Abstract
Feed additives such as ractopamine and salbutamol are pharmacologically active compounds, acting primarily as β-adrenergic agonists. This study was designed to investigate whether the sulfation of ractopamine and salbutamol may occur under the metabolic conditions and to identify the human cytosolic sulfotransferases (SULTs) that are capable of sulfating two major feed additive compounds, ractopamine and salbutamol. A metabolic labelling study showed the generation and release of [35S]sulfated ractopamine and salbutamol by HepG2 human hepatoma cells labelled with [35S]sulfate in the presence of these two compounds. A systematic analysis using 11 purified human SULTs revealed SULT1A3 as the major SULT responsible for the sulfation of ractopamine and salbutamol. The pH dependence and kinetic parameters were analyzed. Moreover, the inhibitory effects of ractopamine and salbutamol on SULT1A3-mediated dopamine sulfation were investigated. Cytosol or S9 fractions of human lung, liver, kidney and small intestine were examined to verify the presence of ractopamine-/salbutamol-sulfating activity in vivo. Of the four human organs, the small intestine displayed the highest activity towards both compounds. Collectively, these results imply that the sulfation mediated by SULT1A3 may play an important role in the metabolism and detoxification of ractopamine and salbutamol.
Keywords: feed additive, ractopamine, salbutamol, sulfation, SULT
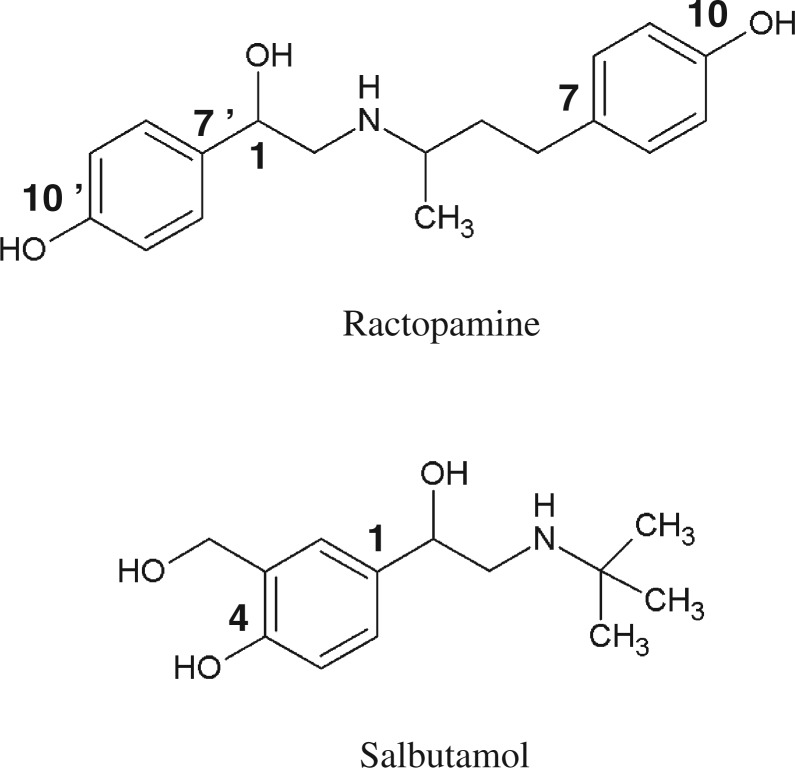
In livestock industry, certain feed additives are commonly used to improve the efficiency of feed utilization and to enhance the meat leanness (1, 2). Prominent among these feed additives are ractopamine (3), clenbuterol (4) and salbutamol (5). These feed additives are pharmacologically active compounds, acting primarily as β-adrenergic agonists (6, 7). Therefore, although these compounds are useful in serving their intended purposes, they may pose a potential health risk to humans when residual feed additives enter the body through dietary meat consumption. Therefore, an important question is whether the human body is equipped with mechanism(s) for protection against these potentially harmful feed additives. β-Adrenergic agonists, such as dobutamine, metaproterenol and terbutaline, are known to be primarily metabolized by conjugation reactions, particularly sulfation and glucuronidation, at their constituent phenolic hydroxyl group(s) (8–12). Like other β-adrenergic agonists, ractopamine and salbutamol also contain phenolic hydroxyl group(s) (Fig. 1). Previous studies have indeed demonstrated the generation of conjugate metabolites of these feed additives in livestock, including cattle, turkey and swine (13, 14). Although glucuronide conjugation of β-adrenergic agonists is a major metabolic pathway in livestock, sulfate conjugation predominates in the metabolism of β-adrenergic agonists in humans (8–14). Therefore, it is possible that sulfate conjugation by cytosolic sulfotransferase(s) (SULT(s)) may play a significant role in humans in the metabolism/detoxification of residual feed additives present in the dietary meat.
Fig. 1.
Chemical structures of ractopamine and salbutamol.
In humans and other mammals, sulfate conjugation catalyzed by the SULTs is known to be involved in the biotransformation and excretion of xenobiotics (15–17). The SULTs catalyze the transfer of a sulfonate group from the active sulfate, 3′-phosphoadenosine 5′-phosphosulfate (PAPS), to an acceptor substrate compound containing either hydroxyl or amino group (18). Sulfate conjugation by the SULT enzymes generally leads to the inactivation of biologically active compounds and/or the increase in their water solubility, thereby facilitating their removal from the body (15–17). In humans, 11 SULTs that are categorized into three distinct gene families have been identified and characterized. Seven of the 11 human SULTs that belong to the SULT1 gene family are SULT1A1 and SULT1A2 (both believed to be the general detoxifying enzymes) (19, 20), SULT1A3 (dopamine/catecholamine sulfotransferase) (21), SULT1B1 (thyroid hormone sulfotransferase) (22), SULT1C2 and SULT1C4 (hydroxyarylamine sulfotransferases) (23, 24) and SULT1E1 (estrogen sulfotransferase) (25). Three that belong to the SULT2 gene family are SULT2A1 (dehydroepiandrosterone sulfotransferase) (26, 27), SULT2B1a (pregnenolone sulfotransferase) (28, 29) and SULT2B1b (cholesterol sulfotransferase) (28, 29). Finally, neuronal/brain sulfotransferase belongs to the SULT4 gene family (30, 31).
We report in this study the generation and release of [35S]sulfated derivatives of ractopamine and salbutamol, two representative feed additive compounds, by HepG2 human hepatoma cells, labelled with [35S]sulfate in the presence of different concentrations of these two feed additive compounds. A systematic investigation of the sulfating activity of all 11 human SULTs towards ractopamine and salbutamol was performed. pH dependence of the major ractopamine-/salbutamol-sulfating SULT, SULT1A3, was examined, and the kinetic parameters of the sulfation of these two feed additive compounds were determined. Moreover, the inhibitory effects of ractopamine and salbutamol on SULT1A3-mediated dopamine sulfation were investigated. Cytosol or S9 fractions of human lung, liver, kidney and small intestine were examined to verify the presence of ractopamine-/salbutamol-sulfating activity in vivo.
