Abstract
Mutations in peroxisome biogenesis proteins (peroxins) can lead to developmental deficiencies in various eukaryotes. PEX14 and PEX13 are peroxins involved in docking cargo-receptor complexes at the peroxisomal membrane, thus aiding in the transport of the cargo into the peroxisomal matrix. Genetic screens have revealed numerous Arabidopsis thaliana peroxins acting in peroxisomal matrix protein import; the viable alleles isolated through these screens are generally partial loss-of-function alleles, whereas null mutations that disrupt delivery of matrix proteins to peroxisomes can confer embryonic lethality. In this study, we used forward and reverse genetics in Arabidopsis to isolate four pex14 alleles. We found that all four alleles conferred reduced PEX14 mRNA levels and displayed physiological and molecular defects suggesting reduced but not abolished peroxisomal matrix protein import. The least severe pex14 allele, pex14-3, accumulated low levels of a C-terminally truncated PEX14 product that retained partial function. Surprisingly, even the severe pex14-2 allele, which lacked detectable PEX14 mRNA and PEX14 protein, was viable, fertile, and displayed residual peroxisome matrix protein import. As pex14 plants matured, import improved. Together, our data indicate that PEX14 facilitates, but is not essential for peroxisomal matrix protein import in plants.
Keywords: Peroxin, Peroxisome import, Organelle targeting, Arabidopsis molecular genetics
Introduction
Eukaryotic organelles employ distinct mechanisms to ensure targeting of proteins synthesized in the cytosol to appropriate subcellular destinations. Proteins housed in peroxisomes are imported through the action of peroxin (PEX) proteins. The PEX5 and PEX7 receptors recognize cytosolic cargo proteins with type 1 or type 2 peroxisomal targeting signals (PTS), respectively. PTS-bearing proteins are translocated into the peroxisome upon receptor binding to a docking complex, which includes the interacting membrane peroxins PEX13 and PEX14 in mammals, plants, and yeasts (reviewed in Lanyon-Hogg et al. 2010). Recent in vitro evidence suggests that yeast PEX5, when recruited to the peroxisomal membrane by PEX14, forms the pore through which PTS1 proteins enter the peroxisome (Meinecke et al. 2010). However, much remains unknown about how proteins enter peroxisomes and the extent to which peroxisomal import mechanisms have diverged during eukaryotic evolution.
In yeast, PEX5-dependent PTS1 import and PEX7-dependent PTS2 import converge at the PEX14-PEX13 docking complex; mutants defective in either docking peroxin exhibit both PTS1 and PTS2 import defects (Elgersma et al. 1996; Albertini et al. 1997). In Saccharomyces cerevisiae, PEX14 and PEX13 each can bind directly to PEX5 and PEX7 and to each other (Elgersma et al. 1996; Albertini et al. 1997; Stein et al. 2002; Niederhoff et al. 2005). The in vitro binding of Pichia pastoris PEX5 to PEX14 is enhanced by PTS1 cargo, whereas PEX5-PEX13 binding is diminished by PTS1 cargo (Urquhart et al. 2000), suggesting that PEX14 is the initial docking site for PEX5-PTS1 cargo complexes and that PEX13 acts after cargo release. Conversely, recent studies in S. cerevisiae suggest that the PEX7-PTS2 import pathway uses PEX13 as the initial docking site followed by interaction with PEX14 (Grunau et al. 2009).
In humans, PEX14 lesions underlie a subset of peroxisome biogenesis disorders (Shimozawa et al. 2004; Huybrechts et al. 2008). As in yeast, mammalian PEX14 interacts with PEX5, PEX7, and PEX13 (reviewed in Azevedo and Schliebs 2006). In contrast to yeast, where PEX5 and PEX7 can independently interact with the docking peroxins and deliver PTS1 and PTS2 cargo, respectively, mammalian PTS2 cargo proteins require a long version of PEX5 (PEX5L) to enter peroxisomes (Braverman et al. 1998; Matsumura et al. 2000; Dodt et al. 2001). PEX5L facilitates the in vitro binding of PEX7-PTS2 cargo complexes to PEX14 (Otera et al. 2000). Although mammalian PEX14 can interact with PEX7 in vitro (Shimizu et al. 1999), immunoprecipitation assays demonstrate that PEX7 primarily associates with PEX13, whereas PEX5L primarily associates with PEX14 (Miyata et al. 2009).
Similar to mammals, Arabidopsis PTS2 protein import depends not only on PEX7, but also on PEX5, the PTS1 receptor (Hayashi et al. 2005; Woodward and Bartel 2005), and Arabidopsis PEX7 binds PEX5 in yeast two-hybrid assays (Nito et al. 2002; Ramón and Bartel 2010). The Arabidopsis pex5-1 missense allele (Zolman et al. 2000), which alters a conserved amino acid residue that in mammalian PEX5L is essential for PEX7 binding and PTS2 import (Matsumura et al. 2000), impairs PTS2 but not PTS1 import (Woodward and Bartel 2005). In Arabidopsis, PEX5 levels and PTS1 import depend on PEX7 function in certain growth conditions (Ramón and Bartel 2010), suggesting that PEX5 is stabilized in the presence of PEX7. As in other eukaryotes (Schliebs et al. 1999; Otera et al. 2002), the N-terminal region of Arabidopsis PEX14 binds PEX5 (Nito et al. 2002), however, direct PEX14-PEX7 interactions have not been reported in plants, and Arabidopsis PEX14 fails to bind PEX7 in yeast two-hybrid assays (Nito et al. 2002). PEX14-blocking antibodies prevent in vitro binding of both PTS1- and PTS2-cargo to sunflower peroxisomes (Lopez-Huertas et al. 1999), and Arabidopsis mutants defective in either PEX14 (Hayashi et al. 2000) or PEX13 (Mano et al. 2006) have deficiencies in both PTS1 and PTS2 import, implying that PEX5 and PEX7 require both docking peroxins for efficient cargo translocation into the peroxisome. Moreover, the N-terminal portion of Arabidopsis PEX13 interacts with PEX7, but not PEX5, in a yeast two-hybrid assay (Mano et al. 2006). These interaction studies suggest that plants, like mammals (Miyata et al. 2009), maintain distinct PEX14-PEX5 and PEX13-PEX7 relationships despite the interdependence of PTS1 and PTS2 pathways in plants (Ramón and Bartel 2010).
Notwithstanding the demonstrated importance of PEX14 and PEX13 in both PTS1 and PTS2 import, Arabidopsis mutants defective in either docking complex peroxin have disparate phenotypes. A PEX13 null allele confers lethal gametophytic defects and completely blocks peroxisomal import of a PTS1-tagged reporter protein in pollen (Boisson-Dernier et al. 2008). In contrast, the single characterized pex14 mutant, peroxisome defective2 (ped2), displays more mild defects despite harboring a nonsense mutation midway through the PEX14 coding sequence (Hayashi et al. 2000). Although ped2 peroxisomes are shrunken and ped2 plants are small and display photorespiration defects, light-grown ped2 root cells display only partial GFP-PTS1 import defects, and dark-grown ped2 seedlings exhibit only partial PTS2 processing defects (Hayashi et al. 2000). Moreover, PTS2 processing is restored as ped2 seedlings mature (Hayashi et al. 2000). These partial and transient matrix protein import defects suggest either that the ped2 allele does not completely eliminate PEX14 function or that PEX14 is not absolutely required for matrix protein import. Moreover, the ped2 accession (Landsberg) precludes direct phenotypic comparison to other Arabidopsis pex mutants, which have been isolated in the Columbia accession. Here we describe the physiological, molecular, and cell biological defects in an allelic series of Arabidopsis pex14 mutants in the Columbia accession. Our analysis of these mutants, including an apparent null allele, suggests that PEX14 facilitates, but is not absolutely required for matrix protein import into Arabidopsis peroxisomes.
Results
An allelic series of pex14 mutants
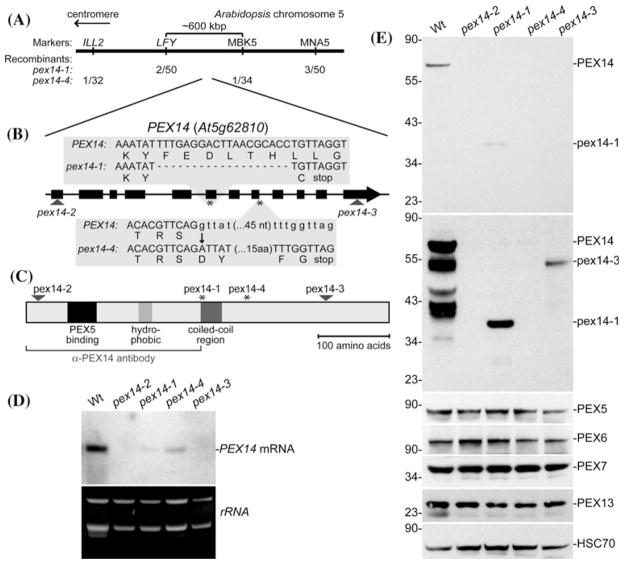
Plant peroxisomes house processes essential for growth and development, including fatty acid β-oxidation, which fuels early seedling growth (Baker et al. 2006), and the related process of indole-3-butyric acid (IBA) conversion to the active auxin indole-3-acetic acid (IAA) (Zolman et al. 2000; Strader et al. 2010; Strader and Bartel 2011). As a result, screens for resistance to IBA or synthetic auxin precursors have facilitated the isolation and characterization of Arabidopsis mutants with defective peroxisomes (Hayashi et al. 1998; Zolman et al. 2000). We isolated four Arabidopsis pex14 alleles, two in forward genetic screens for reduced peroxisomal function and two from a T-DNA insertion collection. pex14-1 was isolated as an IBA-response mutant from the progeny of plants transformed with a T-DNA based cDNA library (LeClere and Bartel 2001). Because the mutant lesion was not linked to the T-DNA, we used recombination mapping to localize the causal mutation near the bottom of chromosome 5 (Fig. 1a). The PEX14 (At5g62810) gene is in this interval, and because the pex14 mutant peroxisome defective2 (ped2, Hayashi et al. 2000) has a similar phenotype (see below), we sequenced PEX14 from the mutant genomic DNA. We found a 19-bp deletion beginning at position 1722 (where 1 is the A of the initiator ATG) that resulted in one out-of-frame amino acid followed by an early stop codon (Fig. 1b).
