Abstract
Melanoma incidence and associated mortality continue to increase worldwide. The lack of treatments with durable responses for stage IV melanoma may be due, at least in part, to an incomplete understanding of the molecular mechanisms that regulate tumor initiation and/or progression to metastasis. Recent evidence supports miRNA dysregulation in melanoma impacting several well-known pathways such as the PI3K/AKT or RAS/MAPK pathways, but also underexplored cellular processes like protein glycosylation and immune modulation. There is also increasing evidence that miRNA can improve patient prognostic classification over the classical staging system and provide new therapeutic opportunities. The integration of this recently acquired knowledge with known molecular alterations in protein coding genes characteristic of these tumors (i.e., BRAF and NRAS mutations, CDKN2A inactivation) is critical for a complete understanding of melanoma pathogenesis. Here, we compile the evidence of the functional roles of miRNAs in melanomagenesis and progression, and of their clinical utility as biomarkers, prognostic tools and potential therapeutic targets. Characterization of miRNA alterations in melanoma may provide new angles for therapeutic intervention, help to decipher mechanisms of drug resistance, and improve patient classification for disease surveillance and clinical benefit.
Introduction
Melanoma is the most aggressive form of skin cancer, and its incidence keeps increasing worldwide (1). There is no curative therapy for advanced stages of melanoma (2); therefore, new indicators of prognosis and therapeutic targets are demanded. MicroRNAs (miRNAs) are small non-coding RNAs that modulate gene expression by repressing protein translation and inducing messenger RNA (mRNA) degradation. Since they bind to the 3ʹUTR of their mRNA targets with imperfect sequence complementarity, each miRNA has the potential of modifying the expression of dozens or perhaps hundreds of genes. Since the discovery of lin-4 in C. elegans in 1993 [3] and the subsequent expansion of miRNAs from a curious phenomenon in worms to a widespread biological regulatory system with the identification of the highly conserved let-7 [4], miRNAs have attracted a strong interest in the scientific community. As a new class of biological molecules, they have added an astounding and unexpected level of regulation of gene expression and have contributed to explain apparent discrepancies between mRNA and protein levels [3]. Over the course of the last decade, miRNAs have been shown to be essential in a variety of normal biological processes necessary for organism development and survival [5].
In humans, more than a 1000 miRNAs have been identified to date, and they regulate the expression of a third of the human genome. MiRNA alterations, resulting from genetic mutations, chromosomal aberrations or epigenetic changes, have been shown to contribute to various developmental defects and diseases, including cancer. The study of miRNA changes in cancer subtypes has yielded several interesting and unexpected findings. First, miRNAs have the ability to subclassify tumor types more robustly than mRNAs [6]. This property is attributed to lineage-specific expression of some miRNAs, whose expression is retained in the corresponding tumors. As discussed later, this ability of miRNAs to evoke the tumor tissue-of-origin may have important diagnostic implications. Second, miRNAs are important modulators of classical oncogenes and tumor suppressors, which turn them into putative tumor suppressors or oncogenes, respectively. Importantly, such behavior is exquisitely context dependent, as the same miRNA can be oncogenic or oncosuppressive in different tumor types, presumed to result from the expression of distinct pools of target genes. Third, miRNAs are extremely stable molecules both in archived tissues and body fluids, a property that in combination with their tissue specificity suggests them as attractive candidate biomarkers.
The first evidence of miRNA alterations in melanoma came from the analysis of genomic alterations characteristic of these tumors, which were significantly enriched in miRNA genes [7]. Moreover, several miRNA profiles of melanoma cell lines or tissues revealed altered patterns of expression compared to normal melanocytes or nevi, respectively (reviewed in [8]). Functional in vitro and/or in vivo testing of some dysregulated miRNAs in melanoma has demonstrated the important contribution of specific miRNAs to the molecular complexity of these tumors.
Here we present an overview of the advances made in the discovery of miRNAs with a functional role in cutaneous melanoma pathogenesis and of preliminary attempts to exploit the diagnostic, prognostic and therapeutic potential held by these small RNAs in melanoma. Table 1 summarizes miRNAs that have been shown to alter some of the oncogenic properties of melanoma cells. We analyze the role of these miRNAs in the context of four cell processes that dictate the genesis and progression of melanoma:
Table I.
Functional miRNA deregulated in melanoma
| Name | ↑or↓ | Source | Discovery | Direct targets | Ref. | |
|---|---|---|---|---|---|---|
| Proliferation | Let-7b | Down | FFPE tissues (157) | qPCR Taqman array | Cyclin D1, D3, CDK4 | [10] |
| miR-193b | Down | FFPE tissues (8) | Agilent miRNA array | Cyclin D1 | [13] | |
| miR-145 | Down | Frozen tissue (7) | qPCR t.a. | c-MYC | [15] | |
| miR-221/222 | Up | Cell lines (9)14;(27)16 | qPCR t.a.14;miRNA array16 | p27, PTEN, TIMP3, c-Kit | [17,18] | |
| miR-205 | Down | Frozen tissues (15)20, (52)21 | Agilent20,Illumina21 miRNA array, | E2F1, E2F5, ERBB3 | [22,23] | |
| miR-9 | Down | FFPE tissues (20) | qPCR t.a. | NFkB1 | [25] | |
| miR-155 | Down | Cell lines (17) | qPCR array | Not Determined | [26] | |
| miR-34a/c | Down | Cell lines (2) | qPCR array | c-MET | [27] | |
| Invasion | Let-7b | Down | Cell lines (1) | qPCR t.a. | BSG | [29] |
| Let-7a | Down | Cell lines (9) | qPCR t.a. | Integrin β3 | [11] | |
| miR-214 | Up | Cell lines (4) | miRNA profile n/p | TFAP2C, ITGA3 | [31] | |
| miR-9 | Down | FFPE tissues (20) | qPCR t.a. | NFkB1 | [25] | |
| miR-34a/c | Down | Cell lines (2) | qPCR t.a. | c-MET | [35] | |
| miR-199* | Down | Cell lines (2) | qPCR t.a. | c-MET | [35] | |
| miR-30b/d | Up | FFPE tisues (59) | Rosetta miRNA array | GALNT7, GALNT1 | [36] | |
| miR-200a/c | Up | Cell lines (11) | qPCR t.a. | MARCKS | [39] | |
| miR-375 | Down | Cell lines (3) | TILDA, NCode miRNA platform | Not Determined | [40] | |
| miR-145 | Down | Frozen tissue (7) | qPCR t.a. | FSCN1 | [15] | |
| miR-182 | Up | Cell lines (14) & FFPE tissues (80) | qPCR t.a. & ISH | MITF-M, FOXO3 | [46] | |
| miR-211 | Down | Cell lines (3)48; (1)49; (51)50 | TILDA/ NCode48 miRNA platform; functional screening49; Agilent miRNA array50 | BRN2, NFAT5, TGFBR2, KCNMA1 | [50–52] | |
| miR-196a | Down | Cell lines (8)51,52 | qPCR t.a.51,52 | HOXB7, HOX-C8 | [53–54] | |
| Cell survival | miR-155 | Down | Cell lines (17) | qPCR array | Not Determined | [26] |
| miR-125b | Down | FFPE tissues (28) | Exiqon miRNA array | Not Determined | [55] | |
| miR-15b | Up | Cell lines (10) | qPCR t.a. | Not Determined | [57] | |
| miR-214 | Up | Cell lines (4) | miRNA profile n/p | Not Determined | [31] | |
| miR-506/514 | Up | Tissue biopsy (33) | qPCR Taqman array | Not Determined | [58] | |
| miR-182 | Up | Cell lines (14)/ FFPE tissues (80) | qPCR t.a. / ISH | FOXO3 | [46] | |
| miR-21 | Up | Cell lines (1)58; Frozen tissues (96)59 | qPCR t.a 58;qPCR Taqman array59 | PTEN,PDCD4, BTG2 | [60,61] | |
| miR-149* | Up | Cell lines (7) | qPCR t.a. | GSK3β | [62] | |
| miR-193b | Down | FFPE tissues (8) | Agilent miRNA array | Mcl-1 | [13] | |
| Immune response | miR-34a/c | Down | Cell lines (8) | qPCR t.a. | ULBP2 | [65] |
| miR-30b/d | Up | FFPE tisues (59) | Rosetta miRNA array | GALNT7, GALNT1 | [36] | |
t.a.: targeted approach. (#): Number of samples analyzed. ISH: In situ hybridization. n/p not published
Regulators of cell cycle/proliferation
Uncontrolled proliferation is an essential step on the path to transformation of cells, of which melanocytes are no exception. Figure 1 summarizes the impact of miRNA deregulation in proliferation pathways. The cell cycle is controlled by the modulation of the expression of cyclins, and their capacity to activate specific cyclin-dependent kinases (CDK). Similar to other malignancies, increased expression of Cyclins D1 and D3 has been reported in melanoma [9]. Several miRNAs have been shown to target them directly, including Let-7 family members, which are consistently downregulated in primary melanomas compared with nevi [10,11], as well as in other tumors [12]. Let-7b ectopic expression directly represses the expression of Cyclins D1, D3 and CDK4, resulting in reduced cell-cycle progression and anchorage-independent growth [10]. In addition, miR-193b, which targets Cyclin D1, has reduced expression in metastatic melanomas versus nevi [13].