Materials and Methods
Materials
Ractopamine, adenosine 5′-triphosphate (ATP), 3′-phosphoadenosine-5′-phosphosulfate (PAPS), 2-morpholinoethanesulfonic acid (MES), 3-(N-morpholino)propanesulfonic acid (MOPS), N-2-hydroxylpiperazine-N′-2-ethanesulfonic acid (HEPES), 3-[N-tris-(hydroxymethyl)methylamino]-propanesulfonic acid (TAPS), 2-(cyclohexylamino)ethanesulfonic acid (CHES), 3-(cyclohexylamino)-1-propanesulfonic acid (CAPS), Trizma base, dithiothreitol (DTT), minimum essential medium (MEM), fetal bovine serum (FBS), penicillin G, streptomycin sulfate and silica gel thin-layer chromatography (TLC) plates were products of Sigma Chemical Company (St. Louis, MO). Salbutamol, carrier-free sodium [35S]sulfate and Ecolume scintillation cocktail were purchased from MP Biomedicals (Irvine, CA). Ultrafree-MC 5000 NMWL filter units were products of Millipore (Bedford, MA). HepG2 human hepatoma cell line (ATCC HB-8065) was from American Type Culture Collection (Manassas, VA). Pooled human lung S9 fraction from a mixed-gender group of 4 donors (Lot No. 0 710 281), liver cytosol from 50 donors (Lot No. 09 103 970), small intestine (duodenum and jejunum) S9 fraction from 9 donors (Lot No. 0 710 351) and kidney S9 fraction from 8 donors (Lot No. 0 510 093) were purchased from XenoTech, LLC (Lenexa, KS). Other chemicals were of the highest grade commercially available.
Metabolic labelling of HepG2 human hepatoma cells
HepG2 cells were routinely maintained, under a 5% CO2 atmosphere at 37°C, in MEM supplemented with 10% FBS, penicillin G (30 µg/ml) and streptomycin sulfate (50 µg/ml). Confluent cells grown in a 24-well culture plate, pre-incubated in sulfate-free (prepared by omitting streptomycin sulfate and replacing magnesium sulfate with magnesium chloride) MEM without FBS for 4 h, were labelled with 0.25 ml aliquots of the same medium containing [35S]sulfate (0.3 mCi/ml) and 5 or 50 µM ractopamine or salbutamol. At the end of an 18-h labelling period, the labelling media were collected, spin filtered to remove high-molecular weight [35S]sulfated macromolecules and subjected to silica gel TLC for the analysis of [35S]sulfated ractopamine or salbutamol, using n-butanol/isopropanol (3:1; by volume) as the solvent system.
Preparation of purified human SULTs
Recombinant human P-form (SULT1A1 and SULT1A2) and M-form (SULT1A3) phenol SULTs, thyroid hormone SULT (SULT1B1), two SULT1Cs (SULT1C2 and SULT1C4), estrogen SULT (SULT1E1), dehydroepiandrosterone (DHEA) SULT (SULT2A1), two SULT2B1s (SULT2B1a and SULT2B1b) and a neuronal SULT (SULT4A1), expressed using pGEX-2TK or pET23c prokaryotic expression system, were prepared as described previously (24, 31–34).
SULT assay
The sulfating activity of the recombinant human SULTs was assayed using PAP[35S] as the sulfate group donor. The standard assay mixture, in a final volume of 20 µl, contained 50 mM of MOPS buffer at pH 7.0, 1 mM DTT and 14 µM PAP[35S]. Stock solutions of the substrates, prepared in dimethyl sulfoxide, were used in the enzymatic assay. The substrate, at 10 times the final concentration (1, 10 and 50 µM) in the assay mixture, was added after MOPS buffer and PAP[35S]. The reaction was started by the addition of the SULT enzyme, allowed to proceed for 10 min at 37°C and terminated by placing the thin-walled tube containing the assay mixture on a heating block, pre-heated to 100°C, for 2 min. The precipitates were cleared by centrifugation at 13,000 × g for 3 min, and the supernatant was subjected to the analysis of [35S]sulfated product using the TLC procedure with n-butanol:isopropanol:88% formic acid:water (8:2:1:1; by volume) for ractopamine or n-butanol:isopropanol (1:6; by volume) for salbutamol as the solvent system. On completion of TLC, the TLC plate was air dried and autoradiographed by using an X-ray film. The radioactive spot corresponding to the sulfated product was located, cut out and eluted in 0.5 ml water in a glass vial. A total of 4.5 ml of Ecolume scintillation liquid was added to each vial, mixed thoroughly and the radioactivity therein was counted by using a liquid scintillation counter. To examine the pH dependence of sulfation of ractopamine and salbutamol by human SULT1A3, 50 mM different buffers (sodium acetate at 4.5 or 5.5; MES at 5.5, 6.0 or 6.5; MOPS at 6.5, 7.0 or 7.5; HEPES at 7.0, 7.5 or 8.0; TAPS at 8.0, 8.5 or 9.0; CHES at 9.0, 9.5 or 10.0 and CAPS at 10.0, 10.5, 11.0 and 11.5), instead of 50 mM MOPS (pH 7.0), were used in individual reactions. To examine the inhibitory effects of ractopamine and salbutamol on the sulfation of dopamine by human SULT1A3, enzymatic assays were performed using 5 µM dopamine as the substrate in the presence of varying concentrations of ractopamine (0.1–1,000 µM) or salbutamol (0.1–4,000 µM). Each experiment was performed in triplicate, together with a control without enzyme. The results obtained were calculated and expressed in nanomoles of sulfated product formed/min/mg purified enzyme. To assay for ractopamine-/salbutamol-sulfating activity of human tissue cytosol or S9 fraction, the reaction mixture was supplemented with 50 mM NaF (a phosphatase inhibitor). The reaction was started by the addition of the cytosol or S9 fraction and allowed to proceed for 20 min, followed by the TLC analysis for [35S]sulfated product as described earlier.
Kinetic analysis
In the kinetic studies on the sulfation of ractopamine and salbutamol, the sulfation assays were carried out using varying concentrations of these substrate compounds and 50 mM MOPS at pH 7.0 according to the procedure described earlier. Data obtained were analyzed based on (i) Michaelis–Menten kinetics, (ii) substrate inhibition kinetics and (iii) Hill kinetics using GraphPad Prism5 software and non-linear regression. Hill and Eadie-Hofstee plots were concomitantly analyzed. The kinetic parameters were calculated based on the following equations:
| (1) |
| (2) |
| (3) |
where Ki is the substrate inhibition constant, S50 is the substrate concentration of resulting in half of Vmax and h is the Hill coefficient.
Miscellaneous methods
PAP[35S] was synthesized from ATP and carrier-free [35S]sulfate using the bifunctional human ATP sulfurylase/adenosine 5′-phosphosulfate kinase, and its purity was determined as described previously (35). The PAP[35S] synthesized was adjusted to the required concentration and a specific activity of 15 Ci/mmol at 1.4 mM by the addition of cold PAPS. Protein determination was based on the method of Bradford with bovine serum albumin as the standard (36).
Results
Generation and release of [35S]sulfated products by HepG2 cells labelled with [35S]sulfate in the presence of ractopamine or salbutamol
HepG2 human hepatoma cells were used to investigate whether sulfation of ractopamine and salbutamol may occur under metabolic conditions. Confluent HepG2 cells grown in a 24-well plate were labelled with [35S]sulfate in sulfate-free medium containing different concentrations of ractopamine or salbutamol. TLC analysis of the labelling media collected at the end of an 18-h labelling period revealed indeed the presence of [35S]sulfated derivatives of ractopamine and salbutamol in a concentration-dependent manner (Fig. 2). These results clearly indicated that sulfation of ractopamine and salbutamol may occur in cells under metabolic conditions.
Fig. 2.
