Abstract
Disruption of distaless homeobox 5 and 6 (Dlx5/6) in mice results in brain, craniofacial, genital, ear and limb defects. In humans, chromosomal aberrations in the DLX5/6 region, some of which do not encompass DLX5/6, are associated with split hand/foot malformation 1 (SHFM1) as well as intellectual disability, craniofacial anomalies and hearing loss, suggesting that the disruption of DLX5/6 regulatory elements could lead to these abnormalities. Here, we characterized enhancers in the DLX5/6 locus whose tissue-specific expression and genomic location along with previously characterized enhancers correlate with phenotypes observed in individuals with chromosomal abnormalities. By analyzing chromosomal aberrations at 7q21, we refined the minimal SHFM1 critical region and used comparative genomics to select 26 evolutionary conserved non-coding sequences in this critical region for zebrafish enhancer assays. Eight of these sequences were shown to function as brain, olfactory bulb, branchial arch, otic vesicle and fin enhancers, recapitulating dlx5a/6a expression. Using a mouse enhancer assay, several of these zebrafish enhancers showed comparable expression patterns in the branchial arch, otic vesicle, forebrain and/or limb at embryonic day 11.5. Examination of the coordinates of various chromosomal rearrangements in conjunction with the genomic location of these tissue-specific enhancers showed a correlation with the observed clinical abnormalities. Our findings suggest that chromosomal abnormalities that disrupt the function of these tissue-specific enhancers could be the cause of SHFM1 and its associated phenotypes. In addition, they highlight specific enhancers in which mutations could lead to non-syndromic hearing loss, craniofacial defects or limb malformations.
INTRODUCTION
In vertebrates, the homeobox transcription factors distaless homeobox 5 and 6 (Dlx5/6) are expressed in the developing brain, olfactory bulb, craniofacial primordia, genital tubercle, otic vesicle and limbs (Supplementary Material, Fig. S1). Mouse models lacking either Dlx5 or Dlx6 are lethal and result in brain, craniofacial, urogenital, bone and inner ear defects but do not exhibit limb abnormalities (1–3). Disruption of both genes in mice leads to a split hand/foot phenocopy (4). In humans, chromosomal aberrations in the DLX5/6 region, some of which do not encompass the coding sequences of DLX5/6 (Fig. 1), cause split hand/foot malformation 1 (SHFM1; OMIM #183600). In addition, SHFM1 is associated with intellectual disability in 33% of patients, craniofacial malformations in more than 35% of patients and deafness in 35% of patients (SHFM1D, OMIM #220600) (5). Except for an autosomal recessive DLX5 missense mutation in a family with severe SHFM and hearing impairment (6), no other coding mutation has been identified in individuals with SHFM1. Altogether, these data suggest that the disruption of DLX5/6 regulatory elements could lead to SHFM1-associated phenotypes.
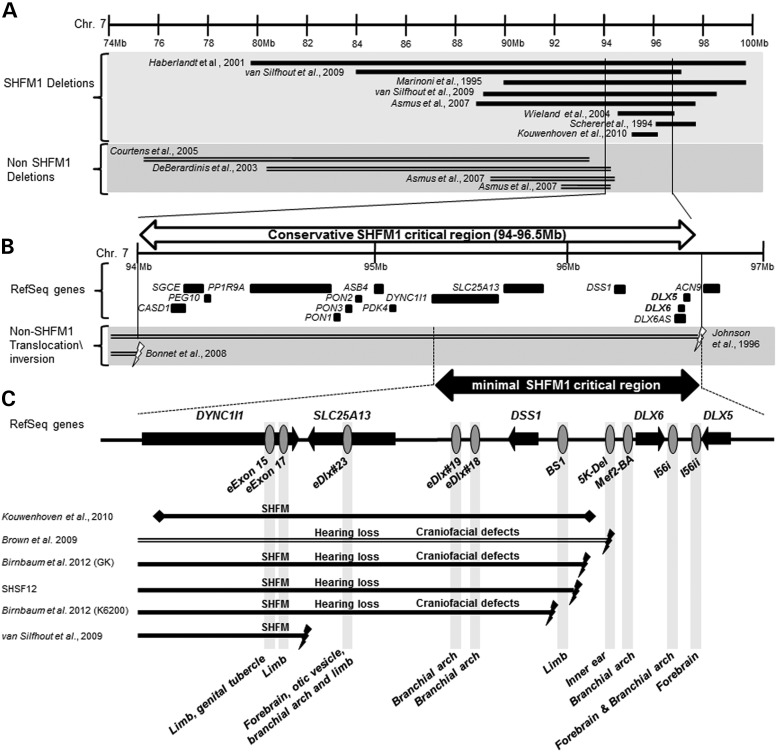
Figure 1.
The SHFM1 critical region defined by 7q21-23 chromosomal abnormalities. (A) Chromosome 7q11-q22 deletions (chromosome 7: 74–100 Mb; hg18) found in humans with SHFM1 and myoclonus-dystonia syndrome (OMIM #159900) (10,16–18,20,21,24–26). (B) Two translocations that are not associated with SHFM1 were used to define the conservative SHFM1 critical region of 2.5 Mb (chr 7: 94 000 000–96 500 000 bp; hg18) (24,25), which contains 15 genes including DLX5/6. (C) The minimal SHFM1 critical region (chromosome 7: 95.3–96.5 Mb; hg18) was defined using the most proximal translocation breakpoint in an individual with isolated SHFM ∼900 kb away from DLX5/6 (23) and a microdeletion that does not include DLX5/6 (10) (both at the centromeric side). Gene orientation is depicted by black arrows and identified mouse tissue-specific enhancers using gray ovals. Black lines represent individuals with a split hand/foot phenotype and unfilled lines represent individuals without a split hand/foot phenotype. Lightning bolts represent translocation and inversion breakpoints and diamonds represent deletions.
The transcriptional regulation of Dlx5/6 is complex and involves tissue-specific enhancers (7–10) and non-coding RNA (11). DLX5/6 are organized in a convergently transcribed bigene cluster and have a similar expression pattern in vertebrates and mammals (Fig. 2G–J), suggesting that evolutionary conserved enhancers could regulate their expression. Using comparative genomics in combination with mouse transgenic assays, two enhancers, I56i and I56ii, that reside in the Dlx5/6 intergenic region (Fig. 1C) were found to be active in the developing forebrain and are thought to regulate Dlx5/6 expression in specific GABAergic interneurons (7,12). Furthermore, a nucleotide variant found in an autistic individual affects the binding of DLX to I56i and reduces DLX5/6 expression in these GABAergic interneurons (12). The I56i enhancer is also active in the branchial arch and its activity is thought to be mediated by heart and neural crest derivatives expressed 2 (Hand2), a gene that down-regulates Dlx5/6 expression in the distal mandibular arch and is crucial for craniofacial development (13). Dlx5/6 branchial arch expression is also thought to be regulated by another enhancer, the MEF2-dependent DLX5/6 branchial arch enhancer that is ∼1.5 kb upstream of the Dlx6 transcription start site (TSS) (Fig. 1C) and is synergistically regulated by Mef2c and Dlx5 (9). A potential DLX5/6 inner ear enhancer located 85 kb proximal to DLX5 was identified due to a 5 kb deletion in a family with an inversion at 7q21.3 that exhibits hearing loss and craniofacial defects (8) (Fig. 1C). In the developing limb, both Dlx5/6 are thought to be regulated by the transcription factor tumor protein p63 (Tp63) (14). In humans, mutations in TP63 were found to cause SHFM4 (OMIM #605289). Using chromatin immunoprecipitation (ChIP) followed by next-generation sequencing for p63, an apical ectodermal ridge (AER) enhancer, named BS1, was identified 300 kb away from DLX5 (Fig. 1C) (10). In addition, we have recently characterized two additional limb enhancers, dynein cytoplasmic 1 intermediate chain 1 (DYNC1I1) eExon 15 and 17, that reside 900 kb proximal to DLX5/6 in coding exons of the DYNC1I1 gene (Fig. 1C) (15) and are thus termed eExons (exonic enhancers). DYNC1I1 eExon 15 has enhancer activity in the AER and DYNC1I1 eExon 17 in the anterior mesenchyme, both features of Dlx5/6 limb expression. Despite the identification of these enhancers, the examination of chromosomal rearrangements in individuals with SHFM1 suggests that additional, yet undetected, proximal DLX5/6 enhancers could be involved in these SHFM1-associated phenotypes (Fig. 1). Thus, a comprehensive enhancer screen is required to elucidate the gene regulatory landscape of the DLX5/6 locus.
