Abstract
Colon cancer growth requires growth-promoting interactions between malignant colonocytes and stromal cells. Epidermal growth factor receptors (EGFR) are expressed on colonocytes and many stromal cells. Furthermore, EGFR is required for efficient tumorigenesis in experimental colon cancer models. To dissect the cell-specific role of EGFR, we manipulated receptor function on stromal cells and cancer cells. To assess the role of stromal EGFR, HCT116 human colon cancer cells were implanted into immunodeficient mice expressing dominant negative (DN) EgfrVelvet/+ or Egfr+/+. To assess the role of cancer cell EGFR, HCT116 transfectants expressing inducible DN-Egfr were implanted into immunodeficient mice. To dissect EGFR signals in vitro, we examined colon cancer cells in monoculture or in cocultures with fibroblasts for EGFR transactivation and prostaglandin synthase 2 (PTGS2) induction. EGFR signals were determined by blotting, immunostaining and real-time PCR. Tumor xenografts in EgfrVelvet/+ mice were significantly smaller than tumors in Egfr+/+ mice, with decreased proliferation (Ki67) and increased apoptosis (cleaved caspase-3) in cancer cells and decreased stromal blood vessels. Mouse stromal transforming growth factor alpha (TGFA), amphiregulin (AREG), PTGS2 and Il1b and interleukin-1 receptor 1 (Il1r1) transcripts and cancer cell beta catenin (CTNNB1) and cyclin D1 (CCND1) were significantly lower in tumors obtained from EgfrVelvet/+ mice. DN-EGFR HCT116 transfectants also formed significantly smaller tumors with reduced mouse Areg, Ptgs2, Il1b and Il1r1 transcripts. Coculture increased Caco-2 phospho-active ERBB (pERBB2), whereas DN-EGFR in Caco-2 cells suppressed fibroblast PTGS2 and prostaglandin E2 (PGE2). In monoculture, interleukin 1 beta (IL1B) transactivated EGFR in HCT116 cells. Stromal cell and colonocyte EGFRs are required for robust EGFR signals and efficient tumor growth, which involve EGFR–interleukin-1 crosstalk.
Introduction
Colon cancer growth is driven by cell–cell and cell–matrix physical interactions and paracrine and autocrine signals involving malignant colonocytes and supporting stromal cells. Colon cancer stroma is increasingly recognized as playing an active role in colonic tumor development (1,2). The stroma includes fibroblasts, immune cells, endothelial cells and the extracellular matrix, which communicate stimulatory and inhibitory cues to tumor epithelial cells via complex networks (1,2). Growth factors, cytokines, chemokines, prostanoids, integrins and other bioactive molecules mediate these bidirectional signals. Among the growth factor signals, the epidermal growth factor receptors (EGFR) and several of their ligands are upregulated in colon cancer (3,4). The receptors are expressed on both malignant colonocytes and several stromal cell types, including fibroblasts and endothelial cells (5,6). In addition, colonic epithelial cells, fibroblasts, endothelial cells and macrophage cells release EGFR ligands (5,7,8). EGFR is also implicated in colonic stem cell regulation and is dysregulated in experimental models of colon cancer (9,10). In prior studies, we showed that EGFR promotes experimental colonic tumorigenesis and tumor progression (11–14). We also identified the proto-oncogenes cyclin D1 (CCND1) and prostaglandin synthase 2 (PTGS2) as important mediators of EGFR in colon cancer development (11,12,14). CCND1, a key regulator of G1 → S cell cycle progression, is upregulated by EGFR in transformed colonocytes (11,12,14). PTGS2, the rate-limiting enzyme for prostaglandin biosynthesis, is also controlled by EGFR in experimental colonic tumorigenesis and is initially increased in stromal myofibroblasts in human colonic adenomas (11,12,14,15).
In prior studies of colonic tumorigenesis, we blocked EGFR in vivo using global pharmacological inhibitors or Egfr germ line mutations that reduced EGFR signals in all cells (11–14). These studies did not determine, however, whether CCND1 and PTGS2 required EGFR signals in colonocytes or stromal cells, respectively. Recent studies, moreover, suggest that the stroma may be important for tumor resistance to EGFR antagonists (16–18). To address the contributions of colonocyte and stromal cell EGFR to tumor growth, we employed in vivo tumor xenograft models and in vitro coculture models to dissect cell-specific roles of EGFR. For in vivo studies, we used parental HCT116 colon cancer cells and exploited a mouse expressing Egfr Velvet, a dominant negative (DN) mutant EGFR, in order to abrogate EGFR signals in the tumor stroma (19,20). To dissect the in vivo contribution of colon cancer cell EGFR to tumor xenograft growth, we bioengineered HCT116 cells to express a dominant negative EGFR (DN-EGFR) under doxycycline-inducible (rtTA) regulation. Unlike Egfr Velvet, this DN-EGFR lacks a cytoplasmic domain. We used these systems to independently inhibit EGFR signals in vivo in stromal cells or colon cancer cells to dissect cell- or compartment-specific EGFR contributions to cell signals and tumor xenograft growth. For these studies, we also examined the effects of stromal cell and colon cancer cell EGFR on pro-inflammatory interleukin 1 beta (IL1B) that is upregulated in colon cancer and has been shown to induce EGFR ligands in colonic fibroblasts (5,21–23).
To dissect in vitro how EGFR and IL1B signals interact and crosstalk between cancer cells and stromal cells, we employed mono- and coculture models. To determine how colon cancer cells modulate PTGS2 expression in stromal fibroblast cells, we employed a novel strategy involving fibroblasts cocultured with colon cancer cells that expressed an inducible DN-EGFR. For fibroblast cells, we utilized CCD-18Co cells, a human embryonic colonic fibroblast cell line (24). In the case of colon cancer cells, we transfected Caco-2 cells with complementary DNA (cDNA) coding for DN-EGFR controlled by an inducible eukaryotic expression system (25). With these systems, we showed that IL1B transactivated EGFR in colon cancer cells and uncovered an important role for colonocyte EGFR in the control of fibroblast PTGS2 expression. These studies have identified EGFR–IL1B crosstalk between stromal cells and cancer cells, which probably contributes to the increases in PTGS2 observed in colon cancers.
Further support for the role of EGFR in colon cancer growth comes from the clinical use of cetuximab, an EGFR-neutralizing antibody. Cetuximab is widely used in combination with cytotoxic therapies to treat metastatic colon cancers that possess wild-type KRAS (26). It is generally believed that the cancer cell is the target of this antibody. Recent studies suggesting that colonic tumors with mutations of the KRAS codon 13 are sensitive to this antibody raise the question of the target cell for this therapy (27). KRAS mutations would theoretically be predicted to make cancer cells resistant to EGFR blockade. Results of the current study suggest that stromal cell EGFR may be an important target of such therapies.