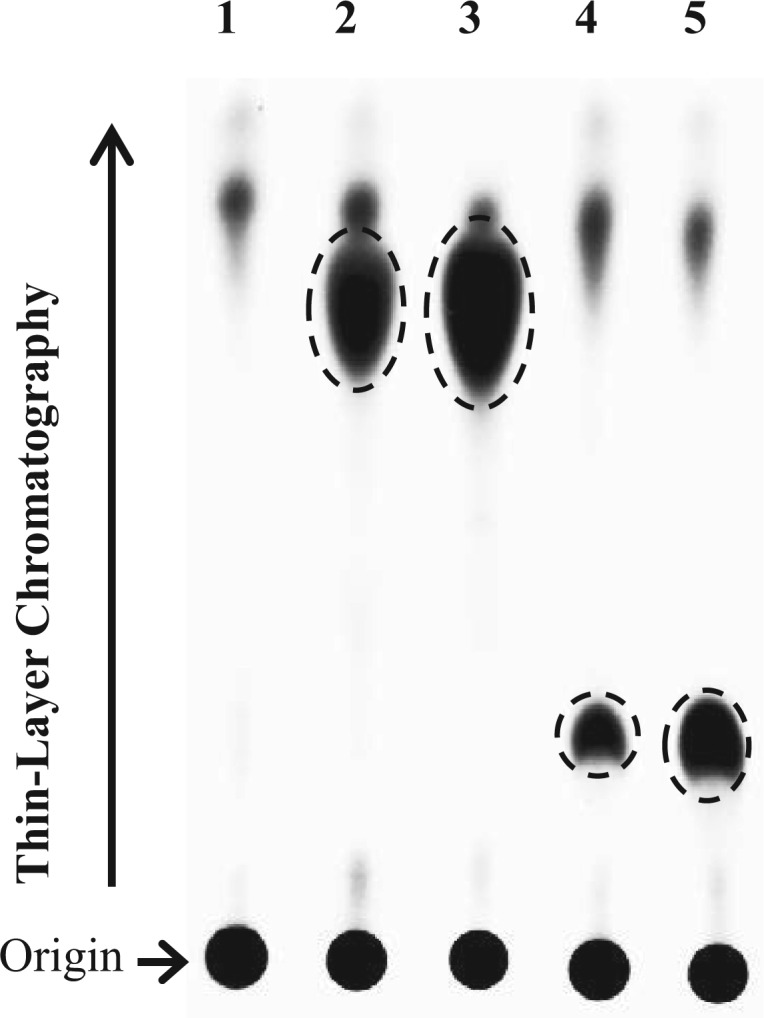
Analysis of the [35S]sulfated products generated and released by HepG2 human hepatoma cells labelled with [35S]sulfate in the presence of ractopamine or salbutamol. The autoradiograph taken from the TLC plate used for TLC analysis of the labelling media is shown. Confluent HepG2 cells were labelled with [35S]sulfate for 18 h in the presence of different concentrations (5 or 50 µM) of ractopamine and salbutamol. Lane 1 shows the control labelling medium without added compound. Lanes 2–5 correspond to the labelling media containing 5 or 50 µM ractopamine and 5 or 50 µM salbutamol, respectively. The figure is representative of three independent experiments.
Differential sulfating activities of the human SULTs towards ractopamine and salbutamol
To identify the enzyme(s) that is(are) responsible for the sulfation of ractopamine and salbutamol, 11 purified human SULTs were examined for sulfating activity with 3 different concentrations (1, 10 and 50 µM) of ractopamine and salbutamol as substrates. Results obtained showed that seven (SULT1A2, SULT1C2, SULT1E1, SULT2A1, SULT2B1a, SULT2B1b and SULT4A1) of the 11 SULTs displayed no detectable activities. Of the other four SULTs, SULT1A3 exhibited considerably stronger activities than the other three towards ractopamine and salbutamol for all concentrations tested (Table I). SULT1A1, SULT1B1 and SULT1C4 displayed sulfating activity towards only ractopamine, but not salbutamol. These results indicated that SULT1A3 is likely the major enzyme responsible for sulfating the two feed additive compounds in the body.
Table I.
Specific activities of the human SULT1A1, SULT1A3, SULT1B1 and SULT1C4 with ractopamine and salbutamol as substratesa.
| Specific activity (nmol/min/mg) |
||||
|---|---|---|---|---|
| Substrate | SULT1A1 | SULT1A3 | SULT1B1 | SULT1C4 |
| Ractopamine | ||||
| 1 µM | NDb | 5.41 ± 0.05 | ND | ND |
| 10 µM | 0.37 ± 0.06 | 41.36 ± 0.37 | ND | 0.60 ± 0.02 |
| 50 µM | 1.36 ± 0.03 | 91.86 ± 3.50 | 0.26 ± 0.03 | 3.61 ± 0.12 |
| Salbutamol | ||||
| 1 µM | ND | 0.27 ± 0.01 | ND | ND |
| 10 µM | ND | 2.01 ± 0.03 | ND | ND |
| 50 µM | ND | 9.93 ± 0.53 | ND | ND |
aSpecific activity refers to nmol substrate sulfated/min/mg purified enzyme. Data represent mean ± standard deviation derived from three determinations. The concentrations of the substrate used in the assay mixtures were 1, 10, or 50 µM.
bND refers to activity not detected. Specific activity determined was lower than the detection limit (estimated to be ∼0.01 nmol/min/mg protein).
Characterization of the ractopamine- and salbutamol-sulfating activity of human SULT1A3
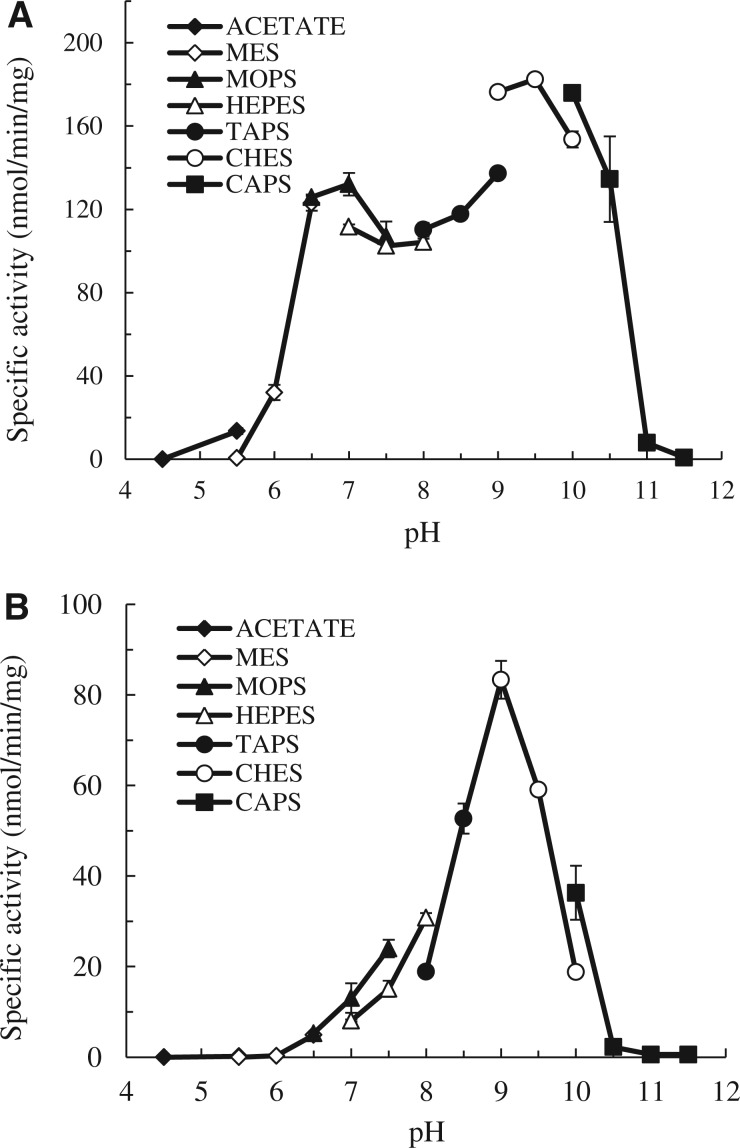
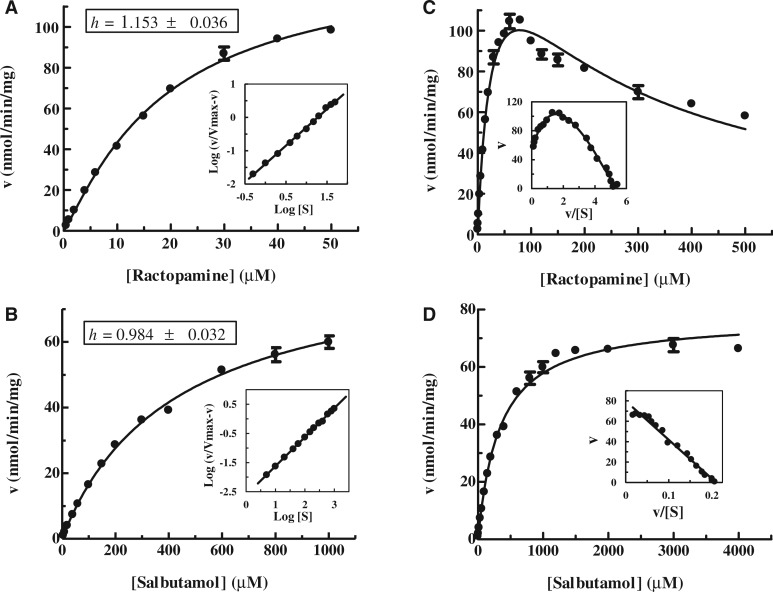
To investigate further the sulfation of ractopamine and salbutamol by human SULT1A3, the pH dependence and kinetic parameters of SULT1A3-mediated sulfation of ractopamine or salbutamol were analyzed. As shown in Fig. 3, a pH optimum at 9.5 and a smaller peak activity at pH 7.0 were observed with ractopamine as substrate, whereas a distinct pH optimum at 9.0 was detected with salbutamol as substrate. The distinct pH-dependence profiles observed may reflect the differential substrate recognition of SULT1A3 towards ractopamine and salbutamol. The kinetics of the sulfation of ractopamine or salbutamol by SULT1A3 was subsequently analyzed using varying concentrations of these two compounds at pH 7.0. Initial rates of the sulfation of ractopamine or salbutamol analyzed using Hill-fitting (sigmoidal) curve showed the hyperbolic (non-sigmoidal) kinetic curves for the sulfation of these two compounds with Hill coefficients (h) of 1.153 ± 0.036 and 0.984 ± 