Fig. 1.
A series of pex14 alleles. a Recombination mapping of pex14-1 and pex14-4. Mapping with the PCR-based markers ILL2 (Davies et al. 1999), LFY (Konieczny and Ausubel 1993), MBK5 (Zolman et al. 2001), and MNA5 (Zolman et al. 2001) localized the defects to a region on the bottom of chromosome 5 that contains the PEX14/PED2 gene. b PEX14 has 12 exons (thick boxes) separated by 11 introns (lines). The positions of the pex14-2 and pex14-3 T-DNA insertions are indicated with triangles. The positions of the pex14-1 and pex14-4 lesions are marked with asterisks. pex14-1 has a 19-bp deletion beginning at position 1722 (where 1 is the A of the initiator ATG) resulting in one out-of-frame amino acid followed by an early stop codon. pex14-4 has a G-to-A mutation at position 2220 which alters the 5′ splice site of the eighth intron, resulting in 19 out-of-frame amino acids followed by an early stop codon. c Schematic showing putative domains in PEX14 and the locations of the defects in the four pex14 alleles. The bracket indicates the protein fragment used to generate the α-PEX14 antibody. d The four pex14 alleles display reduced (pex14-1, pex14-4) or undetectable (pex14-2, pex14-3) levels of full-length PEX14 mRNA. RNA extracted from 10-day-old light-grown seedlings was subjected to RNA gel-blot analysis probed with PEX14 (top panel). The bottom panel shows the ethidium bromide-stained gel prior to transfer. e The four pex14 alleles lack detectable full-length PEX14 protein, but pex14-1 and pex14-3 accumulate pex14 truncation products. Immunoblot of proteins from 8-day-old light-grown seedlings from the indicated lines probed with α-PEX14, α-PEX5, α-PEX6, α-PEX7, α-PEX13, and α-HSC70 (loading control) antibodies. The first and second panels are different exposures of the α-PEX14 panel to show the presence of the lower molecular mass pex14-1 and pex14-3 products in the longer (second) exposure. Positions of molecular mass markers (in kDa) are indicated on the left
We isolated pex14-4 in a screen of ethyl methanesulfonate-mutagenized pools for seedlings that required sucrose for early seedling development and that failed to efficiently produce lateral roots in response to IBA, two hallmarks of peroxisome-defective mutants (Zolman et al. 2000). This mutant was mapped to a similar interval as pex14-1 near the bottom of chromosome 5 (Fig. 1a). Sequencing PEX14 from mutant DNA revealed a G-to-A mutation at position 2,220 that altered the 5′ splice site of the eighth intron. Skipping this intron would result in 19 out-of-frame amino acids followed by an early stop codon (Fig. 1b).
To expand our allelic series, we obtained two additional pex14 alleles from the Salk Institute collection of sequence-tagged lines (Alonso et al. 2003). pex14-2 has a T-DNA insertion 41-bp downstream of the PEX14 start codon, and pex14-3 has a T-DNA insertion 267-bp upstream of the PEX14 stop codon (Fig. 1b).
Arabidopsis PEX14 is a 507-amino acid protein that migrates atm ~75 kDa in SDS–PAGE (Hayashi et al. 2000) and resembles PEX14 from other organisms, with an N-terminal PEX5-binding domain, a hydrophobic region that may anchor the protein in the peroxisomal membrane, and a predicted coiled-coil region (reviewed in Azevedo and Schliebs 2006). The pex14 alleles are distributed throughout the coding sequence, with the pex14-2 allele disrupted upstream of the identified domains, pex14-1 and pex14-4 after the hydrophobic region near the middle of the protein, and pex14-3 near the C-terminus (Fig. 1c). We used RNA gel-blot analysis to examine seedling PEX14 mRNA levels and did not detect full-length PEX14 transcript in either T-DNA allele (pex14-2 and pex14-3; Fig. 1d). Moreover, pex14-1 and pex14-4 displayed reduced PEX14 transcript levels compared to wild type (Fig. 1d), consistent with the possibility that the early stop codons in these alleles (Fig. 1b) led to nonsense-mediated mRNA decay (Stalder and Muhlemann 2008). To determine the consequences of the pex14 mutations on PEX14 protein accumulation, we examined extracts from pex14 plants using an antibody generated against the N-terminal 243 amino acids of Arabidopsis PEX14 (Lingard and Bartel 2009), which we expected would detect any accumulated pex14-1 protein, because the pex14-1 premature stop codon occurs one amino acid residue following Tyr241 (Fig. 1b), as well as pex14-3, and pex14-4 proteins, which would be larger than pex14-1 (Fig. 1c). As expected, none of the pex14 mutants accumulated detectable full-length PEX14 (Fig. 1e). However, we did detect reduced levels of truncated versions of PEX14 in the pex14-1 and pex14-3 mutants (Fig. 1e) migrating at sizes consistent with the location of the mutant lesions (Fig. 1c). Our observation that the truncated PEX14 products did not accumulate to wild-type PEX14 levels is consistent with the reduced pex14 transcript levels in these mutants (Fig. 1d). Our finding that detectable PEX14 mRNA or PEX14 protein did not accumulate in the pex14-2 T-DNA allele (Fig. 1d, e), in combination with the position of the insertion (Fig. 1b), strongly suggests that pex14-2 is a null allele.
We also used immunoblotting to determine whether reduced PEX14 levels impacted levels of other peroxins. Certain pex mutants, including pex6-1 (Zolman and Bartel 2004) and pex7-2 (Ramón and Bartel 2010) accumulate less PEX5 protein than wild type. We detected similar levels of PEX5, PEX6, PEX7, and PEX13 in 8-day-old pex14 and wild-type seedlings (Fig. 1e), suggesting that these peroxins do not require PEX14 for stability. Conversely, PEX14 accumulation is similar in wild type and in pex4, pex5, pex6, and pex7 mutants (Ratzel et al. 2011), suggesting that PEX14 does not depend on the corresponding peroxins for stability.
Reduced IBA responsiveness in pex14 mutants
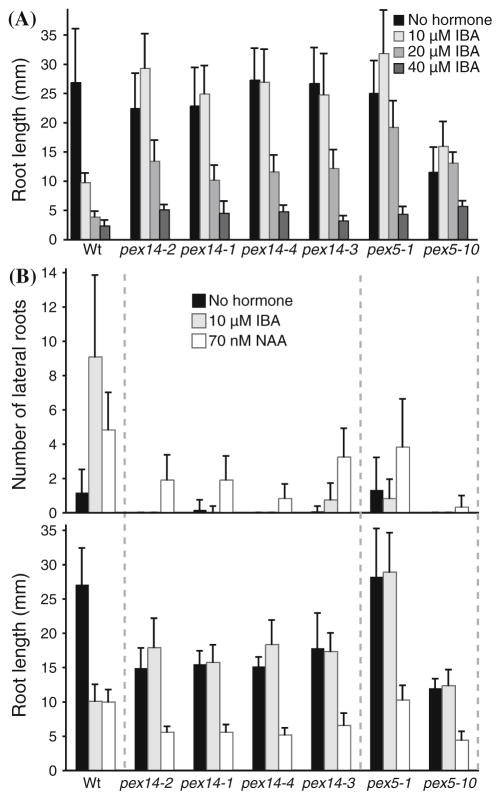
We assayed IBA responsiveness to compare the physiological severity of the pex14 alleles to two previously characterized PTS1 receptor mutants, the pex5-1 missense allele (Zolman et al. 2000; Woodward and Bartel 2005) and the pex5-10 T-DNA insertion allele (Zolman et al. 2005; Khan and Zolman 2010; Ramón and Bartel 2010). Genetic and biochemical data suggest that IBA is β-oxidized to IAA in peroxisomes and that this conversion is necessary for response to exogenous IBA (Zolman et al. 2000, 2007, 2008; Strader et al. 2010, 2011). Consequently, IBA responses, such as the inhibition of primary root elongation and the promotion of lateral root formation, can be used to assess peroxisome function (Zolman et al. 2000). We found that the pex14 mutants were slightly less IBA resistant than pex5-1 and pex5-10 in root elongation assays. Like pex5-1 and pex5-10, the four pex14 mutants displayed complete resistance to the inhibitory effects of 10 μM IBA on root elongation. However, pex14 mutants were only partially resistant to 20 μM IBA, whereas both pex5 mutants still showed substantial resistance at this concentration (Fig. 2a). All of the mutants responded to 40 μM IBA (Fig. 2a).
Fig. 2.
pex14 mutant seedlings are IBA resistant. a Root elongation on IBA. After 8 days of growth under yellow-filtered light on medium supplemented with the indicated concentration of IBA, seedlings were removed from the agar, and the length of the primary root was measured. Error bars represent standard deviations of the means (n ≥ 12). b Lateral root initiation. Lateral roots emerged from the primary root were counted (upper panel) and the lengths of the primary roots were measured (lower panel) 4 days after transfer of 4-day-old seedlings to unsupplemented medium or medium supplemented with the indicated concentration of IBA or NAA. Error bars represent standard deviations of the means (n ≥ 8)
pex14 mutants also displayed lateral root production defects in response to IBA. We counted lateral roots of 8-day-old seedlings grown for 4 days on medium without hormone followed by 4 days on medium supplemented with either IBA or the synthetic auxin 1-naphthaleneacetic acid (NAA), which does not require peroxisomal chain shortening for activity. Wild-type seedlings had few lateral roots without auxin treatment and increased numbers of lateral roots following either IBA or NAA treatment (Fig. 2b). Like wild-type seedlings, the pex14 mutants responded to NAA by forming lateral roots (Fig. 2b). In response to IBA, however, the pex14 mutants displayed dramatically fewer lateral roots than wild type (Fig. 2b). This phenotype is consistent with the lateral root deficiency of other pex mutants (Zolman et al. 2000, 2005; Zolman and Bartel 2004; Woodward and Bartel 2005; Ramón and Bartel 2010), including pex5-1 and pex5-10 (Fig. 2b).