Fig. 1.
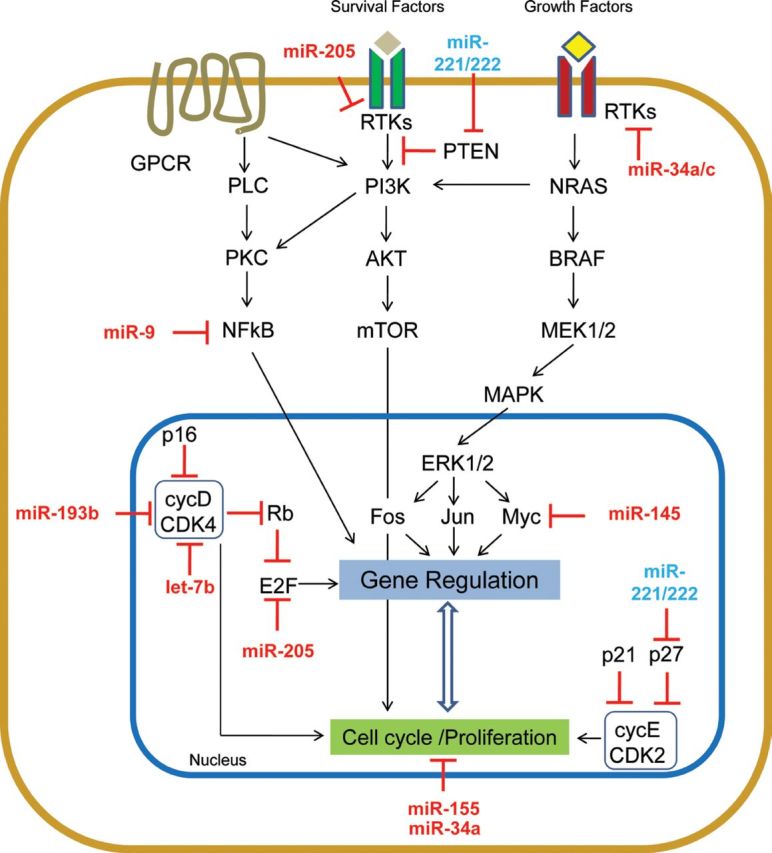
miRNA that intersect with cell cycle/proliferation pathways in melanoma.
Increased levels of Cyclins can also result from transcriptional activation of c-MYC, an oncogene altered in many tumors, including melanoma [14]. Noguchi et al. reported that ectopic expression of miR-145, which is lost in canine and human melanoma, inhibited cellular growth, an effect that was partially mediated by reduction of c-MYC protein [15].
The activity of Cyclin-CDK complexes is, in addition, regulated by other proteins. For instance, p27 (CDKN1B) binds and inhibits the function of CyclinD1-CDK4 and thus acts as a critical regulator of the G1-S transition. Low levels of p27 or its phosphorylation by AKT result in uncontrolled cell-cycle progression into S phase. P27 is a direct target of the miR-221/222 cluster. This cluster is almost undetectable in human melanocytes but its expression increases stepwise through progression of melanoma. Overexpression of miR-221/222 in melanoma cells yielded an increased proliferative rate, whereas its inhibition strongly reduced cell growth, invasion and foci formation in vitro and impaired in vivo tumor growth [16]. Moreover, the miR-221/222 cluster can also modulate other regulators of proliferation, including the tumor suppressors PTEN and TIMP3 [17]. Interestingly, miR-221/222 also target the oncogenic tyrosine kinase receptor c-Kit [18], which is shown to be repressed in certain types of melanoma [19].
Other critical cell-cycle regulators are the E2F transcription factors, whose activity may result in increased proliferation or apoptosis. Frequent genomic amplification of the E2F1 locus leads to increased expression of this transcription factor in melanoma [20]. Dar and colleagues reported that miR-205 loss in melanoma results in increased proliferation due to activation of pathways triggered by receptor tyrosine kinases (RTKs). MiR-205 is able to directly target E2F1 and E2F5 in melanoma and their expression inversely correlates during melanoma progression. miR-205 is located in a region (Chr. 1q) of frequent deletion in melanoma [21]. Restoration of miR-205 expression reduced the phosphorylation of AKT, decreasing melanoma cell proliferation in vitro and in vivo [22]. Furthermore, miR-205 targets ERBB3 [23], a membrane-bound RTK that interacts with and participates in EGF receptor signaling.
Another important pathway associated with melanoma proliferation is NF-kB. Elevated levels and increased phosphorylation of c-REL, a key NF-kB transcription factor, has been identified in clinical melanoma specimens [24]. Liu and others proposed that the loss of miR-9 in metastatic melanoma is causal of higher expression of NF-kB components and subsequent increases in proliferative capacity. The authors clearly demonstrate that overexpression of miR-9 negatively modulates NFkB1 protein levels, reducing cell proliferation [25].
Other miRNAs have been shown to modulate melanoma proliferation in vitro, but the underlying mechanisms remain unclear. For instance, ectopic expression of miR-155, a miRNA downregulated in several melanoma cell lines relative to melanocytes, suppresses proliferation [26], but no targets or downstream modulators have been reported. In another recent study, restoration of miR-34a levels resulted in inhibition of cell proliferation in vitro and tumor growth in vivo [27].
In conclusion, a series of miRNAs that regulate cell-proliferation genes have been identified as dysregulated in melanoma. Excluding miR-221/222, the expression of most of these miRNAs is reduced in melanoma, suggesting a broader role as negative regulators of cell proliferation.
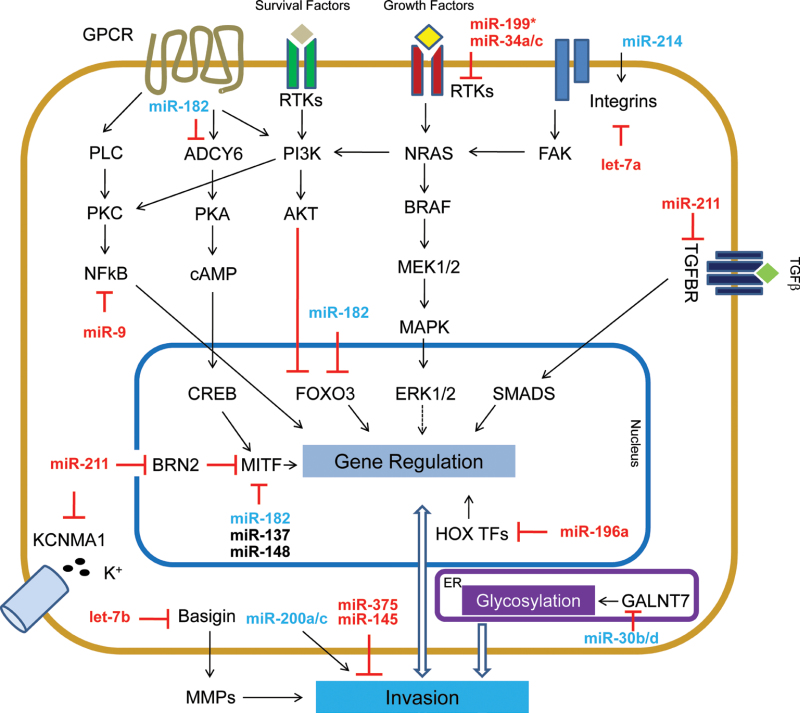
Regulators of migration/invasion
The progression to metastasis of tumor cells is thought to require the acquisition of several properties, including migratory capacity and the ability to invade through surrounding tissues to enter blood or lymphatic circulation [28]. Numerous studies report modulation of the migratory/invasive properties of melanoma cells by miRNAs, suggesting this as a common occurrence (see Figure 2). Several of the miRNAs described below directly target known regulators of invasion/migration, while others impact migration and invasion through previously unlinked cellular processes.