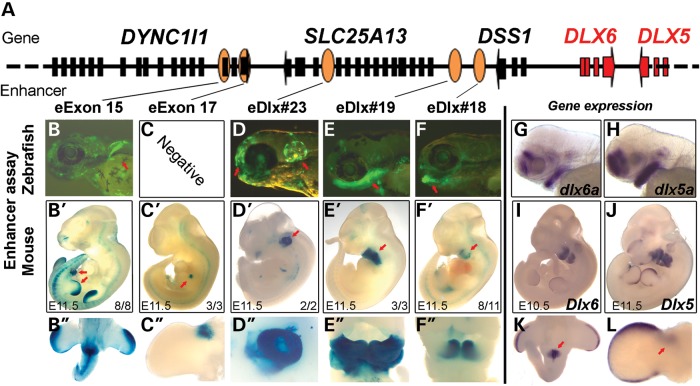
Figure 2.
Functional enhancers in the minimal SHFM1 critical region characterized using zebrafish and mice. (A) A schematic of the minimal SHFM1 region. Black and red boxes represent coding exons and the orange ovals represent sequences that have enhancer activity. (B–F) Tissue-specific enhancers in zebrafish at 72 hpf. Tissue-specific enhancer expression is indicated by the red arrows: (B) pectoral fin, (D) otic vesicle and forebrain and (E and F) branchial arches. (B′–F″) Mouse enhancer expression at E11.5. (B′ and B″) DYNC1I1 eExon 15 shows the AER, limb mesenchyme and genital tubercle enhancer activity. (C′ and C″) DYNC1I1 eExon 17 shows the anterior limb bud mesenchyme enhancer activity. (D′ and D″) eDlx#23 shows the otic vesicle, forebrain, branchial arch and limb bud mesenchyme enhancer activity. (E′–F″) eDlx#18 and eDlx#19 show branchial arch enhancer activity. The red arrows highlight the tissue-specific expression and the numbers in the bottom right of the embryos indicate the number of embryos showing this expression pattern/total LacZ stained embryos. (G and H) Zebrafish whole-mount in situ hybridization of dlx6a and dlx5a at 72 hpf. (I and J) Mouse whole-mount in situ hybridization of Dlx6 and Dlx5. (K) Dlx5 expression in the genital tubercle. (L) Dlx5 is expressed in the limb, both in the AER and in the anterior limb bud (red arrow) similar to DYNC1I1 eExon 15 and 17 expression (B′ and C′), respectively. Images B′, C′, C′, I, J and L were taken with permission from Birnbaum et al. (15).
In this study, we analyzed the chromosomal aberrations of individuals with SHFM1 and defined a minimal SHFM1 critical region. Using a zebrafish transgenic assay, we tested 26 evolutionary conserved non-coding sequences for enhancer activity and found that 8 of them function as brain, olfactory bulb, branchial arch, otic vesicle and/or fin enhancers that recapitulate dlx5a/6a expression. Several of these enhancers showed a similar spatial enhancer activity in mice, supporting their evolutionary importance. Comparison of the location of these enhancers and SHFM1-associated chromosomal rearrangements showed correlation between the clinical phenotypes and the expression patterns of these enhancers. This study suggests that mutations or disruptions of these tissue-specific enhancers could lead to brain, craniofacial, ear and limb defects.
RESULTS
Defining the SHFM1 critical region
In order to identify regulatory elements whose disruption could lead to SHFM1 and its associated phenotypes, we first defined the SHFM1 critical region by analyzing available individuals and previously reported cases with chromosomal aberrations around the 7q21-22 region (Fig. 1). We initially analyzed previously published 7q21-22 chromosomal deletions that either lead to SHFM1 or do not cause SHFM1 (Fig. 1A) (10,16–23). Using these chromosomal deletions along with two translocations in individuals that did not exhibit a SHFM phenotype, we defined a conservative SHFM1 critical region of 2.5 Mb (chr 7: 94 000 000–96 500 000 bp; hg18) that contains 15 genes, including DLX5/6 (Fig. 1B) (24,25).
To narrow down the SHFM1 critical region even further, we fine-mapped the breakpoint coordinates of a family (SHSF12) with autosomal dominant SHFM1 and sensorineural hearing loss with incomplete penetrance and variable phenotypic expression that was previously linked to the DLX5/6 locus (26). The affected SHSF12 family members were found to have inversion breakpoints within chr 7: 29 043 157 and 96 185 954 which is proximal to DLX5/6 (Fig. 1C) (27,28). The inversion was balanced with minimal changes in the normal sequence at each breakpoint and segregated with the SHFM and hearing loss phenotype. We also analyzed two other SHFM1 families, K6200 and GK, that we previously reported (15,26–28). The K6200 family has autosomal dominant SHFM, hearing loss and craniofacial defects with incomplete penetrance and variable phenotypic expression. The affected family members have paracentric inversion breakpoints with chr 7: 96 219 611 and 109 486 136 which is proximal to DLX5/6 (Fig. 1C) (26–28). The GK family has SHFM, hearing loss and craniofacial defects and a chromosomal translocation (7;20)(q22;p13) with the 7q22 breakpoint proximal to DLX5/6 (Fig. 1C) (15). In addition, we referred to two other reported SHFM1 cases: an individual with SHFM1 who has a de novo pericentric inversion of chromosome 7: 46, XY, inv(7) (p22q21.3), with the breakpoint mapped to chromosome 7: 95.53–95.72 Mb (23) and another individual with a split foot phenotype who has an 880 kb microdeletion of 95.39–96.27 Mb (Fig. 1C) (10). Combined, these cases allowed us to define a minimal SHFM1critical region of ∼1.2 Mb (chr 7: 95 300 000–96 500 000; Fig. 1C) where DLX5/6-associated enhancer(s) could reside.