pex14 growth defects
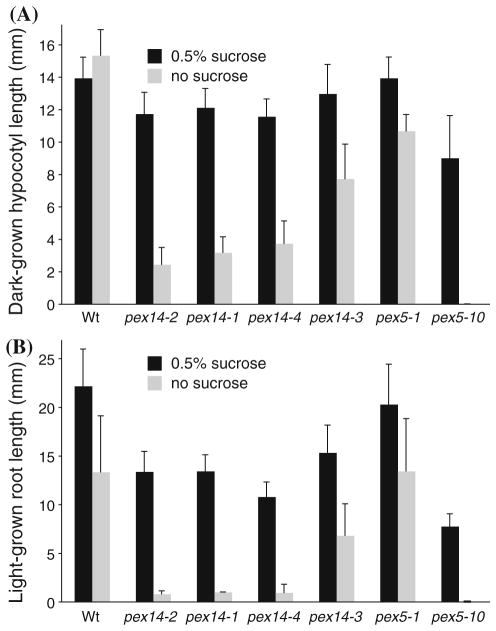
Peroxisomes are the sole organelles housing fatty acid β-oxidation in plants (reviewed in Graham 2008), and mutants defective in β-oxidation of seed storage fatty acids typically display growth defects following germination that can be at least partially restored by exogenous sucrose (Hayashi et al. 1998; Zolman et al. 2000), presumably because this fixed carbon source replaces the energy normally provided by the acetyl-CoA freed during β-oxidation of stored fatty acids. To indirectly assess the ability of pex14 seedlings to metabolize endogenous oil reserves, we monitored seedling growth on media with and without supplemented sucrose. Whereas dark-grown pex14 hypocotyls were of nearly wild-type lengths when seedlings were germinated and grown with 0.5% sucrose, we found dramatic pex14 hypocotyl elongation defects in the absence of sucrose (Fig. 3a). The pex14-2, pex14-1, and pex14-4 mutants were more severely sucrose dependent than pex14-3 (Fig. 3a).
Fig. 3.
pex14 mutant seedlings are sucrose dependent. a Hypocotyl elongation in the dark. Seedlings were grown with or without 0.5% sucrose for 1 day under white light and 5 days in darkness, after which hypocotyl lengths were measured. Error bars represent standard deviations of the means (n ≥ 14). b Light-grown root elongation. Primary roots were measured after 8 days of growth with or without 0.5% sucrose under white light. Error bars represent standard deviations of the means (n ≥ 12)
We also observed sucrose dependence in light-grown pex14 seedlings. The slight root elongation defect of pex14 mutants grown on sucrose in the light was exacerbated in the absence of sucrose (Fig. 3b). The sucrose dependence of light-grown pex14 seedlings was observed not only in root elongation (Figs. 3b, 4a, b), but also in leaf expansion (Fig. 4a, b). Like sucrose dependence in the dark, the root and shoot expansion defects were least severe in the pex14-3 mutant (Fig. 3b).
Fig. 4.
pex14 mutant growth defects. a, b Light-grown 14-day-old wild-type Col-0 (Wt), pex14, and pex5 seedlings on unsupplemented medium (a) or medium supplemented with 0.5% sucrose (b). c 21-day-old plants transferred to soil after 14 days on medium supplemented with 0.5% sucrose. d 28-day-old plants transferred to soil after 14 days on medium supplemented with 0.5% sucrose. e 41-day-old plants transferred to soil after 14 days on medium supplemented with 0.5% sucrose
Even when supplied with sucrose, pex14 mutants were smaller than wild type and pex5-1 (Fig. 4b). Although this defect persisted after plants were transferred to soil (Fig. 4c–e), all of the pex14 mutants were fertile and produced viable seeds. Initially, the pex14 mutants were less delayed than pex5-10 when grown in soil (Fig. 4d), but at maturity, three of the pex14 mutants (pex14-2, pex14-1, and pex14-4) were smaller than pex5-10, with small rosette leaves and reduced stature (Fig. 4d, e). As with other phenotypes, the pex14-3 allele conferred less severe growth defects than the other pex14 lesions (Fig. 4).
In summary, our analyses of pex14 physiological phenotypes revealed growth defects in the presence and absence of sucrose that were generally intermediate between the moderate defects of pex5-1 and the severe defects of pex5-10, both in the light and the dark (Figs. 3, 4a, b). Among the pex14 mutants, several defects appeared less severe in pex14-3 than in pex14-2, pex14-1, or pex14-4 (Figs. 3, 4), suggesting that the pex14-3 protein (Fig. 1e) retained partial PEX14 function and that the other pex14 lesions conferred more complete disruptions.
pex14 defects in PTS2 protein processing
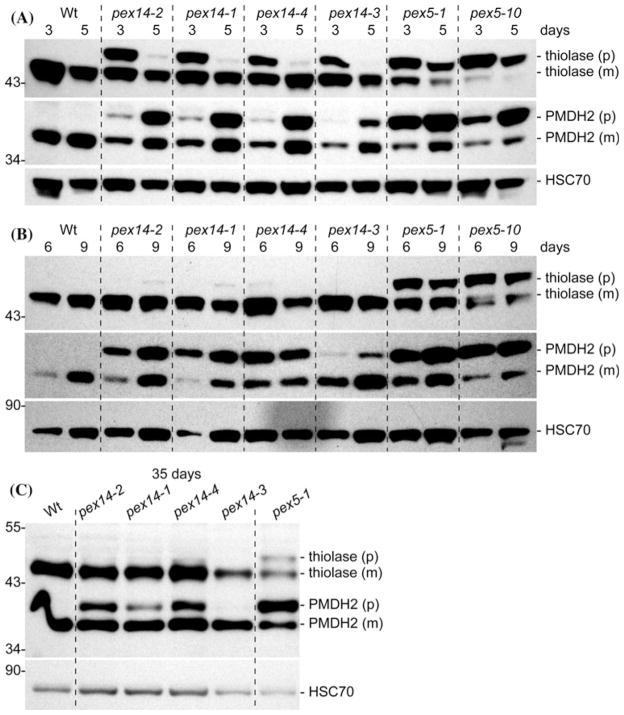
We used immunoblot analyses to determine whether the physiological defects displayed by the pex14 mutants were accompanied by defects in matrix protein import. Because the PTS2-containing signal is removed from PTS2 cargo proteins by the DEG15 peroxisomal protease after cargo enters the peroxisome, and because the DEG15 protease itself is a PTS1 protein (Helm et al. 2007; Schuhmann et al. 2008), monitoring PTS2 removal indirectly assesses per-oxisomal import of PTS1 and/or PTS2 proteins. We analyzed PTS2 processing of 3-ketoacyl-CoA thiolase (thiolase) and peroxisomal malate dehydrogenase (PMDH) in seedlings and leaves of mature plants. As previously reported (Woodward and Bartel 2005; Zolman et al. 2005; Ramón and Bartel 2010), both proteins were fully processed in wild type and displayed marked processing defects in both pex5 mutants (Fig. 5a). All four pex14 mutants displayed PTS2 processing defects; these defects appeared less severe in the pex14-3 mutant. Interestingly, the pex14 PTS2 processing defects were less severe than either pex5-1 or pex5-10 in both 3- and 5-day-old seedlings (Fig. 5a). In particular, unprocessed thiolase was a minor fraction of total thiolase in 5-day-old pex14 seedlings, whereas less than half of the thiolase was processed in either pex5-1 or pex5-10 at 5 days (Fig. 5a). Like thiolase, more than half of the PMDH was processed in 3-day-old pex14 seedlings, whereas the bulk of PMDH was unprocessed in 3-day-old pex5-1 and pex5-10 seedlings (Fig. 5a). In contrast to thiolase processing defects, PMDH processing defects appeared more severe in 5-day-old pex14 seedlings than in 3-day-old seedlings, perhaps because PMDH levels are increasing from 3 to 8 days after sowing while thiolase levels are declining (Lingard et al. 2009). Like thiolase, PMDH processing defects were not as severe in any of the pex14 mutants as the defects detected in either pex5 mutant.
Fig. 5.
pex14 mutants display transient defects in PTS2 protein processing. Immunoblots of proteins prepared from 3- and 5-day-old seedlings (a), 6- and 9-day-old seedlings (b), or 35-day-old rosette leaves (c) probed with α-thiolase and α-PMDH2 antibodies, which recognize precursor (p) and mature (m) polypeptides, and α-HSC70, a loading control. Positions of molecular mass markers (in kDa) are indicated on the left. Upon processing, the N-terminal 4-kDa peptide containing the PTS2 is removed from the 48.5 kDa thiolase precursor and the 37.5 kDa PMDH precursor
The recovery of thiolase PTS2 processing ability as pex14 seedlings matured from 3 to 5 days old (Fig. 5a) was consistent with the previous characterization of the ped2 allele of pex14, which also displays only transient thiolase processing defects (Hayashi et al. 2000). In contrast to thiolase, however, the partial PMDH processing defects were maintained as pex14 seedlings matured from 3 to 9 days old (Fig. 5a, b). In mature plants, however, PMDH processing defects were no longer apparent in the pex14-3 mutant and were reduced in the other pex14 mutants compared to pex5-1, which still processed less than half of the PMDH in 35-day-old plants (Fig. 5c). Thus the recovery of PTS2 processing efficiency seems not to be a specific feature of thiolase, but may reflect a general improvement as pex14 mutants mature in either PTS1 import (including import of the DEG15 PTS2-processing protease), PTS2 import, or both.
pex14 defects in peroxisomal matrix protein import
To examine the effects of pex14 mutations on PTS1 import and to compare the PTS1 import defects of pex14-2 and pex14-1, we used confocal fluorescence microscopy to examine lines carrying a reporter driven by the isocitrate lyase (ICL) promoter in which green fluorescent protein (GFP) is fused to the N-terminus of the PTS1 protein ICL (Lingard et al. 2009). As expected, this reporter was distributed in a punctate pattern in cotyledon cells from 3- and 9-day-old wild-type seedlings (Fig. 6a), consistent with a fully peroxisomal localization. In contrast, GFP-ICL fluorescence appeared largely cytosolic in pex14-1 seedlings, with some punctate foci barely discernable in 9-day-old seedlings (Fig. 6a). In pex14-2 seedlings, GFP-ICL fluorescence appeared fully cytosolic at both 3 and 9 days (Fig. 6a). The more complete mislocalization of GFP-ICL in pex14-2 is consistent with the possibility that the pex14-2 lesion confers a more severe disruption of PEX14 function and PTS1 import than does the pex14-1 lesion.