Fig. 2.
miRNA implicated in cell invasion, as relates to known invasion pathways in melanoma.
Of those miRNAs that target known migration/invasion genes, Fu and collaborators suggest that the loss of let-7b in melanoma cells enhances their ability to metastasize because of an increase of Basigin (BSG) expression. BSG is a known enhancer of extracellular matrix metalloprotease (MMP) production. The authors demonstrate that overexpressing let-7b in a mouse melanoma cell line leads to reduced BSG and MMP-9 protein, resulting in less distant metastasis [29].
In addition to MMPs, other proteins which play key roles in sensing and remodeling the extracellular matrix are altered in melanoma. For example, some adhesion molecules, such as Integrin β3 (ITGB3) have been shown to increase the invasive properties of melanoma cells [30], but the mechanisms that lead to its increased expression remain unknown. Muller et al. show that ITGB3 is a direct target of let-7a, whose loss in melanoma results in higher ITGB3 levels and increased invasive potential [11].
In contrast, another miRNA-regulated integrin, ITGA3, is targeted by miR-214, which has been found to be overexpressed in melanoma [31], suggesting that integrin regulation can have complex functional consequences. In addition to ITGA3, miR-214 also targets TFAP2C, a transcription factor, whose knock down recapitulates miR-214’s effects on invasion. Moreover, along with these two direct targets, miR-214 overexpression modulates a variety of surface molecules involved in cell adhesion and motility [31]. Among them, miR-214 overexpression results in loss of CDH1 (E-cadherin) expression, a common occurrence in cells of a variety of tumors as they become invasive and metastatic. These results nicely show that miRNAs can have pleiotropic effects by targeting several direct and indirect mediators of metastasis. Accordingly, Liu et al. uncover a miRNA that regulates a molecular pathway that converges on E-cadherin downregulation and therefore results in increased migration and invasion. MiR-9, which is frequently lost in metastatic melanoma, contributes to increased expression and activity of its direct target NFkB1, and NFkB activation leads to increased expression of SNAI1 (SNAIL), a well-described repressor of E-cadherin. Restoration of miR-9 levels in melanoma cells caused NFkB1 downregulation, which was accompanied by reduced SNAIL and increased E-cadherin. Together these changes resulted in reduced tumor growth and metastasis [25].
Another key surface protein that has been clearly linked with invasion is the receptor c-MET (MET). MET overexpression is associated with a complex biological program called “invasive growth” that results in cell motility, invasion and protection from apoptosis in many different tumors. In addition, MET is a relevant prognostic factor for overall survival and aggressive behavior in melanoma [32–34]. Recently, some miRNAs have been shown to contribute to the regulation of MET expression. In particular, miR-34b, miR-34c and miR-199a-3p were found to directly target MET causing a reduction in its mRNA and protein levels. Accordingly, inhibiting these miRNAs increased MET protein expression and significantly enhanced migration and cell adhesion in cell lines [35].
MiRNA can also impact cell-surface proteins by interfering with their post-translational modifications. Particularly, miR-30b and miR-30d, which are overexpressed in primary and metastatic melanoma tissues, modify the glycosylation pattern of transmembrane proteins. These changes, mediated by the reduction of GALNT7, a galactosamine transferase, result in higher invasive potential in vitro and in vivo [36]. Aberrant glycosylation may affect cell adhesion and motility by altering the function of a diverse set of proteins, including integrins [37] and E-cadherin [38].
During the metastatic phase, melanoma cells encounter different local microenvironments that demand adaptation to successfully survive the process. Elson-Schwab et al. demonstrate that this plasticity can be modulated by miR-200 family members, which control the switch between the “ameboid-like” and “mesenchymal-like” types of cell migration [39]. Moreover, other miRNAs, including miR-375 [40] and miR-145 [15], have been shown to affect invasion by causing changes in cell morphology, although the mechanisms by which this occurs are unclear.
A key molecule that has been shown to control melanoma plasticity is the microphthalmia-associated transcription factor, MITF. MITF is considered the master regulator of the melanocytic lineage by controlling diverse cellular programs, including differentiation, cellular response mechanisms, cell cycle, cell survival, motility and miRNA biogenesis (reviewed by [41]). Both oncogenic and tumor-suppressive roles have been ascribed to MITF, and recently, several proposed models attempt to reconcile these paradoxical data. Current theories propose that high levels of MITF cause differentiation and cell-cycle arrest, while medium levels will, instead, enhance melanoma cell proliferation. In contrast, low MITF levels will switch cells from a proliferative state to an invasive one [42,43]. Regulation of MITF in melanoma cells is undoubtedly complicated, and regulation by miRNAs has added to this complexity. In melanoma, MITF is directly modulated by several miRNAs, including miR-137 [44], miR-148 [45] and miR-182 [46]. MiR-182 was found to be overexpressed in multiple melanoma cell lines and tissues, and its overexpression enhances melanoma cell invasiveness. These effects were phenocopied by MITF knockdown, and importantly, overexpression of MITF abolished miR-182’s proinvasive effects [46]. Other miRNAs indirectly impact MITF levels. For instance, miR-221/222 repress the expression of the receptor KIT [18,47], which results in reduced MITF activation [48,49] and increased cell invasion.
In addition to its regulation by miRNA, MITF also controls the expression of miRNA, such as miR-211[50]. miR-211 is downregulated in melanoma cell lines [51,52] and tissues [50] relative to normal melanocytic controls. Although its loss of expression appears to be a very early event in melanomagenesis, it was identified as a strong suppressor of invasion in a large-scale miRNA invasion screen. Mechanistically, miR-211 was found to modulate several known players in melanoma metastasis, including IGF2R, TGFBR2 and NFAT5 [51]. Moreover, miR-211 is also able to regulate BRN2, a known repressor of MITF. Loss of miR-211 results in higher levels of BRN2, which in turn, inhibit the expression of MITF, creating a negative feed-back loop that maintains melanoma cells in a dedifferentiated, proinvasive state [52].
In a similar manner, deregulation of HOX transcription factors confers a more undifferentiated and invasive phenotype to cancer cells [53]. Braig et al. found that miR-196a, whose expression is downregulated in melanoma cell lines [54] and tissues [53], directly repress HOXB7 and HOXC8 expression. miR-196a restoration resulted in reduced invasion that correlated with decreased HOXB7 [54] and HOXC8 expression [53].
In summary, miRNAs are involved in the regulation of a diverse set of molecular programs, from cell adhesion and glycosylation at the cell surface to transcriptional regulation in the nucleus, being all important in the complex process of metastasis.