Materials and methods
Materials
EgfrVelvet/wt and Rag1-/- mice were obtained from the Jackson Laboratory and both had C57BL6/J background. EgfrVelvet/wt × Rag1-/- were intercrossed to establish and expand F2 colonies of EgfrVelvet/wt Rag1-/- and Egfrwt/wt Rag1-/-. NOD-Scid IL2Rgamma null mice were also obtained from the Jackson Laboratory. Unsupplemented and doxycycline-supplemented rodent chow was purchased from Harlan Teklad Laboratory (Madison, WI). DNA and RNA extractions were performed using the Qiagen DNeasy kit (#69504) and RNeasy lipid extraction kit (#78404), respectively (Qiagen, Valencia, CA). RNAlater™ RNA storage solution, and DNA-free™ DNase I kit were purchased from Ambion (Austin, TX). TRIzol® RNA–DNA–Protein isolation reagent was obtained from Gibco BRL (Gaithersburg, MD). RiboGreen® RNA quantitation reagent and kit were purchased from Molecular Probes (Eugene, OR). Custom PCR primers were obtained from Integrated DNA Technologies (Coralville, IA). Other PCR reagents, including Moloney murine leukemia virus reverse transcriptase, random hexamers, and SYBR Green were purchased from Applied Biosystems (Foster City, CA). HotStarTaq™ DNA polymerase was supplied by Qiagen (Santa Clarita, CA). SuperScript III Platinum Two-Step qRT-PCR kit was obtained from Invitrogen (Carlsbad, CA). Superfrost Plus slides were purchased from Fisher Scientific (Pittsburgh, PA). Polyclonal antibodies to phospho (active) EGFR (SC-16802), phospho (active) ERBB2 (SC-12352R), phospho-(active) extracellular signal-regulated kinase (ERK)-1 and ERK-2 (SC-7383), and anti-CCND1 (SC-718) and anti-panERBB2 (SC-284) and anti-amphiregulin (SC-5796) antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies to phospho-active EGFR recognize human EGFR phosphorylated on tyrosine 1092 and antibodies to phospho-active ERBB2 recognize human ERBB2 phosphorylated on tyrosine 1248. Polyclonal pan ERK (#9102), pan AKT antibodies (#9271) and pSTAT3 antibodies (#9138) were obtained from Cell Signaling Technology (Beverly, MA). C225 antibodies were provided by Imclone (Bridgewater, NJ). Monoclonal antibodies against Ki67 (clone SP1) were obtained from Neomarkers (Fremont, CA). Monoclonal beta actin (ACTB) antibodies and anti-smooth muscle α-actin antibodies (Clone 1A4, catalogue A 2547) were obtained from Sigma–Aldrich (St. Louis, MO). Rat monoclonal anti-nestin (#556309) and mouse monoclonal anti-beta catenin (CTNNB1) antibodies (#610153) were obtained from BD Pharmingen (Palo Alto, CA). Rabbit anti-PTGS2 antibodies (#160106) were purchased from Cayman Chemicals (Ann Arbor, MI). Anti-cleaved caspase-3 antibodies (CP229A) were purchased from Biocare Medical (Concord, CA). Polyclonal pan EGFR antibodies were obtained from Upstate Biotechnology (Lake Placid, NY). Monoclonal antibodies to transforming growth factor alpha (TGF-α, ab9578) were purchased from Abcam (Cambridge, MA). The antibodies used in this study recognized both human and mouse proteins. Recombinant human interleukin-1B was obtained from R&D Systems (Minneapolis, MN) and recombinant human EGF, from EMD Millipore (Billerica, MA). RC–DC protein assay was from Bio-Rad Labs (Richmond, CA). Wst-1 assay kit was purchased from Roche Applied Science (Indianapolis, IN). Kodak (Rochester, NY) supplied the X-OMAT AR film. Polyvinylidene difluoride membranes (Immobilon-P) were purchased from Millipore (Bedford, MA). Unless otherwise noted, all other reagents were of the highest quality available and were obtained from Sigma–Aldrich.
Cell culture
Low-passage CCD-18Co colonic fibroblasts, Caco-2 cells, and HCT-116 cells were obtained from the American Type Culture Collection and cultured at 37°C in a humidified atmosphere of 5%CO2:95% air as recommended by ATCC and described by scientists in our laboratory (13,28,29). Cells were maintained in supplemented Dulbecco’s Minimum Essential Medium containing 20% fetal bovine serum (Caco-2) or McCoy’s 5A Modified Medium (HCT116), containing 10% serum or Eagle’s minimal essential medium (CCD-18Co cells) containing 15% serum. Media were supplemented with penicillin and streptomycin (50 units/ml). Cells were tested and found to be negative for mycoplasma.
DN-EGFR-expressing HCT116 transfectants
The DN-EGFR-expressing plasmid was obtained from Alex Ulrich (Max Planck Institute, Berlin, Germany). This is a deletion mutant of human EGFR, which encodes a mutant EGFR lacking the intracellular C-terminal residues (30). The encoded DN-EGFR binds EGFR ligands and undergoes dimerization but cannot transactivate wild-type ERBB partners. The DN-EGFR was hemagglutinin epitope (HA) tagged on the C-terminal end and subcloned into pIND-Tight vector (Invitrogen) and the construct was verified by sequencing. HCT116 cells expressing high levels of rtTA doxycycline transactivator (pTet-on system) were described previously (31). These cells were transfected with HA-DN-EGFR-pIND using a standard lipofectamine protocol as described (29) and cotransfected with a 10 µg/ml blasticidin resistance vector for selection. Monoclonal populations were expanded and maintained under G418 (400 µg/ml) and blasticidin (5 µg/ml) selection and screened for doxycycline-inducible (0.5 µg/ml) blockade of receptor signals in EGF-treated cells.
DN-EGFR-expressing Caco-2 transfectants
For in vitro coculture studies, we chose Caco-2 colon cancer cells because these cells express inducible PTGS2 protein, whereas HCT116 cells in cell culture express only PTGS2 transcripts. DN-EGFR was subcloned into the doxycycline pIND-Tight expression vector and the construct was verified by sequencing. Caco-2 cells stably expressing the tTA doxycycline transactivator (pTet off system) were obtained from Dr Jerrold Turner (University of Chicago). Cells were transfected with pIND-DN-EGFR and cotransfected with a 10 µg/ml blasticidin resistance vector for selection. Transfectants were screened for inducible (under doxycycline-free conditions) DN-EGFR expression as assessed by inhibition of EGF-induced phospho-active EGFR (pEGFR). Several monoclonal populations were characterized and shown to have similar inhibitory responses in culture.
HCT116 cell tumor xenografts
To assess the role of stromal cell EGFR on tumor growth, we prepared immunodeficient mice that were wild-type (wt) for Egfr or expressed DN-EgfrVelvet/+ by intercrossing Rag1-/- mice with EgfrVelvet/+ mice, which were both on a C57BL6/J background. The Velvet mutation was genotyped as described (19) and the Rag1 locus was genotyped following the protocol provided by Jackson Laboratory. For tumor xenograft studies, HCT116 parental cells (5 × 106) were injected subcutaneously into the flanks of Rag1 -/- Egfr+/+ or Rag1-/- EgfrVelvet/+ mice. Tumor growth was monitored over time and volumes calculated as length × (width)2/2. Tumors were harvested 3 weeks after cell inoculation. To assess the role of colon cancer cell EGFR, we examined growth of HCT116 DN-EGFR and empty vector (EV) transfectants in Egfr+/+ NOD2-Scid Il2rg null mice. We chose NOD2-Scid Il2rg immunodeficient mice rather than Rag1 null mice for these experiments because we are planning metastasis studies using DN-EGFR transfectants. In preliminary experiments, we found that HCT116 cells metastasized from cecum to the liver by hematogenous spread in NOD-Scid IL2Rgamma null mice, but not in Rag1 null mice (unpublished results). Transfectants (5 × 106) were injected subcutaneously into the flank. After 1 week, tumor nodules were palpable and mice were continued on unsupplemented chow or chow was supplemented with 625mg/kg doxycycline. Tumor dimensions were monitored and volumes calculated as length × (width)2/2. Animals were killed 40 days after implantation and tumor tissue was fixed in 10% formalin or flash-frozen in liquid nitrogen.
Mono- and coculture transwell studies. Parental or transfected Caco-2 human colon cancer cells and CCD-18Co human colonic fibroblasts were plated under identical conditions either in monoculture or were cocultured on opposing sides of transwell membranes. In coculture experiments, the transwell was inverted and CCD-18Co cells plated on the lower surface. Several hours later, when fibroblasts were adherent, the transwell was re-inverted and placed in the transwell chamber. Caco-2 cells were seeded on the upper surface. Parental cells were either untreated (control, C) or treated with EGF (10ng/ml), IL1B (10ng/ml) or C225 anti-EGFR-neutralizing antibodies (20 µg/ml). For cocultures using Caco-2 transfectants, cells were plated overnight in the presence of 25ng/ml doxycycline (DN-EGFR off). After 18h, fresh medium with doxycycline (DN-EGFR silenced) or without doxycycline (DN-EGFR on) was added. Next, 4 or 24h after media change, conditioned media from colon cancer cells and fibroblasts were separately collected and cells were harvested for protein or RNA analysis. To assess the ability of IL1B to transactivate EGFR, parental HCT116 cells were pre-treated with C225 antibodies (20 µg/ml) or vehicle. After 2h, cells were treated with IL1B (10ng/ml) or vehicle. After 5min, whole-cell lysates were prepared and phospho-active ERBB (pERBB2) was assayed. EGF (10ng/ml, 5min) was included in separate wells as a positive control.
Prostaglandin E2 measurements
Conditioned media were assayed for prostaglandin E2 (PGE2) by EIA (Cayman Company) and values were calculated from a standard curve and expressed as means ± SD.
Western blotting
Proteins were extracted in sodium dodecyl sulfate–containing Laemmli buffer, quantified by RC-DC, reducing agent compatible and detergent compatible assay and subjected to western blotting, as described previously (32). Briefly, blots were incubated overnight at 4°C with specific primary antibodies, followed by 1h incubation with appropriate peroxidase-coupled secondary antibodies that were detected by enhanced chemiluminescence using X-OMAT film. Xerograms were digitized with an Epson scanner (San Jose, CA) and band intensity quantified using UN-SCAN-IT software (Silk Scientific Corporation, Orem, UT). Blots were re-probed for ACTB (β-actin) expression to assess loading. Densities were normalized to ACTB loading and expressed as fold increases of control (EV or untreated) samples. For tumor xenografts, values were expressed as fold of proteins in tumors from Egfrwt mice.