Fig. 6.
pex14 mutants display defects in peroxisome import of both PTS1- and PTS2-tagged matrix proteins. a Confocal microscopic images of cotyledon epidermal cells from 3-day- and 9-day-old light-grown seedlings of Col-0 (Wt) and pex14-1 or pex14-2 expressing GFP-ICL from the ICL promoter (Lingard et al. 2009). b, c Confocal microscopic images of cotyledon epidermal cells from 3-day- and 9-day-old light-grown seedlings of Col-0 (Wt) and pex14-1 expressing 35S-GFP-PTS1 (b) (Zolman and Bartel 2004) or 35S-PTS2-GFP (c) (Woodward and Bartel 2005). In a–c, the corresponding 3-day-old wild-type and mutant images were acquired with identical microscope settings and the 9-day-old wild-type and mutant images were acquired with identical microscope settings. Scale bar 20 μm. d–f Immunoblot of proteins prepared from 3- and 10-day-old seedlings probed sequentially with α-GFP, α-HSC70, a loading control (d), α-ICL (e), and α-thiolase (f). Precursor (p) and mature (m) PTS2 proteins (PTS2-GFP and thiolase) are marked on the right, and the position of GFP-PTS1 is marked on the left. An asterisk marks the position of a protein that cross-reacts with the ICL antibody. Positions of molecular mass markers (in kDa) are indicated on the left
ICL functions in the glyoxylate cycle (Eastmond et al. 2000) and is normally degraded a few days after germination (Lingard et al. 2009). We could still detect ICL and GFP-ICL in pex14 mutants after these proteins had been mostly degraded in wild type (Fig. 6a, e). This stabilization is consistent with our previous finding that ICL must be imported into the peroxisome to undergo efficient degradation (Lingard et al. 2009).
To compare the effects of a pex14 mutation on PTS1 and PTS2 matrix protein import, we crossed pex14-1 to transgenic lines containing PTS1- or PTS2-targeted GFP derivatives driven by the cauliflower mosaic virus 35S promoter. In 3- and 9-day-old wild-type cotyledon cells, both GFP-PTS1 (Zolman and Bartel 2004) and PTS2-GFP (Woodward and Bartel 2005) appeared in punctate structures indicative of peroxisomes (Fig. 6b, c). Unlike in wild type, both GFP-PTS1 and PTS2-GFP appeared partially cytosolic in 3-day-old pex14-1 seedlings (Fig. 6b, c). These data confirmed that both PTS1 and PTS2 import is partially defective in pex14-1.
Because we saw a recovery of thiolase PTS2 processing as pex14 seedlings aged (Fig. 5a), we also analyzed import of GFP-PTS1 and PTS2-GFP in older mutant plants. Cotyledon epidermal cells of 9-day-old pex14-1 plants expressing GFP-PTS1 exhibited both punctate and cytosolic fluorescence, similar to that seen in 3-day-old seedlings (Fig. 6b). In contrast, epidermal cells from 9-day-old pex14-1 plants expressing PTS2-GFP exhibited primarily punctate fluorescence, with some cytosolic fluorescence persisting only in stomatal cells. Analysis of PTS2 processing in pex14 transgenic seedlings by immunoblotting confirmed that the PTS2-GFP processing defect became less severe as seedlings aged (Fig. 6d), similar to endogenous thiolase (Figs. 5, 6f). These results suggested that PEX14 might be more important for PTS1 import than for PTS2 import as seedlings mature.
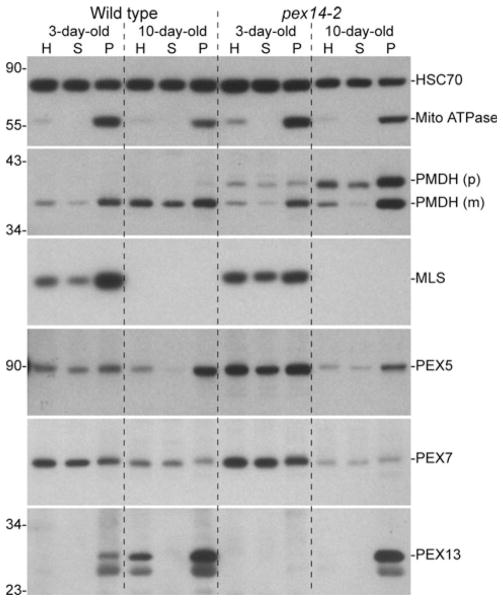
PTS2 processing requires both import of the PTS2 protein into the peroxisome and import of the DEG15 PTS2-processing enzyme, which is a PTS1-containing protein (Helm et al. 2007; Schuhmann et al. 2008). To examine possible causes of pex14 PTS2 processing defects, we fractionated extracts from 3- and 10-day-old seedlings into cytosolic and organellar fractions (Ratzel et al. 2011). As expected, we found the mitochondrial ATPase in the organellar fraction and most of the HSC70 in the cytosolic fraction of both wild type and pex14-2 (Fig. 7). When we examined the PTS2 protein PMDH, we found unprocessed PMDH in both the cytosolic and organellar fractions in both 3- and 10-day-old pex14-2 seedlings (Fig. 7), consistent with the PTS2-GFP import defect in pex14-1 (Fig. 6c). In the cytosolic fractions, which would include both cytosolic proteins and proteins from organelles that ruptured during fractionation, we observed that more than half of the PMDH was unprocessed in pex14-2, whereas less than half of the PMDH was unprocessed in the pex14-2 organellar fractions. We found that the PTS1 protein malate synthase (MLS), a glyoxylate cycle enzyme that only is present in young seedlings (Lingard et al. 2009), appeared more cytosolic in 3-day-old pex14-2 seedlings than in wild type (Fig. 7). The reduced organellar association of MLS is consistent with the reduced punctate fluorescence of PTS1-tagged GFP derivatives (Fig. 6a, b) and suggests that PTS1 import defects may contribute to the PTS2 processing defects of pex14 mutants. Interestingly, however, substantial MLS remained associated with the pex14-2 organellar pellet, suggesting that PTS1 import, like PTS2 import, was not fully blocked even in the pex14-2 presumptive null allele.
Fig. 7.
pex14-2 displays reduced organellar association of PTS1- and PTS2-cargo proteins and delayed PEX13 accumulation. Extracts from 3- and 10-day-old light-grown wild-type and pex14-2 seedlings were separated by centrifugation into soluble and organellar pellet fractions. For each sample, 1% of the total homogenate (H), 1% of the soluble fraction (S), and 25% of the pellet fraction (P) were separated using SDS–PAGE and processed for sequential immuno-blotting using the indicated antibodies. HSC70 and the mitochondrial membrane complex V subunit α (mito ATPase) were used as cytosolic and organellar controls, respectively. Precursor (p) and mature (m) proteins contain or lack the N-terminal PTS2 peptide, respectively. Positions of molecular mass markers (in kDa) are indicated on the left
To determine if the loss of PEX14 was accompanied by altered receptor localization, we examined PEX5 and PEX7 distribution in the fractions. PEX7 was similarly distributed between the cytosolic and organellar fractions in wild type and pex14-2 at both 3 and 10 days, and PEX5 was similarly distributed between the cytosolic and organellar fractions at 3 days (Fig. 7). However, the PEX5 shift to the organellar fraction observed in 10-day-old wild-type seedlings did not appear as complete in 10-day-old pex14-2 seedlings (Fig. 7), suggesting that PEX5 may be targeted to the peroxisome less efficiently in the absence of PEX14.
We also examined the distribution of the docking peroxin PEX13 in pex14-2 seedling extracts. As expected, we found PEX13 in the organellar pellet in wild-type seedlings (Ratzel et al. 2011) and in 10-day-old pex14-2 seedlings (Fig. 7). However, we did not detect PEX13 in 3-day-old pex14-2 seedlings, even in the concentrated organellar fraction, and PEX13 appeared to be less abundant in 10-day-old pex14-2 seedlings than in wild type (Fig. 7), suggesting that PEX14 may stabilize PEX13 in the peroxisome membrane. Alternatively, because PEX13 accumulates as seedlings mature (Fig. 7), the delayed appearance of PEX13 in the pex14 mutant may reflect the slow growth of pex14 seedlings.
Discussion
Most peroxins assist in importing proteins from the cytosol to the peroxisomal matrix, either directly, as cargo-binding receptors (PEX5, PEX7) or docking receptor-cargo complexes at the peroxisome membrane (PEX13, PEX14), or indirectly, by promoting the recycling of receptors back to the cytosol for further import rounds (e.g., PEX1, PEX2, PEX4, PEX6, PEX10, PEX12, PEX22, PEX26). Over the past decade, Arabidopsis mutants have been reported that are defective in nearly all of the conserved peroxins implicated in matrix protein import and receptor recycling, including PEX2 (Hu et al. 2002), PEX4 (Zolman et al. 2005), PEX5 (Zolman et al. 2000; Khan and Zolman 2010), PEX6 (Zolman and Bartel 2004), PEX7 (Woodward and Bartel 2005; Ramón and Bartel 2010), PEX10 (Schumann et al. 2003; Sparkes et al. 2003), PEX12 (Fan et al. 2005; Mano et al. 2006), PEX13 (Mano et al. 2006; Boisson-Dernier et al. 2008; Ratzel et al. 2011), PEX14 (Hayashi et al. 2000), PEX22 (Zolman et al. 2005), and PEX26 (APEM9, Goto et al. 2011). Analysis of these mutants has revealed and confirmed that peroxisomes are important for seedling establishment following germination, lateral root development, photorespiration, jasmonate biosynthesis, and IBA β-oxidation to IAA (reviewed in Hayashi and Nishimura 2006).