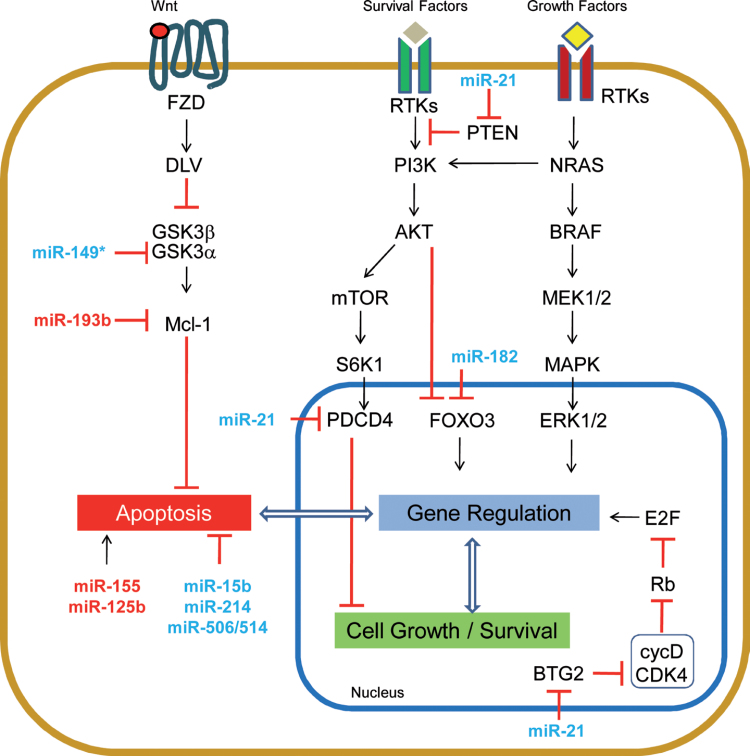
Regulators of cell survival
During both tumor initiation and progression, transformed cells encounter different challenges to their viability. Most directly, cell survival is controlled by cell-death pathways, which are triggered when normal cells go awry. A wealth of evidence has shown that evasion of these cell-death pathways are required for tumor maintenance. A role for miRNAs in the maintenance of cell survival has been proposed in several malignancies, although results in melanoma are still early in their development (summarized in Figure 3). MiR-155 and miR-125b, both of which are less expressed in melanoma than in melanocytes [26,55], are prime examples. MiR-155 restoration caused growth inhibition and apoptotic cell death in multiple melanoma cell lines [26], whereas miR-125b ectopic expression induced cellular senescence. In addition, cells with decreased levels of miR-125b were protected from spontaneous apoptosis [56].
Fig. 3.
miRNA implicated in cell survival pathways in melanoma.
In contrast to apoptosis, there are multiple miRNAs that promote cell survival and are found to be upregulated in melanoma. The inhibition of miR-15b [57], miR-214 [31] and the miR-506/514 cluster [58] resulted in apoptotic cell death through undefined mechanisms. Specific mechanisms have been proposed for miR-182, miR-21 and miR-149*. Inhibition of miR-182 resulted in apoptosis of some melanoma cell lines, likely via upregulation of FOXO3 and its known downstream target BIM, a proapoptotic Bcl-2 family member central to the intrinsic apoptotic pathway [46]. Similarly, the inhibition of miR-21, a well-characterized oncogenic miRNA [59], sensitized mouse melanoma cells to interferon-induced apoptosis, and reduced the metastatic potential of these cells [60]. Moreover, in human melanoma cells, miR-21 inhibition resulted in an increase of its direct target PTEN, with consequent suppression of AKT phosphorylation, reduction of Bcl-2 and increased Bax [61].
MiR-149* is another miRNA impacting the intrinsic apoptotic pathway. This miRNA is expressed in response to p53 activation; however, rather than contributing to p53’s tumor-suppressor functions, miR-149* provides a mechanism to bypass the induction of apoptosis by p53 activation. MiR-149* directly targets glycogen synthase kinase-3α (GSK3α), which leads to the stabilization of MCL1. Inhibition of miR-149* in an in vivo model yielded smaller tumors with concomitant reduction of MCL1, and increased GSK3α and caspase activity [62], suggesting induction of apoptosis as the mechanism by which tumor growth was suppressed. Interestingly, MCL1 is also directly targeted by miR-193b in melanoma. miR-193b, which is lowly expressed in melanoma, inversely correlates with MCL1 levels in melanoma tissues. Restoration of miR-193b decreases MCL1 but is insufficient to induce cell death alone. However, miR-193b does sensitize melanoma cells to apoptotic induction [13]. While both miR-149* and miR-193b converge on MCL1 regulation, these findings suggest that these miRNAs have additional important downstream mediators that exert their phenotypic outputs.
In summary, several miRNAs, which are deregulated in melanoma, have been implicated in the regulation of melanoma cell survival. Further pursuit of these data will unravel new resistance mechanisms that may provide rationale for the design of new therapies.
Modulators of the immune response
Melanoma is a notoriously immunogenic cancer. Numerous antigens and antigen-specific lymphocytes have been isolated from melanoma cells and patients, respectively (reviewed in [63]). As such, to survive and propagate, melanoma cells must acquire the ability to escape immune control, often by downregulating certain antigens and proteins that are essential for immune recognition, or creating a broadly immunosuppressive microenvironment (reviewed in [64]). Recently, miRNAs have been described as modulators of this immune-evasive capability of melanoma cells.
As for modulation of proteins essential for tumor recognition, Heinemann et al. demonstrated that the tumor-suppressive miR-34a/c directly controls ULBP2 expression, a ligand of the natural killer cells (NK) and cytotoxic T lymphocytes (CTL) immunoreceptor NKG2D. They propose that the interaction of transmembranal ULBP2, present on melanoma cells, with the NKG2D receptor activates NK cells and CTLs, thereby creating an immune barrier for tumor progression. Overexpression of miR-34a/c reduced the activation of NK cells and the cytotoxic activity of CTL cells through this mechanism [65].
Our group reported that miR-30b/d overexpression enhances the metastatic potential of melanoma cells, in part through the creation of an immunosuppressive environment. As previously mentioned, the targeting of GALNT7 by miR-30b/d alters the glycosylation landscape of membrane-bound proteins. An unexpected consequence of this alteration is increased secretion of immune suppressive molecules such as IL-10. Interestingly, our group demonstrated that more regulatory T cells (Foxp3+) but fewer CD3+ T cells were present in metastasis-bearing lungs of mice injected with cells overexpressing miR-30b/d relative to control [36].
Immune evasion is an important paradigm in melanoma, and the mechanisms by which this occurs are intensely researched. Thus far, only two studies have been published specifically exploring miRNAs in melanoma-immune evasion, but the results clearly show an important role for miRNAs in this process.
Limitations of functional studies of microRNAs in melanoma
Differences in microRNA expression between cell lines and tumors. miRNA candidates described as dysregulated in melanoma have often been selected from comparisons of melanoma cell lines to cultured melanocytes. It remains unknown whether melanocytes, melanocytic progenitors or stem cells are the cell-of-origin of melanocytic transformation. The cell of origin may be of importance because the basal levels of miRNA and mRNA are likely to change depending on the differentiation state of the initiating cell. In addition, miRNA expression patterns are thought to change significantly from patient tissues to cell lines, with a large portion of miRNAs present in tumor tissues altered on cell-culture adaptation. For these reasons, it is critical to examine the expression of studied miRNAs in patient tissue to definitively show alteration in melanoma. In contrast, studies initiated from miRNA expression in clinical samples must take into consideration the confounding effect of other cell types (e.g., fibroblasts, lymphocytes) that may be present within the specimen. In addition, changes of expression may be strictly a consequence of local microenvironments, making difficult the comparison of, for instance, metastases in one organ site versus another. Several recent studies have begun to use short-term melanoma cultures to perhaps help bridge the gap between these two methodologies.
Lack of in vivo demonstration of miRNA role in melanoma pathogenesis. Many of the studies summarized above have been exclusively conducted in vitro. Further studies in human samples and animal models would greatly inform the relevance of these observations to human disease. In addition, the development of genetic models, which can be combined with established melanoma models, to generate tissue specific, conditional knock-in or knock-outs of oncomiRs will help to further define miRNA roles in melanoma.
Limited scope of microRNA target analyses.miRNAs exert their effects by simultaneously modulating multiple targets and pathways. However, most studies have provided a single miRNA:target model, likely oversimplifying the broad impact of a miRNA on global gene expression.
Outstanding questions and future directions
Comparative analysis of miRNA expression versus activity
The vast majority of studies have analyzed small sets of miRNAs by qRT-PCR or profiled miRNA expression more broadly by qRT-PCR, various array methodologies, or more recently, deep sequencing. While each of these provides information regarding the level or relative level of miRNA expression in the samples analyzed, none provide a measure of miRNA activity. Historically, the assumption has been that expression and activity will strongly correlate, but as with the expression of mRNA with protein, and protein levels with protein function, this may prove untrue. No methods are currently available to explore miRNA activity in patient samples. However, in cell lines, tools such as miRNA sensor reporters can be used to correlate miRNA expression with activity, thereby providing a clearer picture of miRNA functionality in melanoma.