0.032, respectively (Fig. 4A and B). The results of non-linear regression analysis were confirmed by Hill plots, based on log (v/Vmax−v) versus log[S]. To obtain the kinetic parameters, saturation curve analyses were examined using two hyperbolic kinetics, Michaelis–Menten kinetics and substrate inhibition kinetics. As shown in Fig. 4C, the sulfation of ractopamine was fitted to substrate inhibition kinetics, which was further confirmed by an Eadie–Hofstee plot, based on v versus v/[S], with a hook in the upper quadrant (37). Conversely, the sulfation of salbutamol was fitted to Michaelis–Menten kinetics, which was further confirmed by a linear Eadie–Hofstee plot (37) (Fig. 4D). Table II shows the kinetic constants determined for the sulfation of ractopamine by substrate inhibition kinetics and the sulfation of salbutamol by Michaelis–Menten kinetics. The Km value determined with ractopamine (27.52 µM) was an order of magnitude lower than that with salbutamol (332.7 µM), indicating that the affinity of SULT1A3 for ractopamine was much higher than that for salbutamol. The catalytic efficiency of SULT1A3, as reflected by the Vmax/Km (6.20 and 0.23, respectively), was nearly 27 times higher with ractopamine than with salbutamol. These results indicated that SULT1A3 was able to catalyze the sulfation of ractopamine much more efficiently than with salbutamol.
Fig. 3.
pH dependency of the sulfating activity of human SULT1A3 with (A) ractopamine and (B) salbutamol as substrates. Enzymatic assays with a final concentration of 50 µM for each substrate were carried out under standard assay conditions as described in Materials and Methods, using different buffer systems as indicated. Data shown represent calculated mean ± standard deviation derived three experiments.
Fig. 4.
Kinetic analysis for the sulfation of ractopamine and salbutamol by human SULT1A3. (A) and (B) The hyperbolic curve analyses of the sulfation of ractopamine and salbutamol. The fitting curves were generated using sigmoidal program and the Hill coefficients (h) were determined by Hill equation. Hill plots are inserted under each fitting curve. (C) and (D) The saturation curve analyses of the sulfation of ractopamine and salbutamol. The fitting curves were generated using substrate inhibition (C) and Michaelis–Menten kinetics (D). Eadie–Hofstee plots are inserted under each fitting curve. Data shown represent calculated mean ± standard deviation derived three experiments.
Table II.
Kinetic constants of the sulfation of ractopamine and salbutamol by human SULT1A3a.
| Vmax (nmol/min/mg) | Km (µM) | Vmax/Km | Ki (µM) | |
|---|---|---|---|---|
| Ractopamineb | 170.7 ± 6.0 | 27.52 ± 1.91 | 6.20 | 222.3 ± 16.6 |
| Salbutamolc | 77.17 ± 0.81 | 332.7 ± 12.2 | 0.23 | — |
aResults represent means ± standard deviation derived from three determinations.
bKinetic parameters were determined based on the equation for substrate inhibition kinetics.
cKinetic parameters were determined based on the equation for Michaelis–Menten kinetics.
Inhibitory effects of ractopamine and salbutamol on SULT1A3-mediated sulfation of dopamine
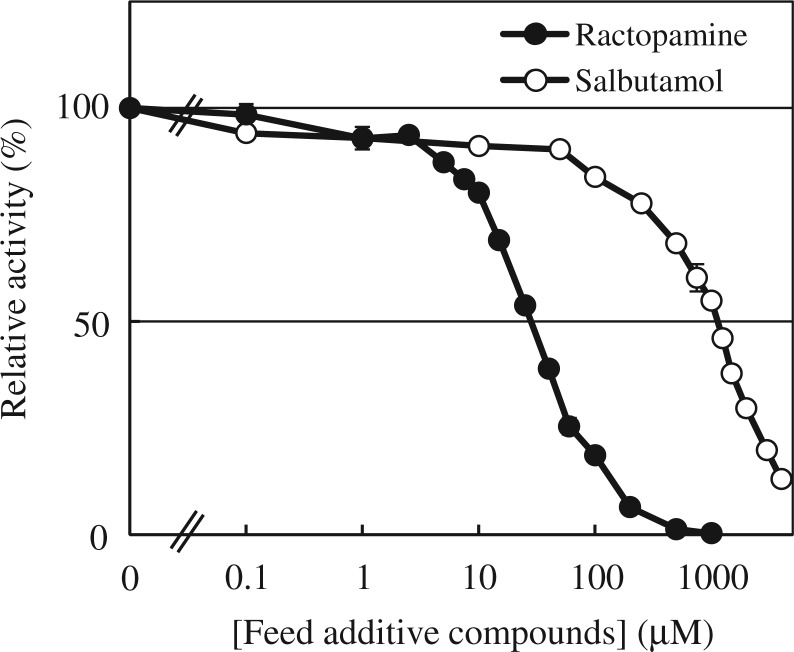
To investigate the inhibitory effects of ractopamine and salbutamol on the sulfation of dopamine by SULT1A3, enzymatic assays were carried out using 5 µM of dopamine as substrate in the presence of increasing concentrations of ractopamine or salbutamol. Activity data obtained showed the inhibition of the sulfation of dopamine by ractopamine or salbutamol in a concentration-dependent manner. Based on the results shown in Fig. 5, the IC50 values determined for ractopamine and salbutamol were 28.02 ± 1.03 and 955.7 ± 1.0 µM, respectively.