Because certain peroxisomal β-oxidation enzymes are required for embryogenesis (Rylott et al. 2003, 2006), any peroxins directly or indirectly needed to import these enzymes into peroxisomes also are expected to be required for embryogenesis. Indeed, null alleles of the ring-finger peroxin genes (PEX2, PEX10, PEX12) and of APEM9, the plant PEX26 equivalent, confer embryonic lethality (Hu et al. 2002; Schumann et al. 2003; Sparkes et al. 2003; Fan et al. 2005; Goto et al. 2011). Combining pex5-1 with pex7-1 confers incompletely penetrant embryonic defects (Woodward and Bartel 2005), and combining pex5-10 or pex5-1 with the stronger pex7-2 allele confers embryonic lethality (Ramón and Bartel 2010). Moreover, a PEX13 null allele confers lethal gametophytic defects (Boisson-Dernier et al. 2008). Whether this early requirement applies to all plant peroxins is not known because surprisingly few Arabidopsis peroxin mutants with confirmed null phenotypes have been described. Other reported T-DNA insertion alleles disrupting PEX genes are either in the 5′ UTR and confer only weak defects, as in pex7-1 (Woodward and Bartel 2005; Ramón and Bartel 2010), pex13-1 (Ratzel et al. 2011), and pex22-1 (Zolman et al. 2005), or do not completely abolish function despite insertion in a central exon, as in pex5-10 (Khan and Zolman 2010; Ramón and Bartel 2010). pex mutants isolated using forward genetic screens for IBA resistance, sucrose dependence, or aberrant peroxisome morphology are of course viable, as are knockdown lines generated using RNAi (Nito et al. 2007), but none are demonstrated null alleles. Consistent with an incomplete disruption, pex2 (ted3 allele, Hu et al. 2002), pex4-1 (Zolman et al. 2005), pex5-1 (Zolman and Bartel 2004), pex6-1 (Zolman and Bartel 2004), pex7-2 (Ramón and Bartel 2010), pex12 (apm4 allele, Mano et al. 2006), and pex26 (apem9-1 allele, Goto et al. 2011) all are missense alleles. Although the apm3 allele of pex13 is a nonsense allele, it occurs late in the coding sequence (codon 263 out of 304) and confers only a partial loss of PEX13 function (Mano et al. 2006). This notable under-representation of nonsense alleles and splice site mutations in the extensive collection of Arabidopsis pex mutants isolated through forward-genetic screens is consistent with the possibility that complete loss of these peroxins, like the ring finger peroxins, PEX13, and PEX26, may confer embryonic or gametophytic lethality. Moreover, the absence of confirmed T-DNA alleles within the coding sequence of many PEX genes hints at an essential role for additional peroxins in gametogenesis, as insertions preventing female gametogenesis may be missing from T-DNA collections (Li et al. 2006).
Our analysis of a pex14 allelic series provides an apparent exception to the pattern of essential PEX genes in Arabidopsis and extends the previous analysis of the ped2 allele of pex14 that was isolated in the Landsberg accession (Hayashi et al. 2000). Our data confirm the demonstrated role for PEX14 in efficient import of both PTS1 and PTS2 cargo into peroxisomes (Fig. 6; Hayashi et al. 2000), but do not reveal an essential role for PEX14 in importing matrix proteins after the early seedling stage nor an essential role for PEX14 in gametophytic or embryonic development. Although none of our pex14 alleles accumulated detectable full-length PEX14 protein (Fig. 1e), the pex14-3 allele displayed the weakest defects in most assays of peroxisomal function (Figs. 2, 3, 4, 5), suggesting that the truncated pex14-3 protein detected in this mutant (Fig. 1e) retained some function. Like the ped2 allele (Hayashi et al. 2000), pex14-1 and pex14-4 are disrupted midway through PEX14. The pex14-2 allele, which is disrupted by a T-DNA insertion early in the PEX14 coding sequence and lacked detectable PEX14 mRNA and PEX14 protein (Fig. 1), conferred defects of similar severity to the pex14-1 and pex14-4 alleles in physiological assays but appeared to have a slightly stronger block than pex14-1 in GFP-ICL import into seedling peroxisomes (Fig. 6a), consistent with the possibility that pex14-1 (and pex14-4) may retain slight residual PEX14 function. Indeed, we detected low levels of a truncated pex14-1 protein product in the pex14-1 mutant (Fig. 1e). Our observations that pex14 PTS2 processing defects (Figs. 5, 6) and PTS2 import defects (Figs. 6, 7) grew less severe as plants matured and accumulated PEX13 (Fig. 7) are consistent with the possibility that residual inefficient import in the absence of PEX14 gradually transfers cargo into pex14 peroxisomes and this efficiency may improve as receptor complexes can shift towards PEX13-based import.
Interestingly, even the most severe pex14 mutants were less IBA resistant (Fig. 2) and displayed more moderate PTS2 processing defects (Fig. 5) than either the pex5-1 missense allele (Zolman et al. 2000) or the pex5-10 T-DNA insertion allele (Zolman et al. 2005), suggesting that loss of PEX14 rendered a less complete block in peroxisome function than partial loss of PEX5 function. The same relative order of severity among the pex5 and pex14 alleles was not observed in all phenotypes, however; the pex14 mutants displayed sucrose dependence intermediate between the strong sucrose dependence of pex5-10 and the weak sucrose dependence of pex5-1 (Fig. 3), and pex14 growth defects were more similar to pex5-10 than to pex5-1, which resembles wild type as a mature plant (Fig. 4; Ramón and Bartel 2010). The pex5-10 allele confers strong physiological and PTS2 processing defects and severely blocks both PTS1 and PTS2 import pathways (Zolman et al. 2005; Khan and Zolman 2010; Ramón and Bartel 2010), whereas pex5-1 displays severe PTS2 import defects but imports PTS1 proteins normally (Woodward and Bartel 2005; Ramón and Bartel 2010). The pex14 mutants displayed severe PTS1 import defects combined with more moderate PTS2 import defects (Fig. 6). The contrasting physiological and molecular defects of pex5-1 and pex14 mutants suggest that one or more PTS2 proteins limit IBA responsiveness, whereas PTS1 import may limit fatty acid β-oxidation and growth in the light.
Together, our physiological and molecular analyses suggest that PEX14 augments but ultimately is dispensable for matrix protein import in Arabidopsis. Supporting this possibility is the observation that pex14 alleles are the most frequently recovered pex mutants in our forward genetic screens for reduced peroxisome function. Moreover, the lesions in these mutants (e.g., premature termination of the PEX14 polypeptide) are suggestive of severe loss of function, whereas other pex mutants that have emerged from these screens are generally missense alleles. Additionally, our most severe allele lacks detectable PEX14 mRNA and PEX14 protein (Fig. 1d, e), consistent with the possibility that pex14-2 is a null allele.
The requirement for PEX14 in matrix protein import appears to vary by organism. PEX14 is reported to be essential for matrix protein import in some organisms, including humans. For example, fibroblasts from a patient homozygous for a nonsense mutation in PEX14 two codons beyond the premature stop in Arabidopsis pex14-1 completely lack PTS1 and PTS2 import, as monitored using GFP reporters (Shimozawa et al. 2004). However, there are indications that PEX14 may not be essential for matrix protein import in all systems. Although a Hansenula polymorpha pex14Δ strain overexpressing PEX5 remains unable to utilize methanol as a carbon source, indicating peroxisome function is not fully restored, PEX5 overexpression partially restores a subset of PTS1 proteins to the peroxisome matrix, indicating that some PTS1 import can occur without PEX14 (Salomons et al. 2000).
In Arabidopsis, PEX14, but not PEX13, binds PEX5, whereas PEX13, but not PEX14, binds PEX7 (Nito et al. 2002; Mano et al. 2006), and PEX5 binds PEX7 (Nito et al. 2002; Ramón and Bartel 2010). The observations that an Arabidopsis pex13 null allele confers gametophyte lethality and completely disrupts GFP-PTS1 import into pollen peroxisomes (Boisson-Dernier et al. 2008) suggests that PEX13 plays an essential role in matrix protein import that cannot be filled by PEX14. Moreover, the IBA resistance, sucrose dependence, and PTS2 processing defects of pex14-2 are all markedly enhanced by a reduction in PEX13 expression that alone does not notably impair peroxisome function (Ratzel et al. 2011). The increased severity of PTS1 and PTS2 import defects in very young versus older pex14 seedlings may reflect the reduced PEX13 levels in younger pex14 seedlings (Fig. 7).
How might matrix proteins be imported without PEX14? It is possible that the interdependence of the PEX5 and PEX7 receptors provides a partially redundant import pathway that reduces the importance of PEX14 versus PEX13 in Arabidopsis compared to organisms such as yeast, in which the receptors independently deliver cargo. Perhaps cargo-laden PEX5-PEX7 complexes normally are targeted to peroxisomes through simultaneous or sequential PEX5-PEX14 and PEX7-PEX13 interactions. When PEX14 is disrupted, PTS2-PEX7 complexes may be inefficiently imported via PEX13. Furthermore, PEX7 docking with PEX13 also may allow PTS1 import via PEX7-PEX5 interactions. Supporting the possibility that PEX7 can contribute to PTS1 import in Arabidopsis, we recently found that PTS1 import is impaired in light-grown pex7 seedlings (Ramón and Bartel 2010). Regardless of the mechanism by which residual peroxisome import is occurring in Arabidopsis pex14 mutants, our results reinforce the idea that although a core set of peroxins are conserved among many eukaryotes, certain aspects of peroxisome biogenesis mechanisms have diversified in particular lineages.
Materials and methods
Plant materials and growth conditions
Plants were grown in soil (Metromix 200; Scotts, Marysville, OH) at 22°C under continuous illumination by cool-white fluorescent bulbs (Sylvania, Danvers, MA). Plants grown aseptically were plated on PN (plant nutrient medium) (Haughn and Somerville 1986) or PNS (plant nutrient medium supplemented with 0.5% [w/v] sucrose) and solidified with 0.6% (w/v) agar, either alone or supplemented with hormones (from 0.1-, 1.0-, 10-, or 100-mM stocks in ethanol) or kanamycin (from a 25-mg/mL stock). Plates were incubated under yellow long-pass filters to slow the breakdown of indolic compounds (Stasinopoulos and Hangarter 1990) unless indicated otherwise.