Improved algorithms and methods of target identification. As the rules that govern miRNA target recognition are further refined, the list of theoretical targets provided by predictive algorithms should become more accurate. Improvements of in silico prediction combined with proteomic or mRNA profilings of cell lines transduced with specific miRNAs should render a more precise landscape of genes modulated by melanoma miRNAs. Moreover, novel tools designed for unbiased direct target identification, such as SILAC [66], HITS-CLIP [67] and PAR-CLIP [68], can provide a deeper understanding of the function of individual miRNAs, but have yet to be implemented in melanoma cell lines.
Investigate the etiology of microRNA alterations in melanoma Numerous ongoing efforts are focused on determining the molecular mechanisms (i.e., transcriptional, genetic, epigenetic) responsible for specific miRNA alterations. The promoter regions of most miRNA genes have not yet been delineated, hindering the study of transcriptional regulators of miRNAs. ChIP-seq mapping of histone marks (i.e., H3K9me3, H3K27me3) will aid in determining the transcription start sites of miRNA genes and in ascertaining whether they are transcriptionally primed/inactive or epigenetically silenced. This information should help spur the study of transcriptional regulation of miRNAs in melanoma.
Determine the contribution of miRNA to melanoma genetic susceptibility or response to therapy. Similar to the reported SNP that modulates miR-221 binding to c-KIT [69], other genetic variants may contribute to genetic susceptibility to melanoma or modulate tumor response to various therapies. To explore these questions, data from genome wide association studies (GWAS), The Cancer Genome Atlas (TCGA) or melanoma-related polymorphisms should be analyzed for predicted miRNA binding sites.
MicroRNAs in the clinical melanoma setting
Prognostic factors such as tumor stage or tumor thickness have limitations in predicting patient outcome. Biologic markers such as growth control genes, extracellular matrix-degrading enzymes, adhesion/signaling molecules, angiogenic factors and immunoregulatory molecules have been proposed as biomarkers for use in melanoma patient care [70–73]. However, none have moved past the preliminary stages of development into a clinically implemented assay, mainly because of difficulties in developing an easily standardized and objectively measured test.
In recent years, the value of miRNAs as potential diagnostic and prognostic biomarkers has been intensively explored as a result of several general properties specific to miRNAs. First, lineage specificity of miRNA expression suggests their potential use as diagnostic tools. Second, miRNA stability in tissues and body fluids allows for longitudinal and retrospective studies. MiRNA profiles or signatures can be useful in detecting melanoma and evaluating its burden and potential for progression. As such, miRNA might become clinical and prognostic biomarkers that can mark early detection of disease, progression and/or response to therapy as well as tumor-type discrimination. Table 2 summarizes the potential clinical utility of miRNAs in melanoma, discussed as follows.
Table II.
MicroRNA with potential clinical applications in melanoma
| Functional category | Name | Source | Discovery type | Correlation | Ref. |
|---|---|---|---|---|---|
| Diagnostic | miR-221 | Serum | Targeted Approach | Higher in MM patients (Stage I to IV) compared to normal subjects | [84] |
| 16 miRNA signature: miR-186, let-7d, miR-18a, miR-145, miR-99a, miR-664, miR-501-5p, miR-378, miR-29c, miR- 1280, miR-365, miR-1249, miR-328, miR-422a, miR-30d, miR-17 | Blood Cells | 866 miRNA array | Classification as melanoma patients | [87] | |
| Prognostic | miR-191 | FFPE Tissues | 470 miRNA array | Higher indicated worse survival | [91] |
| miR-193b | FFPE Tissues | 470 miRNA array | Higher indicated better survival | [91] | |
| miR-221 | Serum | Targeted Approach | Higher with disease burden (pre-operatively and at recurrence compared to post operatively) | [85] | |
| 18 miRNA signature: miR-150, miR-455-3p,miR-145,miR-342-3p,miR-497,miR-155,miR-342-5p,miR-143, miR-193a-3pmiR-146-5p,miR-28-3p,miR-10b,miR-193b,miR-28-5pmiR-143,miR-126,miR-214 | FFPE Tissues | 610 miRNA array | Higher associated with longer post-recurrence survival | [92] | |
| miR-15b | FFPE Tissues | Targeted Approach | Higher predicts lower disease free survival and shorter overall survival | [57] | |
| miR-29c | FFPE Tissues | Targeted Approach | Higher predicts increased overall survival | [93] | |
MicroRNAs as blood-based melanoma markers
miRNAs are remarkably stable in the bloodstream [74,75] since they are thought to be shed from exosomes/microvesicles, which protect them from degradation by endogenous RNAses [75–77]. Several studies examining miRNA in blood samples have indicated the ability of miRNAs to distinguish between disease-free and disease-burdened patients in several types of cancers [74,78–80], offering new hope of miRNA use in early detection or disease surveillance.
In a recently published study, sera samples from melanoma patients (melanoma in-situ through Stage IV) were compared with samples from age- and sex-matched healthy controls for expression of miR-221, a miRNA known to be overexpressed in melanoma [47,81–83]. Serum miR-221 levels were significantly higher in Stage I–IV patients compared to control subjects, and miR-221 levels correlated with tumor thickness in stages I–III. In a different set of eight recurrent patients whose samples were taken at three time points—before primary excision, after excision and at recurrence—miR-221 levels present in sera changed in concordance with disease burden [84]. However, these results are difficult to generalize due to the low proportion (14%) of superficial spreading melanoma cases (which in fact encompasses ~70% of melanomas) included in the cohort studied, compared to relatively high proportion (39%) of acral lentiginous melanoma (which comprises less that 10% of melanomas). In this regard, each melanoma histologic subtype has been shown to exhibit distinct miRNA profiles [85,86] suggesting these studies should be undertaken with specific histologic subtypes or with significantly larger cohorts.
In addition to serum, analysis of blood cells isolated from patients may provide detection markers for non-invasive disease. Leidinger et al. [87] identified a miRNA signature of melanoma comparing the miRNA profiles of blood cells from patients with melanoma (n = 35) versus healthy donors (n = 20). Sixteen significantly deregulated miRNAs were identified that could separate the groups with a classification accuracy of 97.4%, a specificity of 95% and a sensitivity of 98.9% by supervised analysis [87]. These results are particularly promising because most of the patients included in the analysis were diagnosed with early stage melanoma, suggesting that even a small tumor burden significantly impacts the miRNA expression pattern in blood cells. However, no information was provided or discussed regarding whether the miRNA signature is derived from circulating tumor cells or results from the immunologic host response, or both.
Tissue-based MicroRNAs as prognostic markers
Several studies have reported the stability of small RNAs, including miRNAs, in formalin-fixed paraffin embedded (FFPE) tissues [88]. This stability opens the possibility to extract small RNAs from archived samples and conduct retrospective studies. Furthermore, melanoma miRNA profiles obtained from FFPE samples have shown an excellent correlation with those from frozen material [89,90]. This is particularly relevant for the study of primary melanoma, for which conventional gene expression profiles have been difficult to obtain, as the small size of primary melanomas (millimeters) precludes banking of frozen material, leaving only FFPE tissue accessible.
It is promising then that miRNAs, in a variety of contexts, are able to predict disease outcome and prognosis with reasonable accuracy (Table 2). For example, miRNA profiles of 16 melanomas extracted from positive lymph nodes of recurrent melanoma patients with short (<12 months) versus long (>60 months) overall survival revealed clear differences. Low expression levels of miR-191 and high expression of miR-193b were indicative of better survival in this test cohort and in an additional validation cohort of 16 melanomas [91]. Using a similar strategy on a larger specimen collection (n = 59), our group analyzed the miRNA profiles of tumors from patients with metastatic melanoma. We found that higher expression of 18 miRNAs, including again miR-193b, was significantly correlated with longer survival (survival greater than 18 months post-recurrence). A smaller cohort was identified showing six miRNAs (miR-150, miR-342-3p, miR-455-3p, miR-145, miR-155, miR-497) that predict post-recurrence survival in these patients with an estimated accuracy of 80.2% [92].