Fig. 5.
Inhibitory effects of ractopamine or salbutamol on the sulfation of dopamine by SULT1A3. Enzymatic assays using SULT1A3 with 5 µM dopamine as a substrate in the presence of varying concentrations of ractopamine (0.1–1,000 µM) or salbutamol (0.1–4,000 µM) were carried out. Data from three experiments were calculated based on the activity determined in the absence of inhibitor as 100%.
Sulfation of ractopamine and salbutamol by human organ samples
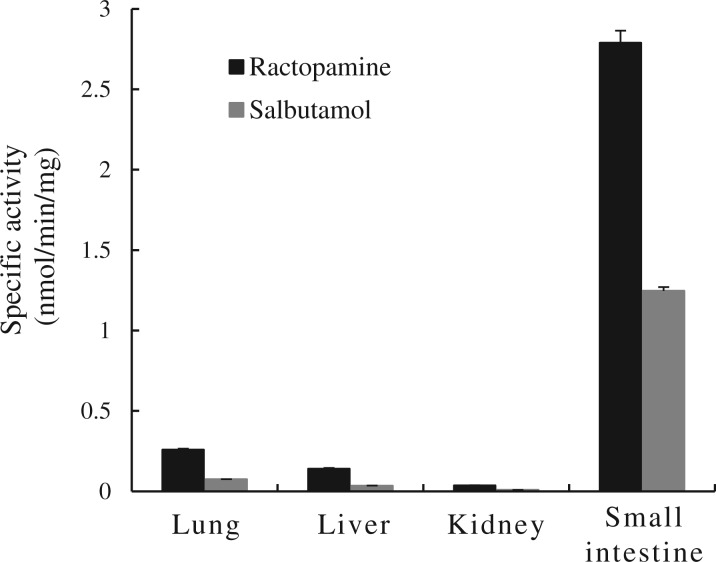
To obtain evidence for the presence of ractopamine- and/or salbutamol-sulfating activity in human tissues, enzymatic assays were performed using cytosol or S9 fraction prepared from human lung, liver, kidney or small intestine. The results showed clearly the presence of sulfating activities towards both ractopamine and salbutamol in samples prepared from all four human organs. Quantitative data shown in Fig. 6 revealed that, compared with the other three human organs, the small intestine exhibited much higher activities (2.79 and 1.25 nmol/min/mg protein, respectively) towards both ractopamine and salbutamol.
Fig. 6.
Sulfating activities of the human lung, liver, kidney and small intestine cytosol or S9 fractions towards ractopamine and salbutamol. Enzymatic assays using human tissues cytosol or S9 fractions based on the procedure described in Materials and Methods. Data shown represent calculated mean ± standard deviation derived three experiments.
Discussion
Considering that the liver is a major detoxification organ in the body, we opted to use the HepG2 human hepatoma cells, a human liver-derived cell line widely used for investigating the metabolism of xenobiotics through the sulfation (38), in this study. Previous studies have demonstrated that most of the SULTs present in the human liver, including SULT1A1, SULT1A2, SULT1A3, SULT1E1, SULT2A1 and SULT2B1, are expressed in HepG2 cells (39–44). A metabolic labelling study initially performed showed the generation and release of sulfated ractopamine and salbutamol, indicating that both of them can be taken up by HepG2 cells and subjected to the sulfation by the SULT enzyme(s) therein. It was noted that for HepG2 cells labelled in the presence of ractopamine or salbutamol, only a single radioactive [35S]sulfated derivative was detected in the labelling medium. In a subsequent study, a systematic survey of the ractopamine-/salbutamol-sulfating activity of the 11 known human SULTs was performed. Of the four SULTs that showed the sulfating activity, SULT1A3 displayed much stronger activities towards both ractopamine and salbutamol. In contrast, SULT1A1, SULT1B1 and SULT1C4 displayed low, yet significant, sulfating activity towards only ractopamine, but not salbutamol. These results indicated that SULT1A3 is the major enzyme responsible for sulfating the two major feed additive compounds. An important issue is with regard to the chemical identity of the sulfated ractopamine and salbutamol. As shown in Fig. 1, two phenolic hydroxyl groups are present in the ractopamine molecule, whereas the salbutamol molecule contains only one phenolic hydroxyl group. The sulfate conjugate of salbutamol at its phenolic hydroxyl group, salbutamol-4-O-sulfate, had been previously identified in humans (45). A sulfate conjugate of ractopamine at the C-10′ phenolic hydroxyl group (cf. Fig. 1) had been reported in the rat (46). Further study is needed to clarify whether human SULT1A3 also catalyzes the sulfation of the C-10′ phenolic hydroxyl group of ractopamine. Another important issue is related to the tissues/organs capable of metabolizing ractopamine and salbutamol through sulfation. SULT1A3 is known to be expressed in many organs including the brain, lung, liver, kidney and gastrointestinal tract (43, 44). Since residual ractopamine and salbutamol, on dietary meat consumption, may enter the body through the gastrointestinal tract, these compounds may first be subjected to the sulfate conjugation in the intestine. Those that escape sulfation in the intestine may then enter the systemic circulation and are transported to other tissues/organs, where they may be sulfated by SULT1A3 and/or other SULT(s) present in local cells. Indeed, ractopamine-/salbutamol-sulfating activities were detected in the cytosol or S9 fractions prepared from the human lung, liver, kidney and small intestine. Interestingly, the small intestine sample showed the highest sulfating activity towards both ractopamine and salbutamol (Fig. 6). This finding is in accord with previous reports on the high level of expression of SULT1A3 in the small intestine (43, 44, 47). It is to be noted that previous studies had also demonstrated the sulfation of salbutamol in the human lung, liver and intestine (48–50). It is noteworthy that although glucuronidation of salbutamol has not been reported in humans, glucuronide metabolites have been detected in other species including bovine, rabbit and rat (14, 45, 51). Although the metabolism and conjugation reactions of ractopamine in humans have not been investigated, both sulfate and glucuronide metabolites of ractopamine have been detected in livestock including turkey and swine (13). The results obtained in this study suggest that SULT1A3-mediated sulfation may play an important role in the metabolism of ractopamine and salbutamol in humans.