Mutants were all in the Columbia (Col-0) accession. pex5-1 (Zolman et al. 2000), pex5-10 (Zolman et al. 2005), pex6-1 (Zolman and Bartel 2004), pex4-1 pex22-1 (Zolman et al. 2005), and pex7-2 (Ramón and Bartel 2010) were described previously. pex14-2 and pex14-3 were isolated from SALK_007441 and SALK_072373 seeds, respectively, generated by the Salk Institute Genomic Analysis Laboratory, La Jolla, CA (Alonso et al. 2003) and obtained from the Arabidopsis Biological Resource Center (Columbus, OH). Segregating pex14-2 and pex14-3 seeds were plated on PNS supplemented with 12 μg/mL kanamycin, and resistant seedlings were transferred to soil. Plants were genotyped by amplifying genomic DNA from pex14-2 plants with the primers PED2-9 (5′-GCTTGCTGAACCTCATTAGCAGGCTTAGTAGCC-3′) and LB1-SALK (5′-CAAACCAGCGTGGACCGCTTGCTGCAACTC-3′), which yields an ~500-bp product, or from pex14-3 plants with primers PED2-6 (5′-GGCAAACCTCATAAAGTATCAATAACCCC-3′) and LB1-SALK, which yields an ~500-bp product. These amplicons were sequenced directly with the LB1-SALK primer to determine the precise location of the T-DNA insert in the mutants.
pex14-1 was isolated from the progeny of Col-0 plants transformed with a T-DNA based cDNA library (LeClere and Bartel 2001) as an IBA-response mutant as previously described (Zolman et al. 2000). Segregation analysis indicated that the T-DNA was not linked to the lesion conferring IBA resistance.
pex14-4 was isolated as a sucrose-dependent IBA-resistant mutant in the Col-0 accession. Twelve pools of ethyl methanesulfonate-mutagenized seed (~72,000 M2 seeds total) were plated on PN and incubated for 1 day in the light followed by 4 days in darkness. Ungerminated seeds and seedlings with short hypocotyls were transferred to PNS plates and incubated under continuous illumination. After 4 days, expanded seedlings (putative sucrose-dependent mutants) were transferred to a PNS plate supplemented with 10 μM IBA. After another 4–10 days under yellow-filtered light, seedlings that failed to produce abundant lateral roots (putative IBA-resistant mutants) were transferred to soil for seed production. Sucrose dependence and IBA resistance were retested in subsequent generations.
pex14-1 was outcrossed to Wassilewskija (Ws) and pex14-4 was outcrossed to Landsberg erecta (Ler) tt4 for recombination mapping. DNA was isolated (Celenza et al. 1995) from IBA-resistant or sucrose-dependent F2 plants, and mutants were mapped using published PCR-based markers (Konieczny and Ausubel 1993; Bell and Ecker 1994; Davies et al. 1999; Zolman et al. 2001). A candidate gene (PEX14/PED2; At5g62810) within the mapping interval was examined for changes by directly sequencing (LoneStar Labs, Houston, TX) the PCR-amplified PEX14 gene. Subsequent determination of the pex14-1 genotype was performed by amplifying genomic DNA with the primers PED2-3 (5′-GTCGTTGGCTGAATATTTTGTTCGGC-3′) and PED2-4 (5′-GTGGCAAGTAAGACCCTAAAGTGAAC-3′), which yields an 1,170-bp product with two BclI sites in Col-0 and three sites in pex14-1. Determination of the pex14-4 genotype was performed by amplifying genomic DNA with the primers PED2-7 (5′-CAGGGCAATCCAACAACATCCCAA-3′) and PED2-8 (5′-TTGAAGGCTTCTCTCCTCTCTGGA-3′), which yields a 601-bp product with one BsaBI site in Col-0 and two sites in pex14-4.
Phenotypic analyses
The pex14 mutants were each backcrossed to Col-0 at least once prior to analysis. All assays were conducted at least twice with similar results. Seeds were surface-sterilized (Last and Fink 1988) and stratified for 24–48 h prior to plating. In root elongation assays on PN or PNS, seedlings were grown for 8 days and removed from the agar, and the length of the primary root was measured. In root elongation assays on IBA, stratified (24 h) seeds were allowed to germinate in one-sixth liquid PN and 0.5% sucrose for 24 h in white light at 22°C prior to plating on PNS plates supplemented with IBA or the equivalent volume of ethanol (mock). In lateral root assays, seedlings were grown on PNS for 4 days, transferred to PNS or PNS supplemented with 10 μM IBA or 70 nM NAA and grown for an additional 4 days, after which the length of the primary root was measured and lateral roots emerged from the main root were counted. For hypocotyl elongation assays, seeds were plated on PN or PNS and incubated for 1 day in white light to induce germination, followed by 5 days in the dark, after which seedlings were removed from the agar and hypocotyl lengths were measured.
RNA gel-blot analysis
RNA was isolated from 10-day-old light-grown seedlings using the RNeasy kit (Qiagen, Valencia, CA) using the standard extraction protocol. Total RNA was subjected to RNA gel-blot analysis as previously described (Dugas and Bartel 2008). Digoxigenin-labeled probes were amplified using a PCR DIG Probe Synthesis Kit (Roche, Indianapolis, IN) according to the manufacturer’s instructions using the primers PEX14-1 (5′-AGAGGCTACTAAGCCTGC-TAATGA-3′) and PEX14-2 (5′-ATGTTGCTGTTCTG TTTCTTCTTG-3′).
Immunoblot analyses
Protein was extracted from seedlings grown under continuous white light on PNS, separated using SDS–PAGE, and electroblotted to HyBond ECL nitrocellulose membranes (Amersham Pharmacia Biotech, Piscataway, NJ) as previously described (Zolman and Bartel 2004). After transfer, the membranes were air dried, blocked for 1 h in 8% non-fat dry milk in TBS-T (blocking buffer), and incubated overnight at 4°C with primary antibodies diluted in blocking buffer: rabbit α-GFP (1:300 dilution, BD Biosciences 8372-2), rabbit α-ICL (1:2,000 dilution, Maeshima et al. 1988), rabbit α-MLS (1:25,000 dilution, Olsen et al. 1993), rabbit α-PEX5 (1:100 dilution, Zolman and Bartel 2004), rabbit α-PEX6 (1:1,000 dilution, Ratzel et al. 2011), rabbit α-PEX7 (1:800 dilution, Ramón and Bartel 2010), rabbit α-PEX13 (1:500 dilution, Mano et al. 2006), rabbit α-PEX14 (1:2,500 dilution, Lingard and Bartel 2009), rabbit α-PMDH2 (1:2,000 dilution, Pracharoenwattana et al. 2007), rabbit α-thiolase (PED1 isoform, 1:10,000 dilution, Lingard et al. 2009), mouse α-complex V subunit α (1:2,000 dilution, MitoScience MS507), or mouse α-HSC70 (1:4,000–1:20,000 dilution, StressGen Bioreagents SPA-817), followed by a 1–4 h incubation with horseradish peroxidase-conjugated α-rabbit or α-mouse secondary antibody (Santa Cruz Biotechnology). Horseradish peroxidase was visualized by incubation with LumiGlo reagent (Cell Signaling Technology, Danvers, MA).
Cell fractionation
Cell fractionation into organellar and cytosolic fractions was as described (Ratzel et al. 2011). Briefly, 500 mg of 3-or 10-day-old light-grown seedlings were homogenized in 1 mL fractionation buffer (150 mM Tris pH 7.6, 10 mM KCl, 1 mM EDTA, 1 mM DTT, 100 mM sucrose, 1× protease inhibitor cocktail [P9559, Sigma]), filtered to give a homogenate (H) fraction, centrifuged at 50×g to remove debris, and then centrifuged at 15,300×g to give a “soluble” (S) supernatant fraction. The pellet was washed once and resuspended in 40 μL of fractionation buffer to give the “pellet” (P) fraction. Cytosolic fractions contained cytosolic proteins as well as contents of lysed organelles, and pellet fractions contained organellar membranes and contents of intact organelles. Following fractionation, 10 μL of the H (1% total homogenate), S (1% total soluble fraction), and P (25% total pellet) fractions were mixed with 10 μL of NuPAGE 2× loading buffer (Invitrogen) and processed for immunoblot analysis as described above.
Microscopy
Col-0 lines expressing 35S-GFP-PTS1 (Zolman and Bartel 2004), 35S-PTS2-GFP (Woodward and Bartel 2005), or ICLp-GFP-ICL (Lingard et al. 2009) were crossed to pex14, and lines homozygous for the mutant and transgene were selected in subsequent generations. Confocal images of cotyledon cells were obtained using a Zeiss multiphoton laser scanning microscope 510 META NLO equipped with a 63× oil-immersion lens. GFP was excited using a 488 nm argon laser. A bandpass emission filter of 500–530 nm was used to detect GFP fluorescence.
Acknowledgments
We thank John Harada (University of California, Davis) for the MLS antibody, Masayoshi Maeshima (Nagoya University, Japan) for the ICL antibody, Shoji Mano and Mikio Nishimura (National Institute for Basic Biology, Okazaki, Japan) for the PEX13 antibody, and Steven Smith and Itsara Pracharoenwattana (University of Western Australia) for the PMDH2 antibody. We thank the Arabidopsis Biological Resource Center at Ohio State University for seeds from Salk Institute insertion lines, and Lucia Strader and Sherry LeClere for EMS- and T-DNA mutagenized Col-0 pools. We are grateful to Lisa Farmer, Wendell Fleming, Jerrad Stoddard, and Lucia Strader for critical comments on the manuscript. This research was supported by the National Science Foundation (MCB-0745122), the National Institutes of Health (R01GM079177), and the Robert A. Welch Foundation (C-1309). N.M.R. was supported in part by a National Institutes of Health predoctoral fellowship (F31-GM081911) and the Rice-Houston Alliance for Graduate Education and the Professoriate Program (NSF HRD-0450363), M.J.L. was supported in part by a postdoctoral fellowship from the USDA (2008-20659), and C.M. was supported by a Howard Hughes Medical Institute Professors Grant (to B.B.).
Contributor Information
Melanie Monroe-Augustus, Department of Biochemistry and Cell Biology, Rice University, 6100 South Main Street, Houston, TX 77005, USA.
Naxhiely Martínez Ramón, Department of Biochemistry and Cell Biology, Rice University, 6100 South Main Street, Houston, TX 77005, USA.