A study analyzing primary melanoma tissues for three miRNAs previously found to be dysregulated in melanoma (miR-15b, miR-210 and miR-34a) showed that high levels of miR-15b represented an independent parameter of low disease-free survival and shorter overall survival in multivariate analyses [57]. In addition, another group compared levels of miR-29a/b/c in stage I through IV tissues. Of these, miR-29c expression was found to be lower in metastatic tissues than in primary tissues. Furthermore, after testing lymph node tissues for miR-29c levels, above median values correlated with significantly improved overall survival [93]. Although the targeted approach used in some of these studies may have limited the discovery space, all miRNAs examined had substantial evidence of dysregulation, correlating with clinical outcome.
Therapeutic potential of microRNA in melanoma
In melanoma, although total vertical thickness of the primary melanoma lesion (e.g., tumor burden or Breslow score) correlates with risk of distant metastasis, 30% of patients with ‘thin’ melanoma ultimately succumb to metastatic disease. No curative treatment exists for Stage IV melanoma, and these patients have a median overall survival of only seven months [94]. Therefore, there is an urgent need to identify patients in early stages of disease that are most likely to progress to stratify them for more rigorous follow-up and aggressive therapy, including sentinel lymph node dissection or systematic adjuvant or neoadjuvant therapies, as these therapies become available [95,96].
Classical treatment options, with limited efficacy, include chemotherapy, immunotherapy, or a combination of both. Although mutant-specific BRAF inhibitors (vemurafenib) or anti-CTLA-4 antibodies (ipilimumab) are showing promising clinical results, emergence of resistance has already been encountered [97,98] and is nearly ubiquitous in vemurafenib-treated patients. In part, this phenomenon is attributed to functional redundancy in tumor cells, which likely buffers the impact of a single gene/target modification on the malignant process. miRNA-based cancer gene therapy (reviewed in [99]) offers the possibility of targeting multiple gene networks that are controlled by an individual miRNA [100].
MiRNAs can be inhibited in vitro with oligonucleotides with specific modifications (i.e., 2ʹ-O-methyl oligonucleotides, locked nucleic acids (LNA)) and delivered in the form of liposomal complexes into adult mice and even non-human primates [101]. These could conceivably be used to negate the translational-regulatory effects of “oncogenic” miRNAs [102,103]. A caveat of most current formulations of miRNA delivery is that they tend to accumulate in the liver with potential toxicity and limiting concentration in other tissues. Nanoparticle-based delivery [104] may improve the specificity and efficacy of miRNA based therapies.
Preliminary in vivo studies have shown therapeutic potential for a few miRNAs in melanoma. Inhibition of miR-221 with antisense oligonucleotides, delivered intratumorally, resulted in reduced tumor growth [47].
Our group showed therapeutic potential of anti-miR-182 oligonucleotides in a spleen-to-liver metastasis model. Mice receiving anti-miR-182 treatment displayed measurably lower miR-182 levels in tumor cells and a reduced burden of liver metastases than controls [105]. Moreover, known targets of miR-182, such as FOXO3 [46] and ADCY6 [106], were upregulated in metastatic lesions of treated mice compared to controls. It is important to note that no changes in weight or any gross histological differences in lung, spleen and brain tissues of mice treated with anti-miR-182 were observed [105].
These preliminary studies offer hope for future melanoma therapies involving miRNAs.
Clinical directions and future perspectives
While ample data has been collected on the ability of specific miRNAs to predict disease presence and burden, more work is necessary to develop melanoma-specific tests that are accurate and reproducible. Blood-based assays using cells or sera offer numerous advantages over conventional tissue testing (i.e., non-invasive sampling method, possibility of longitudinal monitoring), yet more studies with larger cohorts and age- and sex-matched controls are required. In addition, studies should be expanded to larger sample sizes and include a variety of diverse control groups (i.e., blood-related pathologies, systemic inflammatory conditions). In addition, more representative histotype populations are needed to confirm the potential of miRNAs for melanoma surveillance.
In sum, miRNAs may assist in the diagnosis and early detection of melanoma recurrence and predict patient outcome. The development of accurate progression risk biomarkers would greatly enhance the clinical management of melanoma. Currently, anatomic imaging studies (e.g., chest X-ray, MRI, CT, PET) are the mainstays of follow-up care, but the sensitivity of these tests is limited, as tumor cell deposits may grow for months at distant sites before reaching detectable levels [107]. Ultimately, the goal should be to develop easily standardized tests, such as qPCR-based assays, that use a limited number of miRNAs to determine disease burden or predict its outcome.
Moreover, the findings reviewed herein show the enormous potential of miRNAs to guide clinical decisions to sub-classify patients susceptible to novel targeted treatments. Furthermore, functional studies with miRNAs have opened a new avenue to unravel molecular targets that could be exploited therapeutically. However, the evolving field of miRNAs in melanoma still requires large studies with functional data correlated with clinical outcomes before effective targeted approaches can impact patient management.
Funding
Department of Defense (CA093471), Melanoma Research Foundation and NIH/NCI (1R01CA155234-01A1).
Acknowledgments
We thank Avital Gaziel-Sovran, Raffaella Di Micco and Erica Friedman for their critical reading of the manuscript. We apologize to those whose studies could not be mentioned because of space constraints.
References
- 1. Bleyer A., et al. (2006). Cancer in 15- to 29-year-olds by primary site. Oncologist 11 590–601 [DOI] [PubMed] [Google Scholar]
- 2. Kohler B.A., et al. (2011). Annual report to the nation on the status of cancer, 1975-2007, featuring tumors of the brain and other nervous system. J. Natl. Cancer Inst. 103 714–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lee R.C., et al. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75 843–854 [DOI] [PubMed] [Google Scholar]
- 4. Reinhart B.J., et al. (2000). The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403 901–906 [DOI] [PubMed] [Google Scholar]
- 5. Bernstein E., et al. (2003). Dicer is essential for mouse development. Nat. Genet. 35 215–217 [DOI] [PubMed] [Google Scholar]
- 6. Lu J., et al. (2005). MicroRNA expression profiles classify human cancers. Nature 435 834–838 [DOI] [PubMed] [Google Scholar]
- 7. Zhang L., et al. (2006). microRNAs exhibit high frequency genomic alterations in human cancer. Proc. Natl. Acad. Sci. U.S.A. 103 9136–9141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bonazzi V.F., et al. (2012). MicroRNA regulation of melanoma progression. Melanoma Res. 22 101–113 [DOI] [PubMed] [Google Scholar]
- 9. Flørenes V.A., et al. (2000). Levels of cyclin D1 and D3 in malignant melanoma: deregulated cyclin D3 expression is associated with poor clinical outcome in superficial melanoma. Clin. Cancer Res. 6 3614–20 [PubMed] [Google Scholar]
- 10. Schultz J., et al. (2008). MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 18 549–557 [DOI] [PubMed] [Google Scholar]
- 11. Müller D.W., et al. (2008). Integrin beta 3 expression is regulated by let-7a miRNA in malignant melanoma. Oncogene 27 6698–706 [DOI] [PubMed] [Google Scholar]
- 12. Johnson S.M., et al. (2005). RAS is regulated by the let-7 microRNA family. Cell 120 635–647 [DOI] [PubMed] [Google Scholar]
- 13. Chen J., et al. (2011). miR-193b Regulates Mcl-1 in Melanoma. Am. J. Pathol. 179 2162–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Polsky D., et al. (2003). Oncogenes in melanoma. Oncogene 22 3087–3091 [DOI] [PubMed] [Google Scholar]
- 15.Noguchi S, et al. Comparative Study of Anti-Oncogenic MicroRNA-145 in Canine and Human Malignant Melanoma. J Vet Med Sci. (2012) doi: 10.1292/jvms.11-0264. [DOI] [PubMed] [Google Scholar]