In examining the pH dependence of the sulfation of ractopamine by SULT1A3, two distinct pH optima at 7.0 and 9.5, respectively, were detected. In contrast, only one pH optimum at 9.0 was detected with salbutamol as a substrate. These different pH optima may be due to the differences in the chemical structures of the two compounds. Kinetic experiments were performed to gain insight into the mechanism of the sulfation of ractopamine and salbutamol by SULT1A3. Previous studies on the crystal structure of SULT1A1 complexed with p-nitrophenol (pNP) showed two pNP molecules in the substrate-binding pocket (52). The sulfation of pNP by SULT1A1 displayed a slightly positive cooperativity at low concentrations and substrate inhibition profiles at high concentrations (52). For SULT1A3, a molecular modelling analysis suggested the potential superposition of another dopamine molecule in the substrate-binding pocket and kinetic assay showed the substrate inhibition profile at high concentrations of dopamine (53). In our initial kinetic experiments, sigmoidal curve analyses were performed using Hill function at concentration ranges of 0.5–50 µM for ractopamine and 5–1,000 µM for salbutamol to investigate any possible cooperativity of the sulfation of ractopamine or salbutamol by SULT1A3. Hyperbolic curves were observed in the sulfation of both substrates and the Hill coefficient (h) determined indicated a weak positive cooperativity for the sulfation of ractopamine and noncooperativity for the sulfation of salbutamol. Further substrate saturation curve analyses at concentration ranges of 0.5–500 µM for ractopamine and 5–4,000 µM for salbutamol revealed that the sulfation of ractopamine was subjected to substrate inhibition and the sulfation of salbutamol followed that of the Michaelis–Menten kinetics. It was noted that the Km value determined with ractopamine was an order of magnitude lower than that with salbutamol (27.52 versus 332.5 µM), indicating that the affinity of SULT1A3 for ractopamine was much higher than for salbutamol. The catalytic efficiency of SULT1A3, as reflected by the Vmax/Km (6.20 versus 0.23), was 27 times higher with ractopamine than with salbutamol. These results indicated that ractopamine is a much better substrate than salbutamol for SULT1A3 and may be metabolized faster through SULT1A3-mediated sulfation. Nevertheless, it should be noted that previous studies have demonstrated that the Km value of SULT1A3 towards dopamine, a major endogenous substrate of the enzyme, ranged from 2 to 10 µM and the Vmax ranged from 50 to 600 nmol/min/mg enzyme (54–57). Therefore, it seems that ractopamine and salbutamol will have to reach considerably higher concentrations to be used de facto as substrates for SULT1A3, which will then mediate their sulfation just as efficiently as with dopamine.
Since SULT1A3-mediated sulfate conjugation is known to play an important role in the homeostasis of catecholamines (58, 59), an interesting issue is that whether, by serving as substrates for SULT1A3, ractopamine and salbutamol may act as inhibitors for the sulfation of dopamine. Our results showed that both ractopamine and salbutamol indeed exerted inhibitory effects on the sulfation of dopamine in a concentration-dependent manner. With 5 µM of dopamine as a substrate for SULT1A3, the IC50 values of ractopamine and salbutamol were determined to be 28.02 and 955.7 µM, respectively. These IC50 values were in line with the Km values of SULT1A3 for ractopamine and salbutamol (Table II). These results implied that ractopamine or salbutamol, at high concentrations, may interfere with the homeostasis of dopamine and, possibly, other catecholamines as well.
To summarize, this study indicated that sulfation of ractopamine and salbutamol may occur under metabolic conditions and that SULT1A3 is the major human SULT capable of sulfating ractopamine and salbutamol. As substrates for SULT1A3, ractopamine and salbutamol may inhibit the sulfation of dopamine. In line with the tissue distribution of SULT1A3, the highest ractopamine-/salbutamol-sulfating activity was detected in the small intestine, among the four human organ samples tested. More work is warranted to fully elucidate the biochemical/physiological relevance of the sulfation of ractopamine and salbutamol.
Funding
National Institutes of Health (GM085756) and a startup fund from College of Pharmacy, The University of Toledo.
Conflict of interest
None declared.
Glossary
Abbreviations
- ATP
adenosine 5′-triphosphate
- CAPS
3-(cyclohexylamino)-1-propanesulfonic acid
- CHES
2-(cyclohexylamino)ethanesulfonic acid
- DTT
dithiothreitol
- FBS
fetal bovine serum
- HEPES
N-2-hydroxylpiperazine-N′-2-ethanesulfonic acid
- MEM
minimum essential medium
- MES
morpholinoethanesulfonic acid
- MOPS
3-(N-morpholino)propanesulfonic acid
- PAPS
3′-phosphoadenosine 5′-phosphosulfate
- pNP
p-nitrophenol
- SULT
cytosolic sulfotransferase
- TAPS
3-[N-tris-(hydroxymethyl)methylamino]-propanesulfonic acid
- TLC
thin-layer chromatography
References
- 1.Sillence MN. Technologies for the control of fat and lean deposition in livestock. Vet. J. 2004;167:242–257. doi: 10.1016/j.tvjl.2003.10.020. [DOI] [PubMed] [Google Scholar]
- 2.Strydom PE, Frylinck L, Montgomery JL, Smith MF. The comparison of three beta-agonists for growth performance, carcass, characteristics and meat quality of feedlot cattle. Meat. Sci. 2009;81:557–564. doi: 10.1016/j.meatsci.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 3.Mimbs KJ, Pringle TD, Azain MJ, Meers SA, Armstrong TA. Effects of ractopamine on performance and composition of pigs phenotypically sorted into fat and lean groups. J. Anim. Sci. 2005;83:1361–1369. doi: 10.2527/2005.8361361x. [DOI] [PubMed] [Google Scholar]
- 4.Baker PK, Dalrymple RH, Ingle DL, Ricks CA. Use of a beta-adrenergic agonist to alter muscle and fat deposition in lambs. J. Anim. Sci. 1984;59:1256–1261. [Google Scholar]
- 5.Hansen JA, Nelssen JL, Goodband RD, Laurin JL. Interactive effects among porcine somatotropin, the beta-adrenergic agonist salbutamol, and dietary lysine on growth performance and nitrogen balance of finishing swine. J. Anim. Sci. 1994;72:1540–1547. doi: 10.2527/1994.7261540x. [DOI] [PubMed] [Google Scholar]
- 6.Lynch GS, Ryall JG. Role of beta-adrenoceptor signaling in skeletal muscle: implications for muscle wasting and disease. Physiol. Rev. 2008;88:729–767. doi: 10.1152/physrev.00028.2007. [DOI] [PubMed] [Google Scholar]