Identification of enhancer candidates in the minimal SHFM1 critical region using comparative genomics
In order to identify enhancer candidates whose alteration could lead to SHFM1 and its associated phenotypes, we scanned the minimal SHFM1 critical region for non-coding evolutionary conserved regions (ECRs) using the ECR Browser (29). We selected 29 regions with at least 70% identity for ≥100 bp between humans and frogs (Supplementary Material, Table S1). We termed these sequences DLX5/6 enhancer candidates (eDlx). Three of these human–frog non-coding ECRs have been previously shown to function as forebrain [eDlx#1 (I56i), eDlx#2 (I56ii)] and branchial arch [eDlx#1 and eDlx#4 (Mef2-dependent enhancer)] enhancers in mice (9,30). We thus chose the 26 uncharacterized human–frog non-coding ECRs (Supplementary Material, Table S1) for our enhancer screen.
DLX5/6 developmental expression patterns are similar in zebrafish and mice
We next wanted to assess whether zebrafish are a suitable model system for identifying DLX5/6 regulatory elements. We analyzed mRNA expression patterns of dlx5a/6a and Dlx5/6 in both zebrafish and mice, respectively. Using whole-mount in situ hybridization on zebrafish embryos at 72 h post-fertilization (hpf), we observed that dlx5a/6a are expressed in the brain, olfactory bulb, branchial arch and otic vesicle and dlx5a is also expressed in the pectoral fin (Fig. 2G and H). Using a similar assay on mouse embryos at day (E) 11.5, we observed that Dlx5/6 are expressed in the branchial arches, otic vesicle and limb and Dlx5 also shows expression in the forebrain and genital tubercle (Fig. 2I–L, Supplementary Material, Fig. S1). The similar Dlx5/6 expression patterns in both mice and zebrafish suggested that DLX5/6-associated enhancers can initially be identified in zebrafish.
Zebrafish enhancer assays
Since Dlx5/6 developmental expression patterns were comparable in zebrafish and mice and zebrafish enhancer assays are rapid and cost effective, we first tested these 26 enhancer candidates in zebrafish. The enhancer candidate sequences were amplified from human genomic DNA and cloned into a zebrafish enhancer assay vector, containing an E1b minimal promoter followed by the green fluorescent protein (GFP) reporter gene (31). These vectors were microinjected into one-cell stage zebrafish embryos along with the Tol2 transposase to facilitate genomic integration (32). GFP expression was monitored at 24, 48 and 72 hpf. Nine sequences showed consistent GFP expression (≥40% of GFP expressed embryos) in specific tissues (Table 1). While one enhancer showed expression in the somitic muscles (eDlx#8; Supplementary Material, Fig. S2), the other eight enhancers recapitulated dlx5a and dlx6a zebrafish expression patterns. eDlx#14 drove GFP expression in the whole embryo with stronger expression in the olfactory bulb and forebrain (Supplementary Material, Fig. S3A). eDlx#16 showed enhancer expression in the olfactory bulb and forebrain and also showed partial expression in the caudal fin (Supplementary Material, Fig. S3B). eDlx#27 drove GFP expression in the olfactory bulb only at 24 hpf (Supplementary Material, Fig. S3C). eDlx#18 and eDlx#19 drove specific GFP expression in the branchial arch (Fig. 2E and F; Supplementary Material, Fig. S4A and D), and eDlx#19 also drove GFP expression in the pectoral fin at 72 hpf (Supplementary Material, Fig. S4D). eDlx#23 showed enhancer expression in the forebrain, otic vesicle and pectoral fin (Fig. 2D; Supplementary Material, Fig. S5A). eDlx#24 drove GFP expression in the forebrain, pectoral fin and heart. In addition, eDlx#24 drove GFP expression in neurons near the lateral optic tectum at 24 hpf (Supplementary Material, Fig. S5C). eDlx#26 showed enhancer activity in the forebrain, neural tube and heart (Supplementary Material, Fig. S5D). In summary, the zebrafish enhancer assay identified five forebrain enhancers (eDlx#14, eDlx#16, eDlx#23, eDlx#24 and eDlx#26), three olfactory enhancers (eDlx#14, eDlx#16 and eDlx#27), two branchial arch enhancers (eDlx#18 and eDlx#19), an otic vesicle enhancer (eDlx#23) and three pectoral fin enhancers (eDlx#19, eDlx#23 and eDlx#24) that could regulate DLX5/6 expression during development.
Table 1.
Functional enhancers located in the minimal SHFM1 region
| Name | Human genomic position (hg18) | Distance from DLX5 TSS (kb) | Location | Enhancer assay in zebrafish | Enhancer assay in mouse (E11.5) |
|---|---|---|---|---|---|
| I56ii (eDlx#1)a | Chr 7: 96 481 218–96 481 701 | 11 | Intergenic | Forebrain | Forebrain |
| I56i (eDlx#2)a | Chr 7: 96 478 887–96 479 654 | 13 | Intragenic, DLX6AS | Forebrain and branchial arch | Forebrain and branchial arch |
| Mef2-BA (eDlx#4)a | Chr 7: 96 471 500–96 472 022 | 21 | Intragenic, DLX6AS | Not tested | Branchial arch |
| 5K-Dela | Chr 7: 96 401 902–96 407 985 | 90 | Intergenic | Not tested | Inner ear |
| eDlx#8 | Chr 7: 96 301 017–96 301 371 | 191 | Intergenic | Somitic muscle | Not tested |
| BS1a | Chr 7: 96 195 551–96 195 750 | 297 | Intergenic | Forebrain, ear and pectoral fin | Limb (AER) |
| eDlx#14 | Chr 7: 96 063 552–96 063 771 | 429 | Intergenic | Forebrain, olfactory bulb and trunk | Not tested |
| eDlx#16 | Chr 7: 96 059 031–96 059 400 | 433 | Intergenic | Forebrain, olfactory bulb and caudal fin | Negative |
| eDlx#18 | Chr 7: 95 962 912–95 963 295 | 529 | Intergenic | Branchial arch | Branchial arch |
| eDlx#19 | Chr 7: 95 913 505–95 913 684 | 579 | Intergenic | Branchial arch and pectoral fin | Branchial arch |
| eDlx#23 | Chr 7: 95 603 887–95 604 377 | 888 | Intragenic, SLC25A13 | Otic vesicle, forebrain and pectoral fin | Otic vesicle, forebrain, branchial arch and limb |
| DYNC1I1 eExon 17 | Chr 7: 95 564 858–95 565 640 | 927 | DYNC1I1 exon | Negative | Limb |
| DYNC1I1 eExon 15 | Chr 7: 95 543 084–95 543 539 | 949 | DYNC1I1 exon | Pectoral fin, caudal fin and somitic muscle | Limb |
| eDlx#24 | Chr 7: 95 534 652–95 534 810 | 957 | Intragenic, DYNC1I1 | Forebrain, pectoral fin and heart | Negative |
| eDlx#26 | Chr 7: 95 420 675–95 421 193 | 1071 | Intragenic, DYNC1I1 | Forebrain, neural tube and heart | Not tested |
| eDlx#27 | Chr 7: 95 369 635–95 369 750 | 1122 | Intragenic, DYNC1I1 | Olfactory bulb | Not tested |
Mouse enhancer assays