Immunostaining
Immunostaining was carried out as described previously (11). Briefly, 5 µm sections were mounted on Vectabond-coated Superfrost Plus slides. Sections were heated to 60°C for 1h, deparaffinized by three washes for 5min each in xylene, hydrated in a graded series of ethanol washes and rinsed in distilled water. Epitope retrievals were achieved by microwave heating for 15min in 0.01M citrate buffer[pH 6; CTNNB1, cleaved caspase-3, PTGS2, TGF-α, smooth muscle alpha2 actin (SMA)] or steamed in Tris–ethylenediaminetetraacetic acid (EDTA) buffer (pH 9; CCND1, nestin-1 and Ki67), followed by three washes for 2min each in Tris-buffered saline with 0.1% Tween-20 (TBST). Endogenous peroxidase activity was quenched with methanol/H2O2 solution (0.5%). Sections were washed three times in TBST for 2min each and blocked in “Protein Block” for 20min. Sections were incubated with primary antibodies for 1h at room temperature (1:25 dilution for anti-cleaved caspase-3; 1:50 dilution for anti-nestin-1 or anti-CCND1; 1:100 dilution for anti-PTGS2 or anti-TGF-α antibodies; 1:200 dilution for anti-CTNNB1; 1:300 dilution for anti-Ki67; and 1:150 dilution for anti-smooth muscle alpha2 actin antibodies). After three washes in TBST, slides were incubated at room temperature with 1:200 dilution of biotinylated secondary antibodies for 30min. Antigen–antibody complexes were detected using 3,3′-diaminobenzidine as substrate and horseradish peroxidase–labeled DAKO EnVision™+ system or streptavidin–biotin-labeled DAKO LSAB™+ system for nestin-1. For negative controls, sections were incubated with isotype-matched non-immune antibodies and these showed no specific staining. After washing in distilled water, slides were counterstained with Gill’s III hematoxylin, rinsed with water, dehydrated in ethanol and cleared with xylene.
Immunostaining quantitation
Proliferation marker Ki67, apoptosis marker cleaved caspase-3 and angiogenesis marker nestin-1 were quantified by computer-assisted image analysis as described (11). We expressed the nuclear expression of Ki67 as percentage of positive nuclei. We measured cytoplasmic cleaved caspase-3 staining to assess apoptosis and blood vessel density by endothelial nestin-1 staining. Staining levels were quantified by an automated cellular imaging system (ACIS, Clarient San Juan, Capistrano, CA) with settings optimized for nuclear staining, vessel detection or apoptosis, as appropriate, from five representative fields per slide. Proliferation was expressed as percentage of nuclei positive for Ki67 and apoptosis expressed as percentage of cells positive for cleaved caspase-3. Microvessel density was assessed as vessel area normalized to total tissue area. Color-specific thresholds were used to determine brown (positive) and blue (negative) nuclei within the outlined regions of interest and to calculate the ratio of positively stained nuclei to all nuclei and expressed as a percentage (nuclear index). At least five fields per tumor and three tumors per group were scanned for quantitation.
Real-time PCR
Total RNA was extracted using RNeasy Lipid Tissue Mini Kit (Qiagen). Cell lysates were suspended in diethylpyrocarbonate-treated water in 1.5ml tubes. Samples were collected by centrifugation and homogenized by Polytron in 0.5ml QIAzol. Samples were loaded onto an RNA-binding spin column, washed, digested with DNase I and eluted in 30 µl elution buffer. RNA samples were tested using Agilent chip for RNA purity and quantified by RiboGreen. RNA (250ng) was reverse transcribed into cDNA using SuperScript III Platinum Two-Step qRT-PCR kit (Invitrogen) in 20 µL total volume. Incubation conditions were 25°C for 10min, 42°C for 50min and 85°C for 5min. Samples were then incubated with ribonuclease H at 37°C for 20min. The resulting first-strand cDNA was used as template for quantitative PCR in triplicate using SYBR Green QPCR Master Mix kit (Stratagene). Species-specific oligonucleotide PCR primer pairs were designed to cross intron–exon boundaries, where possible, from published sequences in the GenBank database using Primer3 (33). We used species-specific primers to amplify mouse amphiregulin (Areg), Tgfa, Il1b and interleukin-1 receptor 1 (Il1r1) genes. Mouse Areg (NM_009704.3) AREGMF1: 5′–gctattggcatcggc-atc–3′and AREGMR1: 5′–ACA-GTCCCGTTTTCTTGTCG–3′; Mouse Tgfa (NM_031199) TGFAMF1: 5′–TGCTAGCGCTGggtatcc–3′and TGFAMR1: 5′–TGGGCACTTGTTGAAGTGAG–3′; mouse Il1b (NM_008361.3) mIL1bF1: 5′–GCAACTGTTCCTGAACTCAACT–3′; mIL1bR1 5′–ATCTTTTGGGGTCCGTCAACT–3′; mouse Il1r1 (NM_001123382.1) mIL1R1F1: 5′–GTGCTACTGGGGCTCATTTGT–3′and mIL1R1R1: 5′–GGAGTAAGAGGACACTTGCGAAT–3′; mouse Ptgs2 (NM_011198.3) mPtgs2F1: 5′–GACCTGGGTTCACCCGAGGACTG–3′and mPtgs2R1: 5′–CCAAAGACTTCCTGCCCCACAGC–3′. Reverse transcribed cDNA (1 µL of 1:8 dilution) and appropriate primers were mixed with SYBR Green dye I master mixture in 25 µl. Reactants were initially heated to 95°C for 5min, followed by 40 cycles of the following steps: denaturation at 95°C for 10 s, followed by a combined annealing and extension step at 60°C for 30 s. The last cycle was followed by a 7-min extension at 72°C and the thermal denaturation profile was studied to identify the Tm. PCR amplification was verified by melting-curve and electrophoretic analyses of the PCR products on 3% agarose gel. Negative controls (no reverse transcriptase and no template) were included, which yielded no products. The data were analyzed using the comparative Ct method (Delta Ct), and messenger RNA abundance was normalized to ACTB messenger RNA and expressed as fold control.
Statistical methods
Data were expressed as means ± SD. Differences in tumor size, western blotting protein expression, and computer-assisted image analyses were compared by unpaired Student’s t-test. Real-time PCR confidence intervals and significance levels were calculated as described (34). Values of P < 0.05 were considered statistically significant.
Results
Stromal EGFR signals regulate HCT116 tumor xenograft growth
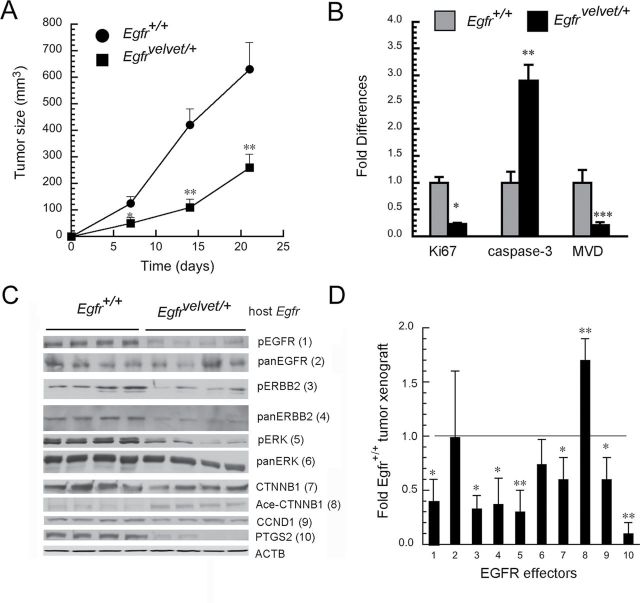
To address the role of stromal EGFR in colon cancer cell growth, we interbred immunodeficient Rag1-/- mice with EgfrVelvet/+ mice that express a DN receptor mutation to produce compound mutant Rag1-/-, EgfrVelvet/+ mice and control Rag-/-, Egfr+/+ mice. We chose parental HCT116 cells because these colon cancer cells form tumor xenografts in immunodeficient mice (35). HCT116 cells possess codon 13 KRAS (G13D) mutations that could potentially make these cells resistant to EGFR blockade. A recent study, however, suggests that cancer cells with codon 13 KRAS mutations remain responsive to EGFR signals because cetuximab anti-EGFR antibodies inhibited their growth in nude mice (27). Although cetuximab blocks EGFR signals in both stromal cells and colon cancer cells, we wanted to dissect the contribution of stromal cell EGFR to tumor growth. As shown in Figure 1A, expression of DN-Egfr Velvet in stromal cells significantly inhibited growth of subcutaneous tumors derived from parental HCT116 cells compared with tumors growing in Egfr+/+ mice. Furthermore, as shown in Figure 1B, compared with tumors in Egfr+/+ mice, proliferation and angiogenesis were reduced by >75%, whereas apoptosis was increased 2.9-fold in tumors growing in EgfrVelvet/+ mice (P < 0.05).