- 7.Yang YT, McElligott MA. Multiple actions of beta-adrenergic agonists on skeletal muscle adipose tissue. Biochem. J. 1989;261:1–10. doi: 10.1042/bj2610001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brès J, Clauzel AM, Pistre MC, Rachmat H, Bressolle F. Metabolism of beta-adrenergic substances. Therapeutic implications. Bull. Eur. Physiopathol. Respir. 1985;21:19s–34s. [PubMed] [Google Scholar]
- 9.Koster AS, Frankhuijzen-Sierevogel AC, Noordhoek J. Glucuronidation of morphine and six beta 2-sympathomimetics in isolated rat intestinal epithelial cells. Drug Metab. Dispos. 1985;13:232–238. [PubMed] [Google Scholar]
- 10.Yan M, Webster LT, Jr, Blumer JL. 3-O-methyldobutamine, a major metabolite of dobutamine in humans. Drug Metab. Dispos. 2002;30:519–524. doi: 10.1124/dmd.30.5.519. [DOI] [PubMed] [Google Scholar]
- 11.MacGregor TR, Nastasi L, Farina PR, Keirns JJ. Isolation and characterization of metaproterenol-3-O-sulfate: a conjugate of metaproterenol in human urine. Drug Metab. Dispos. 1983;11:568–573. [PubMed] [Google Scholar]
- 12.Orlovius AK, Guddat S, Parr MK, Kohler M, Gütschow M, Thevis M, Schänzer W. Terbutaline sulfoconjugate: characterization and urinary excretion monitored by LC/ESI-MS/MS. Drug Test. Anal. 2009;1:568–575. doi: 10.1002/dta.84. [DOI] [PubMed] [Google Scholar]
- 13.Smith DJ. The pharmacokinetics, metabolism, and tissue residues of beta-adrenergic agonists in livestock. J. Anim. Sci. 1998;76:173–194. doi: 10.2527/1998.761173x. [DOI] [PubMed] [Google Scholar]
- 14.Sauer MJ, Dave M, Lake BG, Manchee GR, Howells LC, Coldham NG. Beta2-agonist abuse in food producing animals: use of in vitro liver preparations to assess biotransformation and potential target residues for surveillance. Xenobiotica. 1999;29:483–497. doi: 10.1080/004982599238498. [DOI] [PubMed] [Google Scholar]
- 15.Falany CN, Roth JA. Properties of human cytosolic sulfotransferases involved in the drug metabolism. In: Jeffery HE, editor. Human Drug Metabolism; From Molecular Biology to Man. Boca Raton: CRC Press; 1993. pp. 101–115. [Google Scholar]
- 16.Mulder GJ, Jakoby WB. Sulfation in conjugation reactions. In: Mulder GJ, Jakoby WB, editors. Drug Metabolism. London: Taylor and Francis; 1990. pp. 107–161. [Google Scholar]
- 17.Weinshilboum RM, Otterness DM. Sulfotransferase enzymes in conjugation-deconjugation reactions. In: Kaufman FC, editor. Drug Metabolism and Toxicity. Berlin: Springer-Verlag; 1994. pp. 45–78. [Google Scholar]
- 18.Lipmann F. Biological sulfate activation and transfer. Science. 1958;128:575–580. doi: 10.1126/science.128.3324.575. [DOI] [PubMed] [Google Scholar]
- 19.Veronese ME, Burgess W, Zhu X, McManus ME. Functional characterization of two human sulphotransferase cDNAs that encode monoamine- and phenol-sulphating forms of phenol sulphotransferase: substrate kinetics, thermal-stability and inhibitor-sensitivity studies. Biochem. J. 1994;302:497–502. doi: 10.1042/bj3020497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilborn TW, Comer KA, Dooley TP, Reardon IM, Heinrikson RL, Falany CN. Sequence analysis and expression of the cDNA for the phenol-sulfating form of human liver phenol sulfotransferase. Mol. Pharmacol. 1994;43:70–77. [PubMed] [Google Scholar]
- 21.Wood TC, Aksoy IA, Aksoy S, Weinshilboum RM. Human liver thermolabile phenol sulfotransferase: cDNA cloning, expression and characterization. Biochem. Biophys. Res. Commun. 1994;198:1119–1127. doi: 10.1006/bbrc.1994.1159. [DOI] [PubMed] [Google Scholar]
- 22.Wang J, Falany JL, Falany CN. Expression and characterization of a novel thyroid hormone-sulfating form of cytosolic sulfotransferase from human liver. Mol. Pharmacol. 1998;53:274–282. doi: 10.1124/mol.53.2.274. [DOI] [PubMed] [Google Scholar]
- 23.Her C, Kaur GP, Athwal RS, Weinshilboum RM. Human sulfotransferase SULT1C1: cDNA cloning, tissue-specific expression, and chromosomal localization. Genomics. 1997;41:467–470. doi: 10.1006/geno.1997.4683. [DOI] [PubMed] [Google Scholar]
- 24.Sakakibara Y, Yanagisawa K, Katafuchi J, Ringer DP, Takami Y, Nakayama T, Suiko M, Liu M-C. Molecular cloning, expression, and characterization of novel mouse sulfotransferase that catalyze the sulfonation of N-hydroxy-2-acetylaminofluorene. J. Biol. Chem. 1998;273:33929–33935. doi: 10.1074/jbc.273.51.33929. [DOI] [PubMed] [Google Scholar]
- 25.Aksoy IA, Wood TC, Weinshilboum RM. Human liver estrogen sulfotransferase: identification by cDNA cloning and expression. Biochem. Biophys. Res. Commun. 1994;200:1621–1629. doi: 10.1006/bbrc.1994.1637. [DOI] [PubMed] [Google Scholar]
- 26.Falany CN, Vazquez ME, Kalb JM. Purification and characterization of human liver dehydroepiandrosterone sulphotransferase. Biochem. J. 1989;260:641–646. doi: 10.1042/bj2600641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otterness DM, Wieben ED, Wood TC, Watson WG, Madden BJ, McCormick DJ, Weinshilboum RM. Human liver dehydroepiandrosterone sulfotransferase: molecular cloning and expression of cDNA. Mol. Pharmacol. 1992;41:865–872. [PubMed] [Google Scholar]
- 28.Fuda H, Lee YC, Shimizu C, Javitt NB, Strott CA. Mutational analysis of human hydroxysteroid sulfotransferase SULT2B1 isoforms reveals that exon 1B of the SULT2B1 gene produces cholesterol sulfotransferase, whereas exon 1A yields pregnenolone sulfotransferase. J. Biol. Chem. 2002;277:36161–36166. doi: 10.1074/jbc.M207165200. [DOI] [PubMed] [Google Scholar]
- 29.Her C, Wood TC, Eichler EE, Mohrenweiser HW, Ramagli LS, Siciliano MJ, Weinshilboum RM. Human hydroxysteroid sulfotransferase SULT2B1: two enzymes encoded by a single chromosome 19 gene. Genomics. 1998;53:284–295. doi: 10.1006/geno.1998.5518. [DOI] [PubMed] [Google Scholar]
- 30.Falany CN, Xie X, Wang J, Ferrer J, Falany JL. Molecular cloning and expression of novel sulphotransferase-like cDNAs from human and rat brain. Biochem. J. 2000;346:857–864. [PMC free article] [PubMed] [Google Scholar]
- 31.Sakakibara Y, Suiko M, Pai TG, Nakayama T, Takami Y, Katafuchi J, Liu M-C. Highly conserved mouse and human brain sulfotransferases: molecular cloning, expression, and functional characterization. Gene. 2002;285:39–47. doi: 10.1016/s0378-1119(02)00431-6. [DOI] [PubMed] [Google Scholar]
- 32.Sakakibara Y, Takami Y, Nakayama T, Suiko M, Liu MC. Localization and functional analysis of the substrate specificity/catalytic domains of human M-form and P-form phenol sulfotransferases. J. Biol. Chem. 1998;273:6242–6247. doi: 10.1074/jbc.273.11.6242. [DOI] [PubMed] [Google Scholar]
- 33.Pai TG, Sugahara T, Suiko M, Sakakibara Y, Xu F, Liu M-C. Differential xenoestrogen-sulfating activities of the human cytosolic sulfotransferases: molecular cloning, expression, and purification of human SULT2B1a and SULT2B1b sulfotransferases. Biochim. Biophys. Acta. 2002;1573:165–170. doi: 10.1016/s0304-4165(02)00416-6. [DOI] [PubMed] [Google Scholar]