To test whether these positive zebrafish enhancers have similar expression patterns in mammals, selected enhancers were chosen for a similar transgenic enhancer assay in mice. Since we were interested in SHFM1 and its associated phenotypes, we selected five enhancers with similar dlx5a/6a expression patterns in the branchial arch (eDlx#18 and eDlx#19), otic vesicle (eDlx#23), pectoral fin (eDlx#19, eDlx#23 and eDlx#24) and forebrain, olfactory bulb and caudal fin (eDlx#16) for this assay. The same human sequences tested in zebrafish were cloned into the Hsp68-LacZ vector that contains the heat shock protein 68 minimal promoter followed by a LacZ reporter gene (33). Transgenic mouse embryos were generated, harvested at E11.5 and stained for LacZ. Similar to the zebrafish enhancer results, eDlx#18 (Fig. 2F′ and F″, Supplementary Material, Fig. S4B and C) and eDlx#19 (Fig. 2E′ and E″, Supplementary Material, Fig. S4E and F) drove LacZ expression in the mouse mandibular arch (mbBA1) and eDlx#19 also drove LacZ expression in the second branchial arch (BA2). eDlx#23 drove LacZ expression in the mouse otic vesicle, forebrain, branchial arch and limb (Fig. 2D′ and D″, Supplementary Material, Fig. S5B). It is worth noting that eDlx#23 branchial arch expression was not observed in zebrafish (Fig. 2D) and that its limb expression was different from Dlx5/6 limb expression (Fig. 2D′; Supplementary Material, Fig. S5B). While eDlx#16 (Supplementary Material, Fig. S3B), eDlx#19 (Supplementary Material, Fig. S4D) and eDlx#24 (Supplementary Material, Fig. S5C) showed fin expression in zebrafish, they did not show limb expression in mice (see Supplementary Material, Fig. S4E, for eDlx#19 and data not shown for eDlx#16 and eDlx#24). In addition, we previously characterized two other limb enhancers in this region (15). DYNC1I1 eExon 15 functions as an AER and a limb bud mesenchyme enhancer and is also expressed in the genital tubercle at E11.5 (Fig. 2B′ and B″). DYNC1I1 eExon 17 functions as an anterior limb bud mesenchyme enhancer (Fig. 2C′ and C″). Although both of them function as mouse limb enhancers, only DYNC1I1 eExon 15 drove GFP expression in the pectoral fin (Fig. 2B). Altogether, our current mouse enhancer assay found two specific branchial arch enhancers (eDlx#18 and eDlx#19) and an otic vesicle enhancer that also functions as a forebrain, branchial arch and limb enhancer (eDlx#23). All three enhancers have similar expression patterns to their zebrafish counterparts and largely recapitulate Dlx5/6 expression in mouse E11.5 (Fig. 2I–L).
Mouse mRNA expression analysis of genes in the minimal SHFM1 critical region
In order to determine whether other genes in the minimal SHFM1 critical region, in addition to DLX5/6, could be associated with the phenotypes observed in SHFM1 cases, we analyzed the mouse E11.5 mRNA expression of all known genes (RefSeq, mm9) in this region. Using whole-mount in situ hybridization, the SHFM1 (ectrodactyly) gene (also known as Dss1) showed a similar expression pattern to Dlx5/6 in the branchial arches, otic vesicle, genital tubercle and limb (Supplementary Material, Fig. S1C). Solute carrier family 25 member 13 (Slc25a13) was found to be expressed in the limb and also showed lower expression levels in the branchial arches and otic vesicle compared with Dlx5/6 and Dss1 (Supplementary Material, Fig. S1B). Dync1i1 mRNA expression could not be detected at this time point using this assay (Supplementary Material, Fig. S1A). To further verify the expression of these genes, we also extracted RNA from E11.5 mouse heart, branchial arch, limb and otic vesicle tissues and performed reverse transcriptase–quantitative polymerase chain reaction (RT–qPCR) to measure the tissue-specific expression levels of these genes. Since the expression of Dlx5/6 was not detected in the heart at E11.5 by whole-mount in situ hybridization, the heart tissue served as a negative control. Dlx5/6 were highly expressed in the branchial arch, otic vesicle and limb compared with the heart tissue. Dss1 and Slc25a13 (Supplementary Material, Fig. S1G–J) were expressed in similar levels in all the tested tissues. Dync1i1 was barely detected in the tissues we tested at E11.5 (Supplementary Material, Fig. S1F). While the removal of Dlx5/6 in mice leads to phenotypes similar to those observed in individuals with SHFM1 (1,4,34), these expression results suggest that in addition to the tissue-specific enhancers that we have characterized in this region, these genes could also contribute to the observed phenotypes.
DISCUSSION
In this study, we analyzed chromosomal aberrations of individuals with SHFM1 and its associated phenotypes and defined a minimal SHFM1 critical region (∼1.2 Mb). We characterized the expression pattern of several enhancers in this region using transgenic zebrafish and mice. Each active enhancer recapitulated aspects of Dlx5/6 expression during development. These enhancers drive tissue-specific expression in the brain, olfactory bulb, branchial arches, otic vesicle, genital tubercle and limb, suggesting that mutations within them could cause intellectual disabilities, craniofacial defects, hearing loss, genital abnormalities and limb malformations.
Current mutation analysis studies mainly focus on coding sequences. However, nucleotide changes in gene regulatory elements are increasingly implicated as causes of human disease (12,35–38). Our study supports the longstanding hypothesis that SHFM1 and its associated phenotypes could be due to the disruption of gene regulatory elements leading to the abnormal expression of DLX5/6 during development. Although SHFM1 is associated with the DLX5/6 locus, no coding mutation has been found in individuals with SHFM1, except for an autosomal recessive DLX5 missense mutation in a family with severe SHFM and hearing impairment (6). Along with the observation that knocking out Dlx5/6 in mice leads to embryonic lethality (1,2,4), this could suggest that deleterious coding mutations in DLX5/6 might cause embryonic lethally in humans. In contrast, mutations in DLX5/6 regulatory elements that affect DLX5/6 transcriptional levels in specific tissues could be less deleterious and lead to isolated versions of the phenotypes associated with SHFM1. For example, a nucleotide variant (A>G) found in an autistic individual affects I56i enhancer activity and reduces DLX5/6 expression in specific GABAergic interneurons (12).
Comparison of the genomic location of characterized enhancers and SHFM1-associated chromosomal aberrations showed correlation in several cases between the clinical phenotypes and the tissue-specific enhancer expression patterns (Fig. 1C). For example, in the K6200 family (15) who has autosomal dominant SHFM, hearing loss and craniofacial defects, the paracentric inversion breakpoint coordinates (chr 7: 96 219 611 and 109 486 136) show that several tissue-specific enhancers: DYNC1I1 eExon 15 (AER, genital tubercle), DYNC1I1 eExon 17 (anterior limb mesenchyme), eDlx#23 (forebrain, otic vesicle, branchial arch and limb), eDlx#18 and eDlx#19 (branchial arch) are relocated ∼13 Mb away from DLX5/6. In the GK family (15) who has SHFM, hearing loss and craniofacial defects, the translocation (7;20)(q22;p13) breakpoint at 7q22 relocates the same tissue-specific enhancers: DYNC1I1 eExon 15, DYNC1I1 eExon 17, eDlx#23, eDlx#19, eDlx#18 in addition to the BS1 AER enhancer (10) from the DLX5/6 locus. The findings in both families suggest that the disruption of the interactions between these tissue-specific enhancers and DLX5/6 might lead to limb malformation, hearing loss and craniofacial defects in these families.