Fig. 1.
Stromal cell EGFR controls tumor xenograft growth and EGFR effector signals. HCT116 cells (5 × 106) were implanted in the flanks of Rag1-/- Egfr+/+ and Rag1-/- EgfrVelvet/+ mice that were killed 3 weeks later. (A) Tumor growth; (n = 4 for each genotype, *p < 0.05, **P < 0.005 compared with tumors in Rag1-/- Egfr+/+ mice). (B) Quantitation of Ki67, cleaved caspase-3 and microvessel density (MVD). Tumors were fixed in 10% formalin and stained for Ki67, cleaved caspase-3 and nestin-1. Immunostaining was quantified by computer-assisted image analysis and microvessel density (MVD) as described (11). *P < 0.005; **P < 0.05; ***P < 0.001, compared with tumors in Egfr+/+ mice (n = four tumors for each genotype). (C) Western blots of indicated proteins. Numbers adjacent to proteins refer to bar graph in Figure 1D. (D) Quantitative densitometry of indicated proteins in tumors growing in EgfrVelvet/+ mice expressed as fold proteins in tumors growing in Egfr+/+ mice. 1 = pEGFR; 2 = panEGFR; 3 = pERBB2; 4 = panERBB2, 5 = pERK; 6 = panERK; 7= CTNNB1; 8 = acetylated CTNNB1; 9 = CCND1; and 10 = PTGS2. Horizontal line indicates normalized expression levels of proteins in tumors from Egfr+/+ mice. *P < 0.05, **P < 0.005 compared with proteins from tumors growing in Egfr +/+ mice (n = four tumors for each genotype).
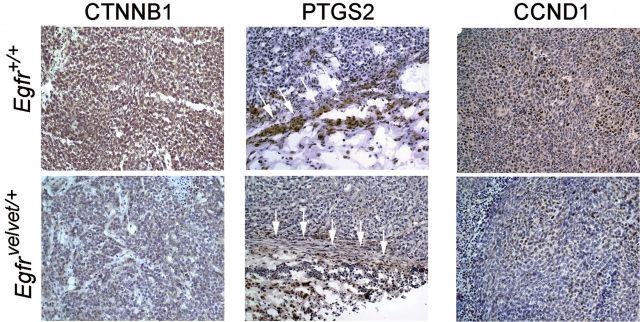
To assess molecular mechanisms that might contribute to these growth differences, we examined CTNNB1 expression and EGFR signals in these tumors. As shown in Figure 1C, EGFR signals and CTNNB1 expression were downregulated in tumors growing in Egfr Velvet/+ mice compared with those in Egfr +/+ mice. EGFR has been reported to stabilize CTNNB1 by inhibiting acetylation on serine 45, which is required for CTNNB1 N-terminal phosphorylation and downregulation (36). In agreement with this, tumors growing in Egfr Velvet/+ mice showed increased CTNNB1 acetylation, which could contribute to reduced CTNNB1 levels in these tumors (Figure 1C). Expression levels of proto-oncogenes CCND1 (cyclin D1) and PTGS2 (Cox-2) were also reduced in tumors growing in Egfr Velvet/+ mice (Figure 1C). We quantified and summarized these effects in Figure 1D. We showed that pEGFR, pERBB2 and pERK, as well as CTNNB1, CCND1 and PTGS2, were all significantly lower in tumors growing in Egfr Velvet/+ mice compared with those in Egfr wt mice. We also assessed tumors for several of these proteins by immunostaining to localize the cell of origin. As shown in Figure 2, CTNNB1 and CCND1 were expressed predominantly in malignant colonocytes, whereas PTGS2 was expressed mainly in stromal cells. In agreement with western blotting results, CTNNB1, PTGS2 and CCND1 were reduced in tumors growing in Egfr Velvet/+ mice compared with the levels in tumors in Egfr +/+ mice (Figure 2).
Fig. 2.
Loss of stromal EGFR reduces expression of CTNNB1 and CCND1 in colon cancer cells and PTGS2 in stromal cells. Tumor xenografts derived from HCT116 cells implanted in Egfr +/+ and Egfr Velvet/+ mice were stained for the indicated proteins as described in Materials and methods. Note decreased CTNNB1 and CCND1 staining in malignant colonocytes and reduced PTGS2 expression in stromal cells in tumors growing in Egfr Velvet/+ mice. For PTGS2, in the middle panel, compare areas indicated by white arrows. PTGS2-positive cells have fibroblast-like appearance and stain for smooth muscle alpha2 actin (SMA; see Figure 3E and 3F). Sections are representative of four tumors stained for the indicated proteins.
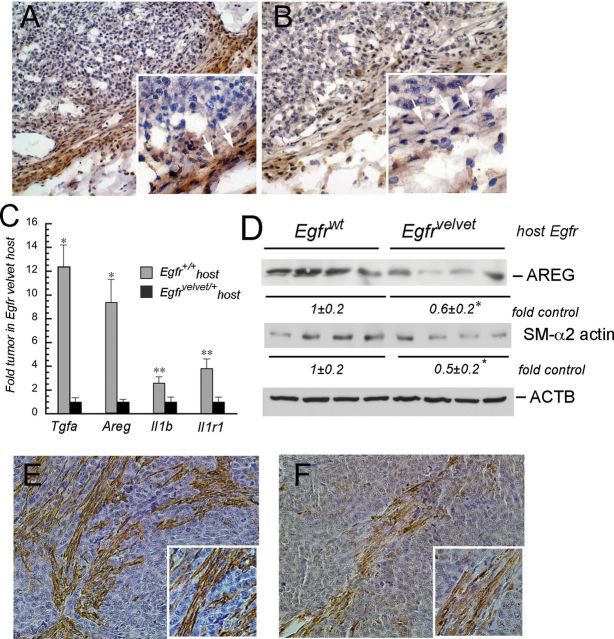
We next examined tumor xenografts for expression of TGF-α (TGFA) and AREG. TGFA was expressed in mouse stromal cells and expression was stronger in Egfr+/+ (Figure 3A), compared with the levels in EgfrVelvet/+ mice (Figure 3B). We also measured transcripts for Tgfa and Areg as well as interleukin-1b (Il1b) and IL1B receptors (Il1r1) using real-time PCR with mouse-specific primers. As shown in Figure 3C, transcript levels for mouse EGFR ligands Tgfa and Areg and for Il1b and Il1r1 were significantly lower in tumors from EgfrVelvet/+ mice. The EGFR ligand AREG was also reduced in tumors in EgfrVelvet mice compared with tumors in Egfr +/+ mice as assessed by western blotting (Figure 3D). Thus, stromal EGFR regulates expression levels of mouse stromal Tgfa and Areg as well as mouse Il1b and Il1r1 transcripts. Transcripts for the human EGFR ligands TGFA, AREG, IL1B and IL1R1, however, were not different in tumors in Egfr+/+ compared with the tumors in EgfrVelvet-recipient mice (data not shown). These results are in agreement with upregulations of EGFR ligands in mouse stroma but not in lung cancer cells, reported in another tumor xenograft model (37).
Fig. 3.
Stromal cell EGFR controls stroma-derived EGFR ligands and Il1b expression in tumor xenografts. (A) TGFA expression in tumor xenograft in Egfr+/+ mice. (B) TGFA expression in tumor xenograft in EgfrVelvet/+ mouse. Note the increased TGFA expression (brown staining) in stromal cells surrounding the tumor in Egfr+/+ mouse compared with stromal cells surrounding the tumor in EgfrVelvet/+ mice. TGFA-expressing cells exhibit a fibroblast-like appearance (compare areas marked with white arrows in insets in A and B). (C) Mouse Tgfa, Areg, Il1b and Il1r1 transcripts are decreased in tumor xenografts growing in EgfrVelvet/+ mice. Real-time PCR was carried out using mouse species-specific primers for the indicated genes. *P < 0.005, **P < 0.05 compared with Egfr+/+ mice (n = four tumors for each genotype). (D) AREG and smooth muscle alpha2 actin (SMA) expression in tumor xenografts in Egfrwt and EgfrVelvet/+ mice. *P < 0.05 compared with tumors in Egfrwt mice. Note that smooth muscle alpha2 actin (SMA) abundance is lower in the tumor growing in EgfrVelvet/+ mouse. E. Smooth muscle alpha2 actin immunostaining in tumor xenograft in Egfrwt mouse. F. Smooth muscle alpha2 actin immunostaining in tumor xenograft in EgfrVelvet mouse. Note the darker brown staining in Figure 3E compared with that in Figure 3F.