Sarah E. Ratzel, Department of Biochemistry and Cell Biology, Rice University, 6100 South Main Street, Houston, TX 77005, USA
Matthew J. Lingard, Department of Biochemistry and Cell Biology, Rice University, 6100 South Main Street, Houston, TX 77005, USA. 700 Chesterfield Parkway, Chesterfield, MO 63017, USA
Sarah E. Christensen, Department of Biochemistry and Cell Biology, Rice University, 6100 South Main Street, Houston, TX 77005, USA
Chaya Murali, Department of Biochemistry and Cell Biology, Rice University, 6100 South Main Street, Houston, TX 77005, USA.
Bonnie Bartel, Email: bartel@rice.edu, Department of Biochemistry and Cell Biology, Rice University, 6100 South Main Street, Houston, TX 77005, USA.
References
- Albertini M, Rehling P, Erdmann R, Girzalsky W, Kiel JA, Veenhuis M, Kunau WH. Pex14p, a peroxisomal membrane protein binding both receptors of the two PTS-dependent import pathways. Cell. 1997;89:83–92. doi: 10.1016/s0092-8674(00)80185-3. [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Azevedo JE, Schliebs W. Pex14p, more than just a docking protein. Biochim Biophys Acta. 2006;1763:1574–1584. doi: 10.1016/j.bbamcr.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Baker A, Graham IA, Holdsworth M, Smith SM, Theodoulou FL. Chewing the fat: β-oxidation in signalling and development. Trends Plant Sci. 2006;11:124–132. doi: 10.1016/j.tplants.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A, Frietsch S, Kim TH, Dizon MB, Schroeder JI. The peroxin loss-of-function mutation abstinence by mutual consent disrupts male-female gametophyte recognition. Curr Biol. 2008;18:63–68. doi: 10.1016/j.cub.2007.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braverman N, Dodt G, Gould SJ, Valle D. An isoform of Pex5p, the human PTS1 receptor, is required for the import of PTS2 proteins into peroxisomes. Hum Mol Genet. 1998;7:1195–1205. doi: 10.1093/hmg/7.8.1195. [DOI] [PubMed] [Google Scholar]
- Celenza JL, Jr, Grisafi PL, Fink GR. A pathway for lateral root formation in Arabidopsis thaliana. Genes Dev. 1995;9:2131–2142. doi: 10.1101/gad.9.17.2131. [DOI] [PubMed] [Google Scholar]
- Davies RT, Goetz DH, Lasswell J, Anderson MN, Bartel B. IAR3 encodes an auxin conjugate hydrolase from Arabidopsis. Plant Cell. 1999;11:365–376. doi: 10.1105/tpc.11.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodt G, Warren D, Becker E, Rehling P, Gould SJ. Domain mapping of human PEX5 reveals functional and structural similarities to Saccharomyces cerevisiae Pex18p and Pex21p. J Biol Chem. 2001;276:41769–41781. doi: 10.1074/jbc.M106932200. [DOI] [PubMed] [Google Scholar]
- Dugas D, Bartel B. Sucrose induction of Arabidopsis miR398 represses two Cu/Zn superoxide dismutases. Plant Mol Biol. 2008;67:403–417. doi: 10.1007/s11103-008-9329-1. [DOI] [PubMed] [Google Scholar]
- Eastmond PJ, Germain V, Lange PR, Bryce JH, Smith SM, Graham IA. Postgerminative growth and lipid catabolism in oilseeds lacking the glyoxylate cycle. Proc Natl Acad Sci USA. 2000;97:5669–5674. doi: 10.1073/pnas.97.10.5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgersma Y, Kwast L, Klein A, Voorn-Brouwer T, van den Berg M, Metzig B, America T, Tabak HF, Distel B. The SH3 domain of the Saccharomyces cerevisiae peroxisomal membrane protein Pex13p functions as a docking site for Pex5p, a mobile receptor for the import PTS1-containing proteins. J Cell Biol. 1996;135:97–109. doi: 10.1083/jcb.135.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan JL, Quan S, Orth T, Awai C, Chory J, Hu JP. The Arabidopsis PEX12 gene is required for peroxisome biogenesis and is essential for development. Plant Physiol. 2005;139:231–239. doi: 10.1104/pp.105.066811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto S, Mano S, Nakamori C, Nishimura M. Arabidopsis ABERRANT PEROXISOME MORPHOLOGY9 is a peroxin that recruits the PEX1-PEX6 complex to peroxisomes. Plant Cell. 2011 doi: 10.1105/tpc.110.080770. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham IA. Seed storage oil mobilization. Annu Rev Plant Biol. 2008;59:115–142. doi: 10.1146/annurev.arplant.59.032607.092938. [DOI] [PubMed] [Google Scholar]
- Grunau S, Schliebs W, Linnepe R, Neufeld C, Cizmowski C, Reinartz B, Meyer HE, Warscheid B, Girzalsky W, Erdmann R. Peroxisomal targeting of PTS2 pre-import complexes in the yeast Saccharomyces cerevisiae. Traffic. 2009;10:451–460. doi: 10.1111/j.1600-0854.2008.00876.x. [DOI] [PubMed] [Google Scholar]
- Haughn GW, Somerville C. Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol Gen Genet. 1986;204:430–434. [Google Scholar]
- Hayashi M, Nishimura M. Arabidopsis thaliana—a model organism to study plant peroxisomes. Biochim Biophys Acta. 2006;1763:1382–1391. doi: 10.1016/j.bbamcr.2006.08.014. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Toriyama K, Kondo M, Nishimura M. 2, 4-dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid β-oxidation. Plant Cell. 1998;10:183–195. doi: 10.1105/tpc.10.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Nito K, Toriyama-Kato K, Kondo M, Yamaya T, Nishimura M. AtPex14p maintains peroxisomal functions by determining protein targeting to three kinds of plant peroxisomes. EMBO J. 2000;19:5701–5710. doi: 10.1093/emboj/19.21.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Yagi M, Nito K, Kamada T, Nishimura M. Differential contribution of two peroxisomal protein receptors to the maintenance of peroxisomal functions in Arabidopsis. J Biol Chem. 2005;280:14829–14835. doi: 10.1074/jbc.M411005200. [DOI] [PubMed] [Google Scholar]
- Helm M, Luck C, Prestele J, Hierl G, Huesgen PF, Frohlich T, Arnold GJ, Adamska I, Gorg A, Lottspeich F, Gietl C. Dual specificities of the glyoxysomal/peroxisomal processing protease Deg15 in higher plants. Proc Natl Acad Sci USA. 2007;104:11501–11506. doi: 10.1073/pnas.0704733104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu JP, Aguirre M, Peto C, Alonso J, Ecker J, Chory J. A role for peroxisomes in photomorphogenesis and development of Arabidopsis. Science. 2002;297:405–409. doi: 10.1126/science.1073633. [DOI] [PubMed] [Google Scholar]
- Huybrechts SJ, Van Veldhoven PP, Hoffman I, Zeevaert R, de Vos R, Demaerel P, Brams M, Jaeken J, Fransen M, Cassiman D. Identification of a novel PEX14 mutation in Zellweger syndrome. J Med Genet. 2008;45:376–383. doi: 10.1136/jmg.2007.056697. [DOI] [PubMed] [Google Scholar]
- Khan BR, Zolman BK. pex5 mutants that differentially disrupt PTS1 and PTS2 peroxisomal matrix protein import in Arabidopsis. Plant Physiol. 2010;154:1602–1615. doi: 10.1104/pp.110.162479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- Lanyon-Hogg T, Warriner SL, Baker A. Getting a camel through the eye of a needle: the import of folded proteins by peroxisomes. Biol Cell. 2010;102:245–263. doi: 10.1042/BC20090159. [DOI] [PubMed] [Google Scholar]
- Last RL, Fink GR. Tryptophan-requiring mutants of the plant Arabidopsis thaliana. Science. 1988;240:305–310. doi: 10.1126/science.240.4850.305. [DOI] [PubMed] [Google Scholar]
- LeClere S, Bartel B. A library of Arabidopsis 35S-cDNA lines for identifying novel mutants. Plant Mol Biol. 2001;46:695–703. doi: 10.1023/a:1011699722052. [DOI] [PubMed] [Google Scholar]
- Li Y, Rosso MG, Ulker B, Weisshaar B. Analysis of T-DNA insertion site distribution patterns in Arabidopsis thaliana reveals special features of genes without insertions. Genomics. 2006;87:645–652. doi: 10.1016/j.ygeno.2005.12.010. [DOI] [PubMed] [Google Scholar]
- Lingard MJ, Bartel B. Arabidopsis LON2 is necessary for peroxisomal function and sustained matrix protein import. Plant Physiol. 2009;151:1354–1365. doi: 10.1104/pp.109.142505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingard MJ, Monroe-Augustus M, Bartel B. Peroxisome-associated matrix protein degradation in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:4561–4566. doi: 10.1073/pnas.0811329106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Huertas E, Oh J, Baker A. Antibodies against pex14p block ATP-independent binding of matrix proteins to peroxisomes in vitro. FEBS Lett. 1999;459:227–229. doi: 10.1016/s0014-5793(99)01239-9. [DOI] [PubMed] [Google Scholar]
- Maeshima M, Yokoi H, Asahi T. Evidence for no proteolytic processing during transport of isocitrate lyase into glyoxysomes in castor bean endosperm. Plant Cell Physiol. 1988;29:381–384. [Google Scholar]
- Mano S, Nakamori C, Nito K, Kondo M, Nishimura M. The Arabidopsis pex12 and pex13 mutants are defective in both PTS1- and PTS2-dependent protein transport to peroxisomes. Plant J. 2006;47:604–618. doi: 10.1111/j.1365-313X.2006.02809.x. [DOI] [PubMed] [Google Scholar]
- Matsumura T, Otera H, Fujiki Y. Disruption of the interaction of the longer isoform of Pex5p, Pex5pL, with Pex7p abolishes peroxisome targeting signal type 2 protein import in mammals. Study with a novel Pex5-impaired Chinese hamster ovary cell mutant. J Biol Chem. 2000;275:21715–21721. doi: 10.1074/jbc.M000721200. [DOI] [PubMed] [Google Scholar]
- Meinecke M, Cizmowski C, Schliebs W, Krüger V, Beck S, Wagner R, Erdmann R. The peroxisomal importomer constitutes a large and highly dynamic pore. Nat Cell Biol. 2010;12:273–277. doi: 10.1038/ncb2027. [DOI] [PubMed] [Google Scholar]