It is worth noting that the correlation between the disruption of functional enhancers and clinical phenotypes was not perfectly observed. In the SHSF12 family, which has autosomal dominant SHFM and hearing loss (27,28), the pericentric inversion breakpoint coordinates show that several tissue-specific enhancers: DYNC1I1 eExon 15, DYNC1I1 eExon 17, BS1, eDlx#23, eDlx#18 and eDlx#19 were relocated ∼67 Mb away from DLX5/6. This inversion likely disrupts the ability of these enhancers to regulate DLX5/6 in the limb and otic vesicle, leading to these phenotypes. However, at least three branchial arch enhancers (eDlx#18, eDlx#19 and eDlx#23) were also relocated in this family, but craniofacial defects were not observed. In addition, Brown et al. (8) reported a three-generation family who has hearing loss and craniofacial defects, but no limb malformations. The affected members of this family were found to have a paracentric inversion at chromosome 7, inv(7)(q21.3q35) that relocates several tissue-specific enhancers: DYNC1I1 eExon 15, DYNC1I1 eExon 17, eDlx#23, eDlx#19, eDlx#18 and BS1 ∼50 Mb away from DLX5/6. In addition to the inversion, a 5 kb deletion was reported at the 7q21.3 breakpoint (chr 7: 96 402 577–96 407 691; hg18) that encompasses a potential ear enhancer (5K-Del). Although several limb-specific enhancers were relocated ∼50 Mb away from DLX5/6, no limb malformations were observed in this family. It is also worth noting that limb malformations are often associated with urogenital developmental abnormalities such as hypospadias (3). Several SHFM-associated genes are expressed in the mouse genital tubercle, including Dlx5/6 (Fig. 2K, Supplementary Material, Fig. S1), and the targeted inactivation of these genes in mice was shown to cause abnormal urethra formation (3). Interestingly, while DYNC1I1 eExon 15 shows enhancer expression in the genital tubercle (Fig. 2B″), SHFM1 patients with chromosomal aberrations that led to the relocation of this enhancer (Fig. 1C) were not reported to have hypospadias or other urogenital abnormalities. One potential explanation for the lack of phenotypes could be enhancer redundancy (39). For example, two other branchial arch enhancers, Mef2-BA and I56i, still remain in the vicinity of DLX5/6 in the SHSF12-affected individuals and could be sufficient for proper branchial arch development. Another explanation could be incomplete penetrance of the phenotype, which was observed in some of these families (27,28).
Previous work has shown that human enhancer sequences can function as active enhancers in zebrafish, even without homologous sequences in zebrafish (40–42). Our results support these findings for most of the enhancers. For example, eDlx#19 and eDlx#23, which do not have homologous sequences in zebrafish, have similar enhancer expression patterns in zebrafish and mice (Fig. 2D and D′; Fig. 2E and E′). However, eDlx#16, eDlx#19 and eDlx#24, which drive fin expression in zebrafish, did not show limb expression in mice. This discrepancy between fin and limb enhancer activity could be attributed to differences in fin versus limb development (43), and further work will be needed in order to evaluate the efficacy of identifying limb enhancers in zebrafish.
In our study, we observed that SLC25A13 and DSS1, in addition to DLX5/6, are also expressed in tissues that could be associated with SHFM1 phenotypes and are also relocated by these various chromosomal aberrations. While the function of DLX5/6 has been well studied, this is not the case for these two additional genes. SLC25A13 encodes a member of the mitochondrial solute carrier family 25 and mutations in SLC25A13 were found to cause citrullinemia type II but not SHFM1-like phenotypes (44,45). In addition, Slc25a13 knockout mice have been reported to develop normally up to 1 year of age and do not show any SHFM1-like phenotypes (46). In this study, we observed that Slc25a13 was expressed in the limb buds, branchial arches and otic vesicle at mouse E11.5 (Supplementary Material, Fig. S1B and G). DSS1 is a subunit of the 26S proteasome and plays a role in ubiquitin-dependent proteolysis (47). In addition, DSS1 interacts with BRCA2 in human cells and contributes to BRCA2 function in homologous recombination-mediated repair and genomic stability (48). DSS1 was named deleted in split hand/split foot 1 region (and also SHFM1), since it was identified within the SHFM1 critical region (49). It was found to have similar expression levels in the branchial arches, otic vesicle and limb buds in mouse E11.5 embryos (Supplementary Material, Fig. S1C and H). While the removal of Dlx5/6 in mice leads to similar phenotypes as those observed in SHFM1 individuals, the observation that Slc25a13 and Dss1 are expressed in SHFM1-associated tissues could suggest that they might have a role in these phenotypes. In addition, the characterized enhancers detected in this and other studies might also be regulating these genes in addition to DLX5/6. Further work will be needed in order to elucidate the function, regulation and involvement of these genes in SHFM1.
MATERIALS AND METHODS
Subjects and chromosomal breakpoint mapping
The SHSF12 family has autosomal dominant SHFM and variable sensorineural hearing loss as reported previously (26). Subsequent studies of this family by pulsed field gel electrophoresis and FISH identified a chromosomal inversion with breakpoints in the SHFM1 critical region (27,28). Southern blot analysis and inverse PCR, as described previously (50), were then used to identify the inversion breakpoints (D.B. Everman, C.T. Morgan, M.E. Laughridge, T. Moss, S. Ladd, B. DuPont, D. Toms, F. Gurrieri, C.E. Schwartz, unpublished data).
Comparative genomics
A 1.2 Mb interval (chr 7: 95 300 000–96 500 000 bp; hg18) encompassing DLX5 and DLX6 was chosen for comparative genomic analysis. Conserved non-coding sequences between human and frogs were selected by the ECR Browser (29) using the default conservation parameters of ≥70% sequence identity for at least 100 bp. These sequences were then screened using the UCSC Genome Browser for those lacking coding sequences and repeats (51). Human–frog ECRs were designated as eDlx#, relevant to their upstream distance from DLX5/6.
Transgenic enhancer assays
Primers were designed to amplify candidate enhancer sequences (Supplementary Material, Table S1). PCR was carried out on human genomic DNA (Roche), and products were cloned into the E1b-GFP-Tol2 enhancer assay vector containing an E1b minimal promoter followed by GFP (31). They were injected following standard procedures (52) into at least 100 embryos per construct along with Tol2 mRNA (32) to facilitate genomic integration. GFP expression was observed and annotated up to 72 hpf. An enhancer was considered positive if 40% of the GFP expressing fish showed a consistent expression pattern. The zebrafish enhancer assay data can be found at: http://zendev.ucsf.edu/index.php. For the mouse enhancer assay, the same human genomic fragments used in zebrafish were transferred into a vector containing the Hsp68 minimal promoter followed by the LacZ reporter gene (53) and sequence verified. Transgenic mice were generated by Cyagen Biosciences using standard procedures (54). Embryos were harvested at E11.5 and stained for LacZ expression as described previously (53). All animal work was approved by the UCSF Institutional Animal Care and Use Committee.