The stromal cells expressing PTGS2 and TGFA possessed fibroblast-like appearance. Because myofibroblasts have been implicated in colon cancer and other tumors, we examined the tumor xenografts for SMA, a marker of myofibroblasts (38,39). As shown in Figure 3E and 3F, there were spindle-shaped fibroblast-appearing cells that stained positive for SMA in tumor xenografts. Surprisingly, tumors growing in Egfr Velvet/+ mice showed lower expression of SMA, suggesting that stromal EGFR directly or indirectly regulates expression of this protein in tumor-associated fibroblasts (Figure 3D).
Colon cancer cell EGFR regulates tumor xenograft growth
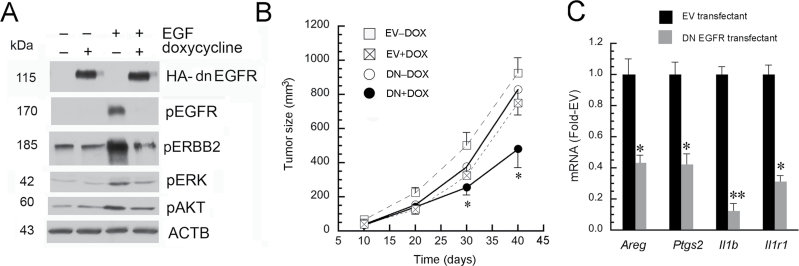
In order to dissect the regulation of tumor xenograft growth by colon cancer cell EGFR, we bioengineered HCT116 cells to express an inducible DN-EGFR under doxycycline regulation (rtTA pTet-on). Unlike Egfr Velvet, this DN-Egfr is a deletion mutant, encoding a receptor that lacks the cytoplasmic domain. We first confirmed that this DN receptor blocked EGFR signals. As shown in Figure 4A, doxycycline upregulated DN-EGFR, as identified with the HA epitope tag. DN-EGFR induction with doxycycline attenuated EGF-induced receptor activation in HCT116 transfectants in culture (Figure 4A).
Fig. 4.
DN-EGFR inhibits EGFR signals in cell culture and tumor xenograft growth of HCT116 transfectants. (A) DN-EGFR inhibits receptor activation and signals in HCT116 transfectants in vitro. Transfectants were plated overnight on six-well plates and then treated with 25ng doxycycline (+) or phosphate-buffered saline (PBS; –). Twelve hours later, cells were stimulated with EGF (+, 10ng/ml) or PBS (–) for 5min. Cells were lysed and indicated proteins were probed by western blotting. Note that doxycycline induced DN-EGFR (HA tag) expression, which blocked EGFR activation. (B) DN-EGFR induction in HCT116 colon cancer cell transfectants inhibits tumor xenograft growth. HCT116 DN-EGFR or EV transfectants were implanted into Egfr +/+ NOD-Scid Il2rg null mice. After one week, mice were provided chow supplemented with 625mg doxycycline/kg chow (+Dox) or continued on standard chow (–Dox). Tumor growth was monitored and tumor size calculated as width2 × length/2. *P < 0.05, compared with DN-Dox mice (n = four tumors/group). Error bars were omitted from EV+Dox group for clarity. (Note EV-Dox and EV+Dox were not significantly different). (C) Induction of DN-EGFR in HCT116 cells reduces mouse stromal Areg, Ptgs2, Il1b and Il1r1 in tumor xenografts. Mice implanted with EV or DN-EGFR transfectants were supplemented with doxycycline as described in Figure 4B (EV+Dox and DN+Dox). RNA was extracted from tumor xenografts at killing. Real-time PCR was carried out using mouse-specific primers, as described in Materials and methods (n = four tumors per group; *P < 0.05, **P < 0.005 compared with tumor xenografts derived from EV cells).
We next implanted HCT116 DN-EGFR or EV transfectants into flanks of Egfr +/+ , NOD-Scid Il2Rgamma null mice. After 1 week, tumors were palpable and mice were continued on standard chow or chow was then supplemented with doxycycline. Tumor size was monitored over time. Animals were killed 40 days after implantation and tumors harvested. Growth curves are summarized in Figure 4B. Doxycycline significantly suppressed growth of HCT116 DN-EGFR but not EV cells in Egfr +/+ mice. To assess the effects of colon cancer cell EGFR on stromal growth factors and cellular markers of inflammation, we examined these tumor xenografts for Areg, Ptgs2, Il1b and Il1r1 by real-time PCR using mouse-specific primers. As shown in Figure 4C, DN-EGFR induction with doxycycline reduced expression of mouse transcripts for Areg, Ptgs2, and Il1b and Il1r1. Taken together, these results showed that EGF receptors on both stromal cells and colon cancer cells are required for efficient HCT116 xenograft growth and robust stromal EGF ligand expression and pro-inflammatory responses.
EGFR signals in mono- and cocultured colon cancer cells and colonic fibroblasts
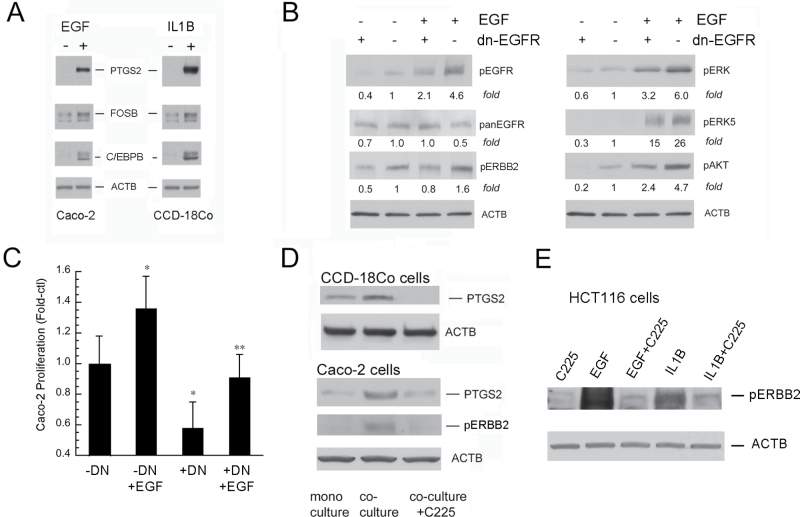
To dissect the autocrine and paracrine cellular signaling pathways in vitro, we examined mono- and cocultured colon cancer cells and stromal cells. For stromal cells, we selected human colonic CCD-18Co fibroblasts that express EGFR (24). For colon cancer cells, we initially selected Caco-2 cells that possess wild-type KRAS and are growth-inhibited by EGFR blockade (40). EGF and IL1B induced PTGS2, C/EBPB and FOSB in monocultured Caco-2 cells and colonic fibroblasts (Figure 5A). We transfected Caco-2 cells with the cytoplasmic-domain-deleted inducible DN-EGFR using a pTet off (tTA) system (25,30). DN-EGFR inhibited EGF-induced signals (Figure 5B) and reduced the growth of Caco-2 transfectants (Figure 5C). We then examined pERBB2 and PTGS2 in Caco-2 cells and CCD-18Co colonic fibroblasts in mono- and coculture conditions. Compared with monoculture, coculture conditions increased pERBB2 in Caco-2 cells and upregulated PTGS2 in colonic fibroblasts and Caco-2 cells (Figure 5D). EGFR controls these coculture-induced increases in pERBB2 and PTGS2 because C225 anti-EGFR antibodies inhibited their increases (Figure 5D). We next examined the effect of IL1B on parental HCT116 cells in monoculture and showed that IL1B transactivated EGFR in these cells (Figure 5E). Because increased pERBB2 was blocked by C225 antibodies, we postulate that IL1B transactivates EGFR by increasing receptor ligands. Increases in stromal IL1B observed in colonic tumors could thereby amplify EGFR signals in stromal cells and colon cancer cells by this mechanism.
Fig. 5.