- Miyata N, Hosoi K, Mukai S, Fujiki Y. In vitro import of peroxisome-targeting signal type 2 (PTS2) receptor Pex7p into peroxisomes. Biochim Biophys Acta. 2009;1793:860–870. doi: 10.1016/j.bbamcr.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Niederhoff K, Meindl-Beinker NM, Kerssen D, Perband U, Schäfer A, Schliebs W, Kunau WH. Yeast Pex14p possesses two functionally distinct Pex5p and one Pex7p binding sites. J Biol Chem. 2005;280:35571–35578. doi: 10.1074/jbc.M502460200. [DOI] [PubMed] [Google Scholar]
- Nito K, Hayashi M, Nishimura M. Direct interaction and determination of binding domains among peroxisomal import factors in Arabidopsis thaliana. Plant Cell Physiol. 2002;43:355–366. doi: 10.1093/pcp/pcf057. [DOI] [PubMed] [Google Scholar]
- Nito K, Kamigaki A, Kondo M, Hayashi M, Nishimura M. Functional classification of Arabidopsis peroxisome biogenesis factors proposed from analyses of knockdown mutants. Plant Cell Physiol. 2007;48:763–774. doi: 10.1093/pcp/pcm053. [DOI] [PubMed] [Google Scholar]
- Olsen LJ, Ettinger WF, Damsz B, Matsudaira K, Webb MA, Harada JJ. Targeting of glyoxysomal proteins to peroxisomes in leaves and roots of a higher plant. Plant Cell. 1993;5:941–952. doi: 10.1105/tpc.5.8.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otera H, Harano T, Honsho M, Ghaedi K, Mukai S, Tanaka A, Kawai A, Shimizu N, Fujiki Y. The mammalian peroxin Pex5pL, the longer isoform of the mobile peroxisome targeting signal (PTS) Type 1 transporter, translocates the Pex7p-PTS2 protein complex into peroxisomes via its initial docking site, Pex14p. J Biol Chem. 2000;275:21703–21714. doi: 10.1074/jbc.M000720200. [DOI] [PubMed] [Google Scholar]
- Otera H, Setoguchi K, Hamasaki M, Kumashiro T, Shimizu N, Fujiki Y. Peroxisomal targeting signal receptor Pex5p interacts with cargoes and import machinery components in a spatiotemporally differentiated manner: conserved Pex5p WXXXF/Y motifs are critical for matrix protein import. Mol Cell Biol. 2002;22:1639–1655. doi: 10.1128/MCB.22.6.1639-1655.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pracharoenwattana I, Cornah JE, Smith SM. Arabidopsis peroxisomal malate dehydrogenase functions in β-oxidation but not in the glyoxylate cycle. Plant J. 2007;50:381–390. doi: 10.1111/j.1365-313X.2007.03055.x. [DOI] [PubMed] [Google Scholar]
- Ramón NM, Bartel B. Interdependence of the peroxisome-targeting receptors in Arabidopsis thaliana: PEX7 facilitates PEX5 accumulation and import of PTS1 cargo into peroxisomes. Mol Biol Cell. 2010;21:1271–1296. doi: 10.1091/mbc.E09-08-0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratzel SE, Lingard MJ, Woodward AW, Bartel B. Reducing PEX13 expression ameliorates physiological defects of late-acting peroxin mutants. Traffic. 2011;12:121–134. doi: 10.1111/j.1600-0854.2010.01136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rylott EL, Rogers CA, Gilday AD, Edgell T, Larson TR, Graham IA. Arabidopsis mutants in short- and medium-chain acyl-CoA oxidase activities accumulate acyl-CoAs and reveal that fatty acid β-oxidation is essential for embryo development. J Biol Chem. 2003;278:21370–21377. doi: 10.1074/jbc.M300826200. [DOI] [PubMed] [Google Scholar]
- Rylott EL, Eastmond PJ, Gilday AD, Slocombe SP, Larson TR, Baker A, Graham IA. The Arabidopsis thaliana multifunctional protein gene (MFP2) of peroxisomal β-oxidation is essential for seedling establishment. Plant J. 2006;45:930–941. doi: 10.1111/j.1365-313X.2005.02650.x. [DOI] [PubMed] [Google Scholar]
- Salomons FA, Kiel JA, Faber KN, Veenhuis M, van der Klei IJ. Overproduction of Pex5p stimulates import of alcohol oxidase and dihydroxyacetone synthase in a Hansenula polymorpha Pex14 null mutant. J Biol Chem. 2000;275:12603–12611. doi: 10.1074/jbc.275.17.12603. [DOI] [PubMed] [Google Scholar]
- Schliebs W, Saidowsky J, Agianian B, Dodt G, Herberg FW, Kunau WH. Recombinant human peroxisomal targeting signal receptor PEX5. Structural basis for interaction of PEX5 with PEX14. J Biol Chem. 1999;274:5666–5673. doi: 10.1074/jbc.274.9.5666. [DOI] [PubMed] [Google Scholar]
- Schuhmann H, Huesgen PF, Gietl C, Adamska I. The DEG15 serine protease cleaves peroxisomal targeting signal 2-containing proteins in Arabidopsis. Plant Physiol. 2008;148:1847–1856. doi: 10.1104/pp.108.125377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann U, Wanner G, Veenhuis M, Schmid M, Gietl C. AthPEX10, a nuclear gene essential for peroxisome and storage organelle formation during Arabidopsis embryogenesis. Proc Natl Acad Sci USA. 2003;100:9626–9631. doi: 10.1073/pnas.1633697100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu N, Itoh R, Hirono Y, Otera H, Ghaedi K, Tateishi K, Tamura S, Okumoto K, Harano T, Mukai S, Fujiki Y. The peroxin Pex14p. cDNA cloning by functional complementation on a Chinese hamster ovary cell mutant, characterization, and functional analysis. J Biol Chem. 1999;274:12593–12604. doi: 10.1074/jbc.274.18.12593. [DOI] [PubMed] [Google Scholar]
- Shimozawa N, Tsukamoto T, Nagase T, Takemoto Y, Koyama N, Suzuki Y, Komori M, Osumi T, Jeannette G, Wanders RJ, Kondo N. Identification of a new complementation group of the peroxisome biogenesis disorders and PEX14 as the mutated gene. Hum Mutat. 2004;23:552–558. doi: 10.1002/humu.20032. [DOI] [PubMed] [Google Scholar]
- Sparkes IA, Brandizzi F, Slocombe SP, El-Shami M, Hawes C, Baker A. An Arabidopsis pex10 null mutant is embryo lethal, implicating peroxisomes in an essential role during plant embryogenesis. Plant Physiol. 2003;133:1809–1819. doi: 10.1104/pp.103.031252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder L, Muhlemann O. The meaning of nonsense. Trends Cell Biol. 2008;18:315–321. doi: 10.1016/j.tcb.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Stasinopoulos TC, Hangarter RP. Preventing photochemistry in culture media by long-pass light filters alters growth of cultured tissues. Plant Physiol. 1990;93:1365–1369. doi: 10.1104/pp.93.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein K, Schell-Steven A, Erdmann R, Rottensteiner H. Interactions of Pex7p and Pex18p/Pex21p with the peroxisomal docking machinery: implications for the first steps in PTS2 protein import. Mol Cell Biol. 2002;22:6056–6069. doi: 10.1128/MCB.22.17.6056-6069.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Bartel B. Transport and metabolism of the endogenous auxin precursor indole-3-butyric acid. Mol Plant. 2011 doi: 10.1093/mp/ssr006. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Culler AH, Cohen JD, Bartel B. Conversion of endogenous indole-3-butyric acid to indole-3-acetic acid drives cell expansion in Arabidopsis seedlings. Plant Physiol. 2010;153:1577–1586. doi: 10.1104/pp.110.157461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader LC, Wheeler DL, Christensen SE, Berens JC, Cohen JD, Rampey RA, Bartel B. Multiple facets of Arabidopsis seedling development require indole-3-butyric acid-derived auxin. Plant Cell. 2011;23:984–999. doi: 10.1105/tpc.111.083071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urquhart AJ, Kennedy D, Gould SJ, Crane DI. Interaction of Pex5p, the type 1 peroxisome targeting signal receptor, with the peroxisomal membrane proteins Pex14p and Pex13p. J Biol Chem. 2000;275:4127–4136. doi: 10.1074/jbc.275.6.4127. [DOI] [PubMed] [Google Scholar]
- Woodward AW, Bartel B. The Arabidopsis peroxisomal targeting signal type 2 receptor PEX7 is necessary for peroxisome function and dependent on PEX5. Mol Biol Cell. 2005;16:573–583. doi: 10.1091/mbc.E04-05-0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Bartel B. An Arabidopsis indole-3-butyric acid-response mutant defective in PEROXIN6, an apparent ATPase implicated in peroxisomal function. Proc Natl Acad Sci USA. 2004;101:1786–1791. doi: 10.1073/pnas.0304368101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Yoder A, Bartel B. Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics. 2000;156:1323–1337. doi: 10.1093/genetics/156.3.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Monroe-Augustus M, Thompson B, Hawes JW, Krukenberg KA, Matsuda SP, Bartel B. chy1, an Arabidopsis mutant with impaired β-oxidation, is defective in a peroxisomal β-hydroxyisobutyryl-CoA hydrolase. J Biol Chem. 2001;276:31037–31046. doi: 10.1074/jbc.M104679200. [DOI] [PubMed] [Google Scholar]
- Zolman BK, Monroe-Augustus M, Silva ID, Bartel B. Identification and functional characterization of Arabidopsis PEROXIN4 and the interacting protein PEROXIN22. Plant Cell. 2005;17:3422–3435. doi: 10.1105/tpc.105.035691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Nyberg M, Bartel B. IBR3, a novel peroxisomal acyl-CoA dehydrogenase-like protein required for indole-3-butyric acid response. Plant Mol Biol. 2007;64:59–72. doi: 10.1007/s11103-007-9134-2. [DOI] [PubMed] [Google Scholar]
- Zolman BK, Martinez N, Millius A, Adham AR, Bartel B. Identification and characterization of Arabidopsis indole-3-butyric acid response mutants defective in novel peroxisomal enzymes. Genetics. 2008;180:237–251. doi: 10.1534/genetics.108.090399. [DOI] [PMC free article] [PubMed] [Google Scholar]