- 16. Felicetti F., et al. (2008). The promyelocytic leukemia zinc finger-microRNA-221/-222 pathway controls melanoma progression through multiple oncogenic mechanisms. Cancer Res. 68 2745–2754 [DOI] [PubMed] [Google Scholar]
- 17. Garofalo M., et al. (2009). miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell 16 498–509 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18. Igoucheva O., et al. (2009). MicroRNA-dependent regulation of cKit in cutaneous melanoma. Biochem. Biophys. Res. Commun. 379 790–794 [DOI] [PubMed] [Google Scholar]
- 19. Curtin J.A., et al. (2006). Somatic activation of KIT in distinct subtypes of melanoma. J. Clin. Oncol. 24 4340–4346 [DOI] [PubMed] [Google Scholar]
- 20. Roberts J.D. (2006). E2F1 amplication and genetic heterogeneity in melanoma. Cancer Biol. Ther. 5 691–692 [DOI] [PubMed] [Google Scholar]
- 21. Limon J., et al. (1988). Chromosome changes in metastatic human melanoma. Cancer Genet. Cytogenet. 30 201–211 [DOI] [PubMed] [Google Scholar]
- 22. Dar A.A., et al. (2011). miRNA-205 suppresses melanoma cell proliferation and induces senescence via regulation of E2F1 protein. J. Biol. Chem. 286 16606–16614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Y, et al. Differential expression of microRNAs during melanoma progression: miR-200c, miR-205 and miR-211 are downregulated in melanoma and act as tumour suppressors. Br. J. Cancer. (2012) doi: 10.1038/bjc.2011.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Amiri K.I., et al. (2005). Role of nuclear factor-kappa B in melanoma. Cancer Metastasis Rev. 24 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu S., et al. (2012). MicroRNA-9 up-regulates E-cadherin through inhibition of NF-KappaB1-Snail1 pathway in melanoma. J. Pathol. 226 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Levati L, et al. (2009). Altered expression of selected microRNAs in melanoma: antiproliferative and proapoptotic activity of miRNA-155 International Journal of Oncology 35 393–400 [PubMed] [Google Scholar]
- 27.Greenberg E., et al. Regulation of cancer aggressive features in melanoma cells by microRNAs. PLoS ONE. (2011);6:e18936. doi: 10.1371/journal.pone.0018936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chiang A.C., et al. (2008). Molecular basis of metastasis. N. Engl. J. Med. 359 2814–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fu T.Y., et al. Let-7b-mediated suppression of basigin expression and metastasis in mouse melanoma cells. Exp. Cell Res. 317 445–51 [DOI] [PubMed] [Google Scholar]
- 30. Li X., et al. (1998). Differential expression of alphav integrins in K1735 melanoma cells. Invasion Metastasis 18 1–14 [DOI] [PubMed] [Google Scholar]
- 31. Penna E., et al. (2011). microRNA-214 contributes to melanoma tumour progression through suppression of TFAP2C. EMBO J. 30 1990–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cruz J., et al. (2003). Expression of c-met tyrosine kinase receptor is biologically and prognostically relevant for primary cutaneous malignant melanomas. Oncology 65 72–82 [DOI] [PubMed] [Google Scholar]
- 33. Otsuka T., et al. (1998). c-Met autocrine activation induces development of malignant melanoma and acquisition of the metastatic phenotype. Cancer Res. 58 5157–5167 [PubMed] [Google Scholar]
- 34.Taghizadeh R., et al. CXCR6, a newly defined biomarker of tissue-specific stem cell asymmetric self-renewal, identifies more aggressive human melanoma cancer stem cells. PLoS ONE. (2010);5:e15183. doi: 10.1371/journal.pone.0015183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Migliore C., et al. (2008). MicroRNAs impair MET-mediated invasive growth. Cancer Res. 68 10128–36 [DOI] [PubMed] [Google Scholar]
- 36. Gaziel-Sovran A., et al. (2011). miR-30b/30d regulation of GalNAc transferases enhances invasion and immunosuppression during metastasis. Cancer Cell 20 1041–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Janik M.E., et al. (2010). Cell migration-the role of integrin glycosylation. Biochim. Biophys. Acta 1800 545–55 [DOI] [PubMed] [Google Scholar]
- 38. Rambaruth N.D., et al. (2011). Cell surface glycan-lectin interactions in tumor metastasis. Acta Histochem. 113 591–600 [DOI] [PubMed] [Google Scholar]
- 39. Elson-Schwab I., et al. MicroRNA-200 family members differentially regulate morphological plasticity and mode of melanoma cell invasion. PLoS ONE. (2010);5 doi: 10.1371/journal.pone.0013176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mazar J., et al. (2011). Epigenetic regulation of microRNA-375 and its role in melanoma development in humans. FEBS Lett. 585 2467–2476 [DOI] [PubMed] [Google Scholar]
- 41. Bell R.E., et al. (2011). The three M’s: melanoma, microphthalmia-associated transcription factor and microRNA. Pigment Cell Melanoma Res. 24 1088–1106 [DOI] [PubMed] [Google Scholar]
- 42. Carreira S., et al. (2006). Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev. 20 3426–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hoek K.S., et al. (2008). In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Res. 68 650–656 [DOI] [PubMed] [Google Scholar]
- 44. Bemis L.T., et al. (2008). MicroRNA-137 targets microphthalmia-associated transcription factor in melanoma cell lines. Cancer Res. 68 1362–8 [DOI] [PubMed] [Google Scholar]
- 45.Haflidadóttir B.S., et al. miR-148 regulates Mitf in melanoma cells. PLoS ONE. (2010);5:e11574. doi: 10.1371/journal.pone.0011574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Segura M.F., et al. (2009). Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc. Natl. Acad. Sci. U.S.A. 106 1814–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Felicetti F., et al. (2008). MicroRNA-221 and -222 pathway controls melanoma progression. Expert Rev. Anticancer Ther. 8 1759–1765 [DOI] [PubMed] [Google Scholar]
- 48. Hou L., et al. (2000). Signaling and transcriptional regulation in the neural crest-derived melanocyte lineage: interactions between KIT and MITF. Development 127 5379–5389 [DOI] [PubMed] [Google Scholar]
- 49.Phung B., et al. C-KIT signaling depends on microphthalmia-associated transcription factor for effects on cell proliferation. PLoS ONE. (2011);6:e24064. doi: 10.1371/journal.pone.0024064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mazar J., et al. The regulation of miRNA-211 expression and its role in melanoma cell invasiveness. PLoS ONE. (2010);5:e13779. doi: 10.1371/journal.pone.0013779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Levy C., et al. (2010). Intronic miR-211 assumes the tumor suppressive function of its host gene in melanoma. Mol. Cell 40 841–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Boyle G.M., et al. (2011). Melanoma cell invasiveness is regulated by miR-211 suppression of the BRN2 transcription factor. Pigment Cell Melanoma Res. 24 525–37 [DOI] [PubMed] [Google Scholar]
- 53. Mueller D.W., et al. (2011). MicroRNA miR-196a controls melanoma-associated genes by regulating HOX-C8 expression. Int. J. Cancer 129 1064–1074 [DOI] [PubMed] [Google Scholar]
- 54. Braig S., et al. (2010). MicroRNA miR-196a is a central regulator of HOX-B7 and BMP4 expression in malignant melanoma. Cell. Mol. Life Sci. 67 353535–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Glud M., et al. (2010). Downregulation of miR-125b in metastatic cutaneous malignant melanoma. Melanoma Res. 20 479–484 [DOI] [PubMed] [Google Scholar]
- 56. Glud M., et al. (2011). MicroRNA miR-125b induces senescence in human melanoma cells. Melanoma Res. 21 253–6 [DOI] [PubMed] [Google Scholar]
- 57. Satzger I, et al. (2010). MicroRNA-15b represents an independent prognostic parameter and is correlated with tumor cell proliferation and apoptosis in malignant melanoma International journal of cancer. Journal international du cancer 126 2553–2562 [DOI] [PubMed] [Google Scholar]
- 58.Streicher K.L., et al. A novel oncogenic role for the miRNA-506-514 cluster in initiating melanocyte transformation and promoting melanoma growth. Oncogene. (2012) doi: 10.1038/onc.2011.345. [DOI] [PubMed] [Google Scholar]
- 59. Pan X., et al. (2011). MicroRNA-21: a novel therapeutic target in human cancer. Cancer Biol. Ther. 10 1224–1232 [DOI] [PubMed] [Google Scholar]