EGF and IL1B activate colon cancer cells and colonic fibroblasts. (A) EGF and IL1B increase PTGS2, FOSB and C/EBPB in Caco-2 cells (left panel) and CCD-18Co cells (right panel). Cells were treated for 4h with vehicle (–) or EGF (+, 10ng/ml) or IL1B (+, 10ng/ml) and lysates were assayed for indicated proteins. (B) DN-EGFR blocks receptor activation in Caco-2 cells. (C) DN-EGFR inhibits basal and EGF-stimulated Caco-2 cell proliferation. Cells were treated with doxycycline (–DN) or media alone (+DN) ; stimulated with EGF (+EGF, 10ng/ml) or vehicle (–EGF); and proliferation was measured 48h later by the Wst-1 assay (*P < 0.05 compared with –DN; **P < 0.05 compared with –DN+EGF). (D) Coculture increases pERBB2 in Caco-2 and PTGS2 in fibroblast and Caco-2 cell by an EGFR-dependent mechanism. Cells were cultured on transwells alone (monoculture) or on opposite sides of the transwells (coculture) for 24h. Where indicated, C225-neutralizing anti-EGFR antibodies (20 µg/ml) were added during coculture. Results represent n = 3 independent platings. (E) IL1B transactivates EGFR in HCT116 colon cancer cells. HCT116 cells were treated with C225 antibodies (20 µg/ml) or vehicle for 2h and then stimulated for 5min with EGF (10ng/ml), or IL1B (10ng/ml). Whole-cell lysates were probed for phospho-active ERBB2 (pERBB2) by western blotting. Results represent n = 2 independent platings.
Colon cancer cell EGFR enhances colonic fibroblast PTGS2 upregulation and PGE2 release
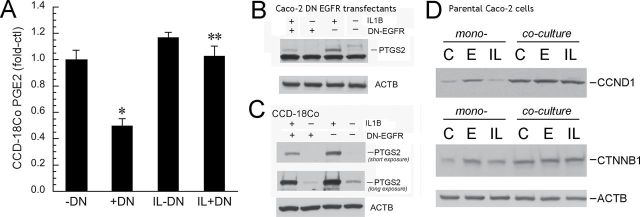
We next examined how colon cancer cell EGFR modulates fibroblast PTGS2 induction by IL1B. This pro-inflammatory cytokine is increased in colon cancer and could play an important role in regulating stromal PTGS2 expression by colon cancer cell EGFR signals (5,21–23). To this end, we cocultured colonic fibroblasts with Caco-2 DN-EGFR transfectants. Induction of DN-EGFR in colon cancer cells suppressed basal and IL1B-stimulated PGE2 secretion from fibroblasts (Figure 6A). DN-EGFR also reduced basal and IL1B-stimulated AREG induction from Caco-2 cells (data not shown). Inhibitory effects of DN-EGFR on PGE2 and AREG were greater on basal compared with IL1B-stimulated expression. DN-EGFR also suppressed basal and IL1B-induced PTGS2 expression in both Caco-2 cells (Figure 6B) and colonic fibroblasts (Figure 6C) under coculture conditions. We also showed that colonic fibroblast coculture increased basal and IL1B-stimulated CTNNB1 and CCND1 expression in Caco-2 colon cancer cells (Figure 6D).
Fig. 6.
EGFR signals in Caco-2 cells regulate fibroblast PTGS2 and PGE2, whereas fibroblasts regulate CTNNB1 and CCND1 expression in Caco-2 cells. (A) Colonic fibroblast PGE2 (pg/ml) by EIA. CCD-18Co colonic fibroblasts and Caco-2 DN-EGFR transfectants were cocultured on transwells. DN-EGFR was suppressed (–DN) or induced (+DN) for 24h, and cells were then treated with IL1B (IL, 10ng/ml) or vehicle for 4h. Fibroblast-conditioned medium was collected for PGE2 analysis and cells were lysed to assess PTGS2 protein expression. *P < 0.05 compared with cells without DN-EGFR induction (–DN), **P < 0.05, compared with IL1B-treated cells without DN-EGFR induction (IL–DN). PGE2 secretion in Caco-2 cells was <10% of CCD-18Co cells (data not shown). (B) PTGS2 expression in Caco-2 cells. Methods were the same as in Figure 6A. (C) PTGS2 expression in CCD-18Co cells. Methods were the same as in Figure 6A. A long exposure is shown in Figure 6C to demonstrate basal PTGS2 in CCD-18Co cells. A short exposure is shown to compare PTGS2 expression in IL1B-treated –DN cells versus IL1B-treated +DN cells. Results represent n = 3 independent platings. (D) Fibroblasts increase basal and EGF- and IL1B-induced CTNNB1 and CCND1 in cocultured Caco-2 cells. Caco-2 cells and CCD-18Co were seeded on transwells and cultured as mono- or cocultures. Cells were treated with vehicle (PBS) or EGF (E, 10ng/ml) or IL1B (IL, 10ng/ml). After 24h, Caco-2 cells were harvested and lysates were probed for CTNNB1 and CCND1. Note that coculture conditions increased expression of these proto-oncogenes in Caco-2 cells under both basal and stimulated conditions. Shown are representative western blots of two independent experiments.
Discussion
Complex growth-promoting interactions between stromal cells and malignant cells contribute to colonic tumor growth (1,2). Prior studies showed that global pharmacological or antibody-mediated EGFR inhibition reduced TGFA expression, cell proliferation, angiogenesis and tumor xenograft growth of colon cancer cells (41,42). In this study, we separated effects of EGFR in the stromal cells and malignant colonocytes to directly examine their respective roles in tumor xenograft growth. Using immunodeficient mice expressing Egfr Velvet, a DN-EGFR with single point mutation, we demonstrated that stromal cell EGFR is required for efficient tumor xenograft growth. In these Egfr Velvet mice, cancer cell proliferation and cell survival were reduced and tumor vasculature decreased in the absence of wild-type EGFR in stromal cells. Conversely, using colon cancer cell transfectants expressing an inducible cytoplasmic-domain-deleted mutant receptor, we showed that cancer cell EGFR also regulates tumor growth. We also showed that stromal EGFR controls tumor-associated myofibroblasts. Stromal myofibroblasts possess EGF receptors and secrete EGFR ligands (5). Other investigators have emphasized the role of tumor-associated myofibroblasts in promoting colonic tumorigenesis and cancer recurrence (43,44).
In some cancers, EGFR is driven by activating mutations or gene amplification. Increases in EGFR signals in colon cancers, however, are generally driven by upregulating receptor and ligand expression without changes in gene copy number. Because in this study stromal EGFR regulated receptor signals in tumor xenografts, we examined the effect of stromal EGFR on receptor expression and ligand abundance. Consistent with reduced EGFR signals in tumors growing in Egfr Velvet/+ mice, protein and transcript levels of the ligands TGFA and AREG were significantly lower in these mice. Thus, upregulated receptor ligands and signals require the presence of functional EGFR in the stroma. It is formally possible that reductions in EGFR signals in malignant colonocytes are critical for growth inhibition observed in tumors growing in Egfr Velvet/+ mice. This might occur, for example, as an epistatic phenomenon with stromal cell EGFR required to maintain functional EGFR in colon cancer cells. Experiments with cancer cells constitutively expressing activated EGFR (e.g. mutant activated EGFR VIII) could address this question. Nevertheless, inhibition of stromal cell EGFR might be a useful strategy for chemoprevention, as also supported by the role of stromal EGFR in mediating resistance to anti-angiogenesis therapy in lung tumor xenograft studies (37).
In the current study, we demonstrated that several oncogenic pathways were inhibited in tumors growing in Egfr-mutant mice. These included reductions in interleukin 1 signals, as assessed by decreases in Il1b and Il1r1 transcripts, and lower levels of EGFR downstream effectors, CCND1 (colonocytes) and PTGS2 (adjacent stroma) in tumors growing in Egfr Velvet/+ mice. PTGS2 is the rate-limiting enzyme for PGE2 biosynthesis. Prostanoids have been shown to play important roles in cancer cell proliferation, apoptotic resistance and angiogenesis (45–47). Because PGE2 can transactivate EGFR and enhance Wnt signaling (48,49), we hypothesize that decreases in PTGS2 contribute to reductions in signals from these key oncogenic pathways in tumors lacking normal stromal EGFR signals.