Whole-mount in situ hybridization
Zebrafish embryos were collected from wild-type matings between 24 and 72 hpf and fixed in 4% paraformaldehyde. Full-length zebrafish dlx5a (MDR1734-96866202, Open Biosystems) and dlx6a (MDR1734-98078461, Open Biosystems) cDNA clones were used to generate digoxygenin-labeled probes. Whole-mount in situ hybridizations were performed according to standard protocols (55). Mouse E11.5 embryos were fixed in 4% paraformaldehyde. Clones containing mouse Dync1i1 (MMM1013-9202215, Open Biosystems), Slc25a13 (MMM1013-65837, Open Biosystems), Shfm1 (Dss1) (MMM1013-7512304, Open Biosystems), Dlx5 (34) and Dlx6 (OMM5895-99863403, Open Biosystems) were used as templates for digoxygenin-labeled probes. Mouse whole-mount in situ hybridizations were performed according to standard procedures (56).
RNA expression analysis
The branchial arch, otic vesicle and whole limbs were carefully dissected from E11.5 mouse embryos. Total RNA was isolated using RNeasy (Qiagen) according to the manufacturer's protocol. qPCR was performed using SsoFast EvaGreen Supermix (Bio-Rad) and run on the Eppendorf Mastercycler 3p realplex 2 real-time PCR. Each RT–qPCR was done using two independent biological experiments with three technical replicates. Specificity and the absence of primer dimers were controlled by denaturation curves. Gene expression was normalized to β-actin and the normalized expression is presented relative to the heart expression. Primer sequences used for amplification are listed in Supplementary Material, Table S2.
SUPPLEMENTARY MATERIAL
FUNDING
This research was supported by the NICHD (R01HD059862) and from the NHGRI (R01HG005058). R.Y.B. is supported in part by the UCSF Program for Breakthrough Biomedical Research postdoctoral award, which is funded in part by the Sandler Foundation. N.A. and R.Y.B. are also supported in part by NINDS (1R01NS079231). N.A. is also supported by NIGMS (GM61390), NHGRI (1R01HG006768) and NIDDK (1R01DK090382). D.B.E and C.E.S. were supported, in part, by a grant from the South Carolina Department of Disabilities and Special Needs and the Genetic Endowment of South Carolina and a previous grant (#8510) from Shriners Hospitals for Children. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH, NICHD, NHGRI, NINDS, NIDDK or the NIGMS.
ACKNOWLEDGEMENTS
We would like to thank members of the Ahituv Lab for helpful comments on the manuscript. We would also like to thank John L.R. Rubenstein for reagents.
Conflict of Interest statement. D.B.E. currently serves as an IPA detailee at NHGRI and the views represented in this article are entirely his own and are unaffiliated with any NIH or NHGRI policy.
REFERENCES
- 1.Acampora D., Merlo G.R., Paleari L., Zerega B., Postiglione M.P., Mantero S., Bober E., Barbieri O., Simeone A., Levi G. Craniofacial, vestibular and bone defects in mice lacking the Distal-less-related gene Dlx5. Development. 1999;126:3795–3809. doi: 10.1242/dev.126.17.3795. [DOI] [PubMed] [Google Scholar]
- 2.Jeong J., Li X., McEvilly R.J., Rosenfeld M.G., Lufkin T., Rubenstein J.L. Dlx genes pattern mammalian jaw primordium by regulating both lower jaw-specific and upper jaw-specific genetic programs. Development. 2008;135:2905–2916. doi: 10.1242/dev.019778. doi:10.1242/dev.019778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suzuki K., Haraguchi R., Ogata T., Barbieri O., Alegria O., Vieux-Rochas M., Nakagata N., Ito M., Mills A.A., Kurita T., et al. Abnormal urethra formation in mouse models of split-hand/split-foot malformation type 1 and type 4. Eur. J. Hum. Genet. 2008;16:36–44. doi: 10.1038/sj.ejhg.5201925. doi:10.1038/sj.ejhg.5201925. [DOI] [PubMed] [Google Scholar]
- 4.Robledo R.F., Rajan L., Li X., Lufkin T. The Dlx5 and Dlx6 homeobox genes are essential for craniofacial, axial, and appendicular skeletal development. Genes Dev. 2002;16:1089–1101. doi: 10.1101/gad.988402. doi:10.1101/gad.988402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elliott A.M., Evans J.A. Genotype-phenotype correlations in mapped split hand foot malformation (SHFM) patients. Am. J. Med. Genet. A. 2006;140:1419–1427. doi: 10.1002/ajmg.a.31244. [DOI] [PubMed] [Google Scholar]
- 6.Shamseldin H.E., Faden M.A., Alashram W., Alkuraya F.S. Identification of a novel DLX5 mutation in a family with autosomal recessive split hand and foot malformation. J. Med. Genet. 2012;49:16–20. doi: 10.1136/jmedgenet-2011-100556. doi:10.1136/jmedgenet-2011-100556. [DOI] [PubMed] [Google Scholar]
- 7.Zerucha T., Stuhmer T., Hatch G., Park B.K., Long Q., Yu G., Gambarotta A., Schultz J.R., Rubenstein J.L., Ekker M. A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain. J. Neurosci. 2000;20:709–721. doi: 10.1523/JNEUROSCI.20-02-00709.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown K.K., Reiss J.A., Crow K., Ferguson H.L., Kelly C., Fritzsch B., Morton C.C. Deletion of an enhancer near DLX5 and DLX6 in a family with hearing loss, craniofacial defects, and an inv(7)(q21.3q35) Hum. Mutat. 2010;127:19–31. doi: 10.1007/s00439-009-0736-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verzi M.P., Agarwal P., Brown C., McCulley D.J., Schwarz J.J., Black B.L. The transcription factor MEF2C is required for craniofacial development. Dev. Cell. 2007;12:645–652. doi: 10.1016/j.devcel.2007.03.007. doi:10.1016/j.devcel.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kouwenhoven E.N., van Heeringen S.J., Tena J.J., Oti M., Dutilh B.E., Alonso M.E., de la Calle-Mustienes E., Smeenk L., Rinne T., Parsaulian L., et al. Genome-wide profiling of p63 DNA-binding sites identifies an element that regulates gene expression during limb development in the 7q21 SHFM1 locus. PLoS Genet. 2010;6:e1001065. doi: 10.1371/journal.pgen.1001065. doi:10.1371/journal.pgen.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng J., Bi C., Clark B.S., Mady R., Shah P., Kohtz J.D. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. doi:10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poitras L., Yu M., Lesage-Pelletier C., Macdonald R.B., Gagne J.P., Hatch G., Kelly I., Hamilton S.P., Rubenstein J.L., Poirier G.G., et al. An SNP in an ultraconserved regulatory element affects Dlx5/Dlx6 regulation in the forebrain. Development. 2010;137:3089–3097. doi: 10.1242/dev.051052. doi:10.1242/dev.051052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barron F., Woods C., Kuhn K., Bishop J., Howard M.J., Clouthier D.E. Downregulation of Dlx5 and Dlx6 expression by Hand2 is essential for initiation of tongue morphogenesis. Development. 2011;138:2249–2259. doi: 10.1242/dev.056929. doi:10.1242/dev.056929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lo Iacono N., Mantero S., Chiarelli A., Garcia E., Mills A.A., Morasso M.I., Costanzo A., Levi G., Guerrini L., Merlo G.R. Regulation of Dlx5 and Dlx6 gene expression by p63 is involved in EEC and SHFM congenital limb defects. Development. 2008;135:1377–1388. doi: 10.1242/dev.011759. doi:10.1242/dev.011759. [DOI] [PubMed] [Google Scholar]