- 34.Suiko M, Sakakibara Y, Liu M-C. Sulfation of environmental estrogen-like chemicals by human cytosolic sulfotransferases. Biochem. Biophys. Res. Commun. 2000;267:80–84. doi: 10.1006/bbrc.1999.1935. [DOI] [PubMed] [Google Scholar]
- 35.Yanagisawa K, Sakakibara Y, Suiko M, Takami Y, Nakayama T, Nakajima H, Takayanagi K, Natori Y, Liu M-C. cDNA cloning, expression, and characterization of the human bifunctional ATP sulfurylase/adenosine 5’-phosphosulfate kinase enzyme. Biosci. Biotech. Biochem. 1998;62:1037–1040. doi: 10.1271/bbb.62.1037. [DOI] [PubMed] [Google Scholar]
- 36.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 37.Hutzler JM, Tracy TS. Atypical kinetic profiles in drug metabolism reactions. Drug Metab. Dispos. 2002;30:355–62. doi: 10.1124/dmd.30.4.355. [DOI] [PubMed] [Google Scholar]
- 38.Shwed JA, Walle UK, Walle T. Hep G2 cell line as a human model for sulphate conjugation of drugs. Xenobiotica. 1992;22:973–982. doi: 10.3109/00498259209049903. [DOI] [PubMed] [Google Scholar]
- 39.Chen X, Baker SM, Chen G. Methotrexate induction of human sulfotransferases in HepG2 and Caco-2 cells. J. Appl. Toxicol. 2005;25:354–360. doi: 10.1002/jat.1071. [DOI] [PubMed] [Google Scholar]
- 40.Miyano J, Yamamoto S, Hanioka N, Narimatsu S, Ishikawa T, Ogura K, Watabe T, Nishimura M, Ueda N, Naito S. Involvement of SULT1A3 in elevated sulfation of 4-hydroxypropranolol in Hep G2 cells pretreated with beta-naphthoflavone. Biochem. Pharmacol. 2005;69:941–950. doi: 10.1016/j.bcp.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 41.Westerink WM, Schoonen WG. Phase II enzyme levels in HepG2 cells and cryopreserved primary human hepatocytes and their induction in HepG2 cells. Toxicol. In Vitro. 2007;21:1592–1602. doi: 10.1016/j.tiv.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 42.Li X, Pandak WM, Erickson SK, Ma Y, Yin L, Hylemon P, Ren S. Biosynthesis of the regulatory oxysterol, 5-cholesten-3beta,25-diol 3-sulfate, in hepatocytes. J. Lipid Res. 2007;48:2587–2596. doi: 10.1194/jlr.M700301-JLR200. [DOI] [PubMed] [Google Scholar]
- 43.Dooley TP, Haldeman-Cahill R, Joiner J, Wilborn TW. Expression profiling of human sulfotransferase and sulfatase gene superfamilies in epithelial tissues and cultured cells. Biochem. Biophys. Res. Commun. 2000;277:236–245. doi: 10.1006/bbrc.2000.3643. [DOI] [PubMed] [Google Scholar]
- 44.Stanley EL, Hume R, Coughtrie MW. Expression profiling of human fetal cytosolic sulfotransferase involved in steroid and thyroid hormone metabolism and in detoxification. J. Mol. Cell Endocrinol. 2005;240:32–42. doi: 10.1016/j.mce.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 45.Lin C, Li Y, McGlotten J, Morton JB, Symchowicz S. Isolation and identification of the major metabolite of albuterol in human urine. Drug Metab. Dispos. 1977;5:234–238. [PubMed] [Google Scholar]
- 46.Smith DJ, Giddings JM, Feil VJ, Paulson GD. Identification of ractopamine hydrochloride metabolites excreted in rat bile. Xenobiotica. 1995;25:511–520. doi: 10.3109/00498259509061870. [DOI] [PubMed] [Google Scholar]
- 47.Riches Z, Stanley EL, Bloomer JC, Coughtrie MW. Quantitative evaluation of the expression and activity of five major sulfotransferases (SULTs) in human tissues: the SULT “pie”. Drug Metab. Dispos. 2009;37:2255–2261. doi: 10.1124/dmd.109.028399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walle UK, Pesola GR, Walle T. Stereoselective sulphate conjugation of salbutamol in humans: comparison of hepatic, intestinal and platelet activity. Br. J. Clin. Pharmacol. 1993;35:413–418. doi: 10.1111/j.1365-2125.1993.tb04159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eaton EA, Walle UK, Wilson HM, Aberg G, Walle T. Stereoselective sulphate conjugation of salbutamol by human lung and bronchial epithelial cells. Br. J. Clin. Pharmacol. 1996;41:201–206. doi: 10.1111/j.1365-2125.1996.tb00183.x. [DOI] [PubMed] [Google Scholar]
- 50.Vietri M, Pietrabissa A, Spisni R, Mosca F, Pacifici GM. Differential inhibition of hepatic and duodenal sulfation of (-)-salbutamol and minoxidil by mefenamic acid. Eur. J. Clin. Pharmacol. 2000;56:477–479. doi: 10.1007/s002280000168. [DOI] [PubMed] [Google Scholar]
- 51.Martin LE, Hobson JC, Page JA, Harrison C. Metabolic studies of Salbutamol-3H: a new bronchodilator, in rat, rabbit, dog and man. Eur. J. Pharmacol. 1971;14:183–199. doi: 10.1016/0014-2999(71)90211-1. [DOI] [PubMed] [Google Scholar]
- 52.Gamage NU, Duggleby RG, Barnett AC, Tresillian M, Latham CF, Liyou NE, McManus ME, Martin JL. Structure of a human carcinogen-converting enzyme, SULT1A1. Structural and kinetic implications of substrate inhibition. J. Biol. Chem. 2003;278:7655–7662. doi: 10.1074/jbc.M207246200. [DOI] [PubMed] [Google Scholar]
- 53.Barnett AC, Tsvetanov S, Gamage N, Martin JL, Duggleby RG, McManus ME. Active site mutations and substrate inhibition in human sulfotransferase 1A1 and 1A3. J. Biol. Chem. 2004;279:18799–18805. doi: 10.1074/jbc.M312253200. [DOI] [PubMed] [Google Scholar]
- 54.Dajani R, Cleasby A, Neu M, Wonacott AJ, Jhoti H, Hood AM, Modi S, Hersey A, Taskinen J, Cooke RM, Manchee GR, Coughtrie MW. X-ray crystal structure of human dopamine sulfotransferase, SULT1A3. Molecular modeling and quantitative structure-activity relationship analysis demonstrate a molecular basis for sulfotransferase substrate specificity. J. Biol. Chem. 1999;274:37862–37868. doi: 10.1074/jbc.274.53.37862. [DOI] [PubMed] [Google Scholar]
- 55.Senggunprai R, Yoshinari K, Yamazoe Y. Inhibitory effects of kynurenic acid, a tryptophan metabolite, and its derivatives on cytosolic sulfotransferases. Biochem. J. 2009;422:455–462. doi: 10.1042/BJ20090168. [DOI] [PubMed] [Google Scholar]
- 56.Yasuda S, Liu M-Y, Suiko M, Sakakibara Y, Liu M-C. Hydroxylated serotonin and dopamine as substrates for human cytosolic SULT1A3. J. Neurochem. 2007;103:2679–2689. doi: 10.1111/j.1471-4159.2007.04948.x. [DOI] [PubMed] [Google Scholar]
- 57.Yasuda S, Yasuda T, Hui Y, Liu M-Y, Suiko M, Sakakibara Y, Liu M-C. Concerted action of the cytosolic sulfotransferase, SULT1A3, and catechol-O-methyltransferase in the metabolism of dopamine in SK-N-MC human neuroblastoma cells. Neurosci. Res. 2009;64:273–279. doi: 10.1016/j.neures.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 58.Strott CA. Sulfonation and molecular action. Endocr. Rev. 2002;23:703–732. doi: 10.1210/er.2001-0040. [DOI] [PubMed] [Google Scholar]
- 59.Eisenhofer G, Kopin IJ, Goldstein DS. Catecholamine metabolism: a contemporary view with implications for physiology and medicine. Pharmacol. Rev. 2004;56:331–349. doi: 10.1124/pr.56.3.1. [DOI] [PubMed] [Google Scholar]