The proto-oncogene CTNNB1 plays a central role in colon cancer development and was also decreased in tumors growing in Egfr Velvet mice. EGFR and PGE2 have been shown to regulate CTNNB1 stability (36,49). Thus, decreases in EGFR signals and PTGS2 expression (and thereby PGE2) would both be predicted to reduce CTNNB1 levels. Consistent with these observations, we showed that CTNNB1 staining was reduced, whereas acetylated (destabilized) CTNNB1 was increased in tumors growing in Egfr Velvet/+ mice. CCND1, a key G1 cell cycle regulator, is controlled by both EGFR and CTNNB1 signals in colon cancers (32,50,51). Decreases in CCND1 in tumors from Egfr Velvet/+ mice presumably reflect reductions in these proto-oncogenes (11,12,14). In this regard, EGFR functions as both an upstream regulator of CTNNB1 (via inhibition of CTNNB1 acetylation and induction of PTGS2) and a downstream effector of CTNNB1 (via, for example, T-cell factor sites in the EGFR promoter and as an activator of CCND1) in colonic tumor growth (14,36,52).
In order to assess the role of colon cancer cell EGFR on tumor xenograft growth, we prepared stable HCT116 transfectants with an inducible deletion mutant DN-EGFR that lacks the cytoplasmic domain (30). This construct has been shown to block EGFR signals in vitro and in vivo (30,53). In the presence of doxycycline, HCT116 transfectants expressed DN-EGFR, which blocked EGF-induced receptor activation and signaling in cell culture. In tumor xenografts, in the absence of doxycycline, growth of DN-EGFR–transfected cells was nearly comparable with EV transfectants in Egfr +/+ mice, indicating low basal expression of the DN-EGFR transgene. In contrast, in the presence of doxycycline, DN-EGFR transfectants grew significantly slower than EV counterparts in Egfr +/+ mice. These results demonstrated that colon cancer cell EGFR, like stromal cell EGFR, plays an important role in tumor xenograft growth. To assess the importance of colon cancer cell EGFR on stromal epidermal growth factors and pro-inflammatory signals, we examined these tumors for mouse Areg, Ptgs2, Il1b and Il1r1 transcripts. These transcripts were significantly reduced in tumor xenografts derived from cells expressing DN-EGFR compared with tumors derived from EV transfectants. Taken together, these results indicate that pro-inflammatory signals and growth factors originating in the stroma require EGFR signals from both stromal cells and cancer cells.
To directly assess the modulation of stromal pro-inflammatory signals by colon cancer cell EGFR, we employed a coculture system with Caco-2 cells and CCD-18Co colonic fibroblasts. We chose Caco-2 cells because they express PTGS2 protein in cell culture, whereas HCT116 cells express only the transcripts (48). We bioengineered these cells to express inducible pTet off (tTA) DN-EGFR. Colonic fibroblasts respond to EGF and IL1B with release of AREG and induction of PTGS2 (5,24). Using this coculture system, we showed that intact EGFR signals in Caco-2 cells enhanced basal and IL1B-stimulated PGE2 release from colonic fibroblasts and AREG expression in Caco-2 cells. As shown in Figure 6D, colonic fibroblasts also increased CTNNB1 and CCND1 levels in Caco-2 cells. Thus, colonic fibroblasts increase cancer cell proliferative and pro-inflammatory signals. Finally, because upregulation of interleukin 1 signals in tumors requires stromal cell EGFR, whereas IL1B can transactivate EGFR on cancer cells, our studies emphasize the importance of EGFR–IL1B signal interaction in driving a pro-inflammatory microenvironment that promotes tumor growth.
In summary, we have shown for the first time that EGFR of both malignant and stromal cells plays critical roles in the growth of tumor xenografts derived from colon cancer cells. Although strategies to target EGFR generally block both compartments, either the stromal or cancer cell component could theoretically be more sensitive to EGFR blockade. In this regard, the presence of EGFRs on endothelial cells was shown to sensitize tumors to the anticancer effects of tyrosine kinase inhibitors in previous studies (54). Stromal targeting of EGFR might explain growth inhibition by anti-EGFR antibodies in tumors without EGFR upregulation (8,55–57). Given the recent demonstration of growth inhibition of colonic tumors with codon 13 KRAS mutations by cetuximab, our results are consistent with suppression being mediated by blockade of stromal cell EGFR (27). The role of stromal cell EGFR blockade in mediating cetuximab growth inhibition, however, will require further study. Studies are in progress to understand how stromal cell EGFR influences growth of isogenic colon cancer cells that possess wild-type KRAS versus those possessing KRAS with codon 12 or codon 13 mutation. Insights into how KRAS mutations might influence stromal EGFR and tumor resistance to EGFR blockade could have important implications for colon cancer chemoprevention and therapy.
Funding
National Institutes of Health [CA36745 to M.B.]; Digestive Disease Research Core Center (P30DK42086); Samuel Freedman GI Cancer Laboratory Fund at the University of Chicago; the University of Chicago Comprehensive Cancer Center 5 P30 CA014599-37; the CR-UK fund CR-UK C5759/A12328.
Acknowledgements
We gratefully acknowledge the generous gift of stable tTA-expressing Caco-2 transfectants from Dr Jerrold Turner (University of Chicago) and the University of Chicago transgenic mouse core facility and Linda Degenstein with assistance in breeding immunodeficient mutant Egfr mice (5P30 CA014599-36). We would also like to thank Dr Maliha Aslam for expert assistance with immunostaining.
Footnotes
Conflict of Interest Statement: None declared.
References
- 1. Li H., et al. (2007). Tumor microenvironment: the role of the tumor stroma in cancer J Cell Biochem., 101 805–815 [DOI] [PubMed] [Google Scholar]
- 2. Inatomi O., et al. (2006). Regulation of amphiregulin and epiregulin expression in human colonic subepithelial myofibroblasts Int. J. Mol. Med. 18 497–503 [PubMed] [Google Scholar]
- 3. Blanchot-Jossic F., et al. (2005). Up-regulated expression of ADAM17 in human colon carcinoma: co-expression with EGFR in neoplastic and endothelial cells J. Pathol. 207 156–163 [DOI] [PubMed] [Google Scholar]
- 4. Barnard J.A., et al. (1995). Epidermal growth factor-related peptides and their relevance to gastrointestinal pathophysiology Gastroenterology 108 564–580 [DOI] [PubMed] [Google Scholar]
- 5. Inatomi O., et al. (2006). Regulation of amphiregulin and epiregulin expression in human colonic subepithelial myofibroblasts Int. J. Mol. Med. 18 497–503 [PubMed] [Google Scholar]
- 6. Hirata A., et al. (2004). Direct inhibition of EGF receptor activation in vascular endothelial cells by gefitinib (‘Iressa’, ZD1839) Cancer Sci. 95 614–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Toyoda H., et al. (1997). Distribution of mRNA for human epiregulin, a differentially expressed member of the epidermal growth factor family Biochem. J. 326 (Pt 1) 69–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. De Luca A., et al. (2008). The role of the EGFR signaling in tumor microenvironment J. Cell. Physiol. 214 559–567 [DOI] [PubMed] [Google Scholar]
- 9. Powell A.E., et al. (2012). The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor Cell 149 146–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Roberts R.B., et al. (2002). Importance of epidermal growth factor receptor signaling in establishment of adenomas and maintenance of carcinomas during intestinal tumorigenesis Proc. Natl. Acad. Sci. U.S.A. 99 1521–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dougherty U., et al. (2008). Epidermal growth factor receptor controls flat dysplastic aberrant crypt foci development and colon cancer progression in the rat azoxymethane model Clin. Cancer Res. 14 2253–2262 [DOI] [PubMed] [Google Scholar]
- 12. Fichera A., et al. (2007). Epidermal growth factor receptor signaling is required for microadenoma formation in the mouse azoxymethane model of colonic carcinogenesis Cancer Res. 67 827–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhu H., et al. (2011). EGFR signals downregulate tumor suppressors miR-143 and miR-145 in Western diet-promoted murine colon cancer: role of G1 regulators Mol. Cancer Res. 9 960–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dougherty U., et al. (2009). Epidermal growth factor receptor is required for colonic tumor promotion by dietary fat in the azoxymethane/dextran sulfate sodium model: roles of transforming growth factor-{alpha} and PTGS2 Clin. Cancer Res. 15 6780–6789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Adegboyega P.A., et al. (2004). Subepithelial myofibroblasts express cyclooxygenase-2 in colorectal tubular adenomas Clin. Cancer Res. 10 5870–5879 [DOI] [PubMed] [Google Scholar]