- 15.Birnbaum R.Y., Clowney E.J., Agamy O., Kim M.J., Zhao J., Yamanaka T., Pappalardo Z., Clarke S.L., Wenger A.M., Nguyen L., et al. Coding exons function as tissue-specific enhancers of nearby genes. Genome Res. 2012;22:1059–1068. doi: 10.1101/gr.133546.111. doi:10.1101/gr.133546.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Courtens W., Vermeulen S., Wuyts W., Messiaen L., Wauters J., Nuytinck L., Peeters N., Storm K., Speleman F., Nothen M.M. An interstitial deletion of chromosome 7 at band q21: a case report and review. Am. J. Med. Genet. A. 2005;134A:12–23. doi: 10.1002/ajmg.a.30106. doi:10.1002/ajmg.a.30106. [DOI] [PubMed] [Google Scholar]
- 17.DeBerardinis R.J., Conforto D., Russell K., Kaplan J., Kollros P.R., Zackai E.H., Emanuel B.S. Myoclonus in a patient with a deletion of the epsilon-sarcoglycan locus on chromosome 7q21. Am. J. Med. Genet. A. 2003;121A:31–36. doi: 10.1002/ajmg.a.20162. doi:10.1002/ajmg.a.20162. [DOI] [PubMed] [Google Scholar]
- 18.Asmus F., Hjermind L.E., Dupont E., Wagenstaller J., Haberlandt E., Munz M., Strom T.M., Gasser T. Genomic deletion size at the epsilon-sarcoglycan locus determines the clinical phenotype. Brain. 2007;130:2736–2745. doi: 10.1093/brain/awm209. doi:10.1093/brain/awm209. [DOI] [PubMed] [Google Scholar]
- 19.Marinoni J.C., Stevenson R.E., Evans J.P., Geshuri D., Phelan M.C., Schwartz C.E. Split foot and developmental retardation associated with a deletion of three microsatellite markers in 7q21.2-q22.1. Clin. Genet. 1995;47:90–95. doi: 10.1111/j.1399-0004.1995.tb03930.x. doi:10.1111/j.1399-0004.1995.tb03930.x. [DOI] [PubMed] [Google Scholar]
- 20.Haberlandt E., Loffler J., Hirst-Stadlmann A., Stockl B., Judmaier W., Fischer H., Heinz-Erian P., Muller T., Utermann G., Smith R.J., et al. Split hand/split foot malformation associated with sensorineural deafness, inner and middle ear malformation, hypodontia, congenital vertical talus, and deletion of eight microsatellite markers in 7q21.1-q21.3. J. Med. Genet. 2001;38:405–409. doi: 10.1136/jmg.38.6.405. doi:10.1136/jmg.38.6.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wieland I., Muschke P., Jakubiczka S., Volleth M., Freigang B., Wieacker P.F. Refinement of the deletion in 7q21.3 associated with split hand/foot malformation type 1 and Mondini dysplasia. J. Med. Genet. 2004;41:e54. doi: 10.1136/jmg.2003.010587. doi:10.1136/jmg.2003.010587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scherer S.W., Poorkaj P., Allen T., Kim J., Geshuri D., Nunes M., Soder S., Stephens K., Pagon R.A., Patton M.A., et al. Fine mapping of the autosomal dominant split hand/split foot locus on chromosome 7, band q21.3-q22.1. Am. J. Hum. Genet. 1994;55:12–20. [PMC free article] [PubMed] [Google Scholar]
- 23.van Silfhout A.T., van den Akker P.C., Dijkhuizen T., Verheij J.B., Olderode-Berends M.J., Kok K., Sikkema-Raddatz B., van Ravenswaaij-Arts C.M. Split hand/foot malformation due to chromosome 7q aberrations(SHFM1): additional support for functional haploinsufficiency as the causative mechanism. Eur. J. Hum. Genet. 2009;17:1432–1438. doi: 10.1038/ejhg.2009.72. doi:10.1038/ejhg.2009.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson E.J., Scherer S.W., Osborne L., Tsui L.C., Oscier D., Mould S., Cotter F.E. Molecular definition of a narrow interval at 7q22.1 associated with myelodysplasia. Blood. 1996;87:3579–3586. [PubMed] [Google Scholar]
- 25.Bonnet C., Gregoire M.J., Vibert M., Raffo E., Leheup B., Jonveaux P. Cryptic 7q21 and 9p23 deletions in a patient with apparently balanced de novo reciprocal translocation t(7;9)(q21;p23) associated with a dystonia-plus syndrome: paternal deletion of the epsilon-sarcoglycan (SGCE) gene. J. Hum. Genet. 2008;53:876–885. doi: 10.1007/s10038-008-0321-z. doi:10.1007/s10038-008-0321-z. [DOI] [PubMed] [Google Scholar]
- 26.Tackels-Horne D., Toburen A., Sangiorgi E., Gurrieri F., de Mollerat X., Fischetto R., Causio F., Clarkson K., Stevenson R.E., Schwartz C.E. Split hand/split foot malformation with hearing loss: first report of families linked to the SHFM1 locus in 7q21. Clin. Genet. 2001;59:28–36. doi: 10.1034/j.1399-0004.2001.590105.x. doi:10.1034/j.1399-0004.2001.590105.x. [DOI] [PubMed] [Google Scholar]
- 27.Everman D., Morgan C., Clarkson K., Gurrieri F., McAuliffe F., Chitayat D., Stevenson R., Schwartz C. Submicroscopic rearrangements involving the SHFM1 locus on chromosome 7q21-22 are associated with split-hand/foot malformation and sensorineural hearing loss. Proc. Greenwood Genet. Center. 2005;24:137. [Google Scholar]
- 28.Everman D., Morgan C., Stevenson R., Schwartz C. Chromosome rearrangements: an emerging theme in the causation of split-hand/foot malformation. Proc. Greenwood Genet. Center. 2006;25:138. [Google Scholar]
- 29.Ovcharenko I., Nobrega M.A., Loots G.G., Stubbs L. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 2004;32:W280–W286. doi: 10.1093/nar/gkh355. doi:10.1093/nar/gkh355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghanem N., Yu M., Poitras L., Rubenstein J.L., Ekker M. Characterization of a distinct subpopulation of striatal projection neurons expressing the Dlx genes in the basal ganglia through the activity of the I56ii enhancer. Dev. Biol. 2008;322:415–424. doi: 10.1016/j.ydbio.2008.07.029. doi:10.1016/j.ydbio.2008.07.029. [DOI] [PubMed] [Google Scholar]
- 31.Li Q., Ritter D., Yang N., Dong Z., Li H., Chuang J.H., Guo S. A systematic approach to identify functional motifs within vertebrate developmental enhancers. Dev. Biol. 2010;337:484–495. doi: 10.1016/j.ydbio.2009.10.019. doi:10.1016/j.ydbio.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawakami K. Transposon tools and methods in zebrafish. Dev. Dyn. 2005;234:244–254. doi: 10.1002/dvdy.20516. doi:10.1002/dvdy.20516. [DOI] [PubMed] [Google Scholar]