- 60. Yang C.H., et al. (2011). MicroRNA miR-21 regulates the metastatic behavior of B16 melanoma cells. J. Biol. Chem. 286 39172–39178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jiang L, et al. The status of microRNA-21 expression and its clinical significance in human cutaneous malignant melanoma. Acta Histochem. doi: 10.1016/j.acthis.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 62. Jin L, et al. (2011). MicroRNA-149*, a p53-responsive microRNA, functions as an oncogenic regulator in human melanoma Proceedings of the National Academy of Sciences of the United States of America 108 15840–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kawakami Y., et al. (2005). Immunological detection of altered signaling molecules involved in melanoma development. Cancer Metastasis Rev. 24 357–366 [DOI] [PubMed] [Google Scholar]
- 64. Márquez-Rodas I., et al. (2011). A new era in the treatment of melanoma: from biology to clinical practice. Clin. Transl. Oncol. 13 787–792 [DOI] [PubMed] [Google Scholar]
- 65. Heinemann A., et al. (2012). Tumor suppressive microRNAs miR-34a/c control cancer cell expression of ULBP2, a stress-induced ligand of the natural killer cell receptor NKG2D. Cancer Res. 72 460–471 [DOI] [PubMed] [Google Scholar]
- 66.Vinther J., et al. Identification of miRNA targets with stable isotope labeling by amino acids in cell culture. Nucleic Acids Res. (2006);34:e107. doi: 10.1093/nar/gkl590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chi S.W., et al. (2009). Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460 479–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hafner M., et al. (2010). Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141 129–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Godshalk S.E., et al. (2011). A Variant in a MicroRNA complementary site in the 3’ UTR of the KIT oncogene increases risk of acral melanoma. Oncogene 30 1542–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rudolph P, et al. (2000). Telomerase activity in melanocytic lesions: A potential marker of tumor biology The American journal of pathology 156 1425–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Polsky D, et al. (2001). HDM2 protein overexpression, but not gene amplification, is related to tumorigenesis of cutaneous melanoma Cancer research 61 7642–6 [PubMed] [Google Scholar]
- 72. Hofmann U.B, et al. (2000). Coexpression of integrin alpha(v)beta3 and matrix metalloproteinase-2 (MMP-2) coincides with MMP-2 activation: correlation with melanoma progression The Journal of investigative dermatology 115 625–32 [DOI] [PubMed] [Google Scholar]
- 73. Berger A.J, et al. (2004). Automated quantitative analysis of HDM2 expression in malignant melanoma shows association with early-stage disease and improved outcome Cancer research 64 8767–8772 [DOI] [PubMed] [Google Scholar]
- 74.Gilad S., et al. Serum microRNAs are promising novel biomarkers. PLoS ONE. (2008);3:e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mitchell P.S., et al. (2008). Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U.S.A. 105 10513–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Valadi H., et al. (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9 654–659 [DOI] [PubMed] [Google Scholar]
- 77. Zomer A., et al. (2010). Exosomes: Fit to deliver small RNA. Commun. Integr. Biol. 3 447–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Resnick K.E, et al. (2009). The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform Gynecologic oncology 112 55–59 [DOI] [PubMed] [Google Scholar]
- 79. Chin L.J., et al. (2008). A SNP in a let-7 microRNA complementary site in the KRAS 3ʹ untranslated region increases non-small cell lung cancer risk. Cancer Res. 68 8535–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Chen X, et al. (2008). Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases Cell research 18 997–1006 [DOI] [PubMed] [Google Scholar]
- 81. Miller B.A., et al. (2008). Cancer incidence and mortality patterns among specific Asian and Pacific Islander populations in the U.S. Cancer Causes Control 19 227–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Fornari F., et al. (2008). MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene 27 5651–5661 [DOI] [PubMed] [Google Scholar]
- 83. Ciafrè S.A., et al. (2005). Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem. Biophys. Res. Commun. 334 1351–8 [DOI] [PubMed] [Google Scholar]
- 84. Kanemaru H, et al. (2011). The circulating microRNA-221 level in patients with malignant melanoma as a new tumor marker Journal of dermatological science 61 187-1–93 [DOI] [PubMed] [Google Scholar]
- 85. Chan E., et al. (2011). MicroRNA signatures differentiate melanoma subtypes. Cell Cycle 10 1845–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Poliseno L, et al. (2011). Distinguishing between nodular and superficial spreading melanoma using specific microRNA alterations Journal of Clinical Oncology 29, (suppl 8540) [Google Scholar]
- 87.Leidinger P., et al. High-throughput miRNA profiling of human melanoma blood samples. BMC Cancer. (2010);10:262. doi: 10.1186/1471-2407-10-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Xi Y., et al. (2007). Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. RNA 13 1668–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Liu A., et al. (2009). MicroRNA expression profiling outperforms mRNA expression profiling in formalin-fixed paraffin-embedded tissues. Int. J. Clin. Exp. Pathol. 2 519–27 [PMC free article] [PubMed] [Google Scholar]
- 90. Glud M., et al. (2009). MicroRNA expression in melanocytic nevi: the usefulness of formalin-fixed, paraffin-embedded material for miRNA microarray profiling. J. Invest. Dermatol. 129 1219–24 [DOI] [PubMed] [Google Scholar]
- 91. Caramuta S., et al. (2010). MicroRNA expression profiles associated with mutational status and survival in malignant melanoma. J. Invest. Dermatol. 130 2062–70 [DOI] [PubMed] [Google Scholar]
- 92. Segura M.F., et al. (2010). Melanoma MicroRNA signature predicts post-recurrence survival. Clin. Cancer Res. 16 1577–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Nguyen T, et al. (2011). Downregulation of microRNA-29c is associated with hypermethylation of tumor-related genes and disease outcome in cutaneous melanoma. Epigenetics: official journal of the DNA Methylation Society 6 388–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Rubin K.M., et al. (2009). Your patient with melanoma: staging, prognosis, and treatment. Oncology (Williston Park) 23 13–21 [PubMed] [Google Scholar]
- 95. Flaherty K.T., et al. (2010). Inhibition of mutated, activated BRAF in metastatic melanoma. N. Engl. J. Med. 363 809–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. O’Day S., et al. (2011). A randomised, phase II study of intetumumab, an anti-av-integrin mAb, alone and with dacarbazine in stage IV melanoma. Br. J. Cancer 105 346–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Johannessen C.M., et al. (2010). COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature 468 968–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Nazarian R., et al. (2010). Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 468 973–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Tong A.W., et al. (2008). Modulation of miRNA activity in human cancer: a new paradigm for cancer gene therapy? Cancer Gene Ther. 15 341–55 [DOI] [PubMed] [Google Scholar]
- 100. van Rooij E., et al. (2007). Control of stress-dependent cardiac growth and gene expression by a microRNA. Science 316 575–9 [DOI] [PubMed] [Google Scholar]
- 101. Elmén J., et al. (2008). LNA-mediated microRNA silencing in non-human primates. Nature 452 896–9 [DOI] [PubMed] [Google Scholar]
- 102. Meister G., et al. (2004). Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA 10 544–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Weiler J., et al. (2006). Anti-miRNA oligonucleotides (AMOs): ammunition to target miRNAs implicated in human disease? Gene Ther. 13 496–502 [DOI] [PubMed] [Google Scholar]
- 104. Davis M.E., et al. (2010). Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature 464 1067–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Huynh C., et al. (2011). Efficient in vivo microRNA targeting of liver metastasis. Oncogene 30 1481–88 [DOI] [PubMed] [Google Scholar]
- 106. Xu S., et al. (2007). MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J. Biol. Chem. 282 25053–66 [DOI] [PubMed] [Google Scholar]
- 107. Schwimmer J, et al. (2000). A review of the literature for whole-body FDG PET in the management of patients with melanoma The quarterly journal of nuclear medicine: official publication of the Italian Association of Nuclear Medicine 44 153-–67 [PubMed] [Google Scholar]