- 16. Wang W., et al. (2009). Crosstalk to stromal fibroblasts induces resistance of lung cancer to epidermal growth factor receptor tyrosine kinase inhibitors Clin. Cancer Res. 15 6630–6638 [DOI] [PubMed] [Google Scholar]
- 17. Mink S.R., et al. (2010). Cancer-associated fibroblasts derived from EGFR-TKI-resistant tumors reverse EGFR pathway inhibition by EGFR-TKIs Mol. Cancer Res. 8 809–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gijsen M., et al. HER2 phosphorylation is maintained by a PKB negative feedback loop in response to anti-HER2 herceptin in breast cancer. PLoS Biol. (2010);8:e1000563. doi: 10.1371/journal.pbio.1000563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Du X., et al. (2004). Velvet, a dominant Egfr mutation that causes wavy hair and defective eyelid development in mice Genetics 166 331–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee D., et al. (2004). Wa5 is a novel ENU-induced antimorphic allele of the epidermal growth factor receptor Mamm. Genome 15 525–536 [DOI] [PubMed] [Google Scholar]
- 21. Lewis A.M., et al. (2006). Interleukin-1 and cancer progression: the emerging role of interleukin-1 receptor antagonist as a novel therapeutic agent in cancer treatment J. Transl. Med. 4 48 doi: 10.1186/1479-5876-4-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Elaraj D.M., et al. (2006). The role of interleukin 1 in growth and metastasis of human cancer xenografts Clin. Cancer Res. 12 1088–1096 [DOI] [PubMed] [Google Scholar]
- 23. Bousserouel S., et al. (2010). Identification of gene expression profiles correlated to tumor progression in a preclinical model of colon carcinogenesis Int. J. Oncol. 36 1485–1490 [DOI] [PubMed] [Google Scholar]
- 24. Kim E.C., et al. (1998). Cytokine-mediated PGE2 expression in human colonic fibroblasts Am. J. Physiol. 275(4 Pt 1)C988–C994 [DOI] [PubMed] [Google Scholar]
- 25. Gossen M., et al. (1992). Tight control of gene expression in mammalian cells by tetracycline-responsive promoters Proc. Natl. Acad. Sci. U.S.A. 89 5547–5551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Giuliani F., et al. (2007). Cetuximab in colon cancer Int. J. Biol. Markers 22(1suppl4)S62–S70 [PubMed] [Google Scholar]
- 27. De Roock W., et al. (2010). Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab JAMA 304 1812–1820 [DOI] [PubMed] [Google Scholar]
- 28. Cerda S.R., et al. (2006). Protein kinase C delta inhibits Caco-2 cell proliferation by selective changes in cell cycle and cell death regulators Oncogene 25 3123–3138 [DOI] [PubMed] [Google Scholar]
- 29. Mustafi R., et al. (2006). Protein Kinase-zeta inhibits collagen I-dependent and anchorage-independent growth and enhances apoptosis of human Caco-2 cells Mol. Cancer Res. 4 683–694 [DOI] [PubMed] [Google Scholar]
- 30. Kashles O., et al. (1991). A dominant negative mutation suppresses the function of normal epidermal growth factor receptors by heterodimerization Mol. Cell. Biol. 11 1454–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Welman A., et al. (2005). Construction and characterization of multiple human colon cancer cell lines for inducibly regulated gene expression J. Cell. Biochem. 94 1148–1162 [DOI] [PubMed] [Google Scholar]
- 32. Bissonnette M., et al. (2000). Mutational and nonmutational activation of p21ras in rat colonic azoxymethane-induced tumors: effects on mitogen-activated protein kinase, cyclooxygenase-2, and cyclin D1 Cancer Res. 60 4602–4609 [PubMed] [Google Scholar]
- 33. Rozen S., et al. (2000). Primer3 on the WWW for general users and for biologist programmersIn Krawetz S., Misener S. (eds.), Bioinformatics Methods and Protocols Humana Press; Totowa, NJ: vol. Methods in Molecular Biology, pp. 365–386 [DOI] [PubMed] [Google Scholar]
- 34. Yuan J.S., et al. (2006). Statistical analysis of real-time PCR data BMC Bioinformatics 7 85–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cascone T., et al. (2011). Upregulated stromal EGFR and vascular remodeling in mouse xenograft models of angiogenesis inhibitor-resistant human lung adenocarcinoma J. Clin. Invest. 121 1313–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li Y., et al. (2008). HDAC6 is required for epidermal growth factor-induced beta-catenin nuclear localization J. Biol. Chem. 283 12686–12690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cascone T., et al. (2011). Upregulated stromal EGFR and vascular remodeling in mouse xenograft models of angiogenesis inhibitor-resistant human lung adenocarcinoma J. Clin. Invest. 121 1313–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. De Wever O., et al. (2008). Stromal myofibroblasts are drivers of invasive cancer growth Int. J. Cancer 123 2229–2238 [DOI] [PubMed] [Google Scholar]
- 39. Gaggioli C., et al. (2007). Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells Nat. Cell Biol. 9 1392–1400 [DOI] [PubMed] [Google Scholar]
- 40. Bishop W.P., et al. (1994). Regulation of Caco-2 cell proliferation by basolateral membrane epidermal growth factor receptors Am. J. Physiol. 267(5 Pt 1)G892–G900 [DOI] [PubMed] [Google Scholar]
- 41. Ciardiello F., et al. (2000). Antiangiogenic and antitumor activity of anti-epidermal growth factor receptor C225 monoclonal antibody in combination with vascular endothelial growth factor antisense oligonucleotide in human GEO colon cancer cells Clin. Cancer Res. 6 3739–3747 [PubMed] [Google Scholar]
- 42. Ciardiello F., et al. (2001). Inhibition of growth factor production and angiogenesis in human cancer cells by ZD1839 (Iressa), a selective epidermal growth factor receptor tyrosine kinase inhibitor Clin. Cancer Res. 7 1459–1465 [PubMed] [Google Scholar]
- 43. Zhu Y., et al. (2003). Cyclooxygenase-2 expression and prostanoid biogenesis reflect clinical phenotype in human colorectal fibroblast strains Cancer Res. 63 522–526 [PubMed] [Google Scholar]
- 44. Tsujino T., et al. (2007). Stromal myofibroblasts predict disease recurrence for colorectal cancer Clin. Cancer Res. 13 2082–2090 [DOI] [PubMed] [Google Scholar]
- 45. Sheng H., et al. (1998). Modulation of apoptosis and Bcl-2 expression by prostaglandin E2 in human colon cancer cells Cancer Res. 58 362–366 [PubMed] [Google Scholar]
- 46. Wang D., et al. (2005). Prostaglandin E2 enhances intestinal adenoma growth via activation of the Ras-mitogen-activated protein kinase cascade Cancer Res. 65 1822–1829 [DOI] [PubMed] [Google Scholar]
- 47. Wang L., et al. (2008). Celecoxib inhibits tumor growth and angiogenesis in an orthotopic implantation tumor model of human colon cancer Exp. Oncol. 30 42–51 [PubMed] [Google Scholar]
- 48. Pai R., et al. (2002). Prostaglandin E2 transactivates EGF receptor: a novel mechanism for promoting colon cancer growth and gastrointestinal hypertrophy Nat. Med. 8 289–293 [DOI] [PubMed] [Google Scholar]
- 49. Castellone M.D., et al. (2005). Prostaglandin E2 promotes colon cancer cell growth through a Gs-axin-beta-catenin signaling axis Science 310 1504–1510 [DOI] [PubMed] [Google Scholar]
- 50. Shtutman M., et al. (1999). The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway Proc. Natl. Acad. Sci. U.S.A. 96 5522–5527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tetsu O., et al. (1999). Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells Nature 398 422–426 [DOI] [PubMed] [Google Scholar]
- 52. Tan X., et al. (2005). Epidermal growth factor receptor: a novel target of the Wnt/beta-catenin pathway in liver Gastroenterology 129 285–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Murillas R., et al. (1995). Expression of a dominant negative mutant of epidermal growth factor receptor in the epidermis of transgenic mice elicits striking alterations in hair follicle development and skin structure EMBO J. 14 5216–5223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kuwai T., et al. (2008). Phosphorylated epidermal growth factor receptor on tumor-associated endothelial cells is a primary target for therapy with tyrosine kinase inhibitors Neoplasia 10 489–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Normanno N., et al. (2003). Target-based agents against ErbB receptors and their ligands: a novel approach to cancer treatment Endocr. Relat. Cancer 10 1–21 [DOI] [PubMed] [Google Scholar]
- 56. Wild R., et al. (2006). Cetuximab preclinical antitumor activity (monotherapy and combination based) is not predicted by relative total or activated epidermal growth factor receptor tumor expression levels Mol. Cancer Ther. 5 104–113 [DOI] [PubMed] [Google Scholar]
- 57. Chung K.Y., et al. (2005). Cetuximab shows activity in colorectal cancer patients with tumors that do not express the epidermal growth factor receptor by immunohistochemistry J. Clin. Oncol. 23 1803–1810 [DOI] [PubMed] [Google Scholar]