- 33.Kothary R., Clapoff S., Brown A., Campbell R., Peterson A., Rossant J. A transgene containing lacZ inserted into the dystonia locus is expressed in neural tube. Nature. 1988;335:435–437. doi: 10.1038/335435a0. doi:10.1038/335435a0. [DOI] [PubMed] [Google Scholar]
- 34.Depew M.J., Liu J.K., Long J.E., Presley R., Meneses J.J., Pedersen R.A., Rubenstein J.L. Dlx5 regulates regional development of the branchial arches and sensory capsules. Development. 1999;126:3831–3846. doi: 10.1242/dev.126.17.3831. [DOI] [PubMed] [Google Scholar]
- 35.Lettice L.A., Hill R.E. Preaxial polydactyly: a model for defective long-range regulation in congenital abnormalities. Curr. Opin Genet. Dev. 2005;15:294–300. doi: 10.1016/j.gde.2005.04.002. doi:10.1016/j.gde.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 36.VanderMeer J.E., Ahituv N. cis-regulatory mutations are a genetic cause of human limb malformations. Dev. Dyn. 2011;240:920–930. doi: 10.1002/dvdy.22535. doi:10.1002/dvdy.22535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smemo S., Campos L.C., Moskowitz I.P., Krieger J.E., Pereira A.C., Nobrega M.A. Regulatory variation in a TBX5 enhancer leads to isolated congenital heart disease. Hum. Mol. Genet. 2012;21:3255–3263. doi: 10.1093/hmg/dds165. doi:10.1093/hmg/dds165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahituv N. Gene Regulatory Sequences and Human Disease. New York: Springer; 2012. [Google Scholar]
- 39.Hong J.W., Hendrix D.A., Levine M.S. Shadow enhancers as a source of evolutionary novelty. Science. 2008;321:1314. doi: 10.1126/science.1160631. doi:10.1126/science.1160631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Navratilova P., Fredman D., Hawkins T.A., Turner K., Lenhard B., Becker T.S. Systematic human/zebrafish comparative identification of cis-regulatory activity around vertebrate developmental transcription factor genes. Dev. Biol. 2009;327:526–540. doi: 10.1016/j.ydbio.2008.10.044. doi:10.1016/j.ydbio.2008.10.044. [DOI] [PubMed] [Google Scholar]
- 41.McGaughey D.M., Vinton R.M., Huynh J., Al-Saif A., Beer M.A., McCallion A.S. Metrics of sequence constraint overlook regulatory sequences in an exhaustive analysis at phox2b. Genome Res. 2008;18:252–260. doi: 10.1101/gr.6929408. doi:10.1101/gr.6929408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fisher S., Grice E.A., Vinton R.M., Bessling S.L., McCallion A.S. Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science. 2006;312:276–279. doi: 10.1126/science.1124070. doi:10.1126/science.1124070. [DOI] [PubMed] [Google Scholar]
- 43.Hall B.K. Fins into Limbs. Chicago: The University of Chicago Press; 2007. [Google Scholar]
- 44.Kikuchi A., Arai-Ichinoi N., Sakamoto O., Matsubara Y., Saheki T., Kobayashi K., Ohura T., Kure S. Simple and rapid genetic testing for citrin deficiency by screening 11 prevalent mutations in SLC25A13. Mol. Genet. Metab. 2012;105:553–558. doi: 10.1016/j.ymgme.2011.12.024. doi:10.1016/j.ymgme.2011.12.024. [DOI] [PubMed] [Google Scholar]
- 45.Kobayashi K., Sinasac D.S., Iijima M., Boright A.P., Begum L., Lee J.R., Yasuda T., Ikeda S., Hirano R., Terazono H., et al. The gene mutated in adult-onset type II citrullinemia encodes a putative mitochondrial carrier protein. Nat. Genet. 1999;22:159–163. doi: 10.1038/9667. doi:10.1038/9667. [DOI] [PubMed] [Google Scholar]
- 46.Sinasac D.S., Moriyama M., Jalil M.A., Begum L., Li M.X., Iijima M., Horiuchi M., Robinson B.H., Kobayashi K., Saheki T., et al. Slc25a13-knockout mice harbor metabolic deficits but fail to display hallmarks of adult-onset type II citrullinemia. Mol. Cell Biol. 2004;24:527–536. doi: 10.1128/MCB.24.2.527-536.2004. doi:10.1128/MCB.24.2.527-536.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sone T., Saeki Y., Toh-e A., Yokosawa H. Sem1p is a novel subunit of the 26 S proteasome from Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:28807–28816. doi: 10.1074/jbc.M403165200. doi:10.1074/jbc.M403165200. [DOI] [PubMed] [Google Scholar]
- 48.Kristensen C.N., Bystol K.M., Li B., Serrano L., Brenneman M.A. Depletion of DSS1 protein disables homologous recombinational repair in human cells. Mutat. Res. 2010;694:60–64. doi: 10.1016/j.mrfmmm.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 49.Crackower M.A., Scherer S.W., Rommens J.M., Hui C.C., Poorkaj P., Soder S., Cobben J.M., Hudgins L., Evans J.P., Tsui L.C. Characterization of the split hand/split foot malformation locus SHFM1 at 7q21.3-q22.1 and analysis of a candidate gene for its expression during limb development. Hum. Mol. Genet. 1996;5:571–579. doi: 10.1093/hmg/5.5.571. doi:10.1093/hmg/5.5.571. [DOI] [PubMed] [Google Scholar]
- 50.Vervoort V.S., Viljoen D., Smart R., Suthers G., DuPont B.R., Abbott A., Schwartz C.E. Sorting nexin 3 (SNX3) is disrupted in a patient with a translocation t(6;13)(q21;q12) and microcephaly, microphthalmia, ectrodactyly, prognathism (MMEP) phenotype. J. Med. Genet. 2002;39:893–899. doi: 10.1136/jmg.39.12.893. doi:10.1136/jmg.39.12.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nusslein-Volhard C., Dahm R. Zebrafish. Oxford: Oxford University Press; 2002. [Google Scholar]
- 53.Pennacchio L.A., Ahituv N., Moses A.M., Prabhakar S., Nobrega M.A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D., et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. doi:10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 54.Andras N., Marina G., Kristina V., Richard B. Manipulating the Mouse Embryo: A Laboratory Manual. (3rd edn) 2003 . Cold Spring Harbor Laboratory Press, Woodbury, NY, USA. [Google Scholar]
- 55.Thisse B., Heyer V., Lux A., Alunni V., Degrave A., Seiliez I., Kirchner J., Parkhill J.P., Thisse C. Spatial and temporal expression of the zebrafish genome by large-scale in situ hybridization screening. Methods Cell Biol. 2004;77:505–519. doi: 10.1016/s0091-679x(04)77027-2. doi:10.1016/S0091-679X(04)77027-2. [DOI] [PubMed] [Google Scholar]
- 56.Hargrave M., Bowles J., Koopman P. In situ hybridization of whole-mount embryos. Methods Mol. Biol. 2006;326:103–113. doi: 10.1385/1-59745-007-3:103. [DOI] [PubMed] [Google Scholar]