7N7 influenza A virus infection. J. Virol. 86:6109–6122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kent WJ, et al. 2002. The human genome browser at UCSC. Genome Res. 12:996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Saleh AD, et al. 2011. Cellular stress induced alterations in microRNA let-7a and let-7b expression are dependent on p53. PLoS One 6:e24429 http://dx.doi.org/10.1371/journal.pone.0024429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Iliopoulos D, Hirsch HA, Struhl K. 2009. An epigenetic switch involving NF-kappaB, Lin28, let-7 microRNA, and IL6 links inflammation to cell transformation. Cell 139:693–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang DJ, Legesse-Miller A, Johnson EL, Coller HA. 2012. Regulation of the let-7a-3 promoter by NF-κB. PLoS One 7:e31240–e31240 http://dx.doi.org/10.1371/journal.pone.0031240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yoboua F, Martel A, Duval A, Mukawera E, Grandvaux N. 2010. Respiratory syncytial virus-mediated NF-kappa B p65 phosphorylation at serine 536 is dependent on RIG-I, TRAF6, and IKK beta. J. Virol. 84:7267–7277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vergoulis T, et al. 2012. Tarbase 6.0: capturing the exponential growth of miRNA targets with experimental support. Nucleic Acids Res. 40:D222–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kumar M, et al. 2011. Let-7 microRNA-mediated regulation of IL-13 and allergic airway inflammation. J. Allergy Clin. Immunol. 128:1077–1085 e1–10 [DOI] [PubMed] [Google Scholar]
- 44. Polikepahad S, et al. 2010. Proinflammatory role for let-7 microRNAs in experimental asthma. J. Biol. Chem. 285:30139–30149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dakhama A, et al. 2005. The enhancement or prevention of airway hyperresponsiveness during reinfection with respiratory syncytial virus is critically dependent on the age at first infection and IL-13 production. J. Immunol. 175:1876–1883 [DOI] [PubMed] [Google Scholar]
- 46. Lukacs NW, et al. 2001. Respiratory syncytial virus predisposes mice to augmented allergic airway responses via IL-13-mediated mechanisms. J. Immunol. 167:1060–1065 [DOI] [PubMed] [Google Scholar]
- 47. Becker Y. 2006. Respiratory syncytial virus (RSV) evades the human adaptive immune system by skewing the Th1/Th2 cytokine balance toward increased levels of Th2 cytokines and IgE, markers of allergy—a review. Virus Genes 33:235–252 [DOI] [PubMed] [Google Scholar]
- 48. Becker S, Quay J, Soukup J. 1991. Cytokine (tumor necrosis factor, IL-6, and IL-8) production by respiratory syncytial virus-infected human alveolar macrophages. J. Immunol. 147:4307–4312 [PubMed] [Google Scholar]
- 49. Chen XM, Splinter PL, O’Hara SP, LaRusso NF. 2007. A cellular micro-RNA, let-7i, regulates Toll-like receptor 4 expression and contributes to cholangiocyte immune responses against Cryptosporidium parvum infection. J. Biol. Chem. 282:28929–28938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tal G, et al. 2004. Association between common Toll-like receptor 4 mutations and severe respiratory syncytial virus disease. J. Infect. Dis. 189:2057–2063 [DOI] [PubMed] [Google Scholar]
- 51. Kurt-Jones EA, et al. 2000. Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus. Nat. Immunol. 1:398–401 [DOI] [PubMed] [Google Scholar]
- 52. Haynes LM, et al. 2001. Involvement of Toll-like receptor 4 in innate immunity to respiratory syncytial virus. J. Virol. 75:10730–10737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jopling CL, Norman KL, Sarnow P. 2006. Positive and negative modulation of viral and cellular mRNAs by liver-specific microRNA miR-122. Cold Spring Harbor Symp. Quant. Biol. 71:369–376 [DOI] [PubMed] [Google Scholar]
- 54. Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 309:1577–1581 [DOI] [PubMed] [Google